Abstract
Closely related species show a high degree of differences in gene expression, but the functional significance of these differences remains unclear. Similarly, stress responses in yeast typically involve differential expression of numerous genes, and it is unclear how many of these are functionally significant. To address these issues, we compared the expression programs of four yeast species under different growth conditions, and found that the response of these species to stress has diverged extensively. On an individual gene basis, most transcriptional responses are not conserved in any pair of species, and there are very limited common responses among all four species. We present evidence that many evolutionary changes in stress responses are compensated either (i) by the response of related genes or (ii) by changes in the basal expression levels of the genes whose responses have diverged. Thus, stress-related genes are often induced upon stress in some species but maintain high levels even in the absence of stress at other species, indicating a transition between induced and constitutive activation. In addition, ∼15% of the stress responses are specific to only one of the four species, with no evidence for compensating effects or stress-related annotations, and these may reflect fortuitous regulation that is unimportant for the stress response (i.e., biological noise). Frequent compensatory changes and biological noise may explain how diverged expression responses support similar physiological responses.
Keywords: evolutionary conservation, functional evolution, gene regulation, biological function
Evolution of gene expression is a central driving force of phenotypic diversity, and has been extensively studied in recent years (1–11). Although, in some particular cases, interspecies differences in gene expression could be linked to phenotypic differences (3, 7, 8), a puzzling observation from the recent flood of genome-wide comparative transcriptome studies, is that many, if not most, genes diverge in expression even among closely related species with very similar physiologies. Although this has led some to conclude that gene expression diverges primarily by neutral drift and that most expression patterns may not be under strong selection pressures (9, 10), this seems to contrast with a rich body of literature that demonstrates the importance of gene regulation. Work on the yeast stress response has presented a similar conundrum (12–14): Induction of some genes is essential for coping with stress, yet most genes induced in a given stress typically do not contribute to fitness in that stress, as their (individual) deletion does not hinder growth in that stress more than in optimal (rich media) conditions.
How can we reconcile the importance of precise gene regulation with the extensive variability of gene expression among related species or growth conditions for which a large proportion is unlikely to have a fitness contribution? One possibility is that, whereas some genes require precise regulation, others could tolerate a large degree of variability in expression without incurring any phenotypic effect, and that these are the primary source of nonadaptive expression differences between species or conditions. Indeed, expression divergence is typically higher for genes that appear to be less important, such as dispensable genes, duplicated genes, and genes with few interactions (4, 15). According to this possibility, functionally important expression patterns could be detected by conservation across species (11, 16). Alternatively, extensive variability could also influence functionally important expression patterns, if these evolve through compensatory changes that alter specific features of gene expression while maintaining the final outcome.
A related issue concerns the distinction between biological function and “biological noise,” which is defined by reproducible events that occur in living cells due to imperfection (i.e., lack of fidelity) of a biological process (17). Transcriptional noise can arise from nonspecific initiation from sites that yield products of no biological significance and/or from chance occurrences of short DNA sequence motifs that bind transcriptional regulatory proteins. When Saccharomyces cerevisiae is subjected to environmental stress, hundreds (and sometimes more than 1,000) genes are differentially expressed, but it is unclear how many of these are important for coping with that stress. Biological noise is likely to evolve faster than biological function and thus evolutionary conservation should provide important information on this issue.
Here, we compared the stress responses of four yeast species (S. cerevisiae, Kluyveromyces lactis, Candida albicans, and Yarrowia lipolytica) that diverged ∼50–300 million years ago and occupy different niches (Fig. S1). We found that the stress response of these species has diverged extensively. However, we also found that this divergence is compensated by two general mechanisms. First, differences in the response of individual genes are often compensated by the response of other functionally related genes, such that the overall response of functional gene sets is highly conserved. Second, a large proportion of the differences in stress responses reflect transitions between a stress-inducible activity and constitutive activity, which does not compromise the final transcriptional output of these genes during stress. We thus propose that flexibility of the regulatory network enables extensive divergence while maintaining the same phenotypic outcome, and consequently that these four species use similar strategies (i.e., sets of genes) to cope with stresses but that these are accomplished by induction of different subsets of genes.
Results
Experimental Strategy.
We measured genome-wide mRNA expression profiles of the four yeasts at standard rich media (YPD) and upon three environmental stresses, including hyper-osmotic shock (Osmo), heat shock (HS), and carbon starvation (Cstarv) (see Materials and Methods). At each of the four conditions, Poly(A)+ mRNA samples of the four species were mixed, and the mixed samples were subjected to Helicos Single Molecule Sequencing (18) with two biological repeats (Fig. S2). The low sequence similarity among these species enabled us to unambiguously assign most reads (∼60%) from the mixed-RNA sample into a particular genomic position without generating a bias against highly conserved genes (Fig. S2), thereby providing a mean for multiplex sequencing that has not yet been available for the Helicos platform.
