Abstract
This work was designed to determine the role of the vascular endothelial growth factor A (VEGF) isoforms during early neuroepithelial development in the mammalian central nervous system (CNS), specifically in the forebrain. An emerging model of interdependence between neural and vascular systems includes VEGF, with its dual roles as a potent angiogenesis factor and neural regulator. Although a number of studies have implicated VEGF in CNS development, little is known about the role that the different VEGF isoforms play in early neurogenesis. We used a mouse model of disrupted VEGF isoform expression that eliminates the predominant brain isoform, VEGF164, and expresses only the diffusible form, VEGF120. We tested the hypothesis that VEGF164 plays a key role in controlling neural precursor populations in developing cortex. We used microarray analysis to compare gene expression differences between wild type and VEGF120 mice at E9.5, the primitive stem cell stage of the neuroepithelium. We quantified changes in PHH3-positive nuclei, neural stem cell markers (Pax6 and nestin) and the Tbr2-positive intermediate progenitors at E11.5 when the neural precursor population is expanding rapidly. Absence of VEGF164 (and VEGF188) leads to reduced proliferation without an apparent effect on the number of Tbr2-positive cells. There is a corresponding reduction in the number of mitotic spindles that are oriented parallel to the ventricular surface relative to those with a vertical or oblique angle. These results support a role for the VEGF isoforms in supporting the neural precursor population of the early neuroepithelium.
Keywords: neural development, forebrain, neurogenesis, VEGF isoforms, telencephalon
Introduction
Key issues in early central nervous system (CNS) development concern how neural stem cell populations contribute to the developing brain and the heterotypic cell-cell interactions that regulate the differentiation and survival processes. Neurogenesis, the formation of new neurons, requires the orchestration of neural stem cell proliferation, initiation of differentiation, and pathway-appropriate neuronal migration and connectivity (reviewed in Nicholls et al., 2001). The coordination of these processes is particularly important in the mammalian cerebrum where precise temporal and spatial organization during development yields the characteristic layering of the cortex (reviewed in Brazel et al., 2003; Fishell and Kriegstein, 2003; Göetz and Sommer, 2005; Mori et al., 2005). Recent evidence has highlighted several transcription factors, including Pax6, Tbr2, neurogenin 2, and Tbr1, that function to sequentially restrict stem cells as they move toward specific fates in the cortex (reviewed in Hevner et al., 2006). Moreover, several signaling pathways, notably notch (Kawaguchi et al., 2008; Mizutani et al., 2007; Shen et al., 2002; Shimojo et al., 2008) and EGF (Sun et al., 2005), have been implicated in cell fate decisions in the CNS partly due to their differential effects on symmetrical versus asymmetrical divisions of neural precursor cells at the ventricular zone.
Neural stem cells give rise to the majority of cell types in the brain in the context of an investing vasculature that is essential to meeting the metabolic demands of the early expanding neuroepithelium. Concomitant with the cellular expansion at the ventricular zones, the vasculature invades from the perineural plexus at the pial surface to establish a secondary plexus that provides a source for additional sprouting vessels (reviewed in Bär, 1980; Lee et al., 2009). A recent study tracking the expression of a series of transcription factors that tightly control vascular investment indicated that the telencephalon was vascularized in a ventral to dorsal and lateral to medial fashion, suggesting that vascular investment is just as stringently regulated as neural epithelial expansion (Vasudevan et al., 2008). A theme of molecular and cellular interdependence is beginning to emerge regarding development of the neural and vascular systems (reviewed in D’Amore and Ng, 2002; Darland and D’Amore, 2001; Greenberg and Jin, 2005; Ruiz de Almodovar et al., 2009; Shima and Mailhos, 2000).
One important factor that has been identified for its potential dual role in the vascular and nervous systems is vascular endothelial growth factor (VEGF), a member of a closely-related family of proteins (VEGF A, B, C, D, E and placental growth factor) (Gale and Yancopoulos, 1999; Neufeld et al., 1999). Here we focus on VEGF-A, hereafter referred to as VEGF, and its role in early forebrain development. Genetic ablation in mice of VEGF or its primary signaling receptor, VEGFR2 (Flk-1/KDR), resulted in early embryonic (E) lethality (E9 to E10) with phenotypes in both vascular and neuronal lineages (Carmeliet et al., 1996; Ferrara et al., 1996; Shalaby et al., 1995). VEGFR2 is expressed throughout the CNS vasculature (Ayadi et al., 2001; Yamaguchi et al., 1993), but on close inspection of these reported results in public database images (EMAGE), VEGFR2 is also present in the forebrain neuroepithelium (EMAGE:882; EMAGE:5807) (Richardson et al., 2009). VEGF is expressed as several isoforms that are generated by alternative splicing of a single gene (Ruiz de Almodovar et al., 2009; Shima et al., 1996). The isoforms differ in their size and in their ability to bind to heparan sulfate proteoglycans on the cell surface and associated with the extracellular matrix (Ferrara, 1999; Houck et al., 1992). The mouse gene encodes three VEGF isoforms that are differentially expressed during development and in different organ systems. VEGF expression in the late developing brain is comprised of ~55–75% VEGF164 and ~20–50% VEGF120; VEGF188 levels are virtually undetectable from E13.5 through to adult, although regional differences in expression were not investigated (Ng et al., 2001).
While VEGF has been shown to be required for vascular development, increasing evidence indicates that VEGF plays a direct role in nervous system development (reviewed in (Rosenstein et al., 2010). CNS expression of VEGFR2 (Flk-1/KDR) was first identified in retinal progenitor cells (Yang and Cepko, 1996) and has since been shown to promote neuronal survival and axon outgrowth in cultured peripheral neurons in vitro (Sondell et al., 1999; Sondell et al., 2000), cortical neurons in vitro and in vivo (Jin et al., 2002; Zhu et al., 2003) and retinal photoreceptors in vitro and in vivo (Saint-Geniez et al., 2008). Recently, expression of VEGF-B has been linked with stimulation of neurogenesis in the adult hippocampus and forebrain (Sun et al., 2006). Moreover, localized expression of VEGF was required for peripheral sensory neurons to act as directional guides for arteriole branching in the skin, although the source of VEGF, neural or glial, was unclear in this study (Mukouyama et al., 2002). VEGF isoforms and cognate receptors have been identified in a neural stem cell population in the subventricular zone of neonatal pig and the expression persists in primary cultures grown as neurospheres (Ara et al., 2010). Examination of VEGF protein in human fetal CNS tissue has demonstrated localized expression throughout the cortical ventricular zones associated with the vasculature, neurons and glia (Sentilhes et al., 2010; Virgintino et al., 2003). A recent study of the role of VEGF in the cerebellum has provided evidence that the VEGF isoforms are differentially expressed in the postnatal cerebellum and are directly involved in granule cell migration mediated by Flk1/VEGFR2 (Ruiz de Almodovar et al., 2010). Therefore, VEGF is uniquely situated to affect both neural and vascular development. The goal of this study was to investigate the role of VEGF isoforms in regulating neurogenesis in the context of the neurovascular interactions that contribute to early forebrain and cortical development.
Materials and Methods
Generation of mouse embryos and genotyping
The VEGF120 mouse line has been previously described (Carmeliet et al., 1999). The VEGF120 homozygous embryos were generated by heterozygous crosses since the homozygous VEGF120 mice die at birth (Carmeliet et al., 1999; Ng et al., 2001). The VEGF-lacZ mice, in which LacZ with a nuclear localization signal has been introduced into the 3′ region of the gene, (Miquerol et al., 1999) were generously provided by Dr. Andras Nagy (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Canada) and are maintained with a heterozygous to wild type cross. Time-pregnant mice (plug date, day 0.5) were used to generate embryos from E7.5, E9.5, and E11.5. Same-stage embryonic mice from different litters were combined as study sets to obtain sufficient wild type and homozygous individuals for statistical confidence. The viability of VEGF120 mice relative to wild type littermates was determined by analyzing genotype yields for E9.5 and E11.5 embryonic time points. A total of 53 E9.5 embryos from 9 litters derived from VEGF120 heterozygous crosses yielded a roughly normal distribution of genotypes with 9 wild-type (18%), 28 heterozygous (54%), and 14 homozygous (27%) embryos. A similar genotypic yield was obtained at E11.5 during neuroepithelial expansion. A total of 42 E11.5 embryos from 13 litters derived from heterozygous crosses yielded 11 wild type (26%), 21 heterozygous (50%), and 10 homozygous (24%) embryos. Genotype was determined using standard PCR of purified genomic DNA (from tail cuts) as described in (Carmeliet et al., 1996; Carmeliet et al., 1999; Saint-Geniez et al., 2006). All animal studies were conducted using NIH recommended guidelines for the care and use of animals in research and approved by the University of North Dakota Institutional Animal Care and Use Committee (#0807-1c).
Immunolabeling
Embryos were fixed in buffered 3.7% paraformaldehyde and gradient equilibrated to 30% sucrose. Cryosectioning of imbedded tissue was done with a Leica HM550 cryostat. Immunolabeling was conducted as previously described (Darland et al., 2003) with modifications for sections. In brief, sections were blocked and permeabilized in 3% donkey serum (Vector Laboratories, Burlingame, CA), 0.1% Triton X-100, 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) 2–3 hours at room temp (RT) or overnight at 4°C. The primary antibody incubation was 1 hour at RT or overnight at 4°C overnight, optimized for each antibody to reduce background. The Pax6 primary antibody was incubated in 1% non-fat milk in Tris-buffered saline solution overnight at 4°C following a previously published method (Nomura et al., 1998). Negative controls were the absence of primary antibody as well as isotype- and species-matched immunoglobulin at concentrations matching the primary antibody. Primary antibodies used were: PAX6 (1:2000 dilution of ascites) and nestin (1: 50 dilution of culture supernatant, Development Studies Hybridoma Bank, Iowa City, IA), phosphohistone H3 (PHH3, 1:200, Upstate Biotechnology, Lake Placid, NY), and Tbr2 (1:300, Abcam, Cambridge, MA). Griffonia simplicifolia lectin B4 conjugated to FITC was used at 1:200 (Vector Laboratories). The fluorochrome-coupled secondary antibodies were incubated one hour at RT (1:200 dilution of cy3 or FITC conjugated secondary donkey or goat antibodies, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Nuclei were labeled with DAPI (Sigma, St. Louis, MO). In cases where signal amplification was required the VectaStain Elite kit (Vector Laboratories) for detection of biotin conjugated-horseradish peroxidase (avidin/biotin complex method) was used. Detection of DNA fragmentation as an indication of apoptosis was done with DEADEnd™ kit (Promega, Madison, WI) according to the manufacturer’s recommended protocol. Slides were permanently mounted with Vectashield mounting medium with or without DAPI (Vector Labs).
