Abstract
Nitrification is a core process in the global nitrogen cycle that is essential for the functioning of many ecosystems. The discovery of autotrophic ammonia-oxidizing archaea (AOA) within the phylum Thaumarchaeota has changed our perception of the microbiology of nitrification, in particular since their numerical dominance over ammonia-oxidizing bacteria (AOB) in many environments has been revealed. These and other data have led to a widely held assumption that all amoA-encoding members of the Thaumarchaeota (AEA) are autotrophic nitrifiers. In this study, 52 municipal and industrial wastewater treatment plants were screened for the presence of AEA and AOB. Thaumarchaeota carrying amoA were detected in high abundance only in four industrial plants. In one plant, thaumarchaeotes closely related to soil group I.1b outnumbered AOB up to 10,000-fold, and their numbers, which can only be explained by active growth in this continuous culture system, were two to three orders of magnitude higher than could be sustained by autotrophic ammonia oxidation. Consistently, 14CO2 fixation could only be detected in AOB but not in AEA in actively nitrifying sludge from this plant via FISH combined with microautoradiography. Furthermore, in situ transcription of archaeal amoA, and very weak in situ labeling of crenarchaeol after addition of 13CO2, was independent of the addition of ammonium. These data demonstrate that some amoA-carrying group I.1b Thaumarchaeota are not obligate chemolithoautotrophs.
Keywords: heterotrophy, physiology, modeling, ammonia monooxygenase
In recent years there have been a number of startling new discoveries in the biogeochemistry of the nitrogen cycle (1–3). Not least of these has been the demonstration that a novel group of archaea, now known to belong to the novel phylum Thaumarchaeota (4, 5), are capable of autotrophic ammonia oxidation (2). This physiology has in the meantime been confirmed for different lineages within this phylum (6–10). The widespread distribution and abundance of these ammonia-oxidizing archaea (AOA) has been shown through metagenomic surveys, targeted retrieval of archaeal 16S rRNA- and ammonia monooxygenase genes, and analysis of characteristic archaeal lipids (11–13). Moreover, a prominent role for AOA relative to ammonia-oxidizing bacteria (AOB) in nitrification in soil, marine, and geothermal systems has been revealed (14–16). This finding has led to the widely held assumption that all amoA-carrying members of the Thaumarchaeota are capable of autotrophic nitrification, although a few reports suggested that some of these organisms might also be able to assimilate organic compounds, like amino acids (17–20). Furthermore, PCR-based studies indicated that certain thaumarchaoetes might not carry amoA genes (20, 21), but these findings might be explained by primer mismatches to certain amoA sequences (22).
Ammonia oxidation as the rate-limiting step of nitrification is also a vital process in engineered biological systems, such as wastewater treatment plants (WWTPs). The microbiology of nitrogen removal in WWTPs has been intensively studied, and the application of molecular tools has led to the identification of the most abundant bacterial nitrifying populations in these systems. The population structure and dynamics of AOB in WWTPs has received particular attention (for a review, see ref. 23). In contrast, there have been very few studies of AEA—the term originally used for amoA-carrying thaumarchaeotes by Dang and colleagues (24, 25)—in engineered biological treatment systems (26–28). When quantitative analyses of AOB and AEA have been conducted in some nitrifying WWTPs, the abundance of AOB has been shown to exceed that of AEA by two to three orders of magnitude. In these cases, AEA were rarely detected at an abundance greater than 103/mL (27, 29, 30). This finding is consistent with the failure to detect thaumarchaeotal sequences in an analysis of archaeal 16S rRNA genes in activated sludge (31). In contrast, higher abundances of AEA were described in some Asian WTTPs and a correlation between AEA abundance and the ammonia concentration in the wastewater was postulated (32, 33). The significance of putative ammonia-oxidizing thaumarchaeota for nitrogen removal in WWTPs therefore remains unclear.
Here we report a survey of the diversity, abundance, and activity of thaumarchaeotes in 52 municipal and industrial WWTPs in Europe and demonstrate that AEA are not widespread in nitrifying reactors. AEA were, however, abundant in a small number of industrial WWTPs. In one of these plants, AEA were two to four orders of magnitude more abundant than AOB, but the measured numbers were far too great to be explained by the amount of ammonia removal occurring in this system. In this treatment plant, AOB assimilated significantly higher amounts of bicarbonate under nitrifying conditions than AEA. This finding, combined with the results from in situ labeling experiments with 13C-inorganic carbon and compound-specific carbon isotope data for archaeal lipids, showed that the AEA in this plant are not obligate chemolithoautotrophs.
Results
Detection of Thaumarchaeota in WWTPs.
