Abstract
Centromere protein A (CENP-A) is a histone H3 variant that marks centromere location on the chromosome. To study the subunit structure and folding of human CENP-A-containing chromatin, we generated a set of nucleosomal arrays with canonical core histones and another set with CENP-A substituted for H3. At the level of quaternary structure and assembly, we find that CENP-A arrays are composed of octameric nucleosomes that assemble in a stepwise mechanism, recapitulating conventional array assembly with canonical histones. At intermediate structural resolution, we find that CENP-A-containing arrays are globally condensed relative to arrays with the canonical histones. At high structural resolution, using hydrogen-deuterium exchange coupled to mass spectrometry (H/DX-MS), we find that the DNA superhelical termini within each nucleosome are loosely connected to CENP-A, and we identify the key amino acid substitution that is largely responsible for this behavior. Also the C terminus of histone H2A undergoes rapid hydrogen exchange relative to canonical arrays and does so in a manner that is independent of nucleosomal array folding. These findings have implications for understanding CENP-A-containing nucleosome structure and higher-order chromatin folding at the centromere.
Keywords: hydrogen exchange, epigenetics
The centromere is the control locus that directs the faithful inheritance of eukaryotic chromosomes at cell division (1, 2). The histone H3 variant, CENP-A, is a highly conserved constituent of all eukaryotic centromeres and is the most attractive candidate for carrying the epigenetic information that specifies the location of the centromere (3). Recent findings have led to several fundamentally different proposals for how CENP-A marks centromere location: (i) CENP-A confers structural and dynamic changes to octameric nucleosomes (4, 5); (ii) CENP-A confers alterations of nucleosomal histone stoichiometry (6, 7), including the incorporation of nonhistone proteins into nucleosome-like structures (8); (iii) CENP-A directs the reversal of handedness of DNA wrapping from left to right (9). The nature and composition of CENP-A-containing nucleosomes remain controversial and areas of intense investigation. The available data regarding their structure, however, could be reconciled by distinct CENP-A-containing complexes existing over the course of a cell cycle-coupled maturation program of newly expressed CENP-A protein that propagates the epigenetic centromere mark (10).
In the bulk chromatin fiber, nucleosome–nucleosome interactions are central to packaging eukaryotic DNA into the nucleus, to compacting chromosomes during mitosis, and to organizing functional subchromosomal domains (11). Although much is known about how chromatin fibers condense in vitro, the extent to which the structured helical histone core of the nucleosome is physically impacted by contact with neighboring nucleosomes in a folded chromatin fiber is not yet known. Eukaryotic centromeres are composed of lengthy arrays of CENP-A-containing nucleosomes. An exception is the budding yeast point centromere that harbors a single CENP-A-containing nucleosome (12, 13). Despite the central role of the specialized CENP-A-containing nucleosomal array in specifying centromere location and directing chromosome inheritance, the internucleosomal interactions of nucleosomal arrays in which CENP-A replaces canonical histone H3 are completely unexplored.
To address the subunit structure and folding of CENP-A-containing nucleosomal arrays, we couple folding measurements using analytical ultracentrifugation (AUC) with mass spectrometry-based hydrogen/deuterium exchange (H/DX-MS). The AUC studies measure the bulk behavior of the arrays, and we find that CENP-A-containing arrays are somewhat more condensed upon folding than canonical arrays. H/DX-MS is an approach capable of measuring the dynamic behavior of the polypeptide backbone of each histone in the nucleosome core. Prior H/DX-MS experiments with subnucleosomal (CENP-A/H4)2 heterotetramers (14) and CENP-A-containing mononucleosomes (4) found that the CENP-A/H4 interface is substantially rigidified by side-chain interactions that restrict transient unfolding of the contacting α-helices (5). In the present study, we demonstrate that the αN-helices of canonical H3-containing nucleosomes are substantially restricted (50- to 100-fold) at their superhelical termini upon nucleosome array folding, indicating an unexpected consequence of nucleosome-nucleosome interactions during chromatin folding. Importantly, both the initial rigidity of CENP-A at its own αN-helix and the rigidity imposed upon chromatin folding are reduced compared to its conventional counterpart containing H3, indicating looseness at the nucleosome superhelical termini.
Results
Stepwise Assembly of CENP-A-containing Polynucleosome Arrays.