Mouse VEGF ELISA
Quantitative comparison of VEGF protein in developing CNS was determined using the VEGF ELISA kit for mouse (R & D Systems, Minneapolis, MN). Neural epithelial tissue was microdissected after a butterfly flat mount of E11.5 embryos and the forebrain and midbrain regions separated. The tissue was triturated in lysis buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing a cocktail of protease and phosphatase inhibitors (Sigma, St. Louis, MO) and passed through a 25-gauge needle to disrupt cells and shred genomic DNA. Samples were stored at −80°C and non-soluble and membranous material separated from the lysed material after a 10-minute centrifugation at 14,000 rpm. Total protein was determined with RCDC- Protein Assay (BioRad Laboratories, Hercules, CA) and 50 μg of total protein was loaded onto the assay plate and compared with a VEGF standard curve, as recommended by the manufacturer.
Quantitative real-time PCR (qPCR)
To generate E7.5 to E11.5 series for RNA analysis, embryos from timed-pregnant mice were saturated in RNALater (Applied Biosystems, Austin, TX) at 4°C and microdissected after at least 24 hours. For the E7.5 samples, the anterior neural ectoderm of three E7.5 embryos was pooled to obtain sufficient RNA for cDNA synthesis. The E9.5 embryo heads were collected with the caudal cut made at the midbrain/hindbrain junction. The E11.5 embryo heads were dissected at the anterior divide between the forebrain and midbrain using the bifurcation of the cerebral artery as the dividing line. Total RNA was purified with PicoPure™ RNA Isolation (Applied Biosystems) and quantified with a Nanodrop spectrophotometer. Two hundred μg of total RNA was converted to cDNA (GeneAmp kit, Applied Biosystems) and real-time PCR quantification of target genes was completed with SybrGreen detection on a AB7300 Thermocycler. Primer pairs were designed for each target gene and optimized using Primer Express Software (Applied Biosystems). The mRNA FASTA sequences were taken from the Entrez Gene website (http://www.ncbi.nlm.nih.gov/sites/entrez) and compared against those found on Ensembl Mouse Gene Viewer (http://www.ensembl.org/Mus_musculus/index.html). Primers were optimized for a GC content of 45–50%, a base pair length of ~20, a melting temperature of 60°C, and an optimal amplicon size of 50–250 bp (see Table I). The amplicons were designed to cross intron/exon boundaries. All primers were checked for minimal hairpins and dimerization using Oligo Analyzer (Integrated DNA Technologies, Coralville, IA). Target product was amplified from E9.5 wild-type neuroepithelium cDNA and subcloned into TOPO-TA-Sequencing (Invitrogen). Cloned products were sequence validated using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on an ABI Prism 3100 Genetic Analyzer. Sequence-validated products were amplified off the plasmid, gel purified (Qiagen) and quantified by nanodrop for use in the standard curve. The analysis approach was based on the previously published protocol (Rhen et al., 2007) and protocols provided following accepted qPCR guidelines (Bustin et al., 2009).
Table I.
qPCR primers, sequence-validated amplicon size, and accession number.
| Target | Primer | Amplicon | Accession # |
|---|---|---|---|
| VEGF120 | For 5′-GCCAGCACATAGGAGAGATGAGC-3′ | 95 bp | NM_001025250.2 |
| Rev 5′-CGGCTTGTCACATTTTTCTGG-3′ | NM_001025257.2 | ||
| VEGF164 | For 5′-GCCAGCACATAGGAGAGATGAGC-3′ | 97 bp | NM_001025250.2 |
| Rev 5′-CAAGGCTCACAGTGATTTTCTGG-3′ | NM_009505.3 | ||
| VEGF188 | For 5′-GCCAGCACATAGGAGAGATGAGC-3′ | 171 bp | NM_001025250.2 |
| Rev 5′-AACAAGGCTCACAGTGAACGCT-3′ | |||
| SSeCKs/Gravin | For 5′-CCGAGAAGAGAAAGGAGCAA-3′ | 147 bp | NM 031185.3 |
| Rev 5′-AAGGCAACTCCACCTTCTCA-3′ | |||
| Pax6 (55a and 56) | For 5′-GCTTGGTGGTGTCTTTGTCA-3′ | 131 bp | NM_013627.3 |
| Rev 5′-TCACACAACCGTTGGATACC-3′ | |||
| nestin | For 5′-CTTTCCTGACCCCAAGCTGA-3′ | 79 bp | NM_016701.3 |
| Rev 5′-TGAGGACAGGGAGCACAGATC-3′ | |||
| PECAM | For 5′-CCGGGAAGTACAAATGCACA-3′ | 150 bp | NM_008816.2 |
| Rev 5′-AGGAACAATTGACCGTCACG-3′ |
The qPCR cycling profile was 2 min. at 50°C, 10 min. at 95°C, then 40 repeated cycles alternating 15 sec. at 95°C with one min. at 60°C. A final dissociation step was included to determine uniformity of product melting temperature. Purified plasmid DNA product for each primer pair was used to generate standard curves ranging from 2ng to 2 × 10−21 (ag) and the inverse log was used to determine DNA concentration in cDNA array samples of wild-type and E9.5 VEGF isoform mice-derived neuroepithelium. Efficiency values for each primer pair were calculated from the slope of the Ct versus DNA concentration curve for the standard using the equation, [log10 (−1/slope)]−1, and final concentrations adjusted accordingly. All samples were quantified in parallel for 18S as a reference gene to allow for variations in cDNA synthesis efficiency between reactions. All cDNA synthesis reactions were quality control checked with standard reverse transcription PCR using glyceraldehyde phosphate dehydrogenase primers prior to being used in the qPCR reactions.
Design-based stereology
We used design-based stereology as an unbiased means of quantifying PHH3- or Tbr2-positive nuclei independent of size or spatial distribution (Schmitz and Hof, 2005). Sections from three or four fixed, frozen brains each of E11.5 wild type or VEGF120 mice were cut at 30μm and collected with gelatin-coated slides (Fisher Scientific). Air dried sections were immunolabeled for PHH3 or Tbr2 as described above with modifications for a biotinylated secondary antibody (Goat anti-rabbit~biotin, 1:200; Jackson) and Vectastain Elite ABC horseradish peroxidase staining according to the manufacturer’s instructions. Immunolabeled sections were counterstained with methyl green solution to label nuclei and permanently mounted after standard alcohol dehydration series and xylene exposure (Vectamount, Vector Labs). Positive and total nuclei were counted using an Olympus BX51WI with motorized XYZ stage. Unbiased quantification of nuclei was conducted using the optional fractionator workflow in StereoInvestigator 9.0 (MicroBrightfield, Inc., Wiliston, VT). For the PHH3 counting, the contour outlined the neuroepithelium within 100μm of the ventricular surface from the forebrain structure to the anterior limit of the diencephalon. For the Tbr2 counting, the contour outlined the same region of the forebrain, however the outer limit of the tracing was taken to the epithelial border beneath the pial vessel surface to ensure inclusion of all positive nuclei within the counting area. Positive nuclei (or total nuclei) were counted in every 10th section in systematically-selected frames based on optical dissector frames and grid sizes determined separately for each counting paradigm to ensure that the coefficient of error was less than 0.1 (10%); details for each are included in the figure legends. The total numbers were estimated with the optical fractionator formula (N=1/ssf.1/asf.1/hsf.ΣQ−) where ssf=section sampling fraction (10), asf=area sampling fraction (area sampled/total area), hsf=height sampling fraction (counting frame height/30 μm), and ΣQ− (total particle count). Neuroepithelial volume was measured based on the total tracing area, the actual z-plane measured section thickness (accounting for tissue shrinkage), and the section interval. For the mitotic spindle counts, a subset of the PHH3-positive nuclei in the anterolateral loop of the telencephalon (bilateral) were counted and mitotic spindles identified and counted relative to their position in relation to the ventricular zone surface (parallel or vertical/oblique).
General statistical analyses
All statistical analyses were completed using GraphPad Prism 4.0c for Macintosh (GraphPad Software, Inc., LaJolla, CA). An F test was used to determine whether the population variances were statistically different or not. Populations with equal variance were analyzed with a two-tailed, unpaired t test with Welch’s correction. Populations with unequal variance were analyzed with a Mann-Whitney test. P values are indicated in the figure legends or on the graph, but significance was determined with α value set to 0.05. Any data transformations (e.g. natural log of ratios) are indicated in the legends. Because of the small sample size, data shown are the mean +/− the standard error of the mean with the n for each sample set indicated in the legend. All embryo data shown were collected from multiple litters to gain sufficient numbers for the assay and to provide as representative a population as possible for comparison.
Microarray and differential gene expression analysis
Microdissected forebrains from E9.5 embryos were collected and total RNA purified as described above. Samples were submitted to Genome Explorations Inc. (Memphis, TN) for further processing and analysis. The RNA concentration was determined from the OD260/280 ratio and quality determined by capillary electrophoresis on the RNA 6000 Nano Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) as per the manufacturers instructions. Following fragmentation, 15 ug of biotinylated cRNA were hybridized for 16 hr at 45°C on GeneChip 430.2 array (Affymetrix; GPL1261). GeneChips were washed and stained with streptavidin-phycoerythrin using the Affymetrix Fluidics Station 450, according to the manufacturer’s recommended protocol. Hierarchical clustering analysis was conducted using the PLIER values (Therneau and Ballman, 2008) for genes that were differentially changed at least 1.5 fold with t-test values ≤ 0.05. The log2 transformed values were mean centered prior to clustering analysis by the farthest neighbor method with Euclidean distance and Pearson Correlation as the similarity metrics. The Affymetrix annotation numbers were uploaded onto the Database for Annotation, Visualization, and Integrated Discovery (DAVID) and the enriched clusters identified with the mouse genome as the population background (Huang et al., 2009a; Huang et al., 2009b). The enrichment score reflects the geometric mean of all the enrichment values for each gene in a given cluster. The Expression Analysis Systematic Explorer (EASE) score (p value) is determined from a modified Fisher’s exact test (Hosack et al., 2003). It reflects the significance of the enrichment score as a representation of a functional gene group relative to the expected representation based on the population background. The Benjamini value corrects for false positives in the enrichment p values with high stringency settings used throughout.
