Abstract
The nucleoprotein (NP) of the influenza virus exists as trimers, and its tail-loop binding pocket has been suggested as a potential target for antiinfluenza therapeutics. The possibility of NP as a drug target was validated by the recent reports that nucleozin and its analogs can inhibit viral replication by inducing aggregation of NP trimers. However, these inhibitors were identified by random screening, and the binding site and inhibition mechanism are unclear. We report a rational approach to target influenza virus with a new mechanism—disruption of NP–NP interaction. Consistent with recent work, E339A, R416A, and deletion mutant Δ402–428 were unable to support viral replication in the absence of WT NP. However, only E339A and R416A could form hetero complex with WT NP, but the complex was unable to bind the RNA polymerase, leading to inhibition of viral replication. These results demonstrate the importance of the E339…R416 salt bridge in viral survival and establish the salt bridge as a sensitive antiinfluenza target. To provide further support, we showed that peptides encompassing R416 can disrupt NP–NP interaction and inhibit viral replication. Finally we performed virtual screening to target E339…R416, and some small molecules identified were shown to disrupt the formation of NP trimers and inhibit replication of WT and nucleozin-resistant strains. This work provides a new approach to design antiinfluenza drugs.
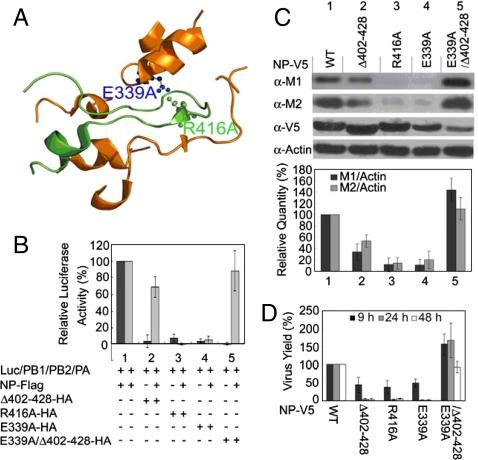
The RNA-dependent RNA polymerase (RDRP) of the influenza A virus is composed of polymerase basic protein 1 (PB1), basic protein 2 (PB2), and acidic protein (PA) (1). The function of RDRP for viral replication requires association with the nucleoprotein (NP) (2) to form the ribonucleoprotein (RNP) complex. Only low resolution structures of the RNP complex are available from cryo-EM studies (2–9), whereas high resolution structures have been reported for some individual components or fragments (10–12). Crystal structures of NP indicate that it exists in trimers (13, 14), with the tail-loop (residues 402–428) region playing an important role in the trimerization (Fig. 1A). Based on the structural information, it was suggested that the tail-loop binding pocket could be a target for antiinfluenza therapeutics (13, 14).
Fig. 1.
Inhibition effect of NP mutants for the RDRP activity and viral replication. (A) Interactions involving the tail-loop (green) and its binding pocket (gold), from Protein Data Bank (PDB) 2IQH with PyMOL. (B) Luciferase-based reporter assays for reconstitution experiments, with 1 μg of each plasmid pPOLI-Luc-RT, pcDNA-PB1, -PB2, -PA, and pClneo-NP-FLAG or -NP mutant-HA. The experiments were performed with and without pClneo-NP-FLAG. (C) The inhibition effects of NP mutant proteins on viral replication. 5 × 105 MDCK cells (WT and mutant stable cell lines) were seeded per well and incubated for 24 h at 37 °C. Then cells were infected with the WSN virus at a multiplicity of infection (MOI) of 0.2. Western blot was performed at 9 h post infection using total cell extract. (D) MDCK cells (WT and mutant stable cell lines) were infected with the WSN virus at MOI of 0.2 as described in (C). The viral titers of the cell culture supernatant for viral replication were determined by plaque assays. The relative virus yields observed 9, 24, and 48 h postinfection are shown. The bar plots are means and SD from three independent experiments.
Disruption of the NP–NP interaction as a strategy for designing antiinfluenza drugs has been further reported. Many mutants of NP, including some tail-loop mutants, lose the ability to support the RDRP activity in reconstitution experiments (2, 15–18). In addition, some of the mutants are shown to exist in monomers instead of trimers. These results support the importance of NP in the RDRP activity and viral replication, and the possibility of NP as a drug target. However, it remains to be shown that molecules capable of disrupting the NP–NP interaction would inhibit viral replication.
