Abstract
Here, we report on the complete genome sequence of the hyperthermophilic Crenarchaeum Thermoproteus tenax (strain Kra1, DSM 2078T) a type strain of the crenarchaeotal order Thermoproteales. Its circular 1.84-megabase genome harbors no extrachromosomal elements and 2,051 open reading frames are identified, covering 90.6% of the complete sequence, which represents a high coding density. Derived from the gene content, T. tenax is a representative member of the Crenarchaeota. The organism is strictly anaerobic and sulfur-dependent with optimal growth at 86°C and pH 5.6. One particular feature is the great metabolic versatility, which is not accompanied by a distinct increase of genome size or information density as compared to other Crenarchaeota. T. tenax is able to grow chemolithoautotrophically (CO2/H2) as well as chemoorganoheterotrophically in presence of various organic substrates. All pathways for synthesizing the 20 proteinogenic amino acids are present. In addition, two presumably complete gene sets for NADH:quinone oxidoreductase (complex I) were identified in the genome and there is evidence that either NADH or reduced ferredoxin might serve as electron donor. Beside the typical archaeal A0A1-ATP synthase, a membrane-bound pyrophosphatase is found, which might contribute to energy conservation. Surprisingly, all genes required for dissimilatory sulfate reduction are present, which is confirmed by growth experiments. Mentionable is furthermore, the presence of two proteins (ParA family ATPase, actin-like protein) that might be involved in cell division in Thermoproteales, where the ESCRT system is absent, and of genes involved in genetic competence (DprA, ComF) that is so far unique within Archaea.
Introduction
Thermoproteus tenax has been the first hyperthermophilic Archaeum described by the pioneering work of Wolfram Zillig and Karl O. Stetter [1], [2]. The strain Kra1 was originally isolated from a solfatare in Iceland [1]. It belongs to the Crenarchaeota and bears important taxonomical meaning for that phylum, representing the type strain of the genus Thermoproteus, which is the type genus of the family Thermoproteaceae [1].
In addition to its hyperthermophilic lifestyle (optimal growth at 86°C and maximal growth at 96°C), the organism is able to grow chemolithoautotrophically in the presence of hydrogen and carbon dioxide [2] as well as chemoorganoheterotrophically on a variety of mono-, di- and polysaccharides, organic acids and alcohols (e.g. glucose, malate, amylase, starch or ethanol) [1]. Less efficient growth has been observed with propionate and casamino acids as substrates. The universal electron acceptor is elemental sulfur, however, polysulfides and thiosulfate are also utilized [1].
In this paper, we describe the complete genome sequence of T. tenax (strain Kra1, DSM 2078T), which gives new insights into the physiological versatility and regulatory potential of this organism.
So far, only 26 crenarchaeotal genomes, of which eleven belong to the genus Sulfolobus and four to the genus Pyrobaculum (according to NCBI, http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi) have been sequenced, versus a total of 53 euryarchaeal genomes, two thaumarchaeal genomes (Cenarchaeum symbiosum, Nitrosopumilus maritimus) [3], [4] and two, not yet validly described and classified strains, i.e. Nanoarchaeum equitans [5], and Candidatus Korarchaeum cryptofilum [6] (www.genomesonline.org) [7]. In addition, the T. tenax genome is of special interest, since it is meanwhile adopted that the related Thermoproteus neutrophilus (strain V24Sta) obviously belongs to the genus Pyrobaculum. Therefore, T. tenax represents the first member of the genus Thermoproteus with available whole genome sequence information. Thus, the present study will not only contribute to unravel unique traits of this organism, but will also contribute to balance the disproportion between the known genomic content of Crenarchaeaota and Euryarchaeota. The here reported detailed genomic analysis, reveals new insights into the physiology as well as genetics and information processing of T. tenax. In addition to the previously suggested reductive TCA cyle [8], [9], all genes encoding enzymes of the novel dicarboxylate/4-hydroxybutyrate cycle [10] were identified, thus, raising questions about the activity of both pathways. In accordance with its autotrophic lifestyle, all pathways for the synthesis of the 20 proteinogenic amino acids were identified in T. tenax. Interestingly, the organism harbors the typical bacterial pathways for the complex branched chain and aromatic amino acid biosynthesis and in addition, archaeal routes, e.g. for proline biosynthesis.