Results
VEGF expression
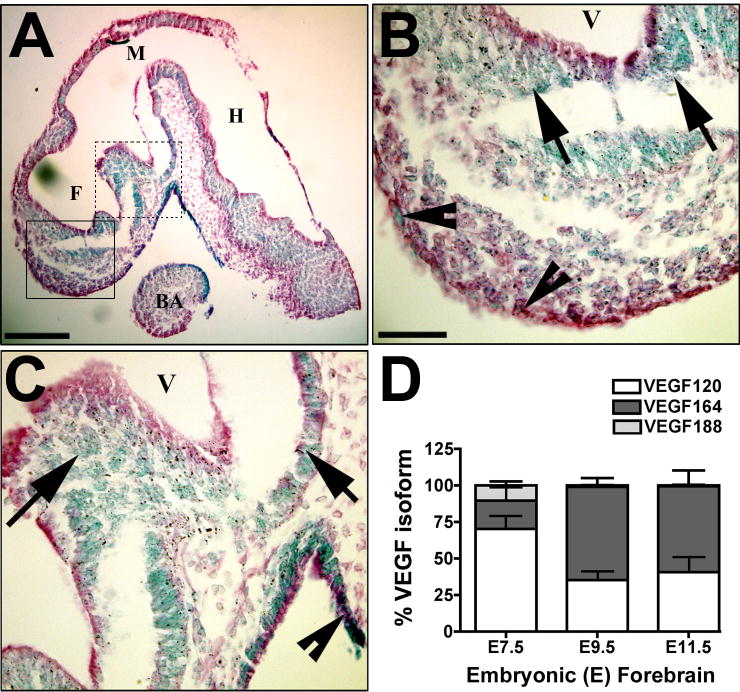
The spatial and temporal pattern of VEGF ligand expression with respect to neurogenesis was determined before evaluating any possible regulatory role in developmental neurogenesis. The developmental window of E9.5 to E11.5 in mouse represents a period of rapid expansion of the neuroepithelium, and vascular investment of the CNS from the perineural plexus occurs in concert, with a temporal delay in the forebrain (reviewed in Bär, 1980; Göetz and Huttner, 2005; Vasudevan et al., 2008). Early in situ hybridization studies demonstrated signal for VEGF mRNA in the forebrain at E10.5 (Ferrara et al., 1996), E12.5 (Dumont et al., 1995), and E17.5 (Breier et al., 1995), although high-resolution images were not available from those studies. In addition, the VEGF isoforms have been detected in brain by RNase protection assay at E13 (Ng et al., 2001) and in the postnatal cerebellum (Ruiz de Almodovar et al., 2010). To determine if VEGF was expressed specifically in the forebrain neuroepithelium, we examined mice expressing the lacZ gene under control of the endogenous VEGF promoter and containing a nuclear localization signal (Miquerol et al., 1999). Mice at E9.5 were examined when the perineural vascular plexus has formed over the neuroepithelium, but vascular invasion into the forebrain has not fully initiated (Figure 1, A–C). Positive nuclei were present throughout the neuroepithelium of the developing mouse brain. Quantification of the three major VEGF isoforms via qPCR from E7.5, E9.5, and E11.5 (Figure 1D) revealed that the VEGF isoform distribution changed proportionally over time and was similar by E11.5 to the distribution previously reported for whole brain at E13 (Ng et al., 2001) and the postnatal cerebellum (Ruiz de Almodovar et al., 2010).
Figure 1. VEGF expression in developing neuroepithelial cells.
(A–D) VEGF-lacZ embryos (E9.5) were stained for β-galactosidase (β-gal) activity, imbedded in paraffin, sectioned in the saggital plane along the midline, and counterstained with eosin. Formation of the secondary vesicles derived from the forebrain (F), midbrain (M) and hindbrain (H) are identified. The first brachial arch mesoderm (BA) is indicated for anatomic reference. The β-gal gene product is targeted to the nucleus (Miquerol et al., 1999), so all VEGF-expressing cells have blue nuclei. Blue nuclei are detected throughout the neuroepithelium. The boxed areas in A are shown in higher magnification in B (solid line) and C (dashed line). Arrows indicate positive nuclei in the neuroepithelium and arrowheads indicate positive cells at the pial surface. The scale bars are 200 μm (A) and 50μm (B–C). (D) Quantitative real-time PCR (qPCR) analysis of the VEGF isoforms, VEGF120, VEGF164, and VEGF188 is graphed as a proportion of the total VEGF mRNA produced for isolated neuroepithelium from E7.5, E9.5 and E11.5 wild-type mice. Data are from 9–11 embryos from 3–5 litters, analyzed at each time point. The mean and standard error of the mean are graphed based on values determined by comparison to an isoform-specific standard curve run in parallel.
Neuroepithelial development in mice lacking VEGF164
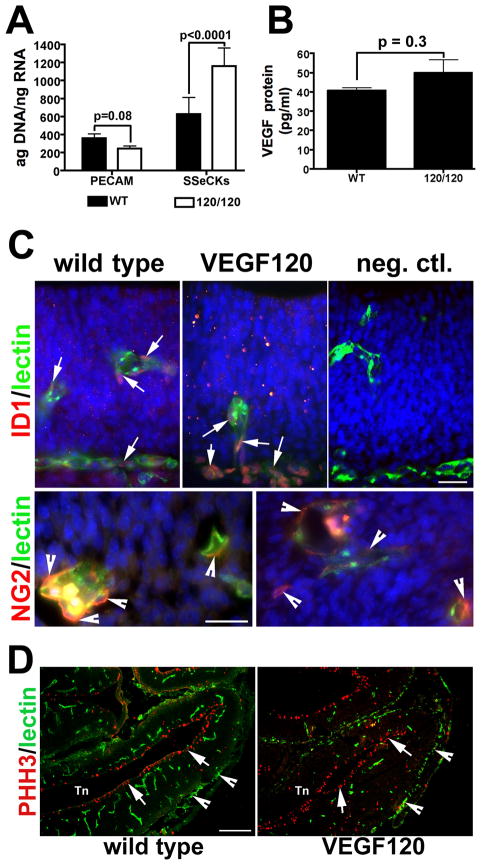
In order to test the hypothesis that VEGF regulates neuroepithelial development, we used a mouse model system of disrupted VEGF isoform expression, the VEGF120 mice. These mice express only a single isoform of VEGF, VEGF120, and display abnormalities in the cardiopulmonary system (Carmeliet et al., 1996; Carmeliet et al., 1999; Ng et al., 2001), the retina (Stalmans et al., 2002), and the granule cells of the cerebellum (Ruiz de Almodovar et al., 2010). These mice lack the predominant brain isoform VEGF164 as well as VEGF188, and express only the diffusible VEGF120 protein, generally displaying early postnatal lethality. To quantify the vascular investment in the VEGF120 mice, we examined expression levels for two genes associated with angiogenesis, platelet endothelial cell adhesion molecule (PECAM) and Src-suppressed C kinase substrate protein (SSeCKS). PECAM is expressed in endothelial cells (EC) and concentrated at the junctions between EC (reviewed in (Woodfin et al., 2007). Expression of SSeCKs has been associated with angiogenesis and blood brain barrier formation in early CNS development (Lee et al., 2003) and reviewed in (Lee et al., 2009). Comparative mRNA was quantified at E9.5, a time point just prior to initiation of vascular ingrowth into the forebrain neuroepithelium. The qPCR analysis showed a 33.3% reduction in mean PECAM levels in the VEGF120 versus wild type mice. In contrast, SSeCKs levels increased by 90% in the VEGF120 animals (Figure 2A). RNase protection has demonstrated that total VEGF mRNA levels are equivalent in the VEGF120 mice relative to wild type (Carmeliet et al., 1999) and we wanted to confirm this at the protein level in the early forebrain. Therefore, we measured levels of total VEGF protein using the VEGF mouse ELISA (Figure 2B). VEGF protein levels in E11.5 forebrain neuroepithelium were not statistically different between VEGF120 and wild type samples (p = 0.32).
Figure 2. Disruption of VEGF isoform expression affects normal vessel formation in developing neuroepithelium.
(A) Determination of relative mRNA expression was quantified by qPCR for PECAM and SSeCKs/Gravin in E9.5 neuroepithelium from wild type and VEGF120 (120/120) mice. Data are from seven embryos collected from at least three litters. Two-way ANOVA analysis of expression indicated that there was a significant source of variation between the genes (45.87%; p<0.0001) with genotype accounting for 5.73% of the variation (p=0.10); the significance from Bonferroni’s post-hoc tests are indicated in the figure. (B) VEGF protein levels in 50 μg total protein were measured in forebrain lysates from E11.5 neuroepithelium using a standard mouse ELISA assay. Data are from seven embryos collected from three litters. Both populations had a normal distribution, but unequal variance (F test, p = 0.003). While the VEGF120 mean value was 20% higher than the wild type mean, the difference was not statistically significant (Mann-Whitney test, p = 0.32). (C) Immunolabeling for Id1 (red) or NG2 (red) in combination with G. simplicifolia lectin~FITC (green) is shown for sections from E11.5 wild type and VEGF120 littermates. Id1-positive endothelial cells were present in the neuroepithelial vessels (arrows) and in the pial vessels in both wild type (left top panel) and VEGF120 (middle top panel). Lectin co-labeling highlighted the differences observed in vessel formation between wild type and VEGF120 mice. Rabbit IgG as a negative control is shown for comparison (right top panel). Scale is 25 μm in the top and bottom panels. NG2-positive pericytes process (arrowheads) with associated lectin-labeled vessels are shown in the bottom panels for wild type (left bottom panel) and VEGF120 (right bottom panel). (D) Immunolabeling for phosphohistone H3 (PHH3) as a marker of mitotic cells is shown for E13.5 dorsolateral telencephalon (Tn) in combination with lectin~FITC. The anterior loop of the forebrain is shown with arrows indicating PHH3-positive cells adjacent to investing blood vessels (green, arrowheads). The scale bar is 200μm.