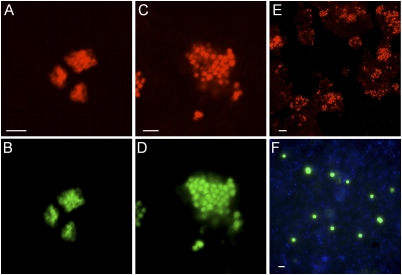
In total, 35 domestic and 17 industrial WWTPs from different geographic locations were screened for the occurrence of AEA. The WWTPs encompassed a wide range of reactor configurations and influent sewage compositions. Of the WWTPs, 46 exhibited high nitrification performance, leading to the removal of at least 90% of the ammonia in the influent (Dataset S1 and ref. 34). Archaeal amoA genes (193 clones) affiliated with group I.1a and group I.1b Thaumarchaeota were detected only in six industrial and one municipal WWTP (Fig. S1). From four of these WWTPs (refinery plants A, D, E, and a tannery plant F) thaumarchaeotal 16S rRNA genes (102 clones) belonging to groups I.1a or I.1b could be retrieved using an archaea-specific PCR assay (Fig. S2). Consistent with this finding was the observation of thaumarchaeote-specific signals with 16S rRNA catalyzed reporter deposition-FISH (CARD-FISH) in only these four industrial WWTPs, using previously published as well as a newly designed probes (Fig. 1, Table 1, and Table S1). The specificity of the CARD-FISH assay was confirmed by double-hybridizations of thaumarchaeotal subgroup-specific probes with the more general probes Cren512 and Arch915. Group I.1b-affiliated Thaumarchaeota in the sludges of plants D and E were detected using a probe specific to cloned 16S rRNA sequences recovered from the WWTPs (Thaum1162). Thaum1162 exclusively hybridized to large, coccoid cells with an average cell diameter of 1 to 2 μm that mainly occurred in aggregates consisting of up to 200 cells. In contrast, AEA detected in plants F and A, which hybridized with the group I.1a-specific probe Cren537, were smaller and formed irregular-shaped colonies with a diameter of 5 to 8 μm that consisted of fewer than 70 cells (Fig. 1). With one exception (brewery WWTP Rapp-Kutzenhausen), AOB were detected by FISH with specific probes in all nitrifying sludges analyzed.
Fig. 1.
Detection of Thaumarchaeota in industrial WWTPs by CARD-FISH. (A) Plant F sludge hybridized with probe Cren512 targeting most Cren- and Thaumarchaeota. (B) The same cells hybridized by group I.1a -specific probe Cren537. (C) Refinery plant D sludge hybridized with the general archaeal probe Arch915. (D) The same cells targeted by the clone-specific probe Thaum1162. (E) Refinery plant D sludge hybridized with probe Arch915. Note the density of AEA colonies reflecting their high relative abundance. (F) Refinery plant E sludge hybridized with probe Thaum1162 after sonication. (Scale bars, 5 μm.)
Table 1.
Measured and modeled abundance of AEA in different samples from the parallel reactors A and B of plant D as determined by qPCR (amoA and 16S rRNA gene), FISH, and modeling
| Log archaeal amoA gene copies/mL |
Log cluster I.1b 16S rRNA gene copies/mL |
Ratio amoA/16S rRNA gene copies |
Modeled log AEA abundance (based on ammonia removal) |
AEA CSAOR (fmol/cell/h)* |
Log FISH counts cells/mL |
||||||
| Refinery D | Reactor A | Reactor B | Reactor A | Reactor B | Reactor A | Reactor B | Reactor A (mean, range) | Reactor B (mean, range) | Reactor A | Reactor B | Reactor A |
| 21.06.2006 | 7.81 ± 0.02 | 7.71 ± 0.07 | 7.72 ± 0.12 | 7.62 ± 0.06 | 1.2 | 1.2 | 5.00 (4.81,5.24) | 5.00 (4.80,5.23) | 0.066 | 0.084 | ND |
| 16.10.2006 | 6.76 ± 0.19 | 7.10 ± 0.30 | 6.85 ± 0.16 | 6.99 ± 0.22 | 0.8 | 1.3 | 4.69 (4.49,4.92) | 4.70 (4.50,4.93) | 0.198 | 0.090 | ND |
| 16.11.2006 | 8.27 ± 0.08 | 8.17 ± 0.09 | 8.00 ± 0.09 | 8.08 ± 0.08 | 1.9 | 1.3 | 4.49 (4.29,4.72) | 4.48 (4.29,4.72) | 0.003 | 0.004 | ND |
| 08.01.2007 | 8.33 ± 0.15 | 8.42 ± 0.14 | 8.01 ± 0.09 | 8.17 ± 0.05 | 2.2 | 1.8 | 4.64 (4.45,4.88) | 4.64 (4.44,4.87) | 0.005 | 0.005 | 7.95 ± 0.05 |
| 07.05.2008 | ND | ND | ND | ND | ND | ND | 8.04 ± 0.08 | ||||
Cell specific ammonia oxidation rates (CSAOR) calculated using the amoA qPCR data are also presented. For quantification of AEA by CARD-FISH, probes Cren1162 and Cren512 were applied. ND, not determined.
*CSAOR calculated from qPCR data assuming that the AEA alone are responsible for ammonia oxidation.
According to the molecular survey data, sludge from the nitrifying refinery plant D contained a single thaumarchaeotal operational taxonomic unit affiliated with group I.1b, which occurred at a relatively high abundance. Because of the low diversity of group I.1b Thaumarchaeota and the co-occurrence of AOB in plant D, the AEA of this plant were selected for subsequent in-depth analyses to assess their role in nitrification.
Quantification of AEA and AOB in Refinery Plant D.
The 16S rRNA gene, as well as archaeal amoA gene-based quantitative PCR data from two parallel reactors of the nitrifying refinery plant D, confirmed the high abundance of AEA in this system. From both reactors four activated sludge samples obtained at different times were analyzed by qPCR and the numbers of AEA [assuming a single 16S rRNA and amoA gene per thaumarchaeotal genome (35)] ranged from 6.76 to 8.42 log10 cells/mL. The ratios of thaumarchaeotal 16S rRNA genes to thaumarchaeotal amoA genes in the eight samples varied between 0.8 and 2.2 (Table 1). In accordance with the qPCR data, quantitative CARD-FISH analysis of two samples from one of the reactors demonstrated that between 7.95 and 8.04 log10 group I.1b thaumarchaeotal cells/mL hybridized with probe Thaum1162 (Table 1), representing up to 5% of the total cell counts in this system. The four samples from both reactors of plant D were also analyzed by qPCR targeting the amoA gene of β-proteobacterial AOB and using FISH targeting AOB 16S rRNA. Bacterial amoA genes were detected in consistent numbers in all samples with an average of 2 × 104 AOB cells/mL (4.3 log10 copies/mL sludge) (Table 2). AOB abundance determined by FISH was somewhat higher than the abundance determined by qPCR (5.6–6.4 log10 copies/mL sludge) (Table 2).
