Methicillin-resistant Staphylococcus aureus (MRSA) colonization is a risk factor for infection in critically ill children. Almost half of children who acquired MRSA colonization in our ICU developed an MRSA infection during their hospitalization or after discharge, highlighting the importance of preventing nosocomial MRSA transmission.
Abstract
Background. Methicillin-resistant Staphylococcus aureus (MRSA) colonization is a predictor of subsequent infection in hospitalized adults. The risk of subsequent MRSA infections in hospitalized children colonized with MRSA is unknown.
Methods. Children admitted to an academic medical center’s pediatric intensive care unit between March 2007 and March 2010 were included in the study. Anterior naris swabs were cultured to identify children with MRSA colonization at admission. Laboratory databases were queried and National Healthcare Safety Network definitions applied to identify patients with MRSA infections during their hospitalization or after discharge.
Results. The MRSA admission prevalence among 3140 children was 4.9%. Overall, 56 children (1.8%) developed an MRSA infection, including 13 (8.5%) colonized on admission and 43 (1.4%) not colonized on admission (relative risk [RR], 5.9; 95% confidence interval [CI], 3.4–10.1). Of those, 10 children (0.3%) developed an MRSA infection during their hospitalization, including 3 of 153 children (1.9%) colonized on admission and 7 of 2987 children (0.2%) not colonized on admission (RR, 8.4; 95% CI, 2.7–25.8). African-Americans and those with public health insurance were more likely to get a subsequent infection (P < .01 and P = .03, respectively). We found that 15 children acquired MRSA colonization in the pediatric intensive care unit, and 7 (47%) developed a subsequent MRSA infection.
Conclusions. MRSA colonization is a risk factor for subsequent MRSA infection in children. Although MRSA colonized children may have lower risks of subsequent infection than adults, children who acquire MRSA in the hospital have similarly high rates of infection. Preventing transmission of MRSA in hospitalized children should remain a priority.
The community prevalence of methicillin-resistant Staphylococcus aureus (MRSA) is increasing due in large part to the epidemic of community-associated MRSA strains [1–3]. More children are being hospitalized with MRSA infections, and transmission of MRSA among children may lead to healthcare-associated infections (HAI) [4–10]. In the United States, MRSA causes 8% of all HAIs, and MRSA infections are associated with an estimated 18650 deaths annually in hospitalized patients [11, 12].
MRSA colonization in adults is a known risk factor for developing an MRSA infection [13, 14]. Adults who acquire MRSA colonization in the hospital have an even greater risk, up to 30%, of developing a subsequent MRSA infection [15]. In an attempt to prevent MRSA transmission and reduce MRSA infections, many hospitals have implemented comprehensive MRSA control programs, including screening high-risk patients for MRSA colonization and placing MRSA carriers in isolation with contact precautions [16]. For children, contact precautions and isolation may hinder family centered care and have unintended and lasting consequences [17]. Despite the increasing number of children colonized or infected with MRSA at time of admission to the hospital, the risk of subsequent infection in hospitalized children colonized with MRSA is unknown. Identifying whether similar risks exist in adults and children would support current pediatric MRSA control programs and inform healthcare workers who counsel families of MRSA-colonized children who worry about their child’s risk of a future infection [18, 19].
Our objective was to measure the risk of developing a subsequent MRSA infection in children colonized with MRSA.
MATERIALS AND METHODS
Setting and Design
The Johns Hopkins Hospital (JHH) is a 920-bed tertiary-care academic medical center with an embedded 175-bed Children’s Hospital. The 26-bed pediatric intensive care unit (PICU) admits ∼1700 medical and surgical patients each year, including hematopoietic stem cell–transplant and organ-transplant patients, as well as cardiac, orthopedic, and neurosurgical patients. Beginning 1 March 2007 naris swabs were performed by nurses to screen patients for MRSA colonization at the time of admission to PICU and weekly thereafter [20]. We retrospectively identified a cohort of patients screened for MRSA colonization on admission to the PICU between 1 March 2007 and 31 March 2010 with a hospital length of stay >2 days. We conducted an observational study to determine the risk of subsequent MRSA infection during each patient’s hospitalization and after discharge, including subsequent hospitalizations. For patients with multiple PICU admissions, the first admission within the study period was considered the patient’s entry into the cohort. The institutional review board approved this study with a waiver of informed consent to review retrospective data.