We then examined the forebrain vasculature during early neuroepithelial expansion at E11.5 (Figure 2C) and compared the patterns of expression of inhibitor of DNA binding-1 (ID1) and nerve-glial antigen 2 (NG2) in E11.5 wild-type and VEGF120 mice (Figure 2C). ID1 is a transcription factor whose expression and nuclear localization are associated with immature and dividing cells, most notably in EC (reviewed in (Benezra et al., 2001; Ruzinova and Benezra, 2003). ID1-positive cells (red) that co-labeled for G. simplicifolia lectin (green) were observed in both pial vessels as well as vessels investing the neuroepithelium, and were particularly prominent in pial vessels of the VEGF120 mice. NG2-positive pericytes were associated with vessels in wild type and VEGF120 neuroepithelium, but the pericyte processes appeared less elaborate in the VEGF120 mice (Figure 2C, bottom panels). We further examined vascular investment two days later (Figure 2D) and observed that blood vessels in the wild type forebrain were well developed and the lateral plexi formed throughout the expanding neuroepithelium at E13.5 (Figure 2D, left panel). In contrast, the VEGF120 mice had compromised vascular investment and limited plexus formation throughout the telencephalon (Figure 2D, right panel).
Comparison of gene expression in wild type and VEGF120 neuroepithelium
Since neural stem cell proliferation at the ventricular surface and in the basal progenitor population is key to successful lamination of the cortex, we examined in parallel the effect of expressing only the diffusible VEGF120 isoform on the pattern of proliferating cells. The phosphorylated form of histone H3 (PHH3) is normally associated with M-phase mitotic cells, although detection as ranged as widely as late G2 to the final stages of anaphase (Hendzel et al., 1997; Nasr and El-Zammar, 2008). PHH3 immunolabeling revealed numerous positive nuclei present along the ventricular zone in the wild type mice (Figure 2D, left panel). PHH3-positive cells were detected in the VEGF120 mice as well, however the mitotic cells were clustered rather than spaced in a continuous line along the ventricular zone (Figure 2D, right panel).
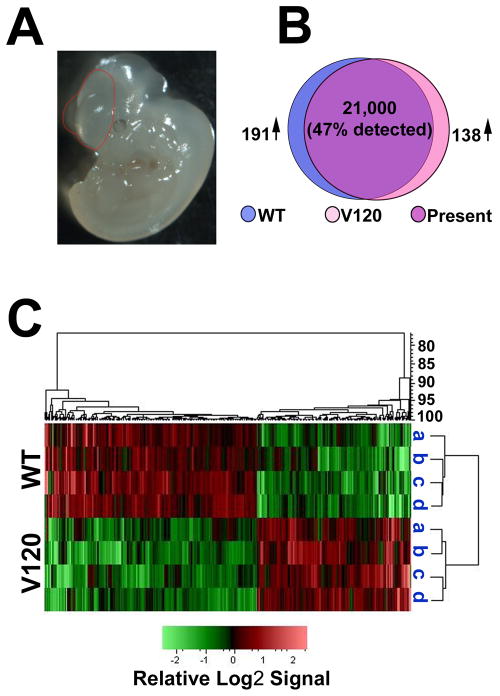
Since we observed differences in vascularization and proliferation pattern in the VEGF120 neuroepithelium from E9.5 to E13.5, we wanted to conduct a genome-wide survey of transcript expression in the VEGF120 mice prior to the period of rapid expansion at E11.5. We speculated that any changes in forebrain gene expression at E9.5 would establish the conditions for subsequent changes in proliferation and neural stem cells at E11.5. Therefore, we used the Affymetrix gene chip platform to compare gene expression profiles at E9.5 for wild type and VEGF120 embryos (Figure 3). We purified total RNA from microdissected wild type and VEGF120 forebrain (Figure 3A) and had four samples of each genotype processed and hybridized to the Affymetric 430.2 mouse gene chip array (Genome Explorations). Of the approximately 28,000 genes represented on 45,000 sites on the chip, 21,000 sites hybridized consistently across all four replicates. Based on filtration criteria of a ≥1.5 fold change in expression with an unpaired t test p value of ≤ 0.05 we identified 329 genes that were differentially expressed (Figure 3B). Cluster analysis of all eight samples showed grouping by genotype, reflecting consistent gene expression changes across litters (Figure 3C).
Figure 3. Differential gene expression in wild type and VEGF120 mice at E9.5.
(A) Representative image of an E9.5 embryo with the microdissected region traced in red. The isolated tissue included the forebrain region with the lateral telencephalon pouches and a caudal border of the ingressing diencephalon. (B) Venn diagram illustrating the number of sites detected on the Affymetrix 430.2 mouse chip comparing four each of wild type (WT, blue) and VEGF120 (V120, pink) samples with the region of common expression indicated as present (purple). The number of differentially expressed genes increased in wild type (191↑) and increased in VEGF120 (138↑) are based on ≥1.5 difference and an unpaired t test p value of ≤0.05. (C) The PLIER signals for the differentially expressed genes were transformed to the base 2 log and row mean centered. The heat map shown was generated via unsupervised (unbranched) hierarchical clustering of the samples with relative Log2 signal intensity indicated below. The dendrogram on the right indicates the distance relationships between the clustered samples while the dendrogram above indicates the relationships among the genes.
We next utilized the DAVID functional annotation database to identify gene groups enriched among the 329 differentially expressed genes relative to the mouse genome. Using the high stringency setting for functional annotation clustering, we identified two major clusters of genes with enrichment scores of 3.3, the egf-like domain (Table II) and the negative regulation of gene expression (Table III) functional groups. We took particular note of the delta-like 1 homolog (Delta) and jagged 2, notch signaling ligands that are both increased in VEGF120 animals in the gene array, 1.78- and 1.5-fold, respectively (Table II). In the latter group, we noted distal-less homeobox 1 (Dll1), whose expression has been linked to control of fate choice in neural precursors via Notch-Delta lateral inhibition (Kawaguchi et al., 2008). The Dll1 gene expression is elevated 1.7-fold in the array. We also noted enrichment of two genes, upregulated in the array, that have been linked to neural precursor differentiation and cell fate choice in the cortex. These include forkhead box P1 (Foxp1), that is normally expressed primarily cortical layers 3–5 as lamination proceeds (Ferland et al., 2003), and Cut-like homeobox 1 (Cux-1) that is associated with upper cortical layers (Nieto et al., 2004) and reelin-positive interneurons (Cubelos et al., 2008). They are upregulated 1.88- and 1.54-fold in the array. While only a small number of genes are differentially expressed in the presence of VEGF120 alone (refer to Figure 3B), the expression changes provide a transcriptional background at E9.5 that precedes and informs the critical neuroepithelial expansion at E11.5.
Table II.
Fold changes in expression of cluster 1 enriched genes are listed for “egf-like domain” with an enrichment score of 3.33 (p=0.00013; Benjamini score=0.031).
| AFFYMETRIX 3PRIME_IVT_ID | GENE NAME (FUNCTIONAL DOMAIN) | FOLD CHANGE ↑ OR ↓ IN VEGF120 | t TEST p VALUE IN ARRAY |
|---|---|---|---|
| 1447352_at | a disintegrin and metallopeptidase domain 8 | 2.24↓ | 0.028 |
| 1421134_at | amphiregulin | 2.83↓ | 0.003 |
| 1427393_at | coagulation factor IX | 2.99↓ | 0.006 |
| 1451759_at | mannan-binding lectin serine peptidase 2 | 1.89↓ | 0.037 |
| 1440173_x_at | selectin, platelet | 1.88↓ | 0.044 |
| 1455978_a_at | matrilin 2 | 1.73↓ | 0.032 |
|
| |||
| 1443131_at | low density lipoprotein-related protein 1B (deleted in tumors) | 3.23↑ | 0.008 |
| 1455499_at | neurexin II | 2.87↑ | 0.026 |
| 1457123_at | neuregulin 4 | 2.01↑ | 0.031 |
| 1447586_at | versican | 1.87↑ | 0.0001 |
| 1449939_s_at | delta-like 1 homolog (Drosophila) | 1.78↑ | 0.032 |
| 1426431_at | jagged 2 | 1.50↑ | 0.037 |
Table III.
Fold changes in expression of cluster 2 enriched genes are listed for “negative regulation of gene expression” with an enrichment score of 3.26 (p=0.00027; Benjamini score=0.046).