Extensive Divergence of Stress Responses Across Four Yeast Species.
Each of the four species responded to each of the three stresses, compared with YPD, by preferentially up-regulating stress genes and down-regulating growth genes (Fig. S3). To examine the conservation of stress responses we compared these genome-wide expression changes (i.e., stress/YPD; Fig. 1 A and B). The highest correlations (r = 0.47 on average) were observed between the responses of the same species to different stresses, consistent with the notion of a general component of the stress response (termed ESR for environmental stress response; refs. 13 and 19–21). Weaker correlations (although still significant, P < 0.05) were observed between the responses of orthologs from different species to the same stresses (r = 0.31 on average), indicating extensive divergence of stress responses. As expected, the lowest correlations were observed between different species responding to different stresses (r = 0.24 on average), but surprisingly, these were only slightly less correlated than the responses of different species to the same stresses, suggesting that the condition-specificity of stress responses has also diverged extensively.
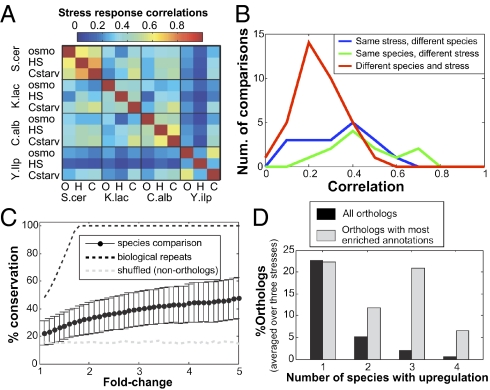
Fig. 1.
Divergence of the yeast stress response. (A) Correlation matrix for the log2-transformed expression responses (YPD/stress) of genes with one-to-one orthologs in each comparison. (B) Distribution of genome-wide correlations for all comparisons of samples from the same species at different conditions (blue), different species at the same stress (green), and different species at different stresses). Each set of correlations is significantly different from zero, and pair of sets is significantly different (P < 0.05). (C) For each threshold (log2 of fold changes), we calculated the percentage of up-regulated genes, by that threshold, in species A that have a similar response (>1.75-fold and P < 0.05) in species B. This analysis was performed for 36 comparisons (12 pairs of species times three stresses) and the plots show the average and SD of the percentages of conserved responses. Black and gray dashed lines reflect a similar analysis of biological repeats (instead of different species) and of a shuffled dataset (such that the comparison is performed between nonorthologs from the same species comparisons), respectively. (D) For the indicated number of species, the percentage of all orthologs (black bars) or orthologs with the most enriched annotations (gray bars) showing up-regulation as averaged over the three stresses.
To further examine the divergence of stress responses, we defined, for each stress and each species, a set of up-regulated and down-regulated genes and asked what fraction of these expression changes are conserved among the species. On average, a gene that is up-regulated (or down-regulated) in one species by at least twofold at a given stress has only ∼30% chance to have a similar response to that stress (>1.75-fold) in another species, and these values vary between ∼10% to ∼50% depending on the exact combination of species and stress condition (Fig. 1C). Notably, approximately half of this apparent conservation is expected simply by chance, as analysis of shuffled gene pairs (i.e., nonorthologous) identifies ∼16% conserved stress responses compared with ∼30% observed for orthologs (gray dashed line, Fig. 1C). The degree of conservation increased for stronger responses, but remained low even for the most highly responsive genes. For example, genes with more than fivefold up-regulation in one species had, on average, less than 50% chance to be up-regulated (>1.75-fold) in another species (Fig. 1C).
The extensive divergence of stress responses is further evident in the very low percentages of stress responding genes whose response is conserved among more than two species. Among the 1,819 genes with one-to-one orthologs among all four species, there are ∼400 genes that are up-regulated in a given condition only in one species, ∼100 genes that respond at two species, and only ∼40 and ∼10 genes whose up-regulation is conserved among three or four species, respectively (Fig. 1D). A list of genes that responds to each stress in at least three of the four yeast species is given in Table S1. As expected, these genes are highly enriched with stress-related functional annotations (e.g., protein folding, autophagy, and carbohydrate metabolism) supporting evolutionary conservation as a method to identify functionally important regulation. However, due to the extensive divergence, these conserved responses reflect only a small minority of the genes with such functional annotations (Fig. 1D), suggesting that a major caveat of the conservation method is that, while it enriches for functionally important responses, it would often exclude many, if not most, of these responses.