Recently Kao et al. (19) and our group (20) reported the use of high throughput screening to identify nucleozin and its analogs as inhibitors that halt viral replication by binding to NP and causing its aggregation. These reports validated that the NP protein is an actual drug target. However, the binding sites of the small molecules remain to be established (though the results of revertants suggest that the binding sites are close to Y52 or Y289), and how the binding induces aggregation is not yet clear.
In this work, we used rational design and virtual screening approaches to demonstrate that deaggregation of NP by disrupting the E339…R416 salt bridge can also inhibit viral replication. We first showed that E339A and R416A mutants form hetero complexes with WT NP and inhibit viral replication. These results established the E339…R416 salt bridge as a sensitive drug target. We then designed peptide inhibitors based on the tail-loop structure to demonstrate the principle—that these peptides can disrupt the NP–NP interaction and inhibit viral replication. Finally we used virtual screening to identify small molecule inhibitors to target the E339…R416 salt bridge. These small molecules were shown to disrupt NP–NP interaction and inhibit viral replication of both the wild-type and the nucleozin-resistant strains.
Results
E339A, R416A, and Δ402–428 Mutants Cannot Support RDRP Activity.
The results in this section are in agreement with other reports published (15, 16) while our work was in progress. Based on the crystal structures, we predicted that E339A, R416A, and the tail-loop deletion (Δ402–428) mutants will show perturbed NP–NP interactions, which could in turn perturb the ability of NP to interact with RDRP productively. Analyses by analytical ultracentrifugation (AUC) indicated that these three mutants exist as monomers in the free form as opposed to trimers for WT NP, and that their ability to bind RNA was significantly perturbed (SI Text and Fig. S1). The luciferase-based reporter assay showed that the three mutants were unable to support the RDRP activity when the mutant was reconstituted with PA, PB1, and PB2 in the absence of WT NP (Fig. 1B, lanes 2–4, black bars), as expected.
E339A and R416A but not Δ402–428 Can Inhibit Viral Replication.
We then performed a competition experiment by including WT NP in the luciferase-based reporter assay of the mutants. As shown in Fig. 1B (gray bars), E339A and R416A were still unable to support the RDRP activity, whereas the deletion mutant restored most of the activity. This result provides a lead for us to propose the following hypothesis: the E339A and R416A mutants, being only slightly perturbed structurally, are still able to form hetero oligomers with WT NP, and that is sufficient to perturb the interactions between NP and RDRP and thus the replication of the virus. On the other hand, the deletion mutant Δ402–428, being more severely perturbed structurally, is less able to interact with the WT NP and affect viral replication. We constructed and tested the “double mutant,” E339A/Δ402–428, which has a greater structural change than the deletion mutant. For the deletion mutant, the remaining E339 may still be able to interact with R416 from WT and cause a small degree of inhibition (gray bar of lane 2), whereas for the double mutant there will be no salt bridge formation with WT NP. As predicted, the double mutant showed nearly full activity in the presence of WT NP (gray bar, last lane of Fig. 1B; the black bar is still near zero as expected also but it is not relevant to this point).
To further test this hypothesis, Madin–Darby Canine Kidney (MDCK) stable cells expressing WT or mutant NP were created and then infected with the influenza A/WSN/33 (H1N1) virus (WSN). The Western blot analyses of the infected cells (Fig. 1C) showed that the cells containing E339A or R416A point mutants can inhibit the replication of the infected virus by up to 90% (lanes 3 and 4). The deletion mutant had a smaller effect (lane 2), and the double mutant showed no inhibition (lane 5). Complementarily, the viral titers of the cell culture supernatant were also determined by plaque assays (Fig. 1D), and the results are fully consistent with the Western blot analyses.
Mechanism for the Inhibition of Viral Replication by E339A and R416A.