Under autotrophic growth conditions T. tenax seems to gain energy by hydrogen oxidation via a single Iron-Nickel hydrogenase and sulfur reductase, which form a short electron transport chain probably mediated by quinones. Energy conservation under heterotrophic growth conditions seems to proceed via a membrane-bound electron transport chain and sulfur has been suggested as final acceptor. Interestingly, two complete operons encoding proteins of complex I (NADH:quinone oxidoreductase) were identified and the genome data give some evidence that either NADH or reduced ferredoxin can serve as electron donator. The presence of the three subunits for NADH binding and oxidation (Nqo1–3 or NuoEFG, NuoG gives ambiguous results) is so far rare for an anaerobic Archaeum. Beside the structurally unusual archaeal A0A1-ATP synthase, a membrane-bound pyrophosphatase seems to be involved in chemiosmosis. The biggest surprise, in respect to physiology, was the identification of all genes required for dissimilatory sulfate reduction and, indeed, growth in the presence of sulfate as terminal electron acceptor could be observed (unpublished data).
Protein transport in T. tenax seems to proceed via the “Sec translocase” secretion pathway as well as the twin arginine translocation (Tat) system. For ion and metabolite transport, as in most Archaea a PEP-dependent phosphotransferase (PTS) system is absent and T. tenax harbors about twice as much secondary transporter compared to ABC transporters. Information processing (i.e. replication, transcription, translation) in T. tenax resembles, like in all Archaea, the respective eukaryal counterparts. Interesting is the finding of four different TFB homologs in T. tenax. Multiplicity of general transcription factors is commonly found in Archaea and a function similar to sigma factors has been proposed previously [11]. In the T. tenax genome no extrachromosomal elements were identified. However, seven clusters of CRISPRs as well as Cas proteins were identified in the genome; the spacer sequences do not show similarity to archaeal viruses and plasmids, which are known to infect or transform T. tenax.
T. tenax harbors the archaeal gene core (157 genes) as well as all 234 Crenarchaeota-specific arCOGs as revealed by comparative genomic analyses. In the Thermoproteales lineage, 19 core gene families have been acquired specifically among those a ParA family ATPase and an actin-like protein. This is of special interest, since the ESCRT system, identified as the major system for cell division in Archaea [12], is missing in Thermoproteales. In addition, six T. tenax specific arCOGS were identified, which are absent in all other crenarchaeal genomes, and among those are genes involved in genetic competence and uptake of DNA (DprA, ComF), which have not been detected in Archaea before.
Results and Discussion
General genome features
The genome of T. tenax consists of a circular chromosome of 1,841,542 bp with an average G+C content of 55.1%. No extra-chromosomal elements remained after the genome sequence assembly. Analysis of the cumulative GC skew of the draft genome sequence was used in search for the origin of replication (http://mips.gsf.de/services/analysis/genskew); the genome sequence was subsequently reorganized, so that the global minimum of the GC skew marks the beginning of the genome sequence (bp 1). However, the only copy of a cdc6 gene, which together with the global minimum of the GC-skew and the ORB-motif is supposed to be a marker for archaeal replication origins [13], is located far away at about 1.6 Mbp (TTX_1848), and the only conserved ORB-motif is located at position 58,820-58,094. Therefore, given the scattered distribution of these three elements, the location of the origin of replication stays uncertain.
Overall 2,051 predicted protein encoding open reading frames (ORFs) remained in the consensus gene set after manual deletion of small, most probably artificial ORFs, covering a total of 90.6% of the genome, which is, as in the closely related Thermofilum pendens (91%) only slightly higher than the values for most other sequenced Crenarchaeota, e.g. Aeropyrum pernix (89.1%), P. aerophilum (88%) or Sulfolobus solfataricus and S. tokodaii (85%). Only one copy for each rRNA gene, 5S (unlinked), 16S, and 23S rRNA, respectively, had been identified in the genome. As common for the Crenarchaeota, many of the 47 annotated transfer RNA genes contain an intron (see below and Table 1). Genes encoding the stable RNA components of RNaseP or the signal recognition particle (7S RNA) are absent, like in most other Thermoproteaceae (according to Rfam database (http://rfam.sanger.ac.uk/) [14]. About 75% of the predicted 2,051 protein coding sequences (1,552 ORFs), could be linked with a putative function, whereas most recent Crenarchaeota annotations name about 60% genes with predicted functions. Twenty-four percent (a total of 497) ORFs were assigned as (conserved) hypothetical or uncharacterized conserved proteins. Totally, 76.6% (1,572) of all predicted proteins were linked to COGs [15] and 95% (1,953) to arCOGs [16], which is slightly above average for crenarchaeotal genomes. Only about 4% (a total of 91) of the 2,051 predicted proteins appear to be unique for T. tenax.
