Abstract
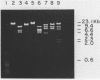
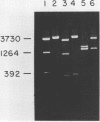
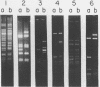
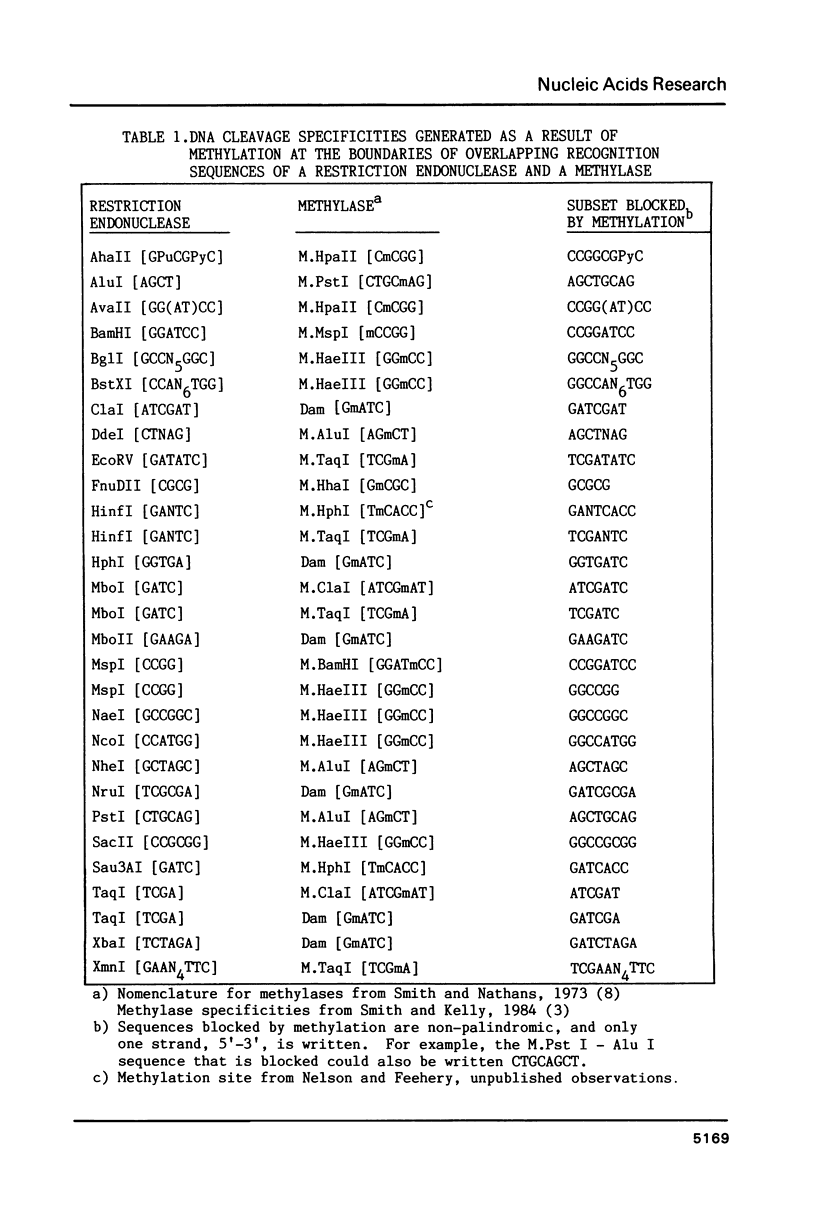
An in vitro method of altering the apparent cleavage specificities of restriction endonucleases was developed using DNA modification methylases. This method was used to reduce the number of cleavage sites for 34 restriction endonucleases. In particular, single-site cleavages were achieved for Nhe I in Adeno-2 DNA and for Acc I and Hinc II in pBR322 DNA by specifically methylating all but one recognition sequence.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K. A cautionary note on the use of certain restriction endonucleases with methylated substrates. Gene. 1980 Oct;11(1-2):169–171. doi: 10.1016/0378-1119(80)90097-9. [DOI] [PubMed] [Google Scholar]
- Brooks J. E., Roberts R. J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982 Feb 11;10(3):913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981 Jun 11;9(11):2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Purification and characterization of two new modification methylases: MClaI from Caryophanon latum L and MTaqI from Thermus aquaticus YTI. Nucleic Acids Res. 1981 Dec 21;9(24):6795–6804. doi: 10.1093/nar/9.24.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. The effect of site specific methylation on restriction endonuclease cleavage (update). Nucleic Acids Res. 1983 Jan 11;11(1):r169–r173. doi: 10.1093/nar/11.1.235-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyer-Weidner M., Jentsch S., Pawlek B., Günthert U., Trautner T. A. Restriction and modification in Bacillus subtilis: DNA methylation potential of the related bacteriophages Z, SPR, SP beta, phi 3T, and rho 11. J Virol. 1983 May;46(2):446–453. doi: 10.1128/jvi.46.2.446-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1984;12 (Suppl):r167–r204. doi: 10.1093/nar/12.suppl.r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]