Definitions
MRSA colonization at the time of PICU admission was defined as having an admission naris surveillance culture grow MRSA or any clinical culture grow MRSA within 3 days of PICU admission [21]. Patients were considered previously colonized if they were not colonized at time of admission, but had an institutional history of MRSA colonization or infection. PICU-acquired MRSA cases became MRSA-colonized or MRSA-infected in the PICU and met the following criteria: (1) had a surveillance or clinical culture obtained >3 days after admission to the PICU growing MRSA [22], (2) had no institutional history of MRSA, and (3) had a previous naris culture that did not grow MRSA during the same PICU admission.
Data Collection and Case Identification
We queried a computerized surveillance support system (Theradoc) to identify cultures from any body site that grew MRSA between 1 March 2007 and 1 October 2010. The follow-up period included a minimum of 6 months following enrollment and ≤3.5 years for patients admitted at the start of the study period. 1 study team member (B. G.) reviewed medical records of patients in whom MRSA grew in a culture sent from JHH inpatient units, the JHH emergency department, and JHH outpatient clinics at the discretion of the patient’s treating clinician. National Healthcare Safety Network’s (NHSN’s) surveillance definitions for HAIs were applied to all cultures to distinguish infection versus colonization [23]. Administrative databases were queried and medical records were reviewed to obtain patient characteristics.
Laboratory Methods
Surveillance cultures were plated on selective agar and evaluated by pulsed-field gel electrophoresis (PFGE) as previously described [20, 24]. DNA was extracted and digested using Sma1 [25, 26]. We used Staphylococcus aureus subspecies NCTC 8325 as a control strain, and all USA PFGE strain types were included for comparison [27]. We considered isolates related if their patterns had ≤3 band differences [27] and isolates unrelated if there were >3 band differences. Strains were compared with USA type strains obtained through the Network of Antimicrobial Resistance in Staphylococcus aureus (NARSA) program supported under NIAID/NIH contract #HHSN272200700055C. CA-MRSA strains included those belonging to the PFGE types USA300 and USA400 [27]. Non–CA-MRSA strains included those belonging to other PFGE types. For isolates not available for strain typing, the original antibiotic susceptibility patterns were reviewed. Strains susceptible to clindamycin and trimethoprim/sulfamethoxazole were considered phenotypic CA-MRSA strains and those resistant to clindamycin or trimethoprim/sulfamethoxazole were considered phenotypic non–CA-MRSA strains based on institutional experience [28, 29].
Statistical Analysis
Data were maintained in Microsoft Access (2007) and analyzed using Stata version 11.0 (Stata Corp). Mean, median, and interquartile ranges (IQR) were calculated for select demographic information. Categorical variables were expressed as numbers and percentages. Comparison of the risk of subsequent infection between those colonized with MRSA at time of first PICU admission, and those not colonized with MRSA were made using a 2-sample test of proportions (Pearson χ2 or Fisher exact test). Kaplan–Meier survival estimates were calculated for patients with and without MRSA colonization, and the log rank test was used to analyze the risk of subsequent MRSA infection during the first hospitalization. A 2-tailed P value of <.05 was considered significant for all statistical tests.
RESULTS
Between 1 March 2007 and 31 March 2010, 3620 patients were admitted to the PICU. Of those admitted, 3140 (87%) were screened for MRSA at the time of admission and were eligible for analysis. The median age was 5.8 years (IQR, 1.0–12.8 years), and 56% were male (see Table 1). Of the 3140 screened, 153 patients (4.9%) were colonized or infected with MRSA at the time of admission to the PICU, including 131 patients (86%) detected by naris surveillance cultures and 22 patients (14%) who had a clinical culture grow MRSA within 3 days of PICU admission. At the time of PICU admission, 43 patients were not colonized with MRSA but had an institutional history of MRSA colonization or infection; they were categorized as not colonized on admission.
Table 1.