Table 2.
Measured and modeled abundance of AOB in different samples from the parallel reactors A and B of plant D as determined by amoA qPCR, FISH, and modeling
| Log bacterial amoA gene copies/mL |
Modeled log AOB abundance (based on ammonia removal) |
AOB CSAOR (fmol/cell/h) |
Log FISH counts cells/mL |
||||
| Refinery D | Reactor A | Reactor B | Reactor A (mean, range) | Reactor B (mean, range) | Reactor A | Reactor B | Reactor A |
| 21.06.2006 | 4.15 ± 0.11 | 4.21 ± 0.16 | 5.88 (5.75,6.03) | 5.87 (5.74,6.02) | 7.2 | 3.7 | 5.92 ± 0.21 |
| 16.10.2006 | 4.31 ± 0.14 | 4.34 ± 0.11 | 5.56 (5.43,5.71) | 5.57 (5.44,5.72) | 6.5 | 1.3 | 5.59 ± 0.49 |
| 16.11.2006 | 4.39 ± 0.19 | 4.45 ± 0.15 | 5.37 (5.23,5.51) | 5.36 (5.23,5.51) | 0.4 | 0.3 | 6.22 ± 0.08 |
| 08.01.2007 | 4.19 ± 0.08 | 4.32 ± 0.12 | 5.52 (5.39,5.67) | 5.51 (5.38,5.66) | 0.9 | 0.3 | 6.35 ± 0.34 |
| 07.05.2008 | ND | ND | (+) | ||||
CSAOR calculated using the quantitative AOB FISH data, assuming that AOB alone are responsible for ammonia oxidation, are also presented. A mix of probes Nso190 and Nso1225 labeled with the same fluorophore was used for detection of AOB by FISH. (+), only occasional signals; ND, not determined.
Consistent with the detection of Thaumarchaeota at high abundance in refinery plant D using nucleic acid based approaches, high amounts of crenarchaeol (GDGT-I) (Fig. S3), a characteristic glycerol dibiphytanyl glycerol tetraether (GDGT) of thaumarchaeotes, was detected (36–38) (Table S2). Reactors A and B of this plant contained 8.8 and 10.9 μg crenarchaeol/g dry sludge, respectively, but a control reactor (Ingolstadt plant), in which no Thaumarchaeota were detected by nucleic acid-based methods, contained only 0.1 μg crenarchaeol/g dry sludge. Furthermore, both reactors of plant D contained 0.4 μg/g dry sludge of the crenarchaeol regioisomer GDGT-VI (Table S2), which is relatively abundant in the group I.1b AOA “Candidatus Nitrososphaera gargensis” (38), and this compound was below the detection limit in the control plant (Table S2). Assuming that all GDGTs are derived from living cells, then it is possible to estimate the number of AEA cells in the sludge based on the lipid concentrations. If we further assume that all AEA cells are spheres with a diameter of 1.5 μm, as is indicated by FISH data, and that 1 μm2 of archaeal cell membrane contains approximately 1.7 × 105 GDGT molecules (39), then the AEA would contain 2.6 fg of GDGT per cell. Using the summed concentrations of thaumarchaeotal GDGTs (Table S2) and a sludge water content of 99% (based on mixed liquor suspended solids measurements from the reactors), we estimate cell numbers in the order of 6 × 107 cells/mL (7.8 log10 cells/mL), which is very similar to our qPCR and FISH based measurements (Table 1).
Modeling the Abundance of Ammonia-Oxidizing Microorganisms in Refinery Plant D.
To determine whether the abundance of AEA in refinery plant D could be explained by autotrophic ammonia oxidation alone, we used the nitrification model developed by Rittman and colleagues (40, 41), to estimate ammonia oxidizer biomass in relation to ammonia removal in wastewater-treatment reactors. To do this determination, we conservatively assumed that 20% of the ammonia was consumed by assimilation and estimated the growth yield of AEA based on the data presented by Könneke et al. (2). The calculated growth yield was 1.2 g per dw/mol N, which compared favorably with the range of growth yields reported for AOB in the literature [0.1–1.4 g per dw/mol N (42)]. This finding is not surprising given the known thermodynamic constraints on growth yields (43, 44). We therefore used the same growth yield and other physiological parameters for AEA that have previously been used to estimate the abundance of AOB in WWTPs based on ammonia removal (40, 41, 45). The possibility that there was an additional contribution to reduced nitrogen in the system from nitrogen fixation was precluded by measurement of nitrogen fixation in sludge samples from refinery plant D by isotope ratio mass spectrometry, which proved negative (Table S3).
