Abstract
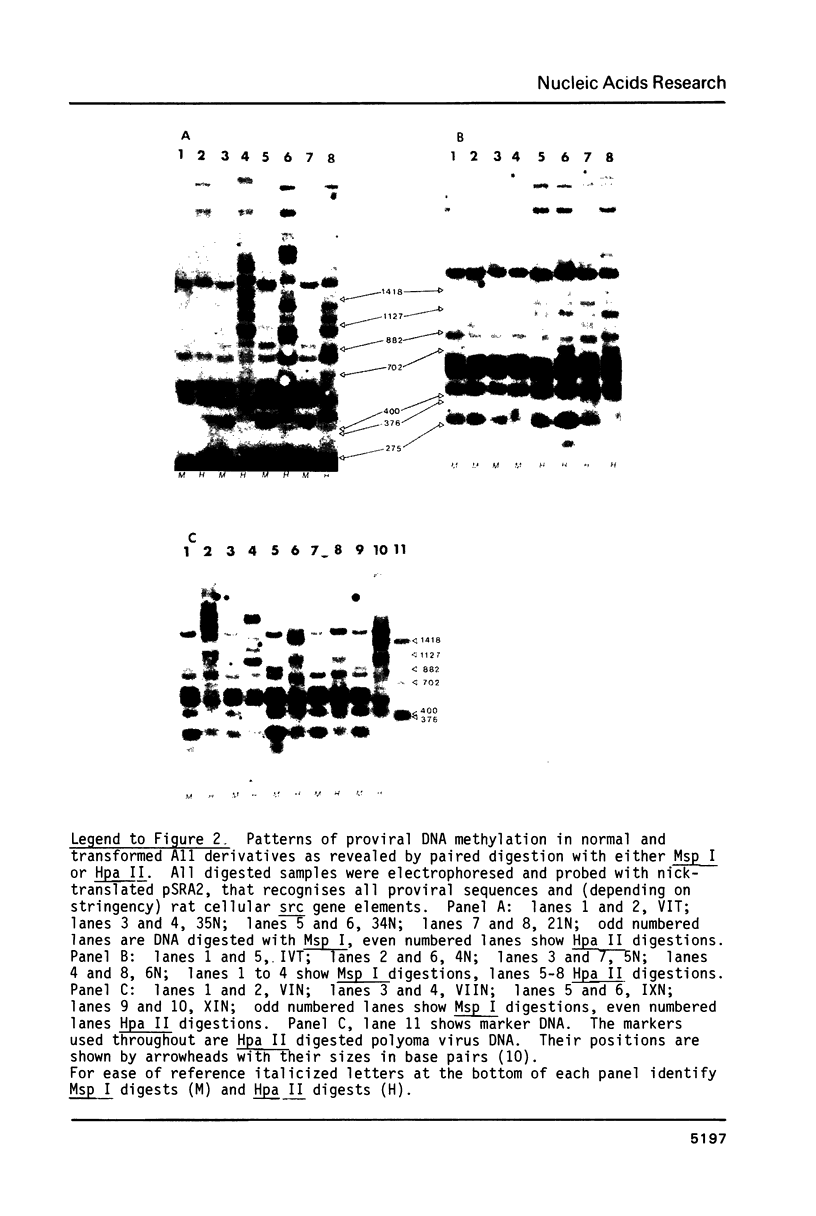
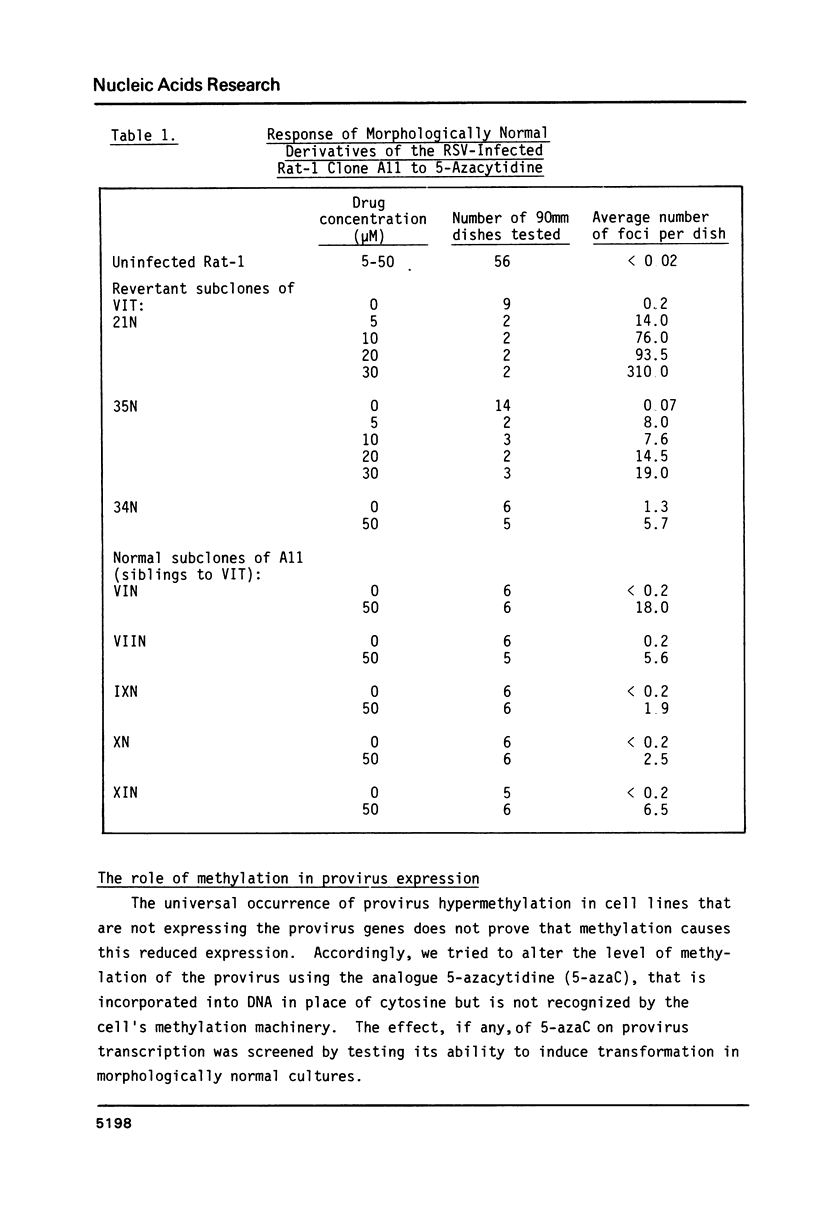
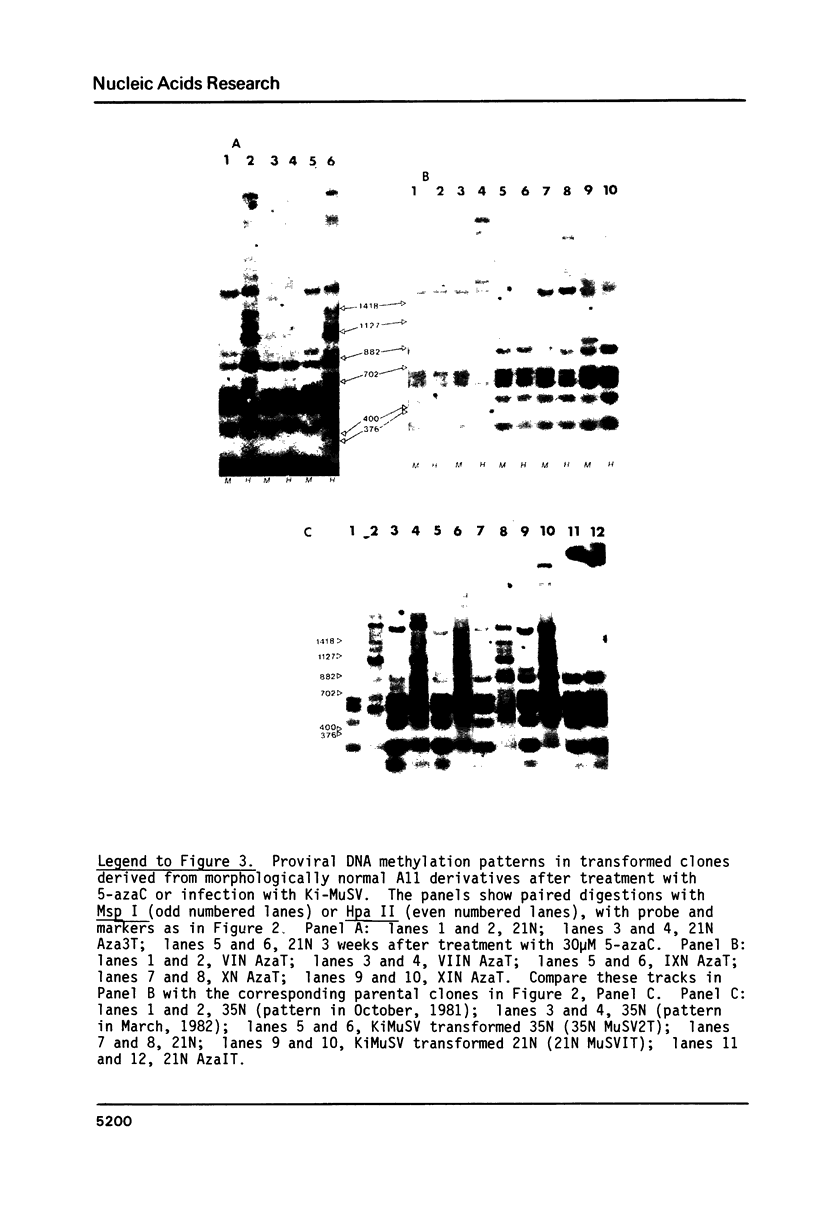
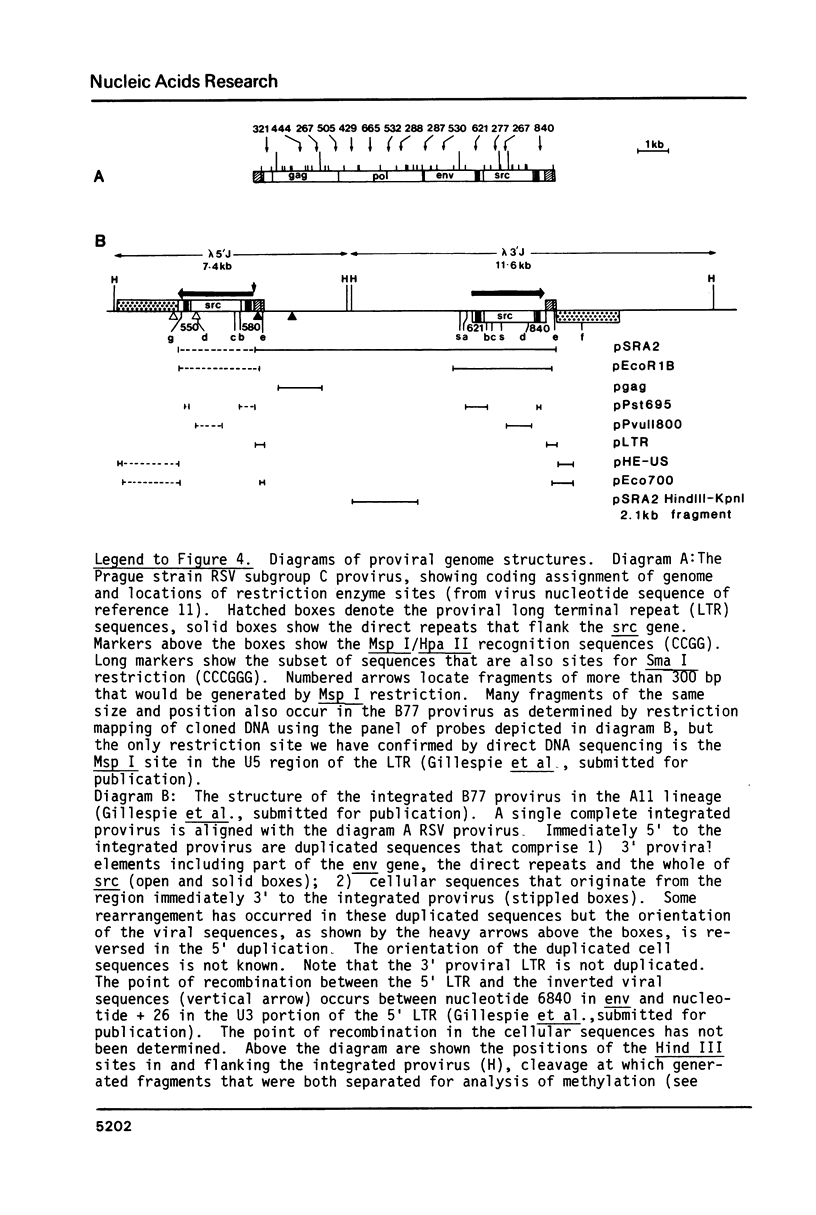
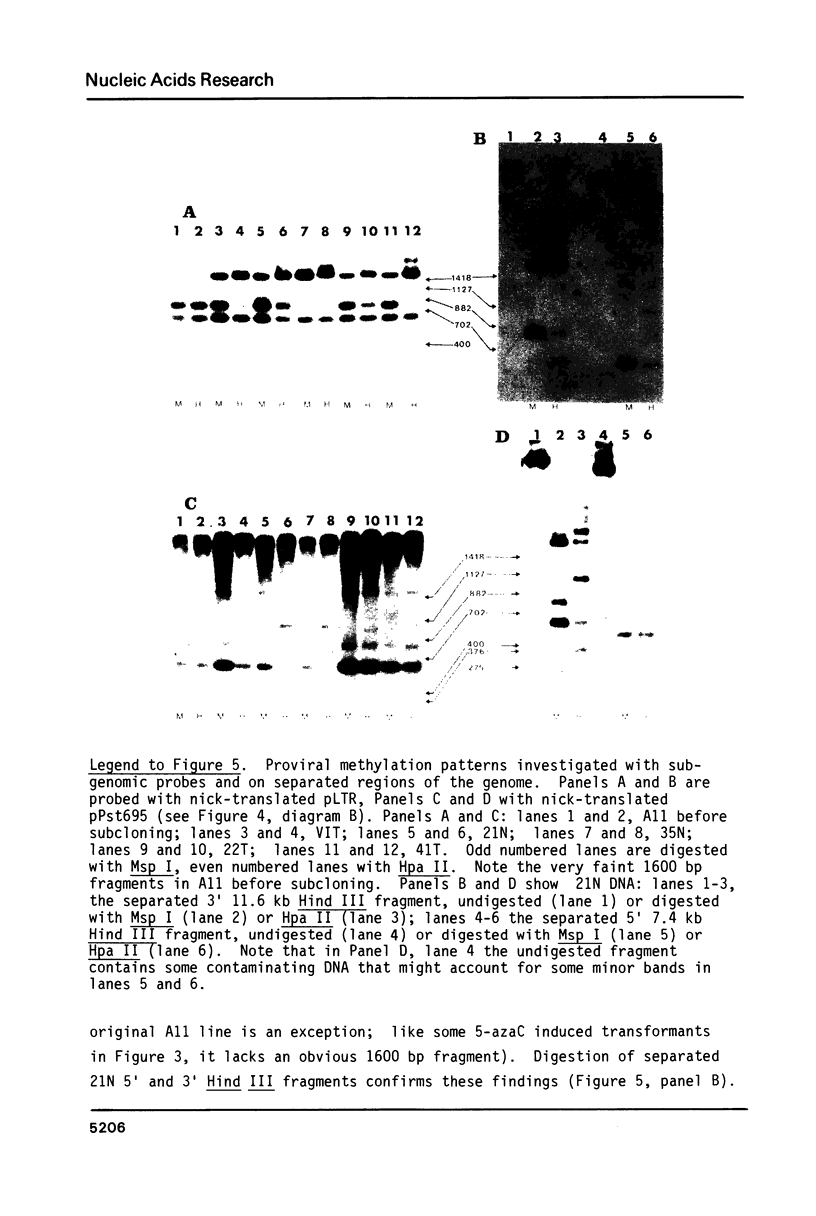
Cells of the A11 lineage of Rat-1 contain a single complete Rous sarcoma provirus. Variation in the activity of this provirus accompanies fluctuations in the lineage between normal and transformed phenotypes. Increased proviral cytosine methylation of the doublet CpG in the tetranucleotide CCGG correlates with transcriptional inactivity and this pattern of cytosine hypermethylation is stable, even when the cells are transformed by another virus. However, transformation can also be induced by 5-azacytidine (but not by other mutagens) and in these transformants reduced proviral cytosine methylation is accompanied by increased proviral transcription. Differences in CCGG methylation between normal and transformed cells are found mainly in the 3' half of the provirus; sites near and within the src gene are heavily methylated only when the provirus is transcriptionally inactive. On the other hand, both transformed and normal A11 derivatives show little, if any, cytosine methylation of CCGG sequences in and flanking the 5' portion of the provirus.