Clinical and Demographic Characteristics of Critically Ill Children Screened for Methicillin-Resistant Staphylococcus aureus Colonization at Time of Intensive Care Unit Admission
| Characteristics | |
| Total number of patients | 3140 |
| Median age, years (IQR) | 5.8 (1.0–12.8) |
| Male sex, no. (%) | 1744 (56) |
| Race, no. (%) | |
| Caucasian | 1637 (52) |
| African-American | 1106 (35) |
| Other | 397 (13) |
| ZIP code of residence, no. (%) | |
| Baltimore City | 989 (32) |
| Other | 2151 (68) |
| Insurance, no. (%) | |
| Private | 1668 (53) |
| Public | 1421 (45) |
| Self-pay | 40 (1) |
| Other | 11 (0.4) |
| MRSA colonized or infected at time of PICU admission, number (%) | 153 (4.9) |
| Previously but not currently MRSA Colonized at time of PICU admission, no. (%) | 43 (1.3) |
| Underlying conditions,a n (%) | |
| Neuromuscular | 1019 (32.4) |
| Cardiovascular | 704 (22.4) |
| Respiratory | 193 (6.1) |
| Renal | 130 (4.1) |
| Gastrointestinal | 75 (2.4) |
| Hematologic deficiencies and immunodeficiencies | 161 (5.1) |
| Metabolic | 139 (4.4) |
| Congenital and genetic | 391 (12.5) |
| Malignancy | 351 (11.2) |
| None | 1155 (36.8) |
| Length of hospital stay, median (IQR), days | 5 (4–11) |
| Length of ICU stay, median (IQR), days | 1.8 (1.0–3.7) |
Abbreviations: ICU, intensive care unit; IQR, interquartile range; MRSA, methicillin-resistant Staphylococcus aureus; PICU, pediatric intensive care unit.
Some patients had >1 underlying condition.
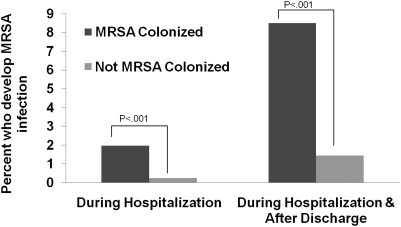
Overall, 56 patients (1.8%) developed a subsequent MRSA infection, including 13 of 153 children colonized on admission (8.5%; 95% confidence interval [CI], 4.5%–14.5%) and 43 of 2987 (1.4%; 95% CI, 1.0–1.9%) not colonized on admission (relative risk [RR] 5.9; 95% CI, 3.4–10.1) [See Figure 1]. Compared with noncolonized children, MRSA-colonized children had an increased risk of respiratory infections (RR 5.3; 95% CI, 2.4–11.8) and nonrespiratory infections (RR 6.5; 95% CI, 3.1–13.6). Of 56 infections, 46 (82%) occurred after hospital discharge, 22 (39%) during subsequent inpatient visits and 24 (43%) during outpatient visits. Of the 42 patients known to be previously colonized with MRSA but not colonized with MRSA at time of admission, 4 (9.5%) developed a MRSA infection, but the risk in this group was not significantly different from that in those colonized at time of PICU admission (RR 1.12; 95% CI, 0.4–3.3).
Figure 1.
Impact of methicillin-resistant Staphylococcus aureus (MRSA) colonization on risk of subsequent infection in critically ill children. The risk of developing a subsequent MRSA infection was compared for children with and without MRSA colonization at time of admission to the pediatric intensive care unit. Patients were monitored for an infection during their hospitalization and after hospital discharge.
Of all the patients, 10 (0.3%) developed a subsequent MRSA infection during their hospitalization, including 3 of 153 (1.9%; 95% CI, 0.4%–5.7%) children colonized on admission and 7 of 2987 (0.2%; 95% CI, .09%–.5%) not colonized on admission (RR 8.4; 95% CI, 2.7–25.8). Of the 42 patients previously colonized with MRSA and not colonized with MRSA at time of admission, none developed an infection during their hospitalization. The incidence rate of subsequent MRSA infections was higher in the colonized group (1.6 per 1000 patient-days) than in the noncolonized group (0.2 per 1000 patient-days) (incidence rate ratio [IRR] 6.9; 95% CI, 1.1–30.1). Patients colonized with MRSA on admission had a shorter time to development of a subsequent MRSA infection compared with those not colonized with MRSA (P < .01 by log-rank test) (Supplementary Figure 1).