Conventional nucleosomes and nucleosomal arrays can be assembled by adding the four core histones at the appropriate molar ratios to defined DNA templates consisting of nucleosome positioning sequences in order to saturate all of the nucleosome binding sites. This is followed by salt dialysis assembly from 2 M to 2.5 mM NaCl (15–17). The stepwise assembly is characterized by the initial binding of the (H3/H4)2 heterotetramer to DNA to form a tetrasome as the [NaCl] is lowered to 1 M, followed by binding of H2A/H2B heterodimers to the tetrasome to complete nucleosome formation as the [NaCl] is lowered to ≤ 0.6 M (16). Whether CENP-A nucleosomes assemble by such a stepwise mechanism has not been tested.
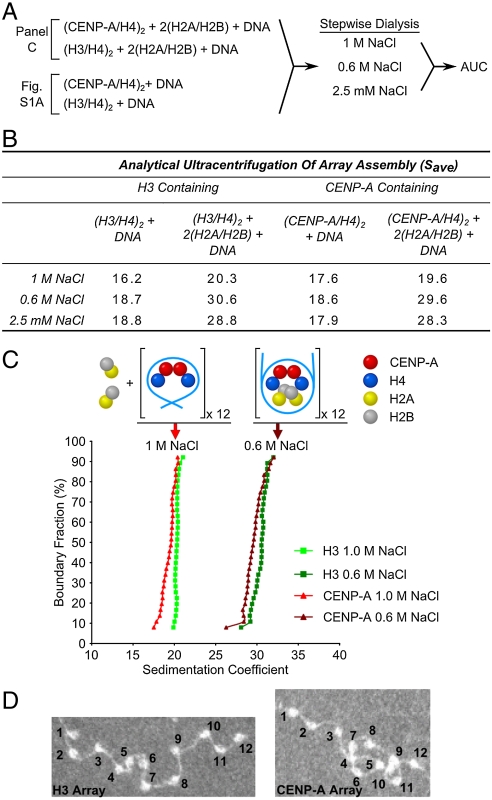
To determine if CENP-A-containing nucleosomal arrays assemble via the same sequential pathway as canonical arrays, samples that contained (CENP-A/H4)2 tetramers, (H2A/H2B) dimers, and a linear DNA template containing twelve tandem copies of the so-called 601 sequence (18, 19) were mixed in 2 M NaCl and then successively dialyzed against buffer containing 1 M NaCl, 0.6 M NaCl and 2.5 mM NaCl. At each dialysis step, the reactions were assayed by AUC (Fig. 1 A–C and Fig. S1 A and B). AUC has been successfully employed in classic sedimentation velocity experiments to measure physical properties of nucleosomal arrays including array composition as well as global changes that accompany intraarray nucleosome–nucleosome interactions and interarray oligomerization (20). The CENP-A-containing samples were compared with otherwise identical samples containing canonical (H3/H4)2 in place of (CENP-A/H4)2, as well as samples that contained only (H3/H4)2 (Fig. 1 A and B). As expected, the canonical H3-containing nucleosomal arrays assembled as previously reported, with the (H3/H4)2 tetramers bound to each DNA repeat at 1 M NaCl to form a 16–20 S 12-mer tetrasomal array (we measured the free 12-mer DNA template sedimenting at approximately 13 S). This is followed by dimer binding at 0.6 M NaCl to generate the completely assembled 28–30 S beads-on-a string species also present in low salt (i.e., 2.5 mM NaCl) (Fig. 1 B and C and Fig. S1 A and B). Similarly, for the centromeric counterpart complexes, the (CENP-A/H4)2 tetramer binds to DNA in 1 M NaCl and forms a 17–20 S tetrasomal array (Fig. 1B and Fig. S1A). In reactions that contain (CENP-A/H4)2 tetramers as well as H2A/H2B dimers the H2A/H2B binding is prevented by the presence of 1 M NaCl and we still observe a 17–20 S array (Fig. 1 B and C). Upon dialysis into 0.6 M NaCl, CENP-A-containing nucleosomal array assembly is completed as H2A/H2B dimers bind to the tetrasomes to form a stable 28–30 S nucleosomal array (Fig. 1 B and C and Fig. S1 A and B). We confirmed by electron microscopy that the 28–30 S CENP-A-containing arrays are saturated with 12 nucleosomes per input linear DNA fragment (Fig. 1D and Fig. S1 D and E). These results indicate that CENP-A arrays form octameric nucleosomes via the same set of intermediate steps as do conventional nucleosomal arrays.
Fig. 1.