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. DNA methylation--how important in gene control? Nature. 1984 Feb 9;307(5951):503–504. doi: 10.1038/307503a0. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Chiswell D. J., Enrietto P. J., Evans S., Quade K., Wyke J. A. Molecular mechanisms involved in morphological variation of avian sarcoma virus-infected rat cells. Virology. 1982 Jan 30;116(2):428–440. doi: 10.1016/0042-6822(82)90137-4. [DOI] [PubMed] [Google Scholar]
- Chiswell D. J., Gillespie D. A., Wyke J. A. The changes in proviral chromatin that accompany morphological variation in avian sarcoma virus-infected rat cells. Nucleic Acids Res. 1982 Jul 10;10(13):3967–3980. doi: 10.1093/nar/10.13.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Lang J., Hayday A., Lania L., Fried M., Chiswell D. J., Wyke J. A. Active viral genes in transformed cells lie close to the nuclear cage. EMBO J. 1982;1(4):447–452. doi: 10.1002/j.1460-2075.1982.tb01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorbe W. J., Luciw P. A., Goodman H. M., Varmus H. E., Bishop J. M. Molecular cloning and characterization of avian sarcoma virus circular DNA molecules. J Virol. 1980 Oct;36(1):50–61. doi: 10.1128/jvi.36.1.50-61.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W., Kruczek I., Eick D., Vardimon L., Kron B. DNA methylation and gene activity: the adenovirus system as a model. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):593–603. doi: 10.1101/sqb.1983.047.01.070. [DOI] [PubMed] [Google Scholar]
- Dyson P. J., Quade K., Wyke J. A. Expression of the ASV src gene in hybrids between normal and virally transformed cells: specific suppression occurs in some hybrids but not others. Cell. 1982 Sep;30(2):491–498. doi: 10.1016/0092-8674(82)90246-x. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Nickol J., Behe M., McGhee J., Jackson D. Methylation and chromatin structure. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):577–584. doi: 10.1101/sqb.1983.047.01.068. [DOI] [PubMed] [Google Scholar]
- Groffen J., Heisterkamp N., Blennerhassett G., Stephenson J. R. Regulation of viral and cellular oncogene expression by cytosine methylation. Virology. 1983 Apr 15;126(1):213–227. doi: 10.1016/0042-6822(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Hoffmann J. W., Steffen D., Gusella J., Tabin C., Bird S., Cowing D., Weinberg R. A. DNA methylation affecting the expression of murine leukemia proviruses. J Virol. 