Table 1. General genome features of T. tenax.
| Genome size | 1,841,542 bp | |
| G+C content | 1,015,210 bp | 55.13% |
| Coding region | 1,669,147 bp | 90.6% |
| CRISPRs (total of 149 repeat modules) | 7 | |
| Stable RNAs | ||
| 23S rRNA | 1 (1,700,676-1,703,699) | |
| 16S rRNA | 1 (1,699,104-1,700,606) | |
| 5S rRNA | 1 (1,298,635-1,298,753) | |
| tRNAs (13 with canonical introns) | 46 (plus one pseudogene) | |
| Predicted protein coding ORFs | 2,051 (1.1 per kb) | |
| Average ORF length | 813 bp | |
| Average intergenic region | 127.6 bp | |
| Predicted functions assigned | 1,552 | 75.7% |
| (Conserved) hypothetical and uncharacterized conserved proteins | 497 | 24.2% |
| No annotation | 2 | <0.1% |
| Assignment to COGs | 1,572 | 76.6% |
| Assignment to arCOGs | 1,953 | 95.2% |
| ORFs unique for T. tenax | 91 | 4.4% |
| ORFs for proteins with signal peptides | 56 | 2.7% |
| ORFs for proteins with transmembrane helices | 412 | 20.1% |
About 2.7% (a total of 56) of the predicted proteins possess a signal peptide. The fraction of transmembrane proteins (20.1%, a total of 412) is normal within the Crenarchaeota. No genes required for the usage of selenocysteine as 21st amino acid were identified. Inteins could not be detected in any of the predicted proteins.
Genes involved in lipopolysaccharide (LPS) synthesis are frequently clustered in regions of microbial genomes that differ significantly from their average G+C content [17]. The function in Archaea is still unclear, since Archaea generally harbor no outer membrane (except Ignicoccus hospitalis; [18]) and LPS, commonly found in Gram-negative Bacteria. The T. tenax genome contains three extended regions of low G+C content (<47%, Table S1). Sixteen of the 23 genes encoded in the largest of these regions (region 3) have functions required for or linked to LPS synthesis, including nine type I/II glycosyltransferases, two polysaccharide biosynthesis proteins, two N-acetyl-glucosaminyl-phosphatidylinositol synthesis proteins, LPS-biosynthesis glycosyltransferase and a membrane protein involved in export of O-antigen. Low G+C region 1 encodes only the three subunits of an ABC transporter that might play a role in the transport of sugar monomers across the periplasm. For the LPS genes encoded in region 3, there is no evidence for a common origin via lateral gene transfer from a donor with low G+C content. Some of the genes in this cluster are most similar to homologs found in a variety of other Archaea, whereas others are most similar to bacterial homologs. Gene duplication in T. tenax as the source of the ten glycosyltransferases in this region can be excluded, because the encoded proteins share a higher degree of sequence similarity with homologs from other organism than between each other.
The largest protein in the genome, encoded by TTX_1887 (2,663 amino acids, corresponding molecular mass of 287 kDa), is a candidate for the S-layer protein, as it shows several of the required features: (i) the protein is rich in serine, threonine, and asparagine as putative glycosylation sites, (ii) it has an N-terminal signal sequence, and (iii) a C-terminal TM helix. Therefore, it is predicted to be anchored in the cytoplasmic membrane facing the environment [19], [20]. When using the NetNGlyc and NetOGlyc servers (http://www.cbs.dtu.dk/services/NetNGlyc/ and http://www.cbs.dtu.dk/services/NetOGlyc/) [21] for glycosylation prediction, five putative O-glycosylation site are predicted and multiple N-glycosylation sites. The genome contains seven (low copy number, 2–5 copies) repeats longer than 300 bp with more than 95% sequence conservation. The longest of these repeats is a pair of cobyrinic acid a,c-diamide synthase genes, cbiA (TTX_0412 and TTX_1195); another pair contains putative cobalamin adenosyltransferases (TTX_0290 and TTX_1504). Five ORFs (TTX_0813, TTX_0867, TTX_1864, TTX_1903, TTX_1904) encoding putative transposases or fragments of inactivated transposases are identified in the T. tenax genome, indicating the rare presence of genetically mobile IS-elements.
In the genome of T. Tenax, seven clusters of regularly interspaced short palindromic repeats (CRISPRs) could be identified (coverage 0.5%; Table S2). In general, Archaea show in comparison to Bacteria very extensive CRISPR clusters and have a highly divergent gene organization of the strictly associated cas genes [22], [23], [24]. The CRISPR/Cas system is supposed to guide antiviral defence by sequence similarity between spacer and phage genome, but also to limit horizontal gene transfer by preventing conjugation and plasmid transformation [25], [26], [27].