CENP-A nucleosome array assembly. (A) Experimental scheme for defining the assembly pathway of CENP-A containing nucleosome arrays. (B) Average sedimentation coefficients (Save is defined at 0.5 boundary fraction) that were measured by AUC of all the different assembly species at varying NaCl concentrations as outlined in A. (CENP-A/H4)2 assembles onto the DNA as a tetramer forming a species sedimenting at approximately 19 S, with complete assembly of an approximately 29 S complex corresponding to a 12-mer octameric nucleosome array that occurs at NaCl concentrations at or below 0.6 M. For corresponding AUC profiles of all 12 conditions shown in B see Fig. S1 A and B. (C) Representative AUC profiles of CENP-A or H3 containing nucleosome arrays analyzed at 1 M and 0.6 M NaCl. All sedimentation coefficients have been corrected for temperature and normalized for water. (D) Electron microscopy (EM) images of CENP-A and H3 containing nucleosome arrays showing that both form similar “beads-on-a-string” 12-mer arrays.
Protection from H/DX Upon Nucleosome Array Folding.
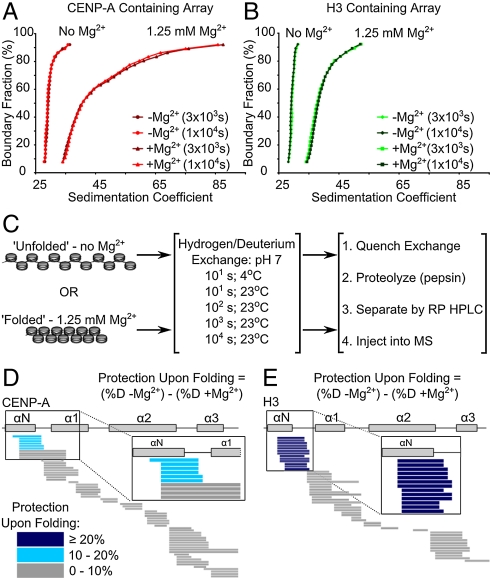
Despite no predicted gross change in their static structures (5, 21), the histone fold domains of (H3/H4)2 and (CENP-A/H4)2 undergo > 1000-fold slowing in H/DX rates upon incorporation into nucleosomes (4). Further protection from H/DX upon nucleosome array folding, however, was previously untested. Nucleosome–nucleosome interactions are restricted in nucleosomal arrays in the absence of cations, most notably Mg2+ (22). Addition of 1–2 mM Mg2+ leads to nucleosomal array folding mediated by internucleosomal contacts that condense the structure so significantly that the 29 S array now sediments at 40–55 S (22) (Fig. 2 A and B). Importantly, such folding behavior is observed in 1.25 mM Mg2+, both with our conventional arrays assembled with canonical recombinant human histones, and with the centromeric counterparts containing CENP-A in place of H3 (Fig. 2 A and B). This indicates that CENP-A does not function by locally decondensing nucleosomal array structure. Indeed, CENP-A-containing nucleosome arrays show a small but highly reproducible shift toward a state that is condensed relative to canonical arrays (Fig. 2 A and B).
Fig. 2.
Protection from H/DX upon nucleosome array folding. (A and B) AUC profiles showing MgCl2 dependent array folding and array stability over the course of the H/DX experiment. (C) Experimental scheme for determining differences in protection from H/DX upon nucleosome array folding. Protection profiles of CENP-A containing nucleosome arrays at 100 s are represented in D, and protection profiles of H3 containing nucleosome arrays at 100 s are represented in E. Protection upon array folding is calculated for each peptide in each array type as the difference of percent deuterium content of the peptide from array in the “unfolded” state minus the percent deuterium content of the same peptide in the “folded” state. Peptides are represented by horizontal bars and color coded based on the difference in protection upon folding.
H/DX measures how polypeptide backbone amide protons are exchanged with deuterons from heavy water in solution. Folded regions (e.g., α-helices and β-sheets) only exchange upon transient unfolding events when amide protons lose main chain hydrogen bonding. Slow exchange can be achieved by many stabilizing interactions (23), including, in the case of DNA-binding proteins, assembly into higher-order complexes with DNA (4, 24, 25). In order to test the extent to which the polypeptide backbone dynamics of the structured histone core of the nucleosomal subunits are altered by array folding, we monitored H/DX exchange behavior of folded and unfolded array fibers (time points at 10, 100, 1,000, and 10,000 s at 23 °C) (Fig. 2C). To potentially detect changes on the most rapidly exchanging regions we also included an additional 10 s time point at 4 °C because such a reduction in temperature leads to a nearly 10-fold slowing in chemical exchange rates at the amide protons that we can measure (23). Throughout this time course, both H3- and CENP-A-containing arrays remain intact and extensively folded as determined by AUC (Fig. 2 A and B). We monitored the H/DX behavior of overlapping peptides spanning the majority of the folded core of the nucleosome with approximately 90% coverage of the histone fold domains and approximately 60% coverage of the total polypeptide length of all histones for both H3- and CENP-A-containing arrays. There is a striking lack of folding-dependent protection at most locations throughout either type of nucleosome core (Fig. S2). However, at peptides spanning much of the αN helix of either H3 or CENP-A, we observed additional protection from H/DX (Fig. 2 D and E and Fig. S2). In the canonical nucleosome structure (21), the αN helix contacts the DNA at the superhelical termini (i.e., the DNA entry/exit site) of the nucleosome.