Of the children with subsequent MRSA infections, the median age was 4 years at time of PICU admission (See Table 2). African-Americans were at increased risk for a subsequent infection compared with non–African-American patients (2.8 vs 1.3%; P < .01). Patients who lived in Baltimore City and those with public insurance were at increased risk for a subsequent MRSA infection (2.5% vs 1.4%; P = .03 and 2.5% vs 1.2%; P = .03, respectively). Most subsequent infections (50%) were respiratory infections, including pneumonia and tracheitis. There was only 1 bloodstream infection identified during the entire follow-up period.
Table 2.
Characteristics of 56 Patients With Subsequent Methicillin-Resistant Staphylococcus aureus Infections
| Characteristic | |
| Age at time of PICU admission, median years (IQR) | 4 (0.8, 12) |
| Race | |
| Caucasian | 21 (38%) |
| African-American | 30 (54%) |
| Other | 5 (8%) |
| MRSA colonized or infected at time of PICU admission, no. (%) | 13 (23.2) |
| Length of hospital stay, median (IQR) days | 9.5 (6–25.5) |
| Length of ICU stay, median (IQR) days | 3.5 (1.7–7.7) |
| Zip code of residence, no. (%) | |
| Baltimore City | 25 (45) |
| Other | 31 (55) |
| Insurance, no. (%) | |
| Private | 20 (36) |
| Public | 36 (64) |
| Time to infection, median (IQR) days | |
| Infections occurring during hospitalization (n = 10) | 8.5 (5–19) |
| Infections occurring during entire follow-up (n = 56) | 195 (37, 511) |
| Site of infection | |
| Pneumonia | 9 (16%) |
| Other respiratorya | 19 (34%) |
| Skin and soft tissue | 13 (23%) |
| Surgical site infection | 10 (18%) |
| Other | 3 (5%) |
| Bloodstream | 1 (2%) |
| Central nervous system | 1 (2%) |
Abbreviations: ICU, intensive care unit; IQR, interquartile range; MRSA, methicillin-resistant Staphylococcus aureus; PICU, pediatric intensive care unit.
Included tracheitis and tracheobronchitis (n = 18) and tonsillitis with abscess (n = 1).
To identify patients who acquired MRSA in the PICU, MRSA surveillance cultures from the naris were sent weekly, and clinical cultures from patients in the PICU for >3 days were monitored. During the study period, there were 15 incident MRSA cases in this cohort. Of these 15 patients, 7 (47%) developed a MRSA infection including 6 (40%) during their hospitalization. Patients who acquired MRSA in the PICU had a higher risk of infection compared with those colonized with MRSA at the time of PICU admission (RR 5.5; 95% CI, 2.5–11.8) and those not colonized with MRSA at the time of PICU admission (RR 32.4; 95% CI, 19.7–53.4). Combining those colonized on admission and those who acquired MRSA in the PICU, 20 of 168 (11.9%) patients known to be colonized during their hospitalization developed a subsequent MRSA infection.
Of the 153 patients colonized on admission, 133 (87%) had isolates available for PFGE analysis. Of these, 72 (61%) isolates were CA-MRSA strains identical to or related to PFGE types USA300, and 61 were non–CA-MRSA strains related to USA100 (n = 20), USA200 (n = 1), USA700 (n = 2), USA1100 (n = 2), or 21 other strains with unique PFGE fingerprints (n = 36). Phenotypic data were available on isolates from patients with a subsequent infection. CA-MRSA strains caused 35 (63%) infections and non CA-MRSA caused 21 (37%). Of the colonized patients who developed a subsequent infection, 7 colonized with USA300-related strains developed an infection with CA-MRSA strains. 5 patients colonized with USA100-related strains became infected, 4 with non–CA-MRSA strains and 1 with a CA-MRSA strain.