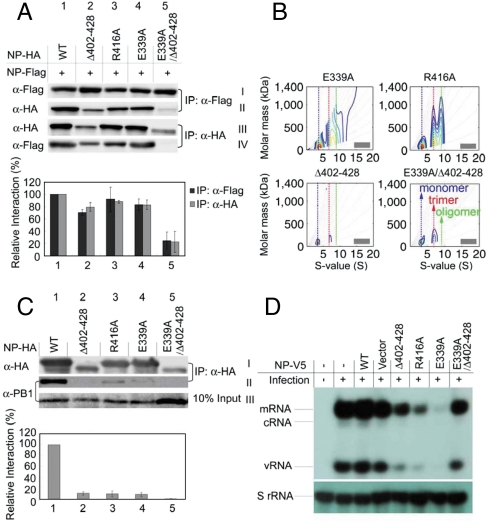
Here we demonstrate the central part of our hypothesis on the mechanism of inhibition—that E339A and R416A, though only slightly perturbed in structure with disruption in the salt bridge, loses their ability to support the function of RDRP. We first examined the binding of NP mutants with WT NP in cells by cotransfection of FLAG-tagged WT NP and HA-tagged mutant NP. As shown in Fig. 2A (rows I and II), the WT NP pulled down R416A (lane 3) and E339A (lane 4) similarly to the control (lane 1), but it pulled down less of the deletion mutant (Δ402–428, lane 2) and nearly none for the double mutant (E339A/Δ402–428, lane 5). Consistently, rows III and IV of Fig. 2A show that the HA-tagged mutants R416A and E339A can pull down the WT NP with good efficiently (lanes 3 and 4), but the double mutant was unable to pull down WT NP (lane 5). The level of expression (row III) was always lower for the deletion mutant and very low for the double mutant, but the trend is clear as shown by the bar plots.
Fig. 2.
IP assays of protein–protein interactions in cells. (A) Interaction of NP-FLAG with NP-HA or mutant-HA. 2 × 106 HEK293T cells were cotransfected with 8 μg of each plasmid. IP analysis was performed 30 h post transfection using total cell extract by anti-FLAG and anti-HA agarose, and visualized by anti-FLAG and anti-HA antibodies. Black bar: row II/row I/row III. Gray bar: row IV/row III/row I. (B) AUC analyses to examine the ability of the NP mutants to mix with WT NP. The complete titration results at 6 different ratios are shown in Fig. S2; only the results of 1∶1 ratio are shown here. Note that different y scales are used in the plots here and in Fig. S2 to show the best comparisons. (C) Pull-down assays for the interaction of NP proteins and PB1. The 2 × 106 HEK293T cells were transfected with 8 μg of each plasmid separately (pClneo-NP-HA or pClneo-mutant-HA) for 24 h, then infected with the WSN virus (MOI = 0.2) for 12 h. IP analysis was performed using total cell extract by anti-HA agarose, and detected by anti-HA and anti-PB1 antibodies. The bar plots represent NP-PB1 interactions: row II/row I/row III. The bar plots are means and SD from three independent experiments. (D) The transcription-replication activity is reduced by NP mutant proteins. MDCK cells were infected with the WSN virus at MOI of 2. RNA was isolated from cells 6 h after infection and analyzed by primer extension assays.
The result of AUC analyses also confirmed that R416A and E339A could bind WT NP to form hetero oligomers, whereas the deletion mutant Δ402–428 is less able to mix with the WT NP and the double mutant is not mixable with WT NP. As mentioned above, pure WT NP exists predominantly as trimers whereas E339A and R416A exist as monomers. As shown in Fig. 2B, the 1∶1 mixture of WT NP with E339A or R416A exists as a mixture of oligomers (evidence for interaction between WT and the mutants), whereas the corresponding mixture with Δ402–428 or in particular E339A/Δ402–428 remains as separate monomer and trimer. Detailed titration experiments with AUC for the interaction between WT and mutant NP proteins are presented in Fig. S2. From Fig. S2D, it is clear that starting from the bottom (pure WT trimer), addition of the mutant monomer caused no mixing (at 1∶1 the trimer:monomer ratio is approximately 1∶1). A small degree of mixing occurred with the deletion mutant (Fig. S2C), and extensive mixing occurred for the point mutants in Fig. S2 A and B.
The interaction of NP mutants with RDRP was next examined in cells infected with the WSN virus. It has been shown previously that NP can pull down PB1, PB2, and PA (21, 22), a property also confirmed by mass spectrometry. We then compared the PB1 binding ability of WT NP and mutants. Fig. 2C shows that, whereas the HA-tagged wild-type NP pulled down PB1 well, all of the NP mutants pulled down little or none. These results support our hypothesis that R416A and E339A could not bind PB1 because the slightly perturbed hetero complexes with WT NP are unable to further interact with RDRP from the infecting virus, whereas the deletion mutant Δ402–428 and the double mutant E339A/Δ402–428 could not bind PB1 because they cannot interact with the WT NP from the infecting virus.