The modeling of data from samples from the two reactors in refinery plant D taken on four separate occasions indicated that the ammonia removal in the reactor would support a population of autotrophic ammonia oxidizers of between 4.48 and 5.00 log10 AEA cells/mL, which is about two to three orders of magnitude lower than the population sizes estimated from quantitative FISH, or qPCR of archaeal amoA, or 16S rRNA genes (Table 1). Predicted AEA numbers are only marginally higher when calculations are based on total Kjeldahl nitrogen (TKN) removal. However, the numbers of autotrophic AOB estimated from the model (5.36–5.88 log10 AOB cells/mL; note that modeled AOB numbers are higher than AOA numbers because of the smaller size of AOB cells in the reactors) are in line with, although higher than, the abundance of AOB amoA genes measured in the same samples (4.15–4.45 log10 AOB cells/mL) and fit well with AOB numbers measured by FISH (5.59–6.35 log10 AOB cells/mL) (Table 2). Quantification of AOB by qPCR may underestimate the true numbers of AOB present in activated sludge because DNA extraction from AOB microcolonies is known to be difficult (46), and thus cell-specific ammonia oxidation rates (CSAOR) were estimated based on the FISH data. Inferred CSAORs of the AOB in the sludge were comparable to values reported for reference strains which range from 0.9 to 53 fmol per cell per hour (47) and CSAORs estimated in situ [0.03–43 fmol per cell per hour (45, 48)] (Table 2). CSAORs based on the abundance of AEA were typically much lower (Table 1). Collectively, these data strongly call into doubt the notion that the amoA-carrying Thaumarchaeota in this system gain most of their energy from chemoautotrophic ammonia oxidation.
Metabolic Activity and Ecophysiology of AEA in Refinery Plant D.
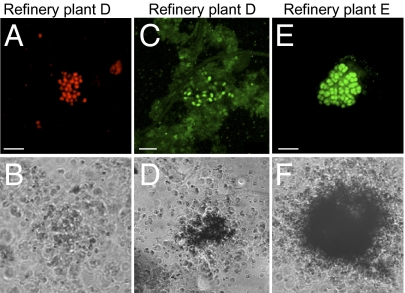
To test whether the sludge from plant D retained its nitrifying capacity during laboratory incubation experiments, live sludge was amended with 0.5 mM NH4Cl and the ammonium concentration was followed for 5 h. In a parallel experiment, 1 μg/mL of diphtheria toxin, an inhibitor of translation in eukarya and archaea (49), was added to the sludge. In both experiments, more than 90% of the added ammonium was removed within 2.5 h (Fig. S4). Furthermore, transcription of amoA genes from group I.1b Thaumarchaeota was detected in incubated sludge from refinery plant D both with and without addition of 2 mM NH4Cl as demonstrated by RT-PCR (Fig. S5). Cloning and sequencing of the amoA RT-PCR product revealed sequences identical to the amoA sequence cluster obtained from genomic DNA of plant D during the survey experiments (Fig. S1, Ref D_11). Subsequently, we combined FISH with microautoradiography (FISH-MAR) (50) to analyze whether the AEA and AOB in plant D incorporated 14C-inorganic carbon in the presence of ammonium. Ammonium chloride (0.5 mM) was added to sludge from plant D (07.05.2008, reactor A) and FISH-MAR was conducted after 6 h of incubation. Unexpectedly, the AEA showed no incorporation of radioactive bicarbonate, although this was strongly assimilated by the AOB (Fig. 2). Absence of autotrophic activity detectable by MAR of the AEA in the presence of ammonia was reproduced in an independent control experiment (09.10.2008, reactor A). In addition, only AOB were shown to fix labeled inorganic carbon in samples from refinery E that, like refinery D, had consistently higher abundance of AEA than AOB (Fig. 2 C–F).
Fig. 2.
FISH-MAR analyses of AOB and AEA in refinery WWTPs under nitrifying conditions in the presence of 14CO2. (A, C, and E) FISH signals obtained with probe Arch915 (A, red signals) and a probe mix (probes Nso190, Nso1225, NEU, NmV, and Nso192) targeting β-proteobacterial AOB (C and E, green signals). (B, D, and F) The corresponding MAR images. (A–D) Samples from refinery D (07.05.2008). (E and F) Samples from Refinery E (26.03.2008). AOB but not AEA showed autotrophic carbon fixation during this experiment. (Scale bars, 5 μm.)
Determination of the δ13C value of the biphytanes released by ether cleavage from GDGTs in the sludge of reactor B of plant D revealed that the crenarchaeol-derived biphytanes (C40:2 and C40:3) (Fig. S3) were depleted by approximately 17‰ compared with the dissolved inorganic carbon (mean δ13C of −27.7‰) of the sludge supernatant (Table S4). Labeling experiments using crenarchaeol as a biomarker for Thaumarchaeota were performed with nitrifying biomass from refinery plant D (reactor A, 9.10.2009). The sludge was incubated with 99 atom% 13C-bicarbonate for 18 h with or without addition of 1 mM NH4Cl. After incubation the crenarchaeol-derived biphytanes C40:2 and C40:3 were very weakly 13C-labeled (Δ δ13C of approximately 7‰) (Table S4), independent of the addition of ammonium.
Discussion
Thaumarchaeota have been shown to play a significant role in nitrification in marine and terrestrial environments (14, 15, 51–53). Despite these findings it is still unclear whether all AEA detected in these environments indeed live from autotrophic ammonia oxidation, and their potential role for nitrogen removal in engineered biological systems has not been clarified as yet (26–28, 33). We report an extensive survey of the distribution, abundance, and activity of AEA in diverse WWTPs. The only occasional occurrence of Thaumarchaeota among 52 plants suggests that they are generally minor contributors to nitrification of most wastewaters. Of 46 nitrifying sludges, only four industrial WWTPs harbored Thaumarchaeota at relative abundances that were above the detection limit of CARD-FISH (Fig. 1). Three of these four sludges originate from petroleum refinery WWTPs, which may reflect a certain habitat preference of the detected AEA. The consistently high abundance of a single Thaumarchaeota operational taxonomic unit throughout the study period in the continuous flow through reactor of refinery plant D demonstrates that they did not originate from allochthonous inflow (i.e., from surrounding terrestrial habitats), but rather accounted for a substantial part of the indigenous actively growing microbial community in this plant. Here, both CARD-FISH cell counts and abundance of amoA and 16S rRNA genes repeatedly revealed thaumarchaeotal cell abundance of 107 to 108 cells/mL. The quantification of crenarchaeol complemented our nucleic acid-based approaches and strongly supported the high abundance of Thaumarchaeota detected by FISH and qPCR in this sludge. In all other sludges tested, in which AEA were not detected by CARD-FISH, β-proteobacterial AOB prevailed (with one exception: Rapp-Kutzenhausen), and were therefore most likely responsible for ammonia oxidation in these plants. In addition, AOB numbers estimated on the basis of a nitrification model were consistent with AOB numbers measured by FISH in the refinery D WWTP, even though they were present at three-to-four orders of magnitude lower abundance than the AEA. Taken together, these data strongly support the contention that β-proteobacterial ammonia oxidizers are the main agents driving the first step of nitrification in most WWTPs (54).