Divergence of the General Stress Response.
We next examined the conservation of the common response to different environmental changes. The general S. cerevisiae ESR has been studied extensively (13, 19–21) and is evident in our data by the high correlations between the genome-wide responses of S. cerevisiae to the three stresses and by the large number of genes that are either down-regulated or up-regulated in all three stresses. However, both of these measures are significantly (P < 0.01) lower in the other three species (Fig. 2A), suggesting that the general component of the stress response (i.e., ESR) is less dominant in these species compared with S. cerevisiae, consistent with a previous report on C. albicans (22).
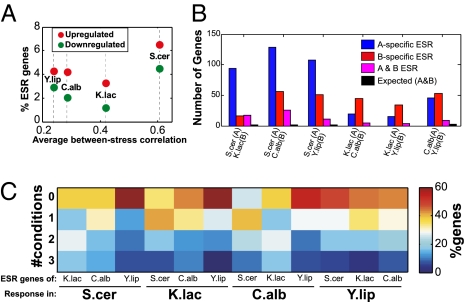
Fig. 2.
Divergence of the general stress response. (A) For each species, the y axis shows the number of predicted ESR genes (up-regulated at all three stresses or down-regulated at all three stresses by at least 1.75-fold), and the x axis shows the average correlation among the expression responses of that species to the three stresses. (B) For each pair of species (A and B), bars show the number of ESR genes specific to A, specific to B, common to A and B, and the expected number of common ESR genes (estimated by random sampling). Each analysis was restricted to the genes with one-to-one orthologs among the pair of species being compared. (C) Percentages of genes up-regulated in zero through three of the stress conditions in the species indicated at the bottom (bold), among the orthologs of the up-regulated ESRgenes in the species indicated above.
ESR genes respond to multiple stresses and therefore might be expected to be most relevant to cope with stress and to have maximal conservation among species. The analysis above demonstrates that the number of ESR genes varies greatly among the species. In addition, we find that the identity of ESR genes is also surprisingly different. For each pair of species (A and B), the number of common ESR genes is significantly smaller than the number of species-specific ESR genes that have one-to-one orthology (P < 0.01, Fig. 2B). Similar results are obtained when we separately analyze up-regulated and down-regulated ESR genes, or when we examine S. cerevisiae ESR genes as previously defined from a larger compendium of stress conditions (13), indicating that the apparent divergence of ESR genes is not due to the use of only three stress conditions in this work (Fig. S4).
We next asked for each pair of species (A and B) whether ESR genes from species A maintain a partial stress response (i.e., respond to a subset of the conditions) in species B. Strikingly, up-regulated ESR genes from one species were most frequently (on average, 45%) responding to zero conditions in another species and least frequently (on average, 9%) responding to three conditions in another species (Fig. 2C). Thus, ESR genes from one species are still more likely (P < 0.01) to respond to stresses in another species than random genes (for which, on average, 82% do not respond to any stress and only 2.5% respond to three stresses), yet they usually lose their responsiveness to most stress conditions. The observed divergence in up-regulated ESR genes therefore does not reflect a threshold effect or the loss/gain of response to individual stresses but rather mostly “all or none” evolutionary transitions. The patterns of down-regulated ESR genes were less uniform among the different pairwise species comparisons (see Fig. S5).
Higher Conservation of the Response of Functional Gene Sets.
The extensive divergence between the stress responses of the four species may indicate that they use different physiological mechanisms to cope with the same stresses, and some specific interspecies may support this possibility. For example, consistent with previous work (8), oxidative phosphorylation genes were up-regulated during stress in S. cerevisiae but were primarily unchanged or down-regulated during stress in the other species (Fig. 3A). Similarly, proteosome subunits were largely unaffected by stress in most species but were specifically up-regulated upon heat shock in C. albicans (Fig. 3A), which may reflect a unique physiology of C. albicans. However, these coherent changes among the stress responses of functionally related genes were extremely rare. In contrast, the average responses of most functional gene sets (based on GO annotations) were highly conserved among the four species. For example, Fig. 3B shows a low correlation between the heat-shock responses of S. cerevisiae and C. albicans across all one-to-one orthologs (Fig. 3B Left), but a much higher correlation for the average response of functional gene sets (Fig. 3B Center). On average, interspecies correlations increased almost twofold, from 0.33 over individual orthologs to 0.62 for functional gene-sets (Fig. S6). These results suggest that the different species have a similar overall response in terms of activating and repressing the same pathways and complexes, yet the finer details of these responses have diverged extensively. We note, however, that the increased correlations of functional gene sets, compared with individual orthologs, may partially also reflect the reduction of experimental noise when averaging over the responses of related genes, as a similar effect, albeit weaker, is observed in comparison of biological repeats (Fig. S6).