Finally, the results of the primer extension assay (Fig. 2D) show that stable expression of E339A or R416A in MDCK cell lines reduced the synthesis of mRNA and vRNA significantly (the cRNA levels are too low to be detected), confirming that the transcription-replication activity of RDRP was inhibited by E339A and R416A. In consistence with the prediction, the deletion mutant Δ402–428 displayed a smaller effect, and the double mutant E339A/Δ402–428 showed relatively minor or no inhibition.
Taken together, the above results suggest that it should be possible to design small molecules or peptides targeting the E339…R416 salt bridge as potential inhibitors of the influenza virus. Furthermore, we predict that these inhibitors should function by disrupting NP–NP interaction, leading to formation of monomers. This mechanism, if confirmed, will be fundamentally different from the mechanism of nucleozin and its analogs that cause severe aggregation of NP trimers to form high-order oligomers as reported recently (19, 20).
Design and Demonstration of Peptides that Disrupt NP Trimers.
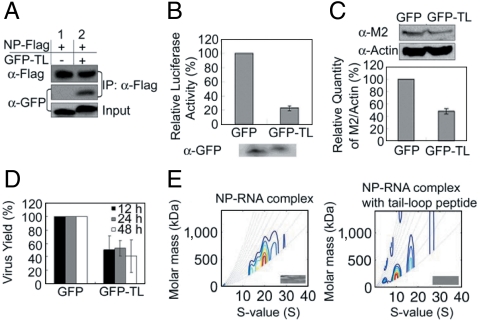
As a proof of principle, we first expressed the tail-loop peptide (residues 402–428) fused to the EGFP in HEK293T cells and showed that it binds FLAG-tagged WT NP by immunoprecipitation (IP) assays (Fig. 3A). Next we used the luciferase-based reporter assay to demonstrate the inhibition effect of the tail-loop peptide on RDRP activity in HEK293T cells (Fig. 3B). Finally the EGFP-fused tail-loop peptide was transfected into HEK293T cells that were subsequently infected with the H1N1 virus. As shown in Fig. 3 C and D, the EGFP-fused tail-loop peptide was able to slow down the replication of the virus by > 50% based on the Western blot analyses and the plaque assays (as described in Fig. 1 C and D). We have also confirmed by AUC analyses that, contrary to the aggregating effect of nucleozin and its analogs (19, 20), binding of the tail-loop peptide causes inhibition of the NP oligomerization in the presence of RNA (Fig. 3E).
Fig. 3.
Effects of the tail-loop peptide 402–428. (A) Interaction of NP WT and EGFP-fused tail-loop peptide. The 2 × 106 HEK293T cells were cotransfected with 8 μg plasmids of pClneo-NP-FLAG in the presence of pEGFP or pEGFP-tail-loop (GFP-TL) plasmid. IP analysis was performed by anti-Flag agarose and visualized by anti-GFP and anti-FLAG antibodies. (B) Luciferase-based reporter assays of the effect of EGFP-tail-loop on the H1N1 polymerase activity. HEK293T cells were cotransfected with pPOLI-Luc-RT, pcDNA-PB1, -PB2, -PA, -NP and with pEGFP (GFP) or GFP-TL plasmid. The expression levels of EGFP and EGFP-tail-loop were determined by Western blot analysis using anti-GFP antibodies. (C) The inhibition effect of EGFP-fused tail-loop peptide (H1N1) on viral replication. HEK293T cells were transfected with pEGFP (GFP) or pEGFP-tail-loop (GFP-TL) plasmid and then infected with the WSN virus.The viral protein M2 and the host protein actin were determined by Western blot analyses that were performed at 9 h post infection at MOI of 0.2. (D) HEK293T cells that were expressed with GFP or GFP-TL were infected with the WSN virus at MOI of 0.2 as described in (C). The viral titers of the cell culture supernatant were determined by plaque assays. The relative virus yields were determined 12, 24, and 48 h post infection. The bar plots are means and SD from three independent experiments. (E) AUC analyses showing that the tail-loop peptide can disrupt the oligomer of the NP–RNA complex (the recombinant NP was purified from Escherichia coli without ribonuclease A treatment). The NP–RNA complex was incubated with synthesized tail-loop peptide at a molar ratio of 1∶1,000.