Our initial assumption was that substantially higher numbers of AEA in refinery WWTP plant D must implicate them as the main agents of ammonia-oxidation in these systems. However, when we modeled the expected abundance of AOA based on ammonia or TKN removal in the plant, the amount of reduced nitrogen oxidized was only sufficient to support a population of autotrophically-growing ammonia-oxidizing microbes of approximately 0.01% to 1% of that observed for the Thaumarchaeota in the system (Table 1). In turn, the predicted numbers were in line with the measured abundance of AOB (Table 2), suggesting that the AOB could very well be the solely responsible agents for nitrification. Moreover CSAORs calculated based on AEA abundance were either vanishingly small or at the lower end of CSAORs estimates from other systems (Table 1) [there is only a single report of a CSAOR lower than 0.22 fmol per cell per hour (47)]. AOB-based CSAORs were by contrast directly in line with what has been measured in pure cultures and the majority of WWTP (45, 47). Of course, the model of ammonia oxidation includes important assumptions regarding the growth yield and endogenous biomass decay terms. However, to obtain predicted numbers, which approach those measured, would require a 10- to 100-fold increase in the yield and a corresponding decrease in endogenous biomass decay. Thus, even if the values used are not completely accurate, it is unlikely that the model is incorrect by the two to four orders of magnitude that the replicated qPCR, CARD-FISH, and archaeal lipid data suggest. One of the most influential factors affecting the modeled cell numbers is the cell biovolume term, where clearly smaller cells would lead to higher cell numbers in the calculations. We addressed this issue by determining AEA biovolume empirically from CARD-FISH images. This method has been further verified by tests with cultured AOA to ensure that the applied CARD-FISH procedure does not lead to significant changes in cell diameter. The predicted numbers would be consistent with the experimentally determined cell counts only if we assume a diameter of the AEA cells in plant D of ca. 0.1 μm [compared with the measured 1.4 μm, which is in line with cell sizes of closely related thaumarchaeotes (55, 56)].
The primary conclusion of the modeling analysis is that the abundance of the Thaumarchaeota, determined independently using CARD-FISH, qPCR, and lipid analysis, is far too great to be explained by chemolithoautotrophic ammonia oxidation alone. Consistent with a nonautotrophic lifestyle, in repeated experiments the AEA in the sludge of plant D (and also in plant E) did not incorporate 14C-bicarbonate in the presence of ammonia at levels detectable by MAR. In contrast, in the same experiments, strong 14C-labeling was observed for the AOB, which were present in this sludge at much lower abundance than the thaumarchaeotes (Fig. 2). Thus, if both AOB and AEA solely lived from autotrophic ammonia oxidation in this continuous flow reactor, the AOB with the much higher bicarbonate fixation rate would be expected to outnumber the AEA. However, as the opposite was observed, AEA must consequently possess a different physiology. In accordance with this conclusion, 13C-bicarbonate incorporation into thaumarchaeotal lipids measured by highly sensitive isotope ratio mass spectrometry was also very limited, and could conceivably have occurred as a result of heterotrophic CO2 fixation (57), and was independent of ammonia addition (Table S4). Likewise, ammonia oxidation in sludge from refinery plant D was not inhibited by diphtheria toxin, an inhibitor of archaeal protein synthesis (Fig. S4) (49), and although expression of archaeal amoA genes was detected in refinery plant D samples, this too was independent of added ammonia (Fig. S5).
A role for Thaumarchaeota in nitrification in the refinery D WWTP was further discounted following failure and subsequent recovery of nitrification in the plant. Following a period of more than 5 y where the AEA were stably present in the plant (Table 1, and Dataset S1), AEA were no longer detectable by CARD-FISH. The loss of the thaumarchaeotal population coincided with an incident at the refinery, which required the deployment of fire-fighting foam that entered the refinery WWTP the day before a sampling expedition. This incident induced a period of poor performance, and when we returned to sample the plant following recovery of the treatment plant performance, despite complete re-establishment of nitrification in the system, no Thaumarchaeota could be detected in the sludge. This finding unambiguously confirmed our conclusion that Thaumarchaeota are not essential for ammonia oxidation in this plant.
Taken together, these data strongly suggest that the AOB present in the refinery D WWTP are responsible for the ammonia oxidation in this system, and that the amoA-carrying Thaumarchaeota are in fact not chemolithoautotrophic ammonia oxidizers. If that is the case, what then is their role in this WWTP where they account for a considerable part of the microbial population? One possibility is that these Thaumarchaeota, which represented a single operational taxonomic unit within the group I.1b, are indeed ammonia oxidizers, but can gain additional energy and carbon from other substrates. This theory, however, is inconsistent with the fact that the levels of ammonia oxidation observed are compatible with the population size of AOB, which were demonstrated to be active in chemolithoautotrophic ammonia oxidation in the reactor (Fig. 2 and Table 2). The amoA-carrying group I.1b Thaumarchaeota might also be heterotrophs using some unknown organic compounds present in the wastewater as a carbon and energy source (19, 20).