The αN Helix of CENP-A is Less Protected than That of H3 Upon Folding of Nucleosomal Arrays.
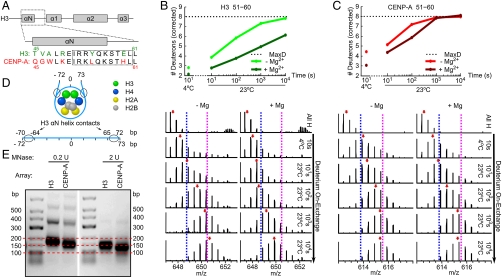
Although peptides spanning the αN helix of CENP-A are additionally protected upon array folding, the magnitude of this protection is less pronounced than for the same region in H3 arrays (Figs. 2 D and E and 3 A–C). The smaller magnitude of protection in this region of CENP-A, relative to H3, was observed in several time points from three independent experiments. H/DX data from several peptides that cover the same region, spanning a portion of the αN helix and following loop in each complex, can be used to compare the local effect of nucleosomal array folding. Representative peptides are shown in Fig. 3 B and C and Fig. S3. While nucleosomal array folding slows the H/DX of the αN helix of H3 by 50–100 times compared to unfolded arrays (Fig. 3B, compare -Mg2+ to +Mg2+), array folding only leads to a 5–10-fold slowing of H/DX in the corresponding region of CENP-A (Fig. 3C). We also noted that in addition to not being as restricted by nucleosomal folding at its αN helix, this region of CENP-A exchanges approximately 10 times faster than the corresponding region in H3 prior to polynucleosome folding (i.e., its beads-on-a-string; compare -Mg2+ in Fig. 3 B and C). Indeed, the αN helix of CENP-A is nearly as flexible in folded chromatin as is the αN helix of H3 in completely unfolded arrays (compare -Mg2+ in Fig. 3B with +Mg2+ in Fig. 3C).
Fig. 3.
Increased flexibility at the αN helix of CENP-A in nucleosomal arrays. (A) Alignment of H3 and CENP-A sequences in the αN and adjacent loop region. Boxed region highlights the sequence of the representative peptides in B and C. (B) H/DX of a representative αN-H3 peptide from nucleosome arrays containing H3. Top panel shows normalized deuterium levels at each time point for arrays that are either “unfolded” (-Mg2+) or “folded” (+Mg2+) and bottom panels show raw peptide data where the dotted blue and pink lines are drawn as guides to visualize differences in H/DX and red arrows indicate peptide centroid values. (C) H/DX of a representative αN-CENP-A peptide from nucleosome arrays containing CENP-A. Top and bottom panels are displayed as in B. (D) Schematic representation of the locations where H3 αN helix contacts nucleosomal DNA. (E) MNase digestion of CENP-A or H3 containing nucleosome arrays.
In the canonical H3-containing nucleosome, the αN helix lies at the DNA entry/exit site (Fig. 3D) (21). Reconstituted CENP-A-containing nucleosomes wrap approximately 150 bp of DNA in a left-handed manner (5). The finding that CENP-A nucleosomal arrays maintain local flexibility after chromatin folding builds on the earlier observation that topologically constrained DNA minicircles containing a single CENP-A nucleosome prefer a more “open” conformation where the exiting DNA strands do not cross (26). In our reconstituted arrays on a repetitive and strongly positioning DNA sequence, digestion with MNase clearly protects less DNA (Fig. 3E). CENP-A-containing arrays have a clear pause site at approximately 150 bp (Fig. 3E, 0.2 U MNase), consistent with a heavily populated steady-state species that is a fully wrapped octameric nucleosome. Further, CENP-A-containing nucleosomes dynamically and transiently release their DNA superhelical termini to allow the digestion of an additional one or two turns of DNA (i.e., 10–20 bp) at either terminus to yield a fragment of approximately 125 bp upon extensive digestion (Fig. 3E, 2 U MNase).