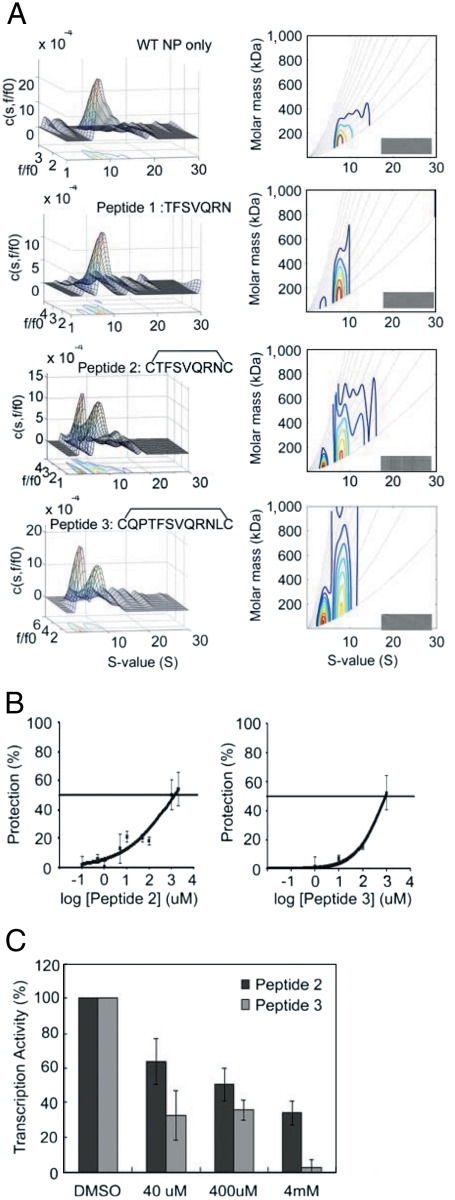
We then tried to improve the effects of the tail-loop peptide by making it shorter and cyclic, and testing their effects by direct addition to cells rather than overexpression in cells. We also used these peptides to show that disruption of the NP trimer formation based on AUC analysis is a feasible predictor for the inhibitory effect. As shown in Fig. 4A, the seven-residue peptide (peptide 1) from residues 411–417 of the tail-loop disrupted WT NP trimerization only slightly, but the effect was substantially enhanced when the peptide is cyclized to restrict its conformation by adding two Cys residues on both ends (peptide 2). A slightly larger cyclic peptide, peptide 3 (residues 409–418), showed a slightly greater effect. Then we analyzed the WSN viral yield reduction by these peptides at different doses. As shown in Fig. 4B, both peptide 2 and peptide 3 were modestly effective in protecting the host cells from viral infection, with the antiviral IC50 values of about 1 mM (Table 1). To verify that the peptides inhibited viral replication by perturbing the RDRP activity, we performed the in vitro transcription assay and the results are shown in Fig. 4C. The results establish that both cyclic peptides 2 and 3 were able to inhibit the influenza viral replication and reduce the viral transcription activity.
Fig. 4.
Effects of short cyclic peptides. (A) Analytical ultracentrifugation (AUC) analyses for NP WT mass distribution free and in the presence of a linear short peptide (peptide 1) and its circularized forms (peptide 2 and 3). The concentrations of NP WT protein and peptides were 3 μM and 300 μM, respectively. The Left panels are c(s,fr) distribution plots and the Right panels are c(s,M) distributions. The insert grayscale bars in the Right panels indicate the residuals bitmap of each fit. Detailed information of AUC data calculation and plot generation are described in SI Materials and Methods. (B) The antiviral dose response curves of the peptides. The MDCK cells were incubated with the peptide (0–2 mM), then inoculated with medium alone or the WSN virus (MOI = 0.001) for 48 h at 35 °C. The number of metabolically viable cells was determined. (C) The inhibition effects in the in vitro transcription assay. The WSN virus was incubated with the peptide (0–4 mM) for 1 h at 25 °C.
Table 1.