Taking this theory into account, one piece of data remains apparently inconsistent with a heterotrophic lifestyle for the thaumarchaeotes from the refinery WWTP. Stable carbon isotope analysis of crenarchaeol demonstrated that this was isotopically depleted by about 17‰ relative to inorganic carbon (Table S4). This pattern is consistent with autotrophic carbon fixation and is observed in marine systems, where the group I.1a Thaumarchaeota are considered to be autotrophic (58, 59). However, typically lipids are 13C depleted relative to the carbon source used by between 5‰ and 20‰ (60). Dissolved inorganic carbon in the refinery treatment plant has a δ13C of −27.7‰ and δ13C sludge biomass was −28.4 ± 0.6‰ (Table S4). This finding is consistent with the main sources of organic carbon in the refinery wastewater being products of crude oil processing, as crude oil δ13C can range from −24‰ to −35‰ depending on oil source, maturity, and migration (61). Thus, archaeal lipids with a δ13C of −46.2 ± 1.9‰ would not be inconsistent with heterotrophic growth of amoA-carrying group I.1b Thaumarchaeota on organic by-products of the crude oil refining process.
This finding being the case, what is the potential role of the ammonia monoxygenase (AMO) homolog carried by nonautotrophic group I.1b Thaumarchaeota? Possibly the amoA gene in AEA is an evolutionary relict that is no longer useful for them, but testifies that these organisms evolved from AOA. A similar scenario has been described for some syntrophic bacteria that, although no longer capable of reducing sulfate, still express their dissimilatory sulfite reductase genes that they inherited from their sulfate-reducing ancestors (62). Such a scenario might have emerged in the AEA if they had lost the downstream detoxification and electron extraction machinery (63) or the enzyme inventory for redox cycling via NO (35) required for gaining energy from ammonia oxidation. However, there are also other equally likely explanations. Bacterial AMO display a high substrate flexibility and are well known for cometabolizing hydrocarbons (64), which is also reflected in the evolutionary relatedness of bacterial ammonia- and methane-monooxygenases (63, 65). This finding is in line with the recent discovery of a novel monooxygenase from the AMO and pMMO family that catalyzes butane oxidation (66). As bacterial and archaeal ammonia monooxygenases are phylogenetically highly divergent, it is plausible that within the diversity of archaeal ammonia monooxygenases, some may in fact use substrates other than ammonia (67). Thus, we hypothesize that the Thaumarchaeota in the petroleum refinery plant D could in fact use hydrocarbons or other compounds present in wastewater from crude oil refining processes that are activated by monooxygenases. To test this theory we conducted a FISH-MAR survey using radiolabeled amino acids, pyruvate, acetate, benzoate, and phenol, none of which were assimilated by the thaumarchaeotes in the sludge. However, as refinery-activated sludge is exposed to a vast complexity of compounds, the lack of incorporation of the tested compounds does not exclude a heterotrophic life mode of these thaumarchaeotes.
Given the known relationship between monooxygenase involved in methane and ammonia oxidation, one further possibility is that the Thaumarchaeota might be methane oxidizers. Indeed we have detected mcrA sequences (Fig. S6) and lipids (Table S2) from methanogens in the sludge and have shown that it has potential for biogenic methane generation at low rates (78 ± 1.2 nmols/mL/d), suggesting that methane might be a carbon source for the Thaumarchaeota we detected. However, the δ13C of the thaumarchaeal lipids were not sufficiently 13C-depleted to support this suggestion.
In summary, our data show that not all amoA-carrying Thaumarchaeota are ammonia-oxidizing autotrophs, and several lines of circumstantial evidence point toward them being heterotrophs, which use organic carbon compounds present in the refinery wastewater. Thus, results from metagenomic, metatranscriptomic, or cloning studies based on the detection of putative archaeal ammonia monooxygenase gene sequences or their transcripts in environmental samples, have to be interpreted with caution, because the retrieved DNA or RNA fragments do not necessarily originate from ammonia oxidizers. In this context it is particularly interesting to note that the group I.1b Thaumarchaeota of refinery plant D are closely related (based on 16S rRNA and amoA) to sequences found in various soils (Figs. S1 and S2). Thus, the role of AOA in carbon sequestration in terrestrial systems might be less prominent than recently suggested (9). As generally group I.1b dominates the Thaumarchaeota communities in soil (68), a metabolism other than ammonia oxidation of some members of this group has important implications for our understanding of the contribution of these organisms to N- and C-cycling in terrestrial ecosystems.
Materials and Methods
Detailed methods are provided in SI Materials and Methods.
WWTPs Sampling.
Samples from 52 WWTPs (Dataset S1) were collected and used for activity measurements, lipid analysis, and DNA extraction or fixed with paraformaldehyde for FISH.
Chemical and Microbiological Analyses.
The wastewater treatment plants were screened for amoA and 16S rRNA genes from AEA and AOB, which were quantified using qPCR, fluorescence in situ hybridization, and lipid analysis. The abundances measured were assessed in relation to nitrogen removal in the plants using a nitrification process model and the activity of the AEA and AOB was measured using FISH combined with MAR with a range of 14C-labeled substrates. In addition, AEA activity was assessed from incorporation of 13C inorganic carbon into archaeal lipids and in situ transcription of archaeal amoA.
Nucleotide Accession Numbers.
Sequences of 16S rRNA and amoA gene fragments determined in this study have been deposited at GenBank, with the following accession numbers: HQ316962–HQ316983 (16S rRNA) and HQ316984–HQ317060 (amoA).