| AFFYMETRIX 3PRIME_IVT_ID | GENE NAME | FOLD CHANGE ↑ OR ↓ IN VEGF120 | t TEST p VALUE IN ARRAY |
|---|---|---|---|
| 1459916_at | paired box gene 5 | 2.13↓ | 0.003 |
| 1421749_at | lin-28 homolog | 1.78↓ | 0.0072 |
| 1434572_at | histone deacetylase 9 | 1.69↓ | 0.0006 |
| 1415996_at | thioredoxin interacting protein | 1.65↓ | 0.017 |
| 1421515_at | nuclear receptor subfamily 6, group A, member 1 | 1.61↓ | 0.031 |
| 1441743_at | paired box gene 3 | 1.54↓ | 0.012 |
|
| |||
| 1438802_at | forkhead box P1 | 1.88↑ | 0.033 |
| 1435022_at | ALX homeobox 1 | 1.83↑ | 0.039 |
| 1449470_at | distal-less homeobox 1 | 1.70↑ | 0.0034 |
| 1419437_at | single-minded homolog 2 | 1.66↑ | 0.033 |
| 1440004_at | suppressor of fused homolog | 1.63↑ | 0.0002 |
| 1420020_at | suppressor of zeste 12 homolog | 1.58↑ | 0.045 |
| 1447564_x_at | piwi-like homolog 4 (Drosophila) | 1.56↑ | 0.042 |
| 1424667_a_at | cut-like homeobox 1 | 1.54↑ | 0.035 |
| 1417302_at | REST corepressor 2 | 1.51↑ | 0.028 |
| 1425528_at | paired related homeobox 1 | 1.51↑ | 0.020 |
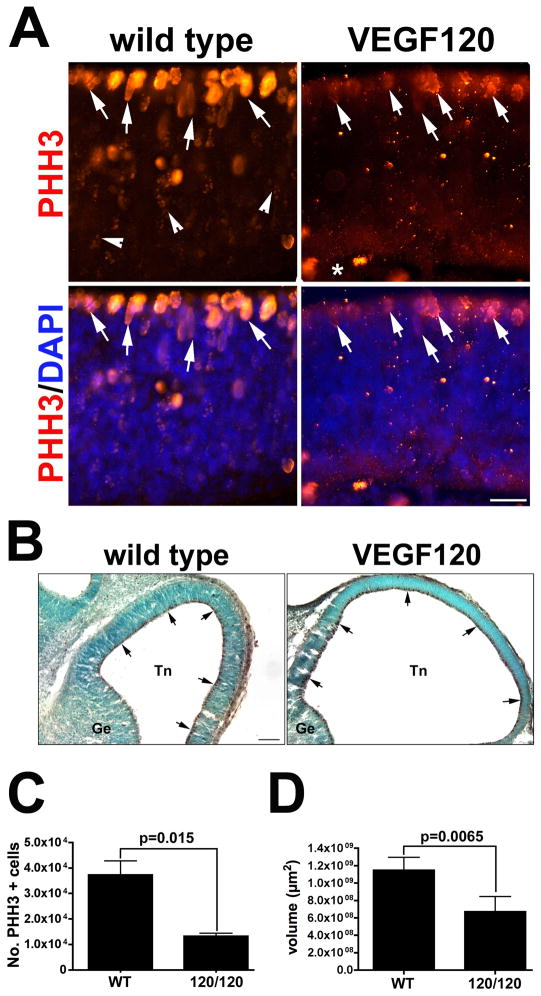
Because a subset of the differentially expressed genes identified by the functional enrichment DAVID analysis had the potential to impact notch signaling or were associated with neural differentiation, we wanted to examine the affects of the altered transcription background on the neural stem populations at E11.5. We first examined PHH3 labeling at E11.5 when early neural stem cell proliferation is at a peak. We observed PHH3-positive nuclei in the ventricular zone in the wild type as well as a secondary population of proliferating cells in the subventricular zone (Figure 4A). In contrast, in VEGF120 mice there was a much more restricted pattern of PHH3-labelled nuclei with the majority of mitotic profiles restricted to the apical surface of the ventricular zone. To more clearly assess the differences in PHH3 labeling, we used design-based stereology to quantify the number of PHH3-positive cells in VEGF120 isoform mice compared to wild type (Figure 4, B–D). We observed a statistically significant drop in the number of PHH3-positive cells and the neural epithelial volume in VEGF120 mice relative to wild type.
Figure 4. Expression of only VEGF120 alters the pattern of proliferating cells in the developing neuroepithelium.
(A) Immunolabeling for in E11.5 neuroepithelium from wild type (left panels) and VEGF120 mice (right panels) is shown with the ventricular zone at the top and the pial surface at the bottom. PHH3-positive cells (red) are localized to the ventricular zone and immediately below. The full area of the neuroepithelium is highlighted with DAPI-positive nuclei (blue) in the two-channel overlay. The white arrows indicate representative PHH3-positive cells. The white arrowheads indicate representative cells that have completed cell division but have retained the characteristic punctate labeling when a portion of the nucleosome still has the antibody epitope available for binding. The scale bar equals 25 μm. This experiment was repeated four times with similar results. The samples shown are from embryonic littermates. (B) Low power images are shown for PHH3 immunolabeling in the lateral telencephalon (Tn) and the ganglionic eminence (Ge) in wild type and VEGF120 E11.5 embryos. Arrows indicate areas of DAB reaction product (brown) localized to the ventricular surface with nuclei counterstained with methyl green. Design-based stereology and unbiased systematic random sampling were used to quantify the number of PHH3-positive cells (C) and the total neural epithelial volume (D). The optical dissector frame was 75 × 75 μm and the counting grid was set at 150 × 200 μm with a counting frame height of 20 μm and 2 μm guard zones on either side. The mean and standard error of the mean are shown for three animals of each genotype. The variances were not significantly different and the p values from an unpaired t test are indicated in the graph.
Because of the altered neuroepithelial proliferation pattern and the shift in gene expression profile in mice with the disrupted VEGF isoform profile, we speculated that there would be a direct impact on the neural stem cell populations. Therefore, we next analyzed Pax6, a transcription factor found in actively proliferating neural stem cells and radial glia (Götz et al., 1998), and nestin, a cytoskeletal protein that has been used as a marker for neural stems (reviewed in (Gilyarov, 2008). Examination of E11.5 mice revealed a Pax6–positive population of cells in the wild type telencephalon near the anterior loop encompassing the majority of the neuroepithelium with a Pax6-negative band of cells adjacent to the pial surface (Figure 5A). In VEGF120 mice the width of the band of Pax6-positive neural epithelial cells appeared reduced with few Pax6-negative cells at the pial surface. Co-labeling for laminin, which localized to the basement membrane at the pial surface and in blood vessels, further highlights the differential distribution of Pax6-positive cells between wild type and VEGF120 mice. Nestin immunolabeling had a radial pattern in the cytoskeleton of cells throughout the expanding neuroepithelium from the ventricular to the pial surface (Figure 5B). Nestin-positive cells were present in the VEGF120 mice, although the radial pattern was disrupted and punctate labeling was observed at the pial surface where the glial end feet normally embed in the basement membrane (Haubst et al., 2006).
Figure 5. Disruption of VEGF isoform expression alters Pax6 and nestin expression in E11.5 neuroepithelium.
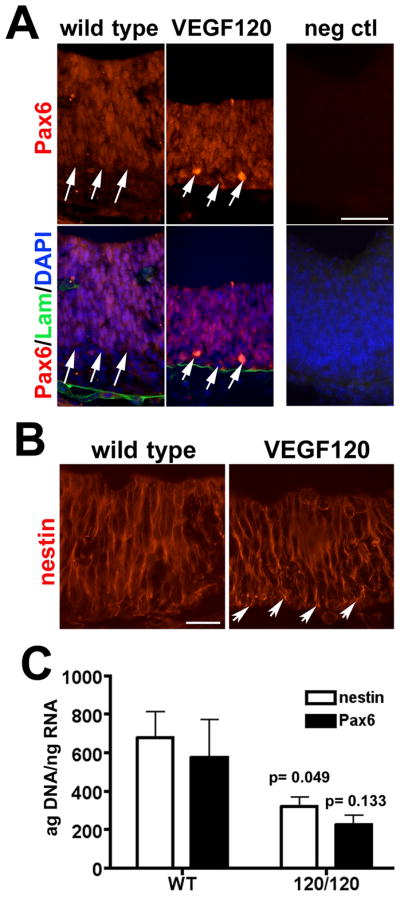
Immunolabeling for (A) Pax6 and (B) nestin is shown in E11.5 neuroepithelium with the ventricular surface at the top and the pial surface at the bottom. (A) The extent of the Pax6-positive domain is indicated by white arrows for wild type and VEGF120 mice and DAPI nuclear stain reveals the cellular population of the neuroepithelium. Labeling for laminin (Lam) in A identifies the limiting basement membrane adjacent to the pial surface and associated with blood vessels. (B) The pattern of nestin filament staining is shown with the white arrows indicating the region of globular nestin expression at the pial surface of VEGF120 embryos. Scale bar is 50 μm in A and 25 μm in B. (C) The qPCR for nestin (white bars) and Pax6 (black bars) are graphed together for comparison, although statistical analyses were conducted separately. RNA from E9.5 wild type and VEGF120 (120/120) neuroepithelium are shown with five or six samples combined from at least three litters. Unpaired t test with Welch’s correction showed that Pax6 expression was reduced by 67% in the VEGF120 compared to wild type, although the observed difference was not significantly different (p = 0.133). Analysis of nestin expression showed a 53% decrease (p = 0.049) in the VEGF120 samples compared to wild type.
In order to quantify the differences observed with the Pax6 and nestin immunolabeling, levels of gene expression in E9.5 wild type and VEGF120 neuroepithelium were measured by qPCR. As expected, Pax6 and nestin mRNA were detected in wild type samples (Figure 5C). Levels of nestin and Pax6 in VEGF120 samples were significantly reduced when compared to the wild type samples; nestin was reduced 53% and Pax6 was reduced 67%. It is important to note that these differences were quantified at E9.5, a time when little to no vascular investment of the forebrain neuroepithelium has occurred, indicating that the differences are due to a direct effect of VEGF120, or the absence of VEGF164 and VEGF188, on the neural stem cell populations.
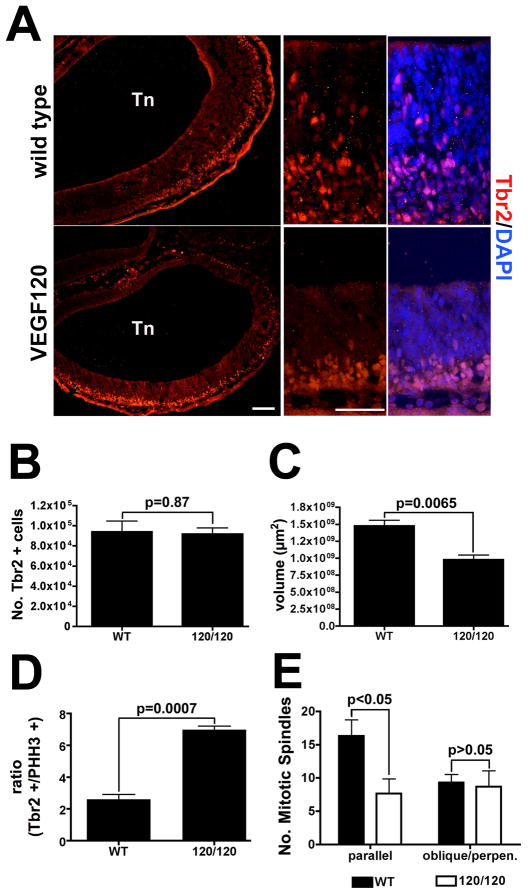
Since the neural stem cell population of the VEGF120 mice was disrupted, we next determined the early consequence for the differentiation process. We examined expression of the T-box transcription factor 2 (Tbr2), that has been identified as a transcriptional marker of intermediate progenitor cells in the developing cortex (reviewed in (Hevner et al., 2006). Tbr2 is normally expressed as stem cells divide and move away from the ventricular zone, but remain in a mitotic state and mark the basal progenitor population of the subventricular zone (Ciruna and Rossant, 1999; Kimura et al., 1999; Sessa et al., 2008). In the wild type mice at E11.5 a layer of Tbr2-positive cells was observed approximately midway between the ventricular and pial surfaces (Figure 6A). Although a Tbr2-positive population was also detected in the VEGF120 mice, the cells were tightly clustered at the primitive cortical plate at the pial surface in a pattern distinct from the wild type.