Antiinfluenza IC50 values (μM) of peptides and small molecule inhibitors
| Peptide 2 | Peptide 3 | Compound 3 | Compound 7 | Compound 12 | Compound 23 | |
| IC50 (μM) | 1,315 | 904 | 2.7 | 37.5 | 39.7 | 118.4 |
| CC50 (μM)* | > 2,000 | > 2,000 | 35.6 | > 100 | > 100 | > 100 |
*CC50 indicated the concentration needed to inhibit 50% growth of MDCK cells in 48 h.
In principle it is possible to design other peptides and peptidomimetics with improved inhibitory effects, but our purpose for the peptide studies was mainly to demonstrate the feasibility of our approach. Our goal was to identify small molecule inhibitors against the E339…R416 target site, as described below.
Virtual Screening for Small Molecules that Can Perturb the E339…R416 Salt Bridge.
The virtual screening method is described in SI Materials and Methods. Twenty-four compounds were selected from a library of 1,775,422 compounds and verified in cell viability antiviral assays (Fig. S3). Of the 24 hits, we tested compounds 3, 5, 7, 12, 20, and 23 (which were readily available commercially) by AUC, and found that four compounds, 3, 7, 12, and 23 were able to disrupt NP trimerization and induce formation of NP monomers (Fig. S4 A–E), suggesting that they likely can hit the target of the E339…R416 salt bridge as predicted. In contrast, we also show that nucleozin (compound 788) and compound 3061 from our previous report (20) caused aggregation of the NP trimer (Fig. S4 F and G).
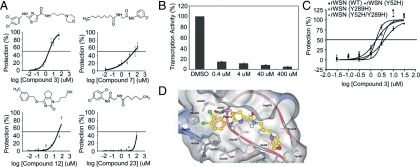
We then further characterized the inhibitory properties of the four compounds. Fig. 5A shows the results of antiviral dose response curves, and Fig. 5B shows the results of antiviral assays. The antiviral IC50 values are shown in Table 1. The results support that the four compounds identified by the virtual screening method are able to inhibit the activity of RDRP and viral replication. Because compound 3 showed the best inhibitory activity, with an antiviral IC50 value as low as 2.7 μM, we focused on compound 3 in the in vitro transcription assay, and confirmed its effect as shown in Fig. 5C.
Fig. 5.
Inhibition effects of small molecule inhibitors. (A) Structures and inhibitory effects of the compounds. The MDCK cells were incubated with compounds (0–100 μM), then inoculated with medium alone or the WSN virus (MOI = 0.001) for 48 h at 35 °C. The number of metabolically viable cells was determined. (B) In vitro transcription assay of compounds. The WSN viruses were incubated with compound 3 (0–400 μM) for 1 h at 25 °C. (C) The antiviral dose response curves of compound 3. The MDCK cells were incubated with the compound (0–100 μM), then inoculated with medium alone, the rWSN virus, nucleozin-resistant rWSN virus (Y289H mutation at NP), compound 3061-resistant rWSN virus (Y52H mutation at NP), or Y52H/Y289H double mutation rWSN virus (MOI = 0.001) at 35 °C. After 48 h, the number of metabolically viable cells was determined. (D) The modeled structure of the NP-compound 3 complex. The ribbon illustrates the NP tail-loop and the amino acids mentioned in the text are colored in pink. The compound 3 is shown in stick-and-ball with the carbon atoms colored in yellow. The NP residues interacting with compound 3 are showed in sticks.
Fig. 5D shows the modeled structure of the NP-compound 3 complex. As illustrated, the dichloro-anilino group of the inhibitor could be docked in a hydrophobic site, formed by Phe304, Trp330, Ala336, Ile347, and Ala387, which is originally occupied by Phe412 from the tail-loop. The aromatic ring of the inhibitor could form aromatic-pi interactions with Phe304 and Trp330 and cation-pi interaction with Arg389. The nitrogen atoms of the anilino-thiazole-carboxamide groups of compound 3 could mimic the tail-loop Arg416 and interact with the carboxylate of Glu339. In addition, the morpholino and propyl moieties of compound 3 could be docked into the pocket that was occupied by Ile408 and Pro419 of the tail-loop.
Comparison with Nucleozin Analogs.