Supplementary Material
Acknowledgments
We thank I. Rijpstra, M. Kienhus, C. Schmied, and C. Baranyi for their excellent technical assistance; R. A. Swainsbury and P. Shawcross for sampling of the United Kingdom sludges; A. Sherry and N. Gray for conducting methanogenesis measurements in the sludge; and C. Schleper, M. Stieglmeier, M. Schmid, and M. Könneke for helpful discussions. This research was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior through a studentship to I.B.; and Austrian Science Fund Grant I44-B06 and the European Science Foundation Contract ERAS-CT-2003-980409 of the European Commission, DG Research, FP6 (to H.D. and I.M.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequences reported in this paper have been deposited in the GenBank database [accession nos: HQ316962–HQ316983 (16S rRNA) and HQ316984–HQ317060 (amoA)].
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106427108/-/DCSupplemental.
References
- 1.Ettwig KF, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 2.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 3.Lam P, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 5.Spang A, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 6.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 7.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pratscher J, Dumont MG, Conrad R. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc Natl Acad Sci USA. 2011;108:4170–4175. doi: 10.1073/pnas.1010981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang LM, et al. Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci USA. 2010;107:17240–17245. doi: 10.1073/pnas.1004947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 12.Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nat Rev Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 13.Hallam SJ, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leininger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 15.Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- 16.Reigstad LJ, et al. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol Ecol. 2008;64:167–174. doi: 10.1111/j.1574-6941.2008.00466.x. [DOI] [PubMed] [Google Scholar]
- 17.Ouverney CC, Fuhrman JA. Marine planktonic archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann M, Saunders AM, Schramm A. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl Environ Microbiol. 2009;75:3127–3136. doi: 10.1128/AEM.02806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herndl GJ, et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agogué H, Brink M, Dinasquet J, Herndl GJ. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–791. doi: 10.1038/nature07535. [DOI] [PubMed] [Google Scholar]
- 21.Muller F, Brissac T, Le Bris N, Felbeck H, Gros O. First description of giant Archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ Microbiol. 2010;12:2371–2383. doi: 10.1111/j.1462-2920.2010.02309.x. [DOI] [PubMed] [Google Scholar]
- 22.Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daims H, Wagner M. The microbiology of nitrogen removal. In: Seviour RJ, Nielsen BH, editors. The Microbiology of Activated Sludge. London, United Kingdom: IWA Publishing; 2010. pp. 259–280. [Google Scholar]
- 24.Dang H, et al. Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J Appl Microbiol. 2009;106:1482–1493. doi: 10.1111/j.1365-2672.2008.04109.x. [DOI] [PubMed] [Google Scholar]
- 25.Dang HY, et al. Diversity, abundance and distribution of amoA-encoding archaea in deep-sea methane seep sediments of the Okhotsk Sea. FEMS Microbiol Ecol. 2010;72:370–385. doi: 10.1111/j.1574-6941.2010.00870.x. [DOI] [PubMed] [Google Scholar]
- 26.Park HD, Wells GF, Bae H, Criddle CS, Francis CA. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol. 2006;72:5643–5647. doi: 10.1128/AEM.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells GF, et al. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: Betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol. 2009;11:2310–2328. doi: 10.1111/j.1462-2920.2009.01958.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang T, et al. Occurrence of ammonia-oxidizing Archaea in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J Appl Microbiol. 2009;107:970–977. doi: 10.1111/j.1365-2672.2009.04283.x. [DOI] [PubMed] [Google Scholar]
- 29.Jin T, Zhang T, Yan QM. Characterization and quantification of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in a nitrogen-removing reactor using T-RFLP and qPCR. Appl Microbiol Biotechnol. 2010;87:1167–1176. doi: 10.1007/s00253-010-2595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao YJ, Bakken LR, Zhao LP, Frostegård A. Functional robustness and gene pools of a wastewater nitrification reactor: comparison of dispersed and intact biofilms when stressed by low oxygen and low pH. FEMS Microbiol Ecol. 2008;66:167–180. doi: 10.1111/j.1574-6941.2008.00532.x. [DOI] [PubMed] [Google Scholar]
- 31.Gray ND, Miskin IP, Kornilova O, Curtis TP, Head IM. Occurrence and activity of Archaea in aerated activated sludge wastewater treatment plants. Environ Microbiol. 2002;4:158–168. doi: 10.1046/j.1462-2920.2002.00280.x. [DOI] [PubMed] [Google Scholar]
- 32.Sonthiphand P, Limpiyakorn T. Change in ammonia-oxidizing microorganisms in enriched nitrifying activated sludge. Appl Microbiol Biotechnol. 2011;89:843–853. doi: 10.1007/s00253-010-2902-y. [DOI] [PubMed] [Google Scholar]
- 33.Limpiyakorn T, Sonthiphand P, Rongsayamanont C, Polprasert C. Abundance of amoA genes of ammonia-oxidizing archaea and bacteria in activated sludge of full-scale wastewater treatment plants. Bioresour Technol. 2011;102:3694–3701. doi: 10.1016/j.biortech.2010.11.085. [DOI] [PubMed] [Google Scholar]
- 34.Pickering RL. United Kingdom: Newcastle University, Newcastle upon Tyne; 2008. How hard is the biomass working?: Can cell specific uptake rates be used to optimise the performance of bacterial biomass in wastewater treatment plants? PhD thesis. [Google Scholar]