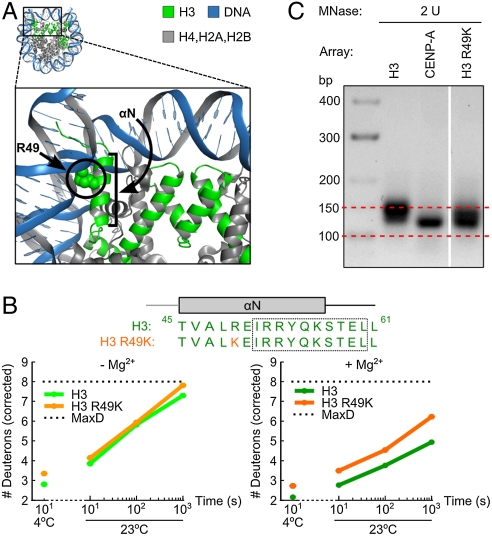
Arg49 of H3 Contributes to Rigidity at the Superhelical Nucleosome Termini in Folded Arrays.
One likely contribution to the increased flexibility in folded CENP-A-containing nucleosomal arrays is the substitution of a lysine at the position corresponding to Arg49 in histone H3. Arg49 of H3 intercalates into the DNA one half turn from the superhelical terminus of the nucleosomal DNA (Fig. 4A) (21). Indeed, with mononucleosomes assembled onto topologically constrained minicircles, the R49K mutation leads to a preference for an open DNA entry/exit arrangement (as opposed to with entering/exiting strands crossed in a closed arrangement) to nearly the same extent as seen with CENP-A mononucleosomes (26). The R49K mutation generates increased local flexibility, measured in six out of six peptides that span at least some portion of the αN helix, including adjacent peptides whose amino acid composition is identical to WT H3 (Fig. 4B and Fig. S4). The increased flexibility in H3 R49K nucleosomes is more pronounced in folded than unfolded arrays. While this substitution does not account for the full extent of the flexibility observed in CENP-A-containing nucleosomal arrays (Fig. 3), the lysine in place of the DNA intercalating arginine is clearly a major contributor. Thus, the R49K mutation in H3 generates an intermediate level of flexibility that also corresponds with an intermediate level of sensitivity to digestion of superhelical DNA termini by MNase (Fig. 4C).
Fig. 4.
Arg49 of H3 contributes to a more rigidified folded fiber. (A) Crystal structure of the human nucleosome particle containing histone H3 (PDB ID code 2CV5) (44) is shown to highlight the location of Arg49 in relation to the αN helix and the DNA entry/exit site. (B) Representative peptides from αN region of H3 containing arrays and mutant H3 R49K arrays. Boxed region highlights the sequence of the representative peptide. (C) MNase digestion of the H3 R49K nucleosome containing arrays.
Beyond the αN helix, we also observed a region of H2A (a.a. 91–134) with 10 out of 11 peptides showing faster exchange rates in CENP-A containing arrays (approximately 10-fold faster than in H3-containing nucleosomes; representative peptides are shown in Fig. S5). This observation was missed in earlier efforts (4) but was readily detected in the present study where many more nucleosome-derived peptides could be monitored due to major technical improvements in peptide resolution. Unlike the αN helices of H3 and CENP-A, the difference in H2A at this location is independent of nucleosomal folding (Fig. S5).
Discussion
CENP-A in the Nucleosomal Array Context.
The measurement of > 100 partially overlapping peptide “probes” that span the majority of each histone subunit in canonical and CENP-A-containing nucleosomal arrays provide a high-resolution view of these structures. Previous high-resolution/site-specific studies of the dynamic and structural impact of CENP-A focused on the subnucleosomal tetramer that it forms with histone H4 (5, 14, 27), mononucleosomes (4, 26), and the ternary complex that CENP-A and H4 forms with the centromeric chromatin assembly protein HJURP (or the HJURP counterpart, Scm3, in yeast) (27–29). The CENP-A targeting domain (CATD), comprised of the L1 and α2-helix of CENP-A, contributes hydrophobic stitches that rigidify the interface between CENP-A and H4 (5, 14). This rigidity is maintained after assembly into mononucleosomes (4). L1, within the CATD, generates a surface on the face of the CENP-A nucleosome that is divergent in shape and electrostatic charge (5) from the corresponding surface on canonical H3-containing nucleosomes (21). These features structurally and dynamically distinguish CENP-A mononucleosomes from conventional ones. The essential role of the CATD in centromere function (30) strongly suggests that these features are key to distinguishing centromeric chromatin from the rest of the chromosome.