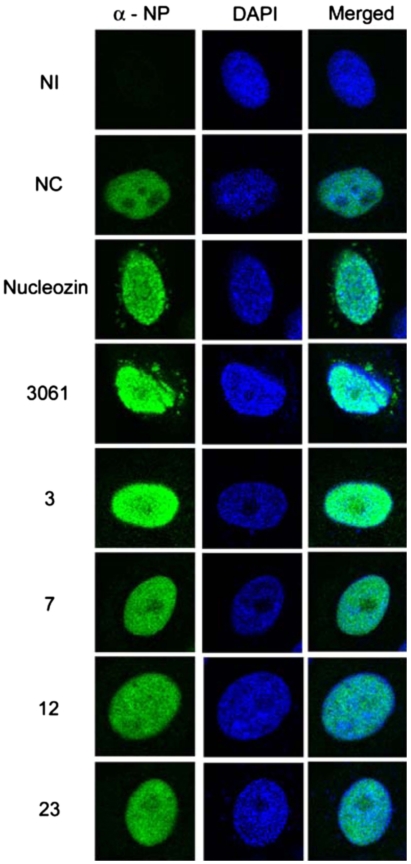
In addition to the AUC analyses showing different mechanism of the interaction with NP between our compounds and nucleozin analogs as described in Fig. S4, the immunostaining data in Fig. 6 also indicate that compounds 3, 7, 12, and 23 did not cause the aggregation that was observed for nucleozin and compound 3061 under in vivo conditions. These results clearly establish that our compounds and nucleozin analogs interact with NP via different mechanisms, and that both interactions can lead to inhibition of the viral replication.
Fig. 6.
Immunofluorescence studies of the compounds. Compounds 3, 7, 12, and 23 did not induce aggregation of NP whereas nucleozin and 3061 did. NI; no infection. NC; no compound. MDCK cells were infected with A/WSN/33 virus (MOI = 1) and incubated with compound nucleozin (5 μM), 3061 (5 μM), 3 (50 μM), 7 (50 μM), 12 (50 μM), and 23 (50 μM) 6 h post infection. Cells were fixed, and immunostaining with anti-NP (mouse) and DAPI (4′,6-diamidino-2-phenylindole) was performed 8 h post infection.
To provide further support to the above statement, we showed that compound 3 is capable of inhibiting nucleozin- and 3061-resistant strains of the virus. Fig. 5C shows the dose response curves, and Table 2 summarizes the IC50 values of the antiviral assay results of these compounds against influenza viruses derived from recombinant WSN (rWSN), nucleozin-resistant rWSN variant (Y289H mutation at NP), and compound 3061-resistant rWSN variant (Y52H mutation at NP) (19, 20), and Y52H/Y289H double mutant. The results indicate that the strains resistant to nucleozin or 3061 are not resistant to compound 3.
Table 2.
Antiinfluenza IC50 values of compound 3 against recombinant WT and mutant strains
| Anti-rWSN IC50 (μM)* | ||||
| Parental | NP Y52H | NP Y289H | NP Y52H/Y289H | |
| Compound 3 | 1.7 | 3.2 | 4.0 | 1.8 |
*Antiinfluenza activities using recombinant virus in WSN background.
Conclusion.
Our results demonstrate that the salt bridge Arg416…Glu339 is a specific target for rational design of new antiinfluenza drugs by disrupting NP–NP interactions. These results complement the recent report by Kao et al. (19) and Su et al. (20) on the use of random screening to identify nucleozin and its analogs that bind NP, cause aggregation, and inhibit viral replication. Together the two approaches reenforce that NP is a highly feasible antiinfluenza drug target.
Even though the four small molecule inhibitors described in this paper are less effective than nucleozin and its analogs in inhibiting the viral replication (the smallest antiviral IC50 obtained from the two studies are 1.7 μM for compound 3 and 0.3 μM for compound 3061), our results are potentially more significant than the two previous reports (19, 20) on the basis of the following considerations: (i) The compounds we identified are only the first generation hits from virtual screening against the target site identified by a rational approach. It is highly likely that these “lead compounds” can be further optimized to become potent inhibitors, particularly because their target site is known. (ii) Our compounds are effective against the viral variants resistant to nucleozin and 3061. (iii) Because the E339…R416 salt bridge is very highly conserved, it is less likely for the virus to develop resistance against the drugs targeting this specific site.