- 35.Walker CB, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Damsté JS, Schouten S, Hopmans EC, van Duin AC, Geenevasen JA. Crenarchaeol: The characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota. J Lipid Res. 2002;43:1641–1651. doi: 10.1194/jlr.m200148-jlr200. [DOI] [PubMed] [Google Scholar]
- 37.Schouten S, et al. Archaeal and bacterial glycerol dialkyl glycerol tetraether lipids in hot springs of Yellowstone National Park. Appl Environ Microbiol. 2007;73:6181–6191. doi: 10.1128/AEM.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitcher A, et al. Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic group I.1b Archaeon. ISME J. 2010;4:542–552. doi: 10.1038/ismej.2009.138. [DOI] [PubMed] [Google Scholar]
- 39.Gabriel JL, Chong PLG. Molecular modeling of archaebacterial bipolar tetraether lipid membranes. Chem Phys Lipids. 2000;105:193–200. doi: 10.1016/s0009-3084(00)00126-2. [DOI] [PubMed] [Google Scholar]
- 40.Furumai H, Rittmann BE. Evaluation of multiple-species biofilm and floc processes using a simplified aggregate model. Water Sci Technol. 1994;29(10-11):439–446. [Google Scholar]
- 41.Rittmann BE, et al. Molecular and modeling analyses of the structure and function of nitrifying activated sludge. Water Sci Technol. 1999;39(1):51–59. [Google Scholar]
- 42.Prosser JI. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1990;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 43.Hoijnen JJ, van Loosdrecht MCM, Tijhuis L. A black box mathematical model to calculate auto- and heterotrophic biomass yields based on Gibbs energy dissipation. Biotechnol Bioeng. 1992;40:1139–1154. doi: 10.1002/bit.260401003. [DOI] [PubMed] [Google Scholar]
- 44.Tijhuis L, Van Loosdrecht MCM, Heijnen JJ. A thermodynamically based correlation for maintenance gibbs energy requirements in aerobic and anaerobic chemotrophic growth. Biotechnol Bioeng. 1993;42:509–519. doi: 10.1002/bit.260420415. [DOI] [PubMed] [Google Scholar]
- 45.Coskuner G, et al. Agreement between theory and measurement in quantification of ammonia-oxidizing bacteria. Appl Environ Microbiol. 2005;71:6325–6334. doi: 10.1128/AEM.71.10.6325-6334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juretschko S, et al. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belser LW. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;33:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 48.Wagner M, Rath G, Amann R, Koops HP, Schleifer KH. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 49.Rohwer F, Azam F. Detection of DNA damage in prokaryotes by terminal deoxyribonucleotide transferase-mediated dUTP nick end labeling. Appl Environ Microbiol. 2000;66:1001–1006. doi: 10.1128/aem.66.3.1001-1006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee N, et al. Combination of fluorescent in situ hybridization and microautoradiography-a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wuchter C, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 53.Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- 54.Koops H-P, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M. The lithoautotrophic ammonia-oxidizing bacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes. New York: Springer; 2006. pp. 778–811. [Google Scholar]
- 55.Simon HM, Dodsworth JA, Goodman RM. Crenarchaeota colonize terrestrial plant roots. Environ Microbiol. 2000;2:495–505. doi: 10.1046/j.1462-2920.2000.00131.x. [DOI] [PubMed] [Google Scholar]
- 56.Simon HM, et al. Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl Environ Microbiol. 2005;71:4751–4760. doi: 10.1128/AEM.71.8.4751-4760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hesselsoe M, Nielsen JL, Roslev P, Nielsen PH. Isotope labeling and microautoradiography of active heterotrophic bacteria on the basis of assimilation of 14CO(2) Appl Environ Microbiol. 2005;71:646–655. doi: 10.1128/AEM.71.2.646-655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoefs MJL, et al. Ether lipids of planktonic archaea in the marine water column. Appl Environ Microbiol. 1997;63:3090–3095. doi: 10.1128/aem.63.8.3090-3095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuypers MMM, et al. Massive expansion of marine archaea during a mid-Cretaceous oceanic anoxic event. Science. 2001;293:92–95. doi: 10.1126/science.1058424. [DOI] [PubMed] [Google Scholar]
- 60.Hayes JM. Fractionation of carbon and hydrogen isotopes in biosynthetic processes. Rev Miner Geochem. 2001;43:225–277. [Google Scholar]
- 61.Stahl WJ. Carbon and nitrogen isotopes in hydrocarbon research and exploration. Chem Geol. 1977;20:121–149. [Google Scholar]
- 62.Imachi H, et al. Non-sulfate-reducing, syntrophic bacteria affiliated with desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl Environ Microbiol. 2006;72:2080–2091. doi: 10.1128/AEM.72.3.2080-2091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tavormina PL, Orphan VJ, Kalyuzhnaya MG, Jetten MSM, Klotz MG. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 2011;3:91–100. doi: 10.1111/j.1758-2229.2010.00192.x. [DOI] [PubMed] [Google Scholar]
- 64.Hyman MR, Murton IB, Arp DJ. Interaction of ammonia monooxygenase from Nitrosomonas europaea with alkanes, alkenes, and alkynes. Appl Environ Microbiol. 1988;54:3187–3190. doi: 10.1128/aem.54.12.3187-3190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 66.Sayavedra-Soto LA, et al. The membrane-associated monooxygenase in the butane-oxidizing Gram-positive bacterium Nocardioides sp. strain CF8 is a novel member of the AMO/PMO family. Environ Microbiol Rep. 2011;3(3):390–396. doi: 10.1111/j.1758-2229.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- 67.Arp DJ, Yeager CM, Hyman MR. Molecular and cellular fundamentals of aerobic cometabolism of trichloroethylene. Biodegradation. 2001;12:81–103. doi: 10.1023/a:1012089908518. [DOI] [PubMed] [Google Scholar]
- 68.Bates ST, et al. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.