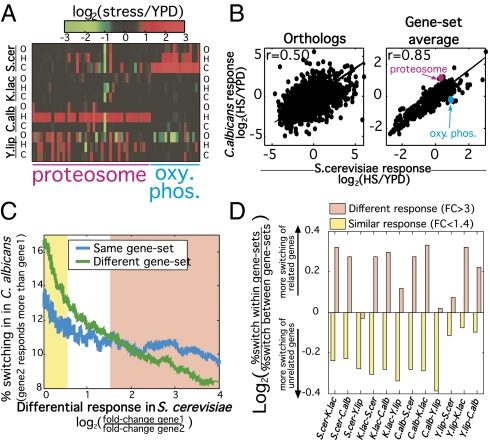
Fig. 3.
Conservation and divergence of functional gene-sets. (A) Heat map of the expression response of genes from two functional gene sets (proteosome and oxidative phosphorylation) at the four species and three stresses. Genes with one-to-one orthologs in at least three species were included, and missing values were given an expression response of zero. (B) Scatterplot of the heat shock expression response of S. cerevisiae and C. albicans over all one-to-one orthologs (Left) and the average response of functional gene-sets (Right). Functional gene sets correspond to GO terms with at least 20 and at most 100 annotated genes in S. cerevisiae. Lines display linear least-square fits. (C) Sliding window analysis of the percentage of gene pairs from the same (blue) or from different (green) functional gene sets, whose difference in response to a given stress switch signs between S. cerevisiae and C. albicans (i.e., if gene 1 has a higher response in S. cerevisiae, then gene 2 has a higher response to the same stress in C. albicans). The gene pairs are ordered by their difference in stress response in S. cerevisiae, defined as log2 ratio of their stress/YPD fold-changes, including each of the individual stresses (i.e., each gene pair was considered three times). Yellow and pink regions mark the gene pairs with a similar (fold-change < 1.4) and different (fold-change > 3) stress responses in S. cerevisiae, respectively. (D) The analysis in C was repeated for all pairwise comparisons. Bars show the log2 ratios of the frequency of switching signs among gene pairs from the same vs. from different functional sets. Yellow and pink bars reflect gene pairs with a similar (fold-change < 1.4) and different (fold-change > 3) stress responses in the first species of the pair (as written in the x axis label).
These results may indicate that genes within a functional set often evolve by compensatory changes in gene expression. This finding is consistent with previous work which suggested that activation of many complexes along the cell cycle is orchestrated by regulation of specific subunits (whereas others maintain a stable expression level) and that the identity of the regulated subunit changes during evolution, whereas the patterns of complex activity is highly conserved (23). Thus, although genes within a functional set (e.g., complex or pathway) tend to be coregulated, there is also ample variability in the responses of individual genes within a functional set that may be subject to extensive divergence. To further test this possibility, we asked whether the variability in stress responses among genes within a functional set is more or less conserved than the variability in stress responses among genes from different functional sets.
We examined the proportion of gene pairs that “switch signs” (the gene with higher response in one species has a lower response in the other species by at least 1.75-fold), for gene pairs from the same or from different functional sets. When examining genes with similar stress responses, we found, as expected, that gene pairs from the same functional set tend to coevolve and thus are less likely to switch signs compared with genes from different functional sets (e.g., Fig. 3C, yellow region). Strikingly, however, an opposite effect is observed for gene pairs with different stress responses (e.g., Fig. 3C, pink region). For these gene pairs, the probability to switch signs is in fact higher if they are from the same functional gene set then if they are from different functional gene sets. Preferential switching of genes within, compared with between, functional sets was significant (P < 0.05) in 10 of the 12 species comparisons (Fig. 3D). Thus, functionally related (but differentially expressed) genes tend to evolve by compensatory changes (i.e., switch signs) and the impact of this tendency is larger than their tendency to coevolve as a result of common regulation.
Evolutionary Transitions Between Regulated and Constitutive Expression.
Interspecies differences in the stress response of some genes may be adaptive and reflect the different physiologies of these species, as appears to be the case for oxidative phosphorylation genes. The stress responses of other genes that differ between species may also be important, but their absence in some species could be compensated. One level of compensation is the response of functionally related genes, as described above. An additional level of compensation may be acting at individual genes if, for example, these genes are regulated by posttranscriptional processes (24, 25). To examine this possibility, we first asked whether genes which are up-regulated upon stress in some species, but not in S. cerevisiae, will be enriched with S. cerevisiae translational up-regulation upon the same stress. Using data on changes in S. cerevisiae ribosome association upon stress (26, 27), we indeed found evidence for such effects in hyperosmotic shock but not in heat shock (Fig. S7).