Materials and Methods
For a complete description of the materials and methods, see SI Materials and Methods. In summary, this section consists of the sources of the compounds and peptides used in the inhibition assays, and the viruses, cells, and plasmids used in various experiments. The section also includes detailed procedures for expression and purification of NP and its mutants, the luciferase-based reporter assay, Western blot analysis, plaque assays, analytical ultracentrifuge analysis, circular dichroism analysis, the virtual screening (Fig. S5), the primer extension assay, the in vitro transcription assay, the preparation of isogenic recombinant influenza viruses, the antiviral assay, the cytotoxicity assay, and the immunofluorescence studies.
Supplementary Material
Acknowledgments.
We thank Shi-Yun Wang for assistance in setting up virus experiments, Dr. Michael M.-C. Lai and K. S. Jeng for providing the plasmids for the luciferase-based report assay, and Dr. Shin-Ru Shih for providing the influenza virus A/WSN/33 (H1N1). We also thank the peptide synthesis labs of Institute of Biological Chemistry (IBC) and Genomics Research Center (GRC), Academia Sinica, Y. M. Shao of IBC, and Dr. R. P. Cheng and Hsiou-Ting Kuo of National Taiwan University for synthesis of peptides. This work was supported by National Science Council Taiwan Grant 95-2745-B-001 to -004 (to M.-D.T.), and by funds from GRC and IBC, Academia Sinica.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113107108/-/DCSupplemental.
References
- 1.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coloma R, et al. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009;5:e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heggeness MH, Smith PR, Ulmanen I, Krug RM, Choppin PW. Studies on the helical nucleocapsid of influenza virus. Virology. 1982;118:466–470. doi: 10.1016/0042-6822(82)90367-1. [DOI] [PubMed] [Google Scholar]
- 4.Jennings PA, Finch JT, Winter G, Robertson JS. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983;34:619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- 5.Pons MW, Schulze IT, Hirst GK, Hauser R. Isolation and characterization of the ribonucleoprotein of influenza virus. Virology. 1969;39:250–259. doi: 10.1016/0042-6822(69)90045-2. [DOI] [PubMed] [Google Scholar]
- 6.Compans RW, Content J, Duesberg PH. Structure of the ribonucleoprotein of influenza virus. J Virol. 1972;10:795–800. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Area E, et al. 3D structure of the influenza virus polymerase complex: Localization of subunit domains. Proc Natl Acad Sci USA. 2004;101:308–313. doi: 10.1073/pnas.0307127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Benito J, et al. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2001;2:313–317. doi: 10.1093/embo-reports/kve063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega J, et al. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J Virol. 2000;74:156–163. doi: 10.1128/jvi.74.1.156-163.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan P, et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 11.He X, et al. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature. 2008;454:1123–1126. doi: 10.1038/nature07120. [DOI] [PubMed] [Google Scholar]
- 12.Tarendeau F, et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol. 2007;14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 13.Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 14.Ng AK, et al. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008;22:3638–3647. doi: 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan WH, et al. Functional analysis of the influenza virus H5N1 nucleoprotein tail loop reveals amino acids that are crucial for oligomerization and ribonucleoprotein activities. J Virol. 2010;84:7337–7345. doi: 10.1128/JVI.02474-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, et al. Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein. J Virol. 2009;83:4153–4162. doi: 10.1128/JVI.02642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mena I, et al. Mutational analysis of influenza A virus nucleoprotein: Identification of mutations that affect RNA replication. J Virol. 1999;73:1186–1194. doi: 10.1128/jvi.73.2.1186-1194.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elton D, Medcalf E, Bishop K, Digard P. Oligomerization of the influenza virus nucleoprotein: Identification of positive and negative sequence elements. Virology. 1999;260:190–200. doi: 10.1006/viro.1999.9818. [DOI] [PubMed] [Google Scholar]
- 19.Kao RY, et al. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol. 2010;28:600–605. doi: 10.1038/nbt.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su CY, et al. High-throughput identification of compounds targeting influenza RNA-dependent RNA polymerase activity. Proc Natl Acad Sci USA. 2010;107:19151–19156. doi: 10.1073/pnas.1013592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas SK, Boutz PL, Nayak DP. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J Virol. 1998;72:5493–5501. doi: 10.1128/jvi.72.7.5493-5501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu WW, Weaver LL, Pante N. Purification and visualization of influenza a viral ribonucleoprotein complexes. J Vis Exp. 2009 doi: 10.3791/1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.