Beyond individual mononucleosomes, each centromere is made up of many CENP-A mononucleosome subunits. CENP-A-containing nucleosomes coalesce on the face of the chromosome to define the location of the mitotic kinetochore assembly, but they are not arranged in a linear context. Instead they are interspersed with conventional H3-containing nucleosomes (31). Whether or not there is a regular geometry to centromeric chromatin organization (31–34), it is very likely that internucleosomal contacts between CENP-A-containing nucleosomes are fundamental in organizing centromeric chromatin. Using H/DX-MS, we have found that a major difference between the subunit structures of folded CENP-A- and H3-containing nucleosomal arrays is increased flexibility at the αN helix of CENP-A that contacts superhelical termini of each nucleosome.
After this work was complete, a crystal structure of an octameric CENP-A-containing nucleosome was reported (35). In the crystal structure, the terminal 13 bp of DNA are not visible (35), consistent with our observations of additional flexibility in the beads-on-a-string configuration (Fig. 3 B and C; -Mg2+). The H/DX studies of nucleosome array folding show the difference at this site between CENP-A- and H3-containing nucleosomes is much greater in folded than unfolded arrays (Fig. 3 B and C). This finding greatly extends our understanding of CENP-A beyond mononucleosomes (4, 5, 35) because it provides the first view of the dynamics and structure of the CENP-A-containing chromatin fiber.
Dynamics at the Superhelical Termini of the Nucleosomal Subunits of Folded Arrays.
For both (H3/H4)2 and (CENP-A/H4)2 heterotetramers, assembly into nucleosomes causes major H/DX protection along their polypeptide backbones (approximately 1,000-fold slower exchange throughout the respective histone fold domains) (4). However, the internucleosomal contacts that accompany nucleosomal array folding apparently remain fluid and do not further restrict the backbone dynamics of the bulk of the octameric histone core of the nucleosome (Fig. 2 D and E and Fig. S2). The observation that the αN helix of H3-containing arrays is substantially affected upon folding (50–100-fold slower H/DX; Fig. 3 B and C and Fig. S3) was not predictable from earlier crystal structures. In both nucleosome (21) and tetranucleosome (36) static structural models, the local structure of histone and DNA are not changed (21, 36). However, it is interesting that rigidity is required for linker DNA and up to 10 bp of terminal nucleosome core DNA in order to make an idealized chromatin fiber model based on the tetranucleosome crystal structure (36). It is also interesting that in the leading models for nucleosome higher-order packing, the nucleosome superhelical termini always face the interior of the fiber (37). Our results suggest that the fiber interior imposes the most rigidity on nucleosome cores.
Our studies also conclusively demonstrate that CENP-A-containing arrays can be readily assembled in a stepwise manner and form condensed fibers in a manner highly similar to canonical nucleosomal arrays (Figs. 1 A–C and 2 A and B). Prior to our present study, the data indicating whether or not CENP-A-containing complexes can assemble nucleosomes in a stepwise manner was not clear. Indeed, it has been reported that after assembly of the (CENP-A/H4)2 heterotetramer a 147 bp DNA template (i.e., the number of base pairs that wraps the canonical nucleosome core particle) resulted in formation of complexes that showed native gel migration consistent with two stacked heterotetramers, not a single one as for canonical (H3/H4)2 heterotetramers assembled onto the same template (26). However, the exact nature of the CENP-A “tetrasomes” was not explored further. Nonetheless, the apparent deviation from the canonical tetrasome (i.e., [H3/H4]2 + 147 bp DNA) behavior (26) was used to strongly question the relevance of subsequently reconstituted octamers to bona fide centromeric chromatin (38).
The histone H3 N-terminal tail is known to participate in array folding (39). We note that the CENP-A N-terminal tail, which is completely divergent in sequence identity from bulk H3 but maintains the strongly basic charge, does not preclude the nucleosome–nucleosome interactions that lead to formation of extensively folded array structures (Fig. 2 A and B). Indeed, we always observe that at the same Mg2+ concentration, CENP-A nucleosomal arrays are somewhat more condensed than canonical arrays (as judged by the right-shifted sedimentation profiles). It is attractive to speculate that the enhanced propensity to fold is related to the loosened superhelical termini of CENP-A nucleosomes, perhaps working in conjunction with the specialized CENP-A N-terminal tail.