DISCUSSION
MRSA-colonized adults are at increased risk of developing a MRSA infection, but data are limited in children. We found that critically ill children colonized with MRSA were at significantly higher risk of subsequent MRSA infection than those not colonized with MRSA. Patients who acquired MRSA in the hospital were at greatest risk of developing a MRSA infection. This highlights the importance of preventing nosocomial MRSA transmission. Importantly, most infections (82%) occurred after hospital discharge. Genotypic and phenotypic data suggest that subsequent infections may be caused by endogenous colonizing strains in most cases.
Our data confirm observations in hospitalized adults that MRSA-colonized patients have an increased risk of subsequent MRSA infection compared with those not colonized with MRSA [13, 14]. Davis and colleagues reported a 9.5-fold increased risk of subsequent infection in critically ill adults colonized with MRSA compared with those not colonized with MRSA [13]. Similarly, we found an 8.4-fold increased risk in those colonized to develop an infection during their hospital admission and a 5.9-fold increased risk during their hospitalization and after discharge. Children who acquired MRSA in our PICU had a significantly greater chance of a subsequent MRSA infection compared with both those colonized and those not colonized at time of ICU admission. This increased risk of infection in those who acquire MRSA colonization in the ICU provides a strong argument for the importance of comprehensive strategies to prevent nosocomial MRSA transmission in children. Traditional MRSA-control strategies including patient isolation have been criticized, as isolated patients may experience more adverse events, be less likely to have vital signs recorded, and have more days without a physician progress note than nonisolated patients [30–32]. Isolation may create a feeling of social isolation and stigma for patients colonized with MRSA [18]. Therefore, given the high risk of infection in patients who acquire MRSA colonization in the hospital, future studies must identify strategies for MRSA prevention that have the least impact on the delivery of care to children.
Similar to hospitalized adults, critically ill children colonized with MRSA have a significant risk of a developing a MRSA infection. Overall, 11.9% of children colonized with MRSA in the ICU developed a MRSA infection, including 8.5% of those colonized on admission and 47% of those who acquired MRSA in the ICU. Subsequent rates of infection in adults have ranged from 19%–30% [13, 15]. Rates of infection may be lower in children who have fewer underlying complex chronic conditions and shorter lengths of ICU stay. Critically ill adults on average have greater severity of illness and a higher device utilization rate, which may increase the risk of infection during hospitalization. This hypothesis is supported by a study of neonates in the ICU, a population with prolonged ICU stays and high device-use rates, in which 26% of colonized neonates developed an MRSA infection [33].
Consistent with previous findings, our data showed that many MRSA infections occur after hospital discharge [11]. Caregivers of children colonized with MRSA frequently have concerns about their child’s risk of subsequent infection [18]. Our data will enable healthcare workers to counsel families about the risks during the child’s hospitalization and after discharge. Strategies to decrease the risk of infection following discharge, such as decolonization, should be considered.
Our data confirm findings from previous studies that have found racial disparities in rates of invasive MRSA disease [3, 34, 35]. Klevens et al found that African-Americans had a higher incidence of invasive MRSA disease than Caucasians and mortality rates that were 80% higher [11]. The reason for these racial disparities is unknown. Previous pediatric studies in hospitalized and healthy children found African-Americans were more likely to have MRSA colonization [20, 36]. Because colonization is a known risk factor for invasive MRSA disease, higher colonization rates in African-Americans may in part explain a disproportionate rate of invasive disease in this group. Findings from our study may help to explain this association. Our data suggest that those with public insurance had an increased risk of infection. Public insurance may represent lower socioeconomic status. Although this study was not designed to identify independent risk factors for infection, after adjusting for insurance status, the association between race and infection was no longer significant (data not shown). Therefore, although African-Americans are more likely to develop an MRSA infection, other important characteristics, such as socioeconomic status, may explain the association between race and infection. Additional large studies are needed to explore racial disparities in MRSA colonization and infection.