1982 Oct;44(1):144–157. doi: 10.1128/jvi.44.1.144-157.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Fanning T. G., Cardiff R. D. Mouse mammary tumor virus: specific methylation patterns of proviral DNA in normal mouse tissues. J Virol. 1984 Jan;49(1):66–71. doi: 10.1128/jvi.49.1.66-71.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathey-Prevot B., Shibuya M., Samarut J., Hanafusa H. Revertants and partial transformants of rat fibroblasts infected with Fujinami sarcoma virus. J Virol. 1984 May;50(2):325–334. doi: 10.1128/jvi.50.2.325-334.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeady M. L., Jhappan C., Ascione R., Vande Woude G. F. In vitro methylation of specific regions of the cloned Moloney sarcoma virus genome inhibits its transforming activity. Mol Cell Biol. 1983 Mar;3(3):305–314. doi: 10.1128/mcb.3.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas R. H., Wright C. A., Cockerill P. N., Wyke J. A., Goodwin G. H. The nuclease sensitivity of active genes. Nucleic Acids Res. 1983 Feb 11;11(3):753–772. doi: 10.1093/nar/11.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Shapiro L. J., Mohandas T. DNA methylation and the control of gene expression on the human X chromosome. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):631–637. doi: 10.1101/sqb.1983.047.01.074. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Stein R., Sciaky-Gallili N., Razin A., Cedar H. Pattern of methylation of two genes coding for housekeeping functions. Proc Natl Acad Sci U S A. 1983 May;80(9):2422–2426. doi: 10.1073/pnas.80.9.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmann H., Jähner D., Jaenisch R. Infectivity and methylation of retroviral genomes is correlated with expression in the animal. Cell. 1981 Oct;26(2 Pt 2):221–232. doi: 10.1016/0092-8674(81)90305-6. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Appella E., Jay G. Developmental activation of the H-2K gene is correlated with an increase in DNA methylation. Cell. 1983 Dec;35(2 Pt 1):457–465. doi: 10.1016/0092-8674(83)90179-4. [DOI] [PubMed] [Google Scholar]
- Wyke J. A., Quade K. Infection of rat cells by avian sarcoma virus: factors affecting transformation and subsequent reversion. Virology. 1980 Oct 30;106(2):217–233. doi: 10.1016/0042-6822(80)90246-9. [DOI] [PubMed] [Google Scholar]