In budding yeast, one of a subset of fungal species where centromere location is determined not by epigenetic information but by a specific 125 bp DNA sequence, the CENP-A counterpart, Cse4p, assembles into an octameric nucleosome that protects only 110–125 bp of DNA with no evidence of further wrapping (40, 41). We note that at the position corresponding to Arg49 in histone H3 (where substitution to the lysine corresponding to human CENP-A leads to increased flexibility; Fig. 4 and Fig. S4), Cse4p has substituted a tyrosine that likely prevents any DNA wrapping by its αN helix. It seems likely that the Arg → Tyr substitution evolved to prevent full 145 bp DNA wrapping and to accommodate the constraints imposed by a system in which the centromere is defined by a particular DNA sequence.
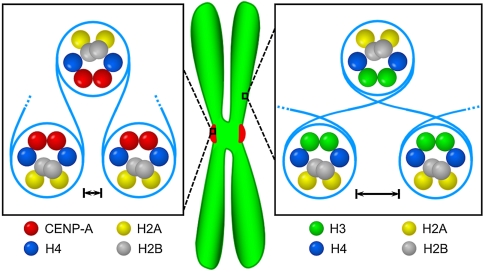
In many eukaryotes, including animals, CENP-A-containing nucleosomes are thought to be arranged in several local clusters of adjacent nucleosomes per centromere interspersed by clusters of H3-containing nucleosomes (31). Although the exact numbers of nucleosomes that make up each cluster is not known, our 12-mer arrays represent a single cluster. Our AUC experiments show that CENP-A-containing arrays are generally more condensed than canonical arrays upon folding, while our H/DX experiments demonstrate local flexibility at the contact point between the CENP-A αN-helix and the nucleosomal terminal DNA. We suggest that both characteristics (Fig. 5) are important for centromeric chromatin. The condensed nature of the array may reflect strong self–self interactions that culminate in the coalescence of discontinuous CENP-A nucleosomes on each centromere into a discrete focus on the surface of the chromosome (31). In addition, the condensed nature of the array may be important to help maintain structural integrity of centromeric chromatin under mitotic spindle stretching forces. The increased flexibility at the DNA entry/exit site may accommodate internucleosomal DNA paths unique to the centromere (31, 33, 34) and/or nucleosomal (or nucleosome-proximal) binding proteins that require increased local access to the DNA and/or histone core. Alternatively, it is formally possible that the increased flexibility at the nucleosomal termini of the human CENP-A protein is a vestige of a common ancestral orthologue that may have lacked any stable DNA binding at the superhelical termini (i.e., similar to the behavior of Cse4 in Saccharomyces cerevisiae) (40, 41).
Fig. 5.
Summary of physical differences identified in this study between CENP-A- and H3-containing nucleosomal arrays. AUC experiments showed that CENP-A-containing arrays are generally more condensed than canonical arrays upon folding (indicated by closer spacing of adjacent CENP-A-containing nucleosomes). At the same time the H/DX experiments measured local relative flexibility at the CENP-A αN-helix, indicating that the DNA at the entry/exit sites is less constrained (indicated by uncrossed internucleosomal DNA for CENP-A-containing arrays). See the text for a discussion of the potential implications of these findings.
Rapid Exchange on the C Terminus of Histone H2A in CENP-A-Containing Nucleosomes.
The region of H2A (a.a. 91–134) that undergoes rapid exchange in CENP-A-containing nucleosomes is juxtaposed to the αN-helix of H3 and is exposed on the surface of the nucleosome in the conventional nucleosome structure (21). Therefore, the increased exchange rates in H2A (a.a. 91–134) is possibly related to the flexibility of the αN-helix of CENP-A. This seems unlikely, however, because there is no correlation with the αN-helix behavior. The H2A H/DX in this region is not substantially slowed in H3-containing arrays upon array folding (Fig. S5) at the same time points where up to a 100-fold slowing is observed in the αN-helix of H3 (Fig. 3C). Another possibility is that the increased H2A H/DX in CENP-A-containing nucleosomes reflects a different orientation of H2A that accompanies rotation of the H2A/H2B dimers. (CENP-A/H4)2 heterotetramers prefer a compact state, either in solution or in crystals, which comes about by rotation at the CENP-A/CENP-A four-helix bundle (5). Upon nucleosome formation, the CENP-A/CENP-A interface may rotate to form a nucleosome of similar shape to the canonical nucleosome (5, 35), or the H2B/H4 four-helix bundles could rotate H2A/H2B dimers away from the central axis of the nucleosome to avoid steric clashes (5). Our H/DX data suggests that the possibility of whether one or both of these two states of CENP-A-containing nucleosomes are substantially populated at centromeres should be the subject of further careful analysis.