To further examine the interplay between transcriptional and posttranscriptional regulation, we next correlated translational efficiency, as estimated by patterns of codon use (28), with mRNA stress response for different pairs of species. Interestingly, we found that genes that respond to stress only in species B tend to have higher translational efficiency in species A than in B, although this effect is weak (Fig. S8). Because codon use presumably reflects translational efficiency at basal conditions (before stress), this result suggests that absence of a stress response may be compensated to some extent by increased basal activity. Therefore, this prompted us to investigate whether there are also compensating mechanism(s) at the level of mRNA. For example, if a gene is required at high mRNA level during some stress, then that level can be achieved either by induction from a low basal mRNA level or by having high basal mRNA level with no induction upon stress.
We compared the interspecies differences in stress responses to those in basal mRNA expression levels (as measured in YPD). To avoid a potential technical artifact whereby errors in the estimation of YPD levels generate a complementary error in the estimation of stress responses [which are computed as log2(stress/YPD)], we used different datasets (biological repeats) to calculate the YPD levels and the stress responses. Indeed, we found that interspecies differences in stress response are strongly associated with differences in basal mRNA levels. For example, among the 183 genes that respond to heat shock in S. cerevisiae but not in K. lactis, 67 genes (37%) have higher basal mRNA levels in K. lactis than in S. cerevisiae (by at least 1.75-fold), whereas only 20 genes (11%) display the opposite pattern (i.e., higher basal levels in S. cerevisiae than in K. lactis; Fig. 4A). Similarly, among the 166 genes that respond to heat shock in K. lactis but not in S. cerevisiae, 56 genes (34%) have higher basal mRNA levels in S. cerevisiae than in K. lactis, whereas only 20 genes (13%) display the opposite pattern. Thus, differences between the response of S. cerevisiae and K. lactis to heat shock are significantly associated with changes in basal mRNA levels (P < 10−4 in both cases) such that genes that are not induced in one species have elevated expression level in that species even without heat shock, thereby compensating for the lack of stress-specific induction. A similar, yet considerably stronger, effect is observed for the carbon starvation response in S. cerevisiae vs. Y. lipolytica, where ∼60% of the genes that respond only in one of the species have higher basal level in the other species, whereas only ∼10% display the opposite pattern (Fig. 4B).
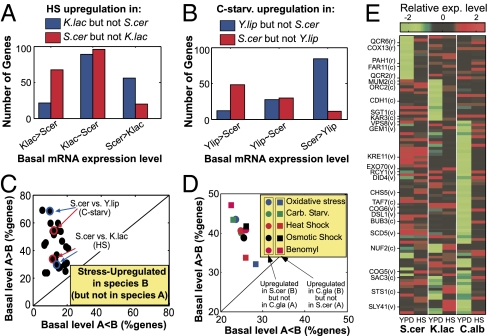
Fig. 4.
Interplay between divergence of stress response and basal levels. (A) Genes up-regulated (>2-fold) during heat shock at K. lactis but not up-regulated (<1.4-fold) at S. cerevisiae (blue bars), and genes up-regulated at S. cerevisiae but not at K. lactis (red bars), were classified according to the difference in basal (YPD) mRNA levels between S. cerevisiae and K. lactis (K.lac∼S.cer refers to a difference smaller than 1.75-fold). (B) Same as A for comparison of S. cerevisiae with Y. lipolytica in response to carbon starvation. (C) For each pair of species (A and B) and at each of the three stresses, we identified genes that respond to that stress in species B (>twofold) but do not respond in species A (<1.4-fold) and calculated the percentage of these genes in which the basal (YPD) expression level is higher in A than in B (y axis) or higher in B than in A (x axis) by at least 1.75-fold. For all 36 comparisons (12 species pairs times 3 stresses), the percentage of A > B was larger than that of A < B (i.e., above the diagonal). The four circles that represent the examples given in (A) and (B) are highlighted. (D) We identified genes that respond to one of four environmental stresses or a chemical stress (Benomyl) either in S. cerevisiae but not in Candida glabrata (circles), or in C. glabrata but not in S. cerevisiae (squares). The percentages of genes in which basal mRNA level is higher in the species without a response (A > B) or in the species with a response (A < B) are shown at the y axis and x axis, respectively. In all 10 cases, the percentage of A > B is higher than that of A < B, and in 7 of the cases this enrichment is statistically significant (P < 0.05). (E) Examples of genes that respond to heat-shock only in a subset of the species but have similar expression levels during heat-shock. The log2 expression levels of each gene are shown for S. cerevisiae, K. lactis, and C. albicans in YPD and heat shock, after normalizing to mean zero. Gene names are shown for genes annotated as respiration (r), cell cycle (c), or vesicle-mediated transport (v). This analysis did not include Y. lipolytica for simplicity, as this species had the most diverged expression patterns.