Figure 6. Tbr2-positive intermediate progenitors are preferentially supported relative to neural stem cells in VEGF120 mice.
(A) Immunolabeling for Tbr2 is shown in E11.5 neuroepithelium from the anterior primitive telencephalon (Tn) with the lateral loop shown in low magnification on the left for wild type and VEGF120 (scale bar is 200 μm). The center panel shows Tbr2-immunopositive nuclei (red) with the ventricular surface at the top and the pial surface at the bottom. Positive cells highlight a secondary population of proliferating precursors intermediate between the ventricular zone and the pial surface in wild-type animals. A restricted band of Tbr2-positive cells is identified in the VEGF120 mice. The same area in overlay with DAPI stained nuclei (blue) is show on the right for comparison (scale bar is 50 μm). The samples shown are from littermates and the immunolabeling was repeated three times with similar results. Design-based stereological quantification of Tbr2-positive cells (B) and neuroepithelial volume (C) in wild type versus VEGF120 mice at E11.5. The optical dissector frame was 75 × 75 μm and the counting grid was set at 80 × 80 μm with a counting frame height of 18 μm and 2 μm guard zones on either side. Values shown are mean and SEM for three samples counted with the investigator masked to genotype. The p values for unpaired t tests are shown in the figures. The number of Tbr2-positive cells was then related to the number of PHH3-positive cells in matched animals and expressed as a ratio (D). The ratios were transformed to the natural log and significance tested with an unpaired t test; p values are indicated in the figure. (E) The number of mitotic spindles in a subset of PHH3-positive cells in the anterolateral telencephalon were quantified as parallel or oblique/perpendicular to the ventricular surface reflecting the orientation of the daughter cells after division. The optical dissector frame was 40 × 40 μm and the counting grid was set at 80 × 80 μm with a counting frame height of 18 μm and 2 μm guard zones on either side. The mean and SEM for three embryos is graphed and significance tested with an unpaired t test; p values are indicated in the figure.
We next quantified the number of Tbr2-positive cells in the E11.5 neuroepithelium to determine the changes in the intermediate progenitor population as a result of altered VEGF isoform profile. Because we saw a decrease in the number of PHH3-positive cells in the VEGF120 mice relative to the wild type, we anticipated seeing a corresponding reduction in the number of Tbr2-positive cells. To our surprise, there was no difference observed in the Tbr2 populations between wild type and VEGF120 (Figure 6B), although the total volume differences measured in this experiment were consistent with that observed for the PHH3-based total volume measurements (Figure 6C). We compared the Tbr2 counts to the PHH3 counts (refer to Figure 4) and expressed these as a ratio (Figure 6D). There was an increase in the number of Tbr2-positive nuclei relative to PHH3-positive nuclei in the VEGF120 mice compared to wild type. In order to assess the mechanism for this apparent shift in the neural stem cell versus intermediate progenitor population, we next quantified the number of total mitotic profiles and their position relative to the ventricular zone surface (Figure 6E). In the PHH3-positive population of cells of the anterolateral telencephalon, we determined that wild type and VEGF120 mice had similar numbers of mitotic spindle orientations that resulted in one daughter cell losing contact with the ventricular zone (90° angle or oblique angle). In contrast, the VEGF120 mice had significantly fewer mitotic spindles where both daughter cells would retain contact with the ventricular surface. Taken together, these results suggest that the VEGF120 supports the intermediate progenitor population at the expense of neural stem cell proliferation.
Discussion
Our results provide evidence for a role for VEGF in early neuroepithelial development in the mammalian cortex. VEGF is expressed temporally and spatially in a manner that is consistent with an effect on both the neural and vascular components of early forebrain expansion during the E9.5 to E11.5 developmental window. The availability of a line of mice that express only VEGF120 has allowed for direct testing of the hypothesis that VEGF164 is required for normal neuroepithelial development. VEGF120 mice, which lack the longer, matrix- and heparan sulfate proteoglycan-binding VEGF isoforms (VEGF164 and VEGF188), displayed reduced neuroepithelial proliferation. Several key genes that are involved in notch signaling (Delta, jagged 2, Dll1), or are associated with neural differentiation (FoxP1, Cux1), were differentially expressed in a microarray analysis of the E9.5 VEGF120 mice. The impact of these changes in gene expression manifested at E11.5 as significant alterations in the neuroepithelium. Levels of nestin mRNA were significantly reduced and there was a restricted area of immunolabeling for Pax6- and nestin-positive stem cell populations relative to wild type controls. However, the number of Tbr2-positive basal progenitors that form a secondary proliferative zone in the developing cortex was unchanged in the VEGF120 mice. Moreover, there was a corresponding reduction in the number of mitotic spindle profiles that would result in both daughter cells retaining contact with the ventricular zone and sustaining an active precursor population. Taken together, these data indicate that while VEGF120 alone is able to mediate some of the roles of VEGF in the early neuroepithelium, VEGF164 and VEGF188 appear to be required to sustain the neural precursor population prior to early cortical lamination.
One of the challenges associated with investigating a potential effect of VEGF on developing neural stem cells is the difficulty in determining whether an observed change is due to a direct effect of VEGF or due to immature or insufficient vascularization. The early forebrain neuroepithelium, which gives rise to many of the anterior brain structures, including the cortex, is vascularized from a neural plexus that invades the expanding neuroepithelium beginning at approximately E9.5. Clear evidence indicates that even as little as a two-fold deviation (increase or decrease) in VEGF levels will lead to abnormal vascularization resulting in increased apoptosis and reduced neuroepithelium (Haigh et al., 2003; Wada et al., 2006). Furthermore, examination of vascularization of the hindbrain of VEGF120 mouse from E10.5 to E13.5 revealed that the vessels in the established vessel network have reduced branching and a larger, distended lumen (Ruhrberg et al., 2002), although overall vascular investment was unaltered. Consistent with these results, the few initial vessels in the forebrain of the VEGF120 mice are grossly similar to those in the wild type embryos, although there are dramatic differences in telencephalic vascularization by E13.5. Thus, regional differences in the timing of vascularization may account for the lack of vessels at E11.5 since the telencephalon is not fully vascularized until E13.5 (Bär, 1980; Vasudevan et al., 2008). Our results suggest that VEGF120 alone is sufficient to support survival of the forebrain neuroepithelial cells, at least to the E11.5 stage where vascular compromise is not yet a significant factor. Studies using a nestin-promoter driven system to control stem cell-specific changes in VEGF expression levels resulted in significant increases in apoptosis that accompanied abnormal vascularization in neural tissues, specifically late stage cortex and retina (Haigh et al., 2003). We do not observe increased apoptosis or overtly abnormal vascularization in the VEGF120 mice at E11.5 (Figure 2 and data not shown), possibly due to the fact that our mouse model represents a shift in isoform profile expression rather than a change in total VEGF protein levels that do not impact vascularization significantly until later in telencephalon development.
One of the most revealing results of our investigations was the functional enrichment of the notch pathway moderating genes and the cortical neuron differentiation-associated genes in our microarray analysis. The DAVID platform proved to be an invaluable tool for mining genes in our microarray whose differential expression might affect neural precursor populations. The notch signaling pathway plays a variety of roles throughout all stages of development in a variety of tissues, particularly the vascular system (Gridley, 2001) and the CNS (Lathia et al., 2008). The genes present in the top two DAVID-enriched clusters collectively reflect a series of transcriptional changes that may be indicative of reduced notch signaling and precocious differentiation. The Dll1 gene, which was upregulated in the VEGF120 mice, has been shown to have differential effects on neural progenitor populations in the telencephalon (Yun et al., 2002) and depending on its expression level, can promote early neurogenesis (Kawaguchi et al., 2008) in the developing cortex. While all four Notch genes (Notch 1–4) were detected in our array, they were not differentially expressed (data not shown). However, the helix-loop-helix transcription factor, single-minded homolog 2 (Sim2) was upregulated in the VEGF120 mice and its expression has been shown to be downregulated by notch signaling induced in MCF10A breast cancer cells (Gustafson et al., 2009).
The possible downregulation of notch signaling would, indeed, lead to precocious differentiation as has been previously reported for several transgenic mouse models that result in disrupted notch-delta signaling (reviewed in (Lathia et al., 2008). Moreover, a background of increased Dll1/Delta/jagged 2 suppressing notch signaling at E9.5 correlates well with early increased expression of FoxP1 and Cux1 that mark cortical layer formation (Cubelos et al., 2008; Ferland et al., 2003; Nieto et al., 2004). We speculate that the marked change in the neuroepithleium at E11.5 that affects neural progenitor proliferation is the result of these early shifts in gene expression. While we observed variation in the total Tbr2 protein levels by western blot in E11.5 neuroepithelium (data not shown), there was no difference in total cell number indicating that absence of VEGF164 and VEGF188 had no significant impact on establishment of the intermediate progenitor population. Analysis at later developmental stages might reveal a greater impact of loss of VEGF164 and VEGF188 on the Tbr2 population or the post-mitotic Tbr1 population and its cortical derivatives. However, it would be difficult to interpret these studies as the expanding neuroepithelium has a growing dependence on vascularization that is increasingly compromised with further development.