CA-MRSA strains, especially PFGE type USA300, continue to be the most common MRSA strain colonizing children admitted to our PICU. We found that 61% of children colonized with MRSA at the time of PICU admission harbored a CA-MRSA strain. The high prevalence of USA300 strains may be multifactorial. First, the community prevalence of MRSA colonization in children is increasing in the Baltimore community and nationwide [1, 2, 20, 28, 37], in large part because of the spread of CA-MRSA strains. Second, children generally have less-frequent exposure to healthcare facilities and traditional HA-MRSA strains. Third, children routinely are in close contact environments where opportunities for hygiene may be limited, such as day care, schools, and sports teams, areas postulated as high-risk environments for MRSA transmission. All patients colonized with CA-MRSA strains developed infections with strains with a CA-MRSA phenotype. Similarly, 80% of children colonized with non–CA-MRSA developed infection with a non–CA-MRSA phenotype. In this small sample of colonized patients who developed subsequent infection, the concordance of colonizing and infecting strains was high.
Several limitations should be considered when interpreting our data. First, only naris cultures were performed to identify asymptomatic MRSA carriers at the time of admission to the PICU. Studies have shown screening extranasal anatomic sites, such as throat, axilla, perineum and stool can increase the detection of MRSA carriers [38, 39], especially those colonized with CA-MRSA strains. Therefore, we may have misclassified colonized patients as noncolonized, including those with a history of MRSA colonization that had a negative naris culture on admission. Thus the infection rate in those not colonized on admission may have been overestimated. Second, we likely underestimated rates of infection after hospital discharge as we relied on inpatient and outpatient cultures processed in our institution’s clinical microbiology laboratory. We did not capture infections that occurred at other healthcare facilities. We do not think that this would have led to a differential bias between children colonized and not colonized on admission to the ICU as a similar percentage of both groups (76% and 79%) visited our institution during the follow-up period. Third, we had relatively few events, so confirming these findings in another cohort would support the conclusions. Finally, we included only patients admitted to the PICU, so our findings may not be generalizable to non-ICU populations.
Overall, our study found that MRSA colonization increased the risk of subsequent MRSA infection in children. Children who acquired MRSA in the ICU were at especially high risk, further emphasizing the need to prevent nosocomial transmission of MRSA. A high risk of infection in select groups of children offers an opportunity to intervene to prevent MRSA infections. MRSA decolonization is not standard practice in hospitalized children. Future studies should address not just interventions to protect the population from MRSA transmission, but interventions such as decolonization to protect the individual child from subsequent infection.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://www.oxfordjournals.org/our_journals/cid/).
Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments.
We thank Sonali Advani and Alicia Budd for their assistance, The JHH Microbiology laboratory staff, the JHH PICU nursing staff, and The JHH Hospital Epidemiology and Infection Control Group for their support of this study.
Financial support.
This work was supported by the National Institute for Allergy and Infectious Diseases at the National Institutes of Health (K23 AI081752-02 to A. M.); and Johns Hopkins School of Medicine Medical Student Research Award (to B. G.).
Potential conflicts of interest.
A. M. and T. P. received grant support from Sage Products. A. M. received grant support from BioMerieux. T. P. received grant support from Merck and is on Advisory Boards for Hospira and BioMerieux. K. C. has received research funds from BD Diagnostics, Akonni, and Microphage, and is on the Scientific Advisory Boards of Quidel and NanoMR. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Creech CB, 2nd, Kernodle DS, Alsentzer A, Wilson C, Edwards KM. Increasing rates of nasal carriage of methicillin-resistant Staphylococcus aureus in healthy children. Pediatr Infect Dis J. 2005;24:617–21. doi: 10.1097/01.inf.0000168746.62226.a4. [DOI] [PubMed] [Google Scholar]