We repeated the above analysis for each of the 12 pairs of species (A and B) and each of the three stresses, and examined the genes that respond to the respective stress in species B but not in species A, and have higher basal level either in A (A > B) or in B (A < B) by at least 1.75-fold. Strikingly, in all comparisons with at least 30 genes ,we found a higher percentage of (A > B) genes, consistent with a compensating effect, and these enrichments were significant (P < 0.05) for 25 of the 27 comparisons. On average (over all comparisons), 43% of the differentially up-regulated genes had higher expression in the species without a response, whereas only 13% had a lower expression in that species, indicating that compensation by basal mRNA levels may account for a large proportion of the interspecies differences in up-regulation. These results could not be accounted by the low basal levels of stress-responsive genes, as control genes with the same basal levels (but no stress response) did not have a similar increase in basal levels at other species (Fig. S9). Similar analysis for the genes that are down-regulated in species B but not in species A showed the same effect, although quantitatively weaker (Fig. S9).
To further test the generality of these results, we sought to analyze interspecies expression differences with other independent datasets. First, we repeated the above analysis when basal mRNA levels of S. cerevisiae, C. albicans, and Y. lipolytica are taken from a previous work (29) and obtained similar results (Fig. S9). Second, we compared the expression programs of S. cerevisiae and C. glabrata using microarray datasets from multiple previous studies, including basal expression levels (29) and response to environmental (13, 30) and chemical (31) stresses. Remarkably, combining these diverse datasets and repeating the above analysis, we again found that interspecies differences in the response to each of these five different stresses are preferentially associated with compensation by basal mRNA levels (Fig. 4D). Taken together, our findings indicate that expression of many yeast stress genes diverge during evolution by altering the degree by which their activation is induced versus constitutive.
Nonconserved Stress Responses May Reflect Biological Noise.
In addition to interspecies differences that are either adaptive or compensated, other nonconserved responses may reflect biological noise, e.g., due to chance occurrences of regulatory sequence motifs that evolved by neutral drift (17). On average, 30% of the genes that are up-regulated in a given species and condition are up-regulated only in that species and not in the three other species (species-specific response, defined as >twofold in one species and <1.4-fold in all others). Of these, approximately half show evidence of compensation, whereas the rest (i.e., ∼15% of all responding genes) are not associated either with compensation or with stress-related functional annotations. Notably, promoters of these species-specific response genes are enriched with transcription factor binding sites that are not conserved even among the closely related Saccharomyces sensu-stricto species, consistent with the possibility that stress responses of many of these genes may reflect biological noise through chance occurrences of sequence motifs (Fig. S10). Taken together, our results suggest that yeast stress responses diverged extensively through compensatory changes and biological noise and thus that the response (or lack thereof) of genes to a given stress should be interpreted with caution.
Discussion
Our comparison of the responses of four yeast species to three stresses revealed extensive divergence. A potential caveat of this and other interspecies studies is that the same conditions might not induce the same responses of different species, as some species are more or less adapted to these conditions. Accordingly, it could be argued that, for some species, YPD is already stressful or one of the stress conditions is not stressful, and thus that we are not capturing their stress responses and interpret this as constitutive activation. However, several evidences argue against this interpretation. First, all species mount a large-scale response to each of the stresses that involves preferential up-regulation of stress genes and down-regulation of growth genes (Fig. S3). Second, we observed an increased conservation when examining the average response of functional gene sets, suggesting that the species do have a similar functional response to these stresses. Third, our results are consistent among many different pairwise species comparisons and among three different stresses. It is difficult to imagine that this differential stress sensitivity affects each pairwise comparison and at each of the conditions. Fourth, in some analyses, we considered the direction of expression divergence and obtained the same results when switching the identity of the species (e.g., in Fig. 4 every species was defined once as A and once as B in each pairwise comparison and for each stress; in every case, we found an enrichment of genes that are induced in B but have constitutively high levels in A). Thus, although there likely is some variability between the species' sensitivity to the tested conditions, the global patterns that we observe are unlikely to reflect such effects.
At face value, the observed divergence may indicate that yeasts deal with stresses differently. However, our results suggest that they may in fact be using a similar set of genes to cope with stresses, but the expression patterns of these genes has diverged extensively through compensatory changes. Such compensation could occur by activation of different genes in the same pathway or complex (23) (Fig. 3), by interplay between transcriptional and posttranscriptional regulation, which we only partially address here (see Figs. S7 and S8), or through a mechanism in which induced genes in some species are constitutively active in other species (Fig. 4).