Methods
H/DX Reactions.
Nucleosome arrays (assembled and characterized by EM, AUC, and MNase digestion as detailed in SI Methods) were incubated for 30 min at 4 °C or 23 °C with either 1 × TEN or 1 × TEN with 1.25 mM MgCl2 prior to starting deuterium on-exchange reactions. Deuterium on-exchange was carried out by adding 5 μL of the array (containing approximately 3.8 μg of array) to 15 μL of deuterium on-exchange buffer (10 mM Tris, pD 7.0, 0.25 mM EDTA, 2.5 mM NaCl in D2O; supplemented with 1.25 mM MgCl2 where indicated) so that the final D2O content was 75%. Reactions were quenched at the indicated time points by withdrawing 20 μL of the reaction volume, mixing in 30 μL ice cold quench buffer (2.5 M GdHCl, 0.8% formic acid, 10% glycerol), and rapidly freezing in liquid nitrogen prior to proteolysis and LC-MS steps (detailed in SI Methods).
H/DX Data Analysis.
MATLAB-based MS data analysis tool ExMS was used for data processing (42). Detailed information regarding the ExMS algorithm is described elsewhere (42) and briefly in SI Methods. The level of H/DX occurring at each time point is expressed as either the number of deuterons or the percentage of exchange within each peptide. In each case, corrections for loss of deuterium label by individual peptides during H/DX-MS analysis (back exchange) were made through measurement of loss of deuterium from reference samples (fully deuterated control, FD) that had been deuterated under denaturing conditions as described elsewhere (43). The loss of deuterium for the FD sample was approximately 10% for most peptides, and the measured centroid values for several of the αN helix-containing peptides are shown as part of Fig. S3. Calculation of deuterium loss correction and other data operations were performed using MATLAB.
Supplementary Material
Acknowledgments.
We thank D.W. Cleveland for plasmids and M.U. Salman for assistance with MATLAB code. This work was supported by National Institutes of Health (NIH) grants GM045916 (J.C.H.) and GM082989 (B.E.B.). T.P. was supported by NIH Grant GM08275 (University of Pennsylvania Structural Biology Training Grant). This work is also supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and a Rita Allen Foundation Scholar Award (B.E.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113621108/-/DCSupplemental.
References
- 1.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panchenko T, Black BE. The epigenetic basis for centromere identity. Prog Mol Subcell Biol. 2009;48:1–32. doi: 10.1007/978-3-642-00182-6_1. [DOI] [PubMed] [Google Scholar]
- 4.Black BE, Brock MA, Bédard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 9.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szerlong HJ, Hansen JC. Nucleosome distribution and linker DNA: Connecting nuclear function to dynamic chromatin structure. Biochem Cell Biol. 2011;89:24–34. doi: 10.1139/O10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 13.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 15.Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: A model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 16.Hansen JC, van Holde KE, Lohr D. The mechanism of nucleosome assembly onto oligomers of the sea urchin 5 S DNA positioning sequence. J Biol Chem. 1991;266:4276–4282. [PubMed] [Google Scholar]
- 17.Hansen JC, Ausio J, Stanik VH, Van Holde KE. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 18.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 19.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 20.Hansen JC. Conformational dynamics of the chromatin fiber in solution: Determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 21.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz PM, Hansen JC. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J Biol Chem. 1994;269:16284–16289. [PubMed] [Google Scholar]
- 23.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalodimos CG, et al. Structure and flexibility adaptation in nonspecific and specific protein–DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 25.Hansen JC, et al. DNA binding restricts the intrinsic conformational flexibility of methyl CpG binding protein 2 (MeCP2) J Biol Chem. 2011;286:18938–18948. doi: 10.1074/jbc.M111.234609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conde e Silva N, et al. CENP-A-containing nucleosomes: Easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 27.Cho U-S, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu H, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Z, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: A repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribeiro SA, et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 36.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 37.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 38.Henikoff S, Furuyama T. Epigenetic inheritance of centromeres. Cold Spring Harb Symp Quant Biol. 2010;75:51–60. doi: 10.1101/sqb.2010.75.001. [DOI] [PubMed] [Google Scholar]
- 39.Zheng C, Lu X, Hansen JC, Hayes JJ. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- 40.Kingston IJ, Yung JSY, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dechassa ML, et al. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kan ZY, Mayne L, Englander SW. ExMS: A data analysis program for HX-MS experiments. J Am Soc Mass Spectrom. 2011. in press. [DOI] [PMC free article] [PubMed]
- 43.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.