- 2.Gorwitz RJ, Kruszon-Moran D, McAllister SK, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis. 2008;197:1226–34. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 3.Moran GJ, Krishnadasan A, Gorwitz RJ, et al. EMERGEncy ID Net Study Group. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–74. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 4.Campbell AL, Bryant KA, Stover B, Marshall GS. Epidemiology of methicillin-resistant Staphylococcus aureus at a children's hospital. Infect Control Hosp Epidemiol. 2003;24:427–30. doi: 10.1086/502226. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan SL, Hulten KG, Gonzalez BE, et al. Three-year surveillance of community-acquired Staphylococcus aureus infections in children. Clin Infect Dis. 2005;40:1785–91. doi: 10.1086/430312. [DOI] [PubMed] [Google Scholar]
- 6.Zaoutis TE, Toltzis P, Chu J, et al. Clinical and molecular epidemiology of community-acquired methicillin-resistant Staphylococcus aureus infections among children with risk factors for health care–associated infection: 2001–2003. Pediatr Infect Dis J. 2006;25:343–8. doi: 10.1097/01.inf.0000207403.67197.cc. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Community-associated methicillin-resistant Staphylococcus aureus infection among healthy newborns–Chicago and Los Angeles County, 2004. MMWR Morb Mortal Wkly Rep. 2006;55:329–32. [PubMed] [Google Scholar]
- 8.Bratu S, Eramo A, Kopec R, et al. Community-associated methicillin-resistant Staphylococcus aureus in hospital nursery and maternity units. Emerg Infect Dis. 2005;11:808–13. doi: 10.3201/eid1106.040885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Healy CM, Hulten KG, Palazzi DL, Campbell JR, Baker CJ. Emergence of new strains of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Clin Infect Dis. 2004;39:1460–6. doi: 10.1086/425321. [DOI] [PubMed] [Google Scholar]
- 10.Seybold U, Kourbatova EV, Johnson JG, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care–associated blood stream infections. Clin Infect Dis. 2006;42:647–56. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 11.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 12.Hidron AI, Edwards JR, Patel J, et al. National Healthcare Safety Network Team. NHSN annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 13.Davis KA, Stewart JJ, Crouch HK, Florez CE, Hospenthal DR. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin Infect Dis. 2004;39:776–82. doi: 10.1086/422997. [DOI] [PubMed] [Google Scholar]
- 14.Corbella X, Dominguez MA, Pujol M, et al. Staphylococcus aureus nasal carriage as a marker for subsequent staphylococcal infections in intensive care unit patients. Eur J Clin Microbiol Infect Dis. 1997;16:351–7. doi: 10.1007/BF01726362. [DOI] [PubMed] [Google Scholar]
- 15.Huang SS, Platt R. Risk of methicillin-resistant Staphylococcus aureus infection after previous infection or colonization. Clin Infect Dis. 2003;36:281–5. doi: 10.1086/345955. [DOI] [PubMed] [Google Scholar]
- 16.Muto CA, Jernigan JA, Ostrowsky BE, et al. Society for Healthcare Epidemiology of America. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect Control Hosp Epidemiol. 2003;24:362–86. doi: 10.1086/502213. [DOI] [PubMed] [Google Scholar]
- 17.Briggs JJ, Milstone AM. Changes over time in caregivers' knowledge, attitudes, and behaviors regarding methicillin-resistant Staphylococcus aureus. J Pediatr. 2011;158:416–421. doi: 10.1016/j.jpeds.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sengupta A, Rand C, Perl TM, Milstone AM. Knowledge, awareness, and attitudes regarding methicillin-resistant Staphylococcus aureus among caregivers of hospitalized children. J Pediatr. 2011;158:416–21. doi: 10.1016/j.jpeds.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koller DF, Nicholas DB, Goldie RS, Gearing R, Selkirk EK. When family-centered care is challenged by infectious disease: pediatric health care delivery during the SARS outbreaks. Qual Health Res. 2006;16:47–60. doi: 10.1177/1049732305284010. [DOI] [PubMed] [Google Scholar]