This last compensatory mechanism is consistent with the possibility that, for many stress-related genes, mRNA levels during stress are constrained whereas their (presumably less functional) basal levels evolve faster, thereby enabling rapid divergence of the expression response to stress without compromising the activity of these genes during stress. For example, Fig. 4E shows the relative expression levels of 95 genes in S. cerevisiae, K. lactis, and C. albicans, at YPD and heat shock. Each of these genes responds to heat shock only in a subset of the species yet has similar expression levels upon heat shock among the three species (and thus differs among the species mostly in basal YPD levels). These genes are enriched with aerobic respiration (“r,” up-regulated primarily in S. cerevisiae), cell cycle regulation (“c,” up-regulated primarily in K. lactis), and vesicle-mediated transport (“v,” up-regulated primarily in C. albicans).
Concomitant changes in a gene's stress response and basal level could most easily be realized by changing only its basal level but without affecting its level upon stress. For example, if a gene is repressed at rich media but this repression is relieved during stress, then mutations that eliminate the repression mechanism would both increase its basal level and decrease its stress response but not affect the level during stress. Such loss of repression mechanisms may account for the interplay that we observe between stress response and basal levels. However, induction of stress-response genes is often accomplished by activating signals, such as transport of stress-related TFs (e.g., MSN2/4) from the cytoplasm to the nucleus or their activation through other means (32). Mutations that affect induction of genes through these stress-specific signals are expected to affect only the stress response of such genes but not their basal levels. Therefore, we expect that many genes evolved by two complementary sets of mutations (those affecting their response to stress and those affecting their basal levels) and that the combined influence of these mutations generates transitions between an inducible stress response and constitutive activation.
Transitions between induced and constitutive expression may reflect physiological differences between these yeast species. For example, although the three other species generate energy primarily by respiration, S. cerevisiae preferentially ferments glucose thus and maintains low levels of respiration genes during rapid growth, which are induced upon stress (8). For other genes, however, such transitions may not reflect physiological differences. For example, a gene that is required only during stress might be stress-induced in one species but constitutively expressed in another species. Widespread nonfunctional basal expression is consistent with recent observations in mammalian cells that numerous genes are expressed in tissues where they have no function (i.e., ectopic expression; refs. 17 and 33–35). Nonfunctional basal expression may reflect expression leakage as a result of neighboring genes or regulatory mutations that accumulated through neutral drift; or it may be beneficial, particularly to microbes that face constantly changing environments, as it relieves the need to “infer” when and to what extent genes should be induced, which might be a complex and error-prone process.
Approximately 15% of the stress-regulated genes behave in a species-specific manner and show no evidence of functional annotations related to stress responses. Regulation of this class of genes could be due to evolutionary adaptations or compensation by as yet unknown mechanisms, but it is also possible that these regulatory events represent biological noise. In this context, biological noise refers to regulatory events in living cells (i.e., are not experimental error) that occur fortuitously and are functionally irrelevant to the stress response. A simple mechanism for such biological noise stems from the fact that DNA binding motifs for transcriptional regulatory factors are short and occur frequently by chance. For example, a typical motif has specificity equivalent to six absolutely defined base pairs and can occur at either one of the DNA strands, and this would occur at a frequency of ∼1/2,000 bp by chance. As binding sites for a given factor should function within a ∼250 bp region corresponding to the promoter, this means that a random gene has a ∼12% chance to be affected by a given transcription factor. Most stress responses are mediated by multiple transcription factors, so the observed value of 15% of species-specific regulation is consistent with biological noise. Although these considerations do not prove that such species-specific regulatory events are indeed due to biological noise, it is a plausible explanation and we suspect that at least some of these regulatory events are indeed functionally insignificant.
Materials and Methods
RNA Sequencing.
Yeast strains (Table S1) were grown at 28 °C in YPD medium until midlog phase (OD600 ∼ 0.6), and then separately subjected to osmotic shock (0.4 M NaCl at 30 °C for 5 min), heat shock (39 °C for 10 min), or carbon starvation (YP medium for 6 h). Equal amounts of poly(A)-containing RNA from different yeasts from the same growth conditions were pooled together before reverse transcription and library preparation and subjected to RNA sequencing with a HeliScope sequencer (Helicos). Raw sequencing data (NCBI accession number SRA029166.1, SRP005689) were mapped against the processed combined reference genome of the four yeasts using HeliSphere software (DGE pipeline) and then assigned to the individual species.
Comparison of Expression Responses.