The five type I repeat clusters are significantly longer than the two type II clusters (Table S2) and show a larger variation in the lengths of the spacer sequences: 37–55 bp versus 41–48 bp. The two types of clusters also differ significantly in the length of their leader sequences. Leader sequences of type I are shorter than those of type II (317–327 bp versus 613–624 bp) [23], and also show a higher degree of sequence similarity between each other. The spacer sequences between the repeat units in CRISPRs are considered to derive from extra-chromosomal elements [24], [28], but homology searches revealed no significant matches between the spacer sequences of T. tenax CRISPRs and archaeal viruses and plasmids, which are known to transform T. tenax (TTV1, TTSV1, and PSV) [29], [30], [31]. The conserved genes cas1 to 5 and the gene of a putative HD-domain superfamily hydrolase (TTX_1254) are clustered between CRISPR 5 and CRISPR 6 and occur near one of the organisms̀ repeat clusters. Thus, the CRISPR organisation of T. tenax corresponds to the A. pernix subtype [22]. Like in other Crenarchaeota, these genes are associated with three genes belonging to COG4343 (TTX_1248), COG1857 (TTX_1251), and COG0640 (TTX_1249).
All general genome features are summarized in Table 1. Table S8 provides all identified T. tenax genes including gene IDs, functional assignments as well as the GenBank GI accession numbers for BLASTP best hits against NCBI Non-redundant database (e-value cutoff 0.001).
Central metabolism
Central carbohydrate metabolism (CCM)
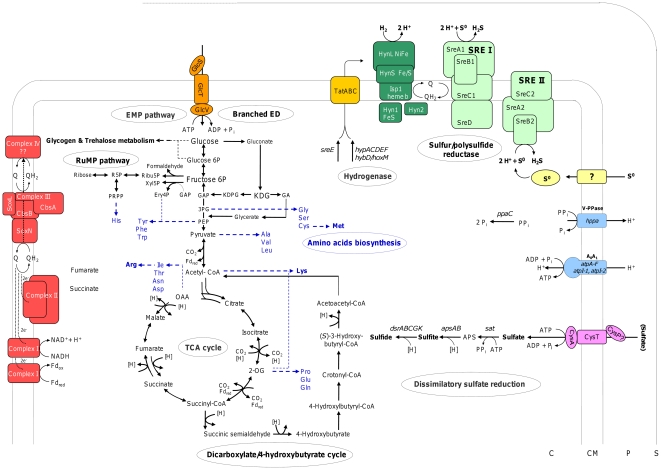
The CCM of T. tenax has been studied in great detail, and genome analysis in combination with biochemical studies revealed the presence of a modified reversible EMP pathway as well as an unusual branched ED pathway for sugar degradation (Figure 1) (for review see [32]). Organic substrates are completely oxidized to CO2 via the oxidative TCA cycle [33], whereas CO2-fixation under autotrophic growth conditions has been assumed to proceed via the reductive TCA cycle (Figure 1) [8], [9], [32]. However, recent studies proposed a novel dicarboxylate/4-hydroxybutyrate cycle for autotrophic CO2 fixation as common CO2 fixation mechanism within autotrophic members of the Thermoproteales [10], [34], [35]. Surprisingly, in the genome of T. tenax all required genes for a functional dicarboxylate/4-hydroxybutyrate cycle could be identified (Figure 1; Table 2). Therefore, experimental analyses have to be awaited in order to elucidate the role of both pathways in CO2 fixation in T. tenax. Recent studies revealed that the conventional oxidative pentose phosphate pathway (OPPP), which is essential for the generation of pentoses, reducing power (NADPH) and erythrose 4-phosphate (E4P) for amino acid biosynthesis, is generally absent in Archaea [36], [37]. Beside the non-oxidative pentose phosphate pathway (NOPPP), the so-called reversed ribulose monophosphate (RuMP) pathway has been shown to provide pentoses for anabolic purposes in most Archaea [36], [37]. The pathway is characterized by the two enzymes 3-hexulose-6-phosphate isomerase (PHI) and 3-hexulose-6-phosphate synthase (HPS) that catalyze the isomerization of fructose 6-phosphate (F6P) to 3-hexulose-6-phosphate and the reversible cleavage into formaldehyde and ribulose 5-phosphate (Ru5P; Figure 1). The HPS-PHI fusion proteins from Pyrococcus horikoshii [38] and Thermococcus kodakaraensis [39] have recently been characterized. In the genome of T. tenax two single ORFs, TTX_1521 and TTX_1049 have been identified, which code for a single HPS and PHI, respectively