Our observations indicate that the full profile of VEGF isoform expression is essential for the establishment and differentiation of the forebrain neuroepithelium and its vasculature. The reduction in overall proliferation and in parallel mitotic spindle profiles, leading daughter cells to maintain stem cell status via apical contact with the ventricular surface, are consistent with an alteration in notch signaling that is predicted by the microarray data. Under normal circumstances, VEGF164 is the predominant isoform in the brain with VEGF188 being expressed in only small amounts. Therefore, the phenotype observed in the VEGF120 mice may be due to absence of the predominant isoform, VEGF164, or due to the absence of any locally-retained VEGF protein (VEGF164 or VEGF188). The VEGF164 isoform may be required for full maintenance of the progenitor population and the VEGF120 alone may be insufficient to sustain normal proliferation and survival. The idea of specific roles for individual isoforms is not necessarily in contradiction to an integrated role for the isoforms based on matrix association versus diffusion. Such an isoform gradient model has been proposed previously for hindbrain and retinal vascularization (Gerhardt et al., 2004; Ishida, 2003; Ruhrberg et al., 2002). The gradient model would lead to the hypothesis that the local retention of VEGF164 and VEGF188 may act as a point source of VEGF at the pial surface while the diffusion of VEGF164 and VEGF120 would establish a gradient to support neural stem cell populations and guide migration of neuronal precursors and post-mitotic neurons as the layered cortex forms. Our results indicate that the absence of the locally-retained isoforms, VEGF164 and VEGF188, leads to reduced proliferation without impact on the number of Tbr2-positive intermediate progenitors. These data contribute to the growing evidence of the multi-faceted role that a traditional “vascular” factor plays during CNS development, contributing to neurovascular development at many levels.
Research Highlights.
VEGF isoforms are differentially expressed in the developing mouse forebrain
Expression of VEGF120 isoform results in reduced proliferation in neuroepithelium
The basal progenitor population is unaltered in VEGF120 mice
Fewer symmetric versus asymmetric mitotic spindle divisions occur in VEGF120 mice
Acknowledgments
The authors thank Dr. Andras Nagy for the use of the VEGF-lacZ mouse line. The authors thank Ken Drees of the UND Biology Department for his technical support with sequencing. The authors wish to acknowledge the help of the faculty startup funds from the University of North Dakota Biology Department, College of Arts and Sciences and the North Dakota EPSCoR program. These studies were supported by National Institutes of Health grants NIH-NINDS R15 NS057807-01/-02 and 3R15NS057807-01S1 (DCD), EY05318 and EY015435 (PAD) with additional support from a predoctoral award from the American Heart Association (AHA0715542Z, JTC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ara J, Fekete S, Zhu A, Frank M. Characterization of Neural Stem/Progenitor Cells Expressing VEGF and its Receptors in the Subventricular Zone of Newborn Piglet Brain. Neurochemical Research. 2010;35:1455–1470. doi: 10.1007/s11064-010-0207-2. [DOI] [PubMed] [Google Scholar]
- Ayadi A, Suelves M, DollÈ P, Wasylyk B. Net, an Ets ternary complex transcription factor, is expressed in sites of vasculogenesis, angiogenesis, and chondrogenesis during mouse development. Mechanisms of Development. 2001;102:205–208. doi: 10.1016/s0925-4773(01)00289-1. [DOI] [PubMed] [Google Scholar]
- Bär T. The vascular system of the cerebral cortex. Advances in Anatomy, Embryology, and Cell Biology. 1980;59:1–62. doi: 10.1007/978-3-642-67432-7. [DOI] [PubMed] [Google Scholar]
- Benezra R, Rafii S, Lyden D. The Id proteins and angiogenesis. Oncogene. 2001;20:8334–8341. doi: 10.1038/sj.onc.1205160. [DOI] [PubMed] [Google Scholar]
- Brazel CY, Romanko MJ, Rothstein RP, Levison SW. Roles of the mammalian subventricular zone in brain development. Progress in Neurobiology. 2003;603:1–21. doi: 10.1016/s0301-0082(03)00002-9. [DOI] [PubMed] [Google Scholar]
- Breier G, Clauss M, Risau W. Coordinate expression of vascular endothelial growth factor receptor-1 (flt-1) and its ligand suggests a paracrine regulation of murine vascular development. Developmental Dynamics. 1995;204:228–239. doi: 10.1002/aja.1002040303. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Ferriera V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Ng Y-S, Nuyen D, Theilmeier G, Brusselmans K, Cornelissen I, Ehler E, Kakkar VV, Stalmans I, Mattot V, Perriard J-C, Dewerchin M, Flameng W, Nagy A, Lupu F, Moons L, Collen D, D’Amore PA, Shima DT. Impaired myocardial angiogenesis and ischemic cardiomyopathy in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Nature Medicine. 1999;5:495–502. doi: 10.1038/8379. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mechanisms of Development. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Sebastián-Serrano A, Kim S, Redondo JM, Walsh C, Nieto M. Cux-1 and Cux-2 control the development of Reelin expressing cortical interneurons. Developmental Neurobiology. 2008;68:917–925. doi: 10.1002/dneu.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amore PA, Ng Y-S. Won’t you be my neighbor? Local induction of arteriogenesis. Cell. 2002;110:289–292. doi: 10.1016/s0092-8674(02)00869-3. [DOI] [PubMed] [Google Scholar]
- Darland DC, D’Amore PA. Cell-cell interactions in vascular development. Current Topics in Developmental Biology. 2001;52:107–149. doi: 10.1016/s0070-2153(01)52010-4. [DOI] [PubMed] [Google Scholar]
- Darland DC, Massingham LJ, Smith SR, Piek E, Saint-Geniez M, D’Amore PA. Pericyte production of cell-associated VEGF is differentiation-dependent and is associated with endothelial survival. Developmental Biology. 2003;264:275–288. doi: 10.1016/j.ydbio.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Dumont DJ, Fong GH, Puri MC, Gradwohl G, Alitalo K, Breitman ML. Vascularization of the mouse embryo: A study of flk-1, tek, tie</I>, and vascular endothelial growth factor expression during development. Developmental Dynamics. 1995;203:80–92. doi: 10.1002/aja.1002030109. [DOI] [PubMed] [Google Scholar]
- EMAGE. Electronic Mouse Atlas of Gene Expression. 2010. [DOI] [PubMed] [Google Scholar]
- Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. Journal of Comparative Neurology. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- Ferrara N. Molecular and biological properties of vascular endothelial growth factor. Journal of Molecular Medicine. 1999;77:527–543. doi: 10.1007/s001099900019. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea K, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Fishell G, Kriegstein AR. Neurons from radial glia: the consequences of asymmetric inheritance. Current Opinion in Neurobiology. 2003;13:34–41. doi: 10.1016/s0959-4388(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Gale NW, Yancopoulos GD. Growth factors acting via endothelial cell-specific receptor tyrosine kinases: VEGFs, angiopoietins, and ephrins in vascular development. Genes & Development. 1999;13:1055–66. doi: 10.1101/gad.13.9.1055. [DOI] [PubMed] [Google Scholar]
- Gerhardt H, Ruhrberg C, Abramsson A, Fujisawa H, Shima D, Betsholtz C. Neuropilin-1 is required for endothelial tip cell guidance in the developing central nervous system. Developmental Dynamics. 2004;231:503–509. doi: 10.1002/dvdy.20148. [DOI] [PubMed] [Google Scholar]
- Gilyarov AV. Nestin in Central Nervous System. Neuroscience and Behavioral Physiology. 2008;38:165–169. doi: 10.1007/s11055-008-0025-z. [DOI] [PubMed] [Google Scholar]
- Göetz M, Huttner WB. The cell biology of neurogenesis. Nature Reviews. Molecular Cell Biology. 2005;6:777–88. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Göetz M, Sommer L. Cortical development: the art of generating cell diversity. Development. 2005;132:3327–3332. doi: 10.1242/dev.01931. [DOI] [PubMed] [Google Scholar]
- Götz M, Stoykova A, Gruss P. Pax6 Controls Radial Glia Differentiation in the Cerebral Cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Greenberg DA, Jin K. From angiogenesis to neuropathology. Nature. 2005;438:954–9. doi: 10.1038/nature04481. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling during vascular development. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5377–5378. doi: 10.1073/pnas.101138098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson TL, Wellberg E, Laffin B, Schilling L, Metz RP, Zahnow CA, Porter WW. Ha-Ras transformation of MCF10A cells leads to repression of Singleminded-2s through NOTCH and C/EBP[beta] Oncogene. 2009;28:1561–1568. doi: 10.1038/onc.2008.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh JJ, Morelli PI, Gerhardt H, Haigh K, Tsien J, Damert A, Miquerol L, Muhlner U, Klein R, Ferrara N, Wagner EF, Betsholtz C, Nagy A. Cortical and retinal defects caused by dosage-dependent reductions in VEGF-A paracrine signaling. Developmental Biology. 2003;262:225–41. doi: 10.1016/s0012-1606(03)00356-7. [DOI] [PubMed] [Google Scholar]
- Haubst N, Georges-Labouesse E, De Arcangelis A, Mayer U, Gotz M. Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal subtypes. Development. 2006;133:3245–3254. doi: 10.1242/dev.02486. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RAM, Englund C. Transcription factors in glutamatergic neurogenesis: Conserved programs in neocortex, cerebellum, and adult hippocampus. Neuroscience Research. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis GJ, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biology. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck KA, Leung DW, Rowland AM, Winer J, Ferrara N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. Journal of Biological Chemistry. 1992;257:26031–26037. [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ishida S, Usui T, Yamashiro K, Kaji Y, Amano S, Ogura Y, Hida T, Oguchi Y, Ambati J, Ng Y-S, D’Amore PA, Shima DT, Adamis AP. VEGF164 -mediated inflammation is required for pathological, but not physiological, ischemia-induced retinal neovascularization. The Journal of Experimental Medicine. 2003;198:483–489. doi: 10.1084/jem.20022027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proceedings of the National Academy of Sciences. 2002;99:11946–11950. doi: 10.1073/pnas.182296499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi D, Yoshimatsu T, Hozumi K, Gotoh Y. Selection of differentiating cells by different levels of delta-like 1 among neural precursor cells in the developing mouse telencephalon. Development. 2008;135:3849–3858. doi: 10.1242/dev.024570. [DOI] [PubMed] [Google Scholar]