- 20.Milstone AM, Carroll KC, Ross T, Shangraw KA, Perl TM. Community-associated methicillin-resistant Staphylococcus aureus strains in pediatric intensive care unit. Emerg Infect Dis. 2010;16:647–55. doi: 10.3201/eid1604.090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren DK, Guth RM, Coopersmith CM, Merz LR, Zack JE, Fraser VJ. Epidemiology of methicillin-resistant Staphylococcus aureus colonization in a surgical intensive care unit. Infect Control Hosp Epidemiol. 2006;27:1032–40. doi: 10.1086/507919. [DOI] [PubMed] [Google Scholar]
- 22.Cohen AL, Calfee D, Fridkin SK, et al. Society for Healthcare Epidemiology of America, Healthcare Infection Control Practices Advisory Committee. Recommendations for metrics for multidrug-resistant organisms in healthcare settings: SHEA/HICPAC Position paper. Infect Control Hosp Epidemiol. 2008;29:901–13. doi: 10.1086/591741. [DOI] [PubMed] [Google Scholar]
- 23.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care–associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–32. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Milstone AM, Song X, Beers C, Berkowitz I, Carroll KC, Perl TM. Unrecognized burden of methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus carriage in the pediatric intensive care unit. Infect Control Hosp Epidemiol. 2008;29:1174–6. doi: 10.1086/592093. [DOI] [PubMed] [Google Scholar]
- 25.Murray BE, Singh KV, Heath JD, Sharma BR, Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–63. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen AE, Goldstein M, Carroll K, Song X, Perl TM, Siberry GK. Evolving epidemiology of pediatric Staphylococcus aureus cutaneous infections in a Baltimore hospital. Pediatr Emerg Care. 2006;22:717–23. doi: 10.1097/01.pec.0000236832.23947.a0. [DOI] [PubMed] [Google Scholar]
- 29.Farley JE, Ross T, Stamper P, Baucom S, Larson E, Carroll KC. Prevalence, risk factors, and molecular epidemiology of methicillin-resistant Staphylococcus aureus among newly arrested men in Baltimore, Maryland. Am J Infect Control. 2008;36:644–50. doi: 10.1016/j.ajic.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stelfox HT, Bates DW, Redelmeier DA. Safety of patients isolated for infection control. JAMA. 2003;290:1899–905. doi: 10.1001/jama.290.14.1899. [DOI] [PubMed] [Google Scholar]
- 31.Evans HL, Shaffer MM, Hughes MG, et al. Contact isolation in surgical patients: A barrier to care? Surgery. 2003;134:180–8. doi: 10.1067/msy.2003.222. [DOI] [PubMed] [Google Scholar]
- 32.Kirkland KB, Weinstein JM. Adverse effects of contact isolation. Lancet. 1999;354:1177–8. doi: 10.1016/S0140-6736(99)04196-3. [DOI] [PubMed] [Google Scholar]
- 33.Huang YC, Chou YH, Su LH, Lien RI, Lin TY. Methicillin-resistant Staphylococcus aureus colonization and its association with infection among infants hospitalized in neonatal intensive care units. Pediatrics. 2006;118:469–74. doi: 10.1542/peds.2006-0254. [DOI] [PubMed] [Google Scholar]
- 34.Fridkin SK, Hageman JC, Morrison M, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–44. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 35.Sattler CA, Mason EO, Jr, Kaplan SL. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr Infect Dis J. 2002;21:910–7. doi: 10.1097/00006454-200210000-00005. [DOI] [PubMed] [Google Scholar]
- 36.Fritz SA, Garbutt J, Elward A, Shannon W, Storch GA. Prevalence of and risk factors for community-acquired methicillin-resistant and methicillin-sensitive staphylococcus aureus colonization in children seen in a practice-based research network. Pediatrics. 2008;121:1090–8. doi: 10.1542/peds.2007-2104. [DOI] [PubMed] [Google Scholar]
- 37.Szczesiul JM, Shermock KM, Murtaza UI, Siberry GK. No decrease in clindamycin susceptibility despite increased use of clindamycin for pediatric community–associated methicillin-resistant Staphylococcus aureus skin infections. Pediatr Infect Dis J. 2007;26:852–4. doi: 10.1097/INF.0b013e318124aa5c. [DOI] [PubMed] [Google Scholar]
- 38.Rohr U, Mueller C, Wilhelm M, Muhr G, Gatermann S. Methicillin-resistant Staphylococcus aureus whole-body decolonization among hospitalized patients with variable site colonization by using mupirocin in combination with octenidine dihydrochloride. J Hosp Infect. 2003;54:305–9. doi: 10.1016/s0195-6701(03)00140-3. [DOI] [PubMed] [Google Scholar]
- 39.Ringberg H, Petersson AC, Walder M, Hugo Johansson PJ. The throat: An important site for MRSA colonization. Scand J Infect Dis. 2006;38:888–93. doi: 10.1080/00365540600740546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.