Expression levels at each condition were averaged over two biological repeats for S. cerevisiae and K. lactis. For C. albicans and Y. lipolytica, where one replicate is of lower quality than the other, we used only the apparent higher-quality dataset in most of analyses, although averaging of the two replicates did not change our main results (Fig. S11). Log2 ratios of expression in each stress with respect to YPD were centered to a mean of zero. Response to a particular stress was defined as a ratio > 1.75-fold (increase or decrease) and P value (binomial test) below 0.05, and ESR genes were defined as those responding to all three stresses in a given species. Species-specific responses were defined as genes with >2-fold (and P < 0.05) response in one species and <1.4-fold in others. Pairwise species comparisons used one-to-one orthologs. Details on the experimental and bioinformatic analyses are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Oliver Rando for insightful comments on the manuscript. This work was supported by National Institutes of Health Grant GM30186 (to K.S.) and a grant from the Binational Science Foundation and the European Research Council (Ideas) (to N.B.). I.T. is supported by the Clore Center of the Weizmann Institute of Science. K.H.W. is supported by the Croucher Foundation Fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the RNA-seq database (NCBI accession nos. SRA029166.1 and SRP005689).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113718108/-/DCSupplemental.
References
- 1.Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- 2.Thompson DA, Regev A. Fungal regulatory evolution: cis and trans in the balance. FEBS Lett. 2009;583:3959–3965. doi: 10.1016/j.febslet.2009.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci USA. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 5.Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- 7.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 8.Ihmels J, et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 9.Yanai I, Graur D, Ophir R. Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. OMICS. 2004;8:15–24. doi: 10.1089/153623104773547462. [DOI] [PubMed] [Google Scholar]
- 10.Khaitovich P, et al. A neutral model of transcriptome evolution. PLoS Biol. 2004;2:E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanai I, Hunter CP. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res. 2009;19:2214–2220. doi: 10.1101/gr.093815.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 13.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JJ, et al. Expression and functional profiling reveal distinct gene classes involved in fatty acid metabolism. Mol Syst Biol. 2006;2:0009. doi: 10.1038/msb4100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tirosh I, Barkai N. Evolution of gene sequence and gene expression are not correlated in yeast. Trends Genet. 2008;24:109–113. doi: 10.1016/j.tig.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 16.McCarroll SA, et al. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 17.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 18.Lipson D, et al. Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol. 2009;27:652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 19.Berry DB, Gasch AP. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell. 2008;19:4580–4587. doi: 10.1091/mbc.E07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasch AP. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast. 2007;24:961–976. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- 21.Gasch AP, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- 22.Enjalbert B, Nantel A, Whiteway M. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol Biol Cell. 2003;14:1460–1467. doi: 10.1091/mbc.E02-08-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen LJ, Jensen TS, de Lichtenberg U, Brunak S, Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- 24.Laurent JM, et al. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics. 2010;10:4209–4212. doi: 10.1002/pmic.201000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leach MD, Stead DA, Argo E, MacCallum DM, Brown AJ. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol Microbiol. 2011;79:1574–1593. doi: 10.1111/j.1365-2958.2011.07542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halbeisen RE, Gerber AP. Stress-Dependent Coordination of Transcriptome and Translatome in Yeast. PLoS Biol. 2009;7:e105. doi: 10.1371/journal.pbio.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preiss T, Baron-Benhamou J, Ansorge W, Hentze MW. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat Struct Biol. 2003;10:1039–1047. doi: 10.1038/nsb1015. [DOI] [PubMed] [Google Scholar]
- 28.Man O, Pilpel Y. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet. 2007;39:415–421. doi: 10.1038/ng1967. [DOI] [PubMed] [Google Scholar]
- 29.Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roetzer A, et al. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol Microbiol. 2008;69:603–620. doi: 10.1111/j.1365-2958.2008.06301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lelandais G, et al. Genome adaptation to chemical stress: clues from comparative transcriptomics in Saccharomyces cerevisiae and Candida glabrata. Genome Biol. 2008;9:R164. doi: 10.1186/gb-2008-9-11-r164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Görner W, et al. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldmesser E, et al. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 2006;7:121. doi: 10.1186/1471-2164-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodríguez-Trelles F, Tarrío R, Ayala FJ. Is ectopic expression caused by deregulatory mutations or due to gene-regulation leaks with evolutionary potential? Bioessays. 2005;27:592–601. doi: 10.1002/bies.20241. [DOI] [PubMed] [Google Scholar]
- 35.Yanai I, et al. Similar gene expression profiles do not imply similar tissue functions. Trends Genet. 2006;22:132–138. doi: 10.1016/j.tig.2006.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.