- Kimura N, Nakashima K, Ueno M, Kiyama H, Taga T. A novel mammalian T-box-containing gene, Tbr2, expressed in mouse developing brain. Developmental Brain Research. 1999;115:183–193. doi: 10.1016/s0165-3806(99)00064-4. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Mattson MP, Cheng A. Notch: from neural development to neurological disorders. Journal of Neurochemistry. 2008;107:1471–1481. doi: 10.1111/j.1471-4159.2008.05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Han J, Bai H-J, Kim K-W. Brain angiogenesis in developmental and pathological processes: regulation, molecular and cellular communication at the neurovascular interface. FEBS Journal. 2009;276:4622–4635. doi: 10.1111/j.1742-4658.2009.07174.x. [DOI] [PubMed] [Google Scholar]
- Lee S-W, Kim WJ, Choi YK, Song HS, Son MJ, Gelman IH, Kim Y-J, Kim K-W. SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nature Medicine. 2003;9:900–906. doi: 10.1038/nm889. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Gertsenstein M, Harpal K, Rossant J, Nagy A. Multiple developmental roles of VEGF suggested by a LacZ-tagged allele. Developmental Biology. 1999;212:307–22. doi: 10.1006/dbio.1999.9355. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–356. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Mori T, Buffo A, Götz M, Gerald PS. Current Topics in Developmental Biology. Vol. 69. Academic Press; 2005. The Novel Roles of Glial Cells Revisited: The Contribution of Radial Glia and Astrocytes to Neurogenesis; pp. 67–99. [DOI] [PubMed] [Google Scholar]
- Mukouyama Y-S, Shin D, Britsch S, Taniguchi M, Anderson DJ. Sensory nerves determine the pattern of arterial differentiation and blood vessel branching in the skin. Cell. 2002;109:693–705. doi: 10.1016/s0092-8674(02)00757-2. [DOI] [PubMed] [Google Scholar]
- Nasr MR, El-Zammar O. Comparison of pHH3, Ki-67, and survivin immunoreactivity in benign and malignant melanocytic lesions. American Journal of Dermatopathology. 2008;30:116–122. doi: 10.1097/DAD.0b013e3181624054. [DOI] [PubMed] [Google Scholar]
- Neufeld GTC, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB Journal. 1999;13:9–22. [PubMed] [Google Scholar]
- Ng Y-S, Rohan R, Sunday M, de Mello DE, D’Amore PA. Differential expression of VEGF isoforms in mouse during development and in the adult. Developmental Dynamics. 2001;220:112–121. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1093>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Nicholls JG, Martin AR, Wallace BG, Fuchs PA. Development of the Nervous System. Sinauer Associates, Inc; Sunderland, Massachusetts: 2001. [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II IV of the cerebral cortex. The Journal of Comparative Neurology. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Nomura T, Kawakami A, Ujisawa FH. Correlation between tectum formation and expression of two PAX family genes, PAX7 and PAX6, in avian brains. Development Growth & Differentiation. 1998;40:485–495. doi: 10.1046/j.1440-169x.1998.t01-3-00003.x. [DOI] [PubMed] [Google Scholar]
- Rasband WS. Image J. U.S. National Institutes of Health; Bethesda, Maryland, USA: 1997–2009. http://rsb.info.nih.gov/ij/ [Google Scholar]
- Rhen T, Metzger K, Schroeder A, Woodward R. Expression of putative sex-determining genes during the thermosensitive period of gonad development in the snapping turtle. Chelydra serpentina. Sexual Development. 2007;1:255–270. doi: 10.1159/000104775. [DOI] [PubMed] [Google Scholar]
- Richardson L, Venkataraman S, Stevenson P, Yang Y, Burton N, Rao J, Fisher M, Baldock RA, Davidson DR, Christiansen JH. EMAGE mouse embryo spatial gene expression database: 2010 update. Nucleic Acids Research. 2009;38:D703–D709. doi: 10.1093/nar/gkp763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein JM, Krum JM, Ruhrberg C. VEGF in the nervous system. Organogenesis. 2010;6:107–114. doi: 10.4161/org.6.2.11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhrberg C, Gerhardt H, Golding M, Watson R, Ioannidou S, Fujisawa H, Betsholtz C, Shima DT. Spatially restricted patterning cues provided by heparin-binding VEGF-A control blood vessel branching morphogenesis. Genes and Development. 2002;16:2684–98. doi: 10.1101/gad.242002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Almodovar C, Coulon C, Salin PA, Knevels E, Chounlamountri N, Poesen K, Hermans K, Lambrechts D, Van Geyte K, Dhondt J, Dresselaers T, Renaud J, Aragones J, Zacchigna S, Geudens I, Gall D, Stroobants S, Mutin M, Dassonville K, Storkebaum E, Jordan BF, Eriksson U, Moons L, D’Hooge R, Haigh JJ, Belin M-F, Schiffmann S, Van Hecke P, Gallez B, Vinckier S, Chédotal A, Honnorat J, Thomasset N, Carmeliet P, Meissirel C. Matrix-binding vascular endothelial growth factor (VEGF) isoforms guide granule cell migration in the cerebellum via VEGF receptor Flk1. The Journal of Neuroscience. 2010;30:15052–15066. doi: 10.1523/JNEUROSCI.0477-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Almodovar C, Lambrechts D, Mazzone M, Carmeliet P. Role and therapeutic potential of VEGF in the nervous system. Physiological Reviews. 2009;89:607–648. doi: 10.1152/physrev.00031.2008. [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends in Cell Biology. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Saint-Geniez M, Maharaj ASR, Walshe TE, Tucker BA, Sekiyama E, Kurihara T, Darland DC, Young MJ, D’Amore PA. Endogenous VEGF is required for visual function: evidence for a survival role on Müller cells and photoreceptors. PLoS ONE. 2008;3:e3554. doi: 10.1371/journal.pone.0003554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Geniez M, Maldonado AE, D’Amore PA. VEGF expression and receptor activation in the choroid during development and in the adult. Invest Ophthalmol Vis Sci. 2006;47:3135–3142. doi: 10.1167/iovs.05-1229. [DOI] [PubMed] [Google Scholar]
- Schmitz C, Hof PR. Design-based stereology in neuroscience. Neuroscience. 2005;130:813–831. doi: 10.1016/j.neuroscience.2004.08.050. [DOI] [PubMed] [Google Scholar]
- Sentilhes L, Michel C, Lecourtois M, Catteau J, Bourgeois P, Laudenbach V, Marret SAL. Vascular endothelial growth factor and its high-affinity receptor (VEGFR-2) are highly expressed in the human forebrain and cerebellum during development. Journal Neuropathology and Experimental Neurology. 2010;69:111–128. doi: 10.1097/NEN.0b013e3181ccc9a9. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao C, Hadjantonakis A-K, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by Indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu X-F, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- Shima DT, Kuroki M, Deutsch U, Ng Y-S, Adamis AP, D’Amore PA. The mouse gene for vascular endothelial growth factor. Genomic structure, definition of the transcriptional unit and characterization of transcriptional and post-transcriptional regulatory sequences. Journal of Biological Chemistry. 1996;271:3877–3883. doi: 10.1074/jbc.271.7.3877. [DOI] [PubMed] [Google Scholar]
- Shima DT, Mailhos C. Vascular developmental biology: getting nervous. Current Opinion Genes & Development. 2000:10. doi: 10.1016/s0959-437x(00)00124-6. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in Notch Signaling Regulate Maintenance of Neural Progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Sondell M, Lundborg G, Kanje M. Vascular endothelial growth factor has neurotrophic activity and stimulates axonal outgrowth, enhancing cell survival and Schwann cell proliferation in the peripheral nervous system. Journal of Neuroscience. 1999;19:5731–40. doi: 10.1523/JNEUROSCI.19-14-05731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondell M, Sundler F, Kanje M. Vascular endothelial growth factor is a neurotrophic factor which stimulates axonal outgrowth through the flk-1 receptor. European Journal of Neuroscience. 2000;12:4243–54. doi: 10.1046/j.0953-816x.2000.01326.x. [DOI] [PubMed] [Google Scholar]
- Stalmans I, Ng YS, Rohan R, Fruttiger M, Bouche A, Yuce A, Fujisawa H, Hermans B, Shani M, Jansen S, Hicklin D, Anderson DJ, Gardiner T, Hammes HP, Moons L, Dewerchin M, Collen D, Carmeliet P, D’Amore PA. Arteriolar and venular patterning in retinas of mice selectively expressing VEGF isoforms. Journal of Clinical Investigation. 2002;109:327–36. doi: 10.1172/JCI14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Goderie SK, Temple S. Asymmetric Distribution of EGFR Receptor during Mitosis Generates Diverse CNS Progenitor Cells. Neuron. 2005;45:873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jin K, Childs JT, Xie L, Mao XO, Greenberg DA. Vascular endothelial growth factor-B (VEGFB) stimulates neurogenesis: evidence from knockout mice and growth factor administration. Developmental Biology. 2006;289:329–335. doi: 10.1016/j.ydbio.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Therneau TM, Ballman KV. What does PLIER really do? Cancer Informatics. 2008;6:423–431. [PMC free article] [PubMed] [Google Scholar]
- Vasudevan A, Long JE, Crandall JE, Rubenstein JLR, Bhide PG. Compartment-specific transcription factors orchestrate angiogenesis gradients in the embryonic brain. Nature Neuroscience. 2008;11:429–439. doi: 10.1038/nn2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgintino D, Errede M, Robertson D, Girolamo F, Masciandaro A, Bertossi M. VEGF expression is developmentally regulated during human brain angiogenesis. Histochemistry & Cell Biology. 2003;119:227–232. doi: 10.1007/s00418-003-0510-y. [DOI] [PubMed] [Google Scholar]
- Wada T, Haigh JJ, Ema M, Hitoshi S, Chaddah R, Rossant J, Nagy A, van der Kooy D. Vascular endothelial growth factor directly inhibits primitive neural stem cell survival but promotes definitive neural stem cell survival. Journal of Neuroscience. 2006;26:6803–6812. doi: 10.1523/JNEUROSCI.0526-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfin A, Voisin M-B, Nourshargh S. PECAM-1: A multi-functional molecule in inflammation and vascular biology. Arteriosclerosis Thrombosis Vascular Biology. 2007;27:2514–2523. doi: 10.1161/ATVBAHA.107.151456. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Dumont DJ, Conlon RA, Breitman ML, Rossant J. flk-1, an flt-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development. 1993;118:489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- Yang K, Cepko CL. Flk-1, a receptor for vascular endothelial growth factor (VEGF), is expressed by retinal progenitor cells. Journal of Neuroscience. 1996;16:6089–99. doi: 10.1523/JNEUROSCI.16-19-06089.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, de Angelis MH, Weinmaster G, Rubenstein JLR. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Jin K, Mao XO, Greenberg DA. Vascular endothelial growth factor promotes proliferation of cortical neuron precursors by regulating E2F expression. FASEB Journal. 2003;17:186–193. doi: 10.1096/fj.02-0515com. [DOI] [PubMed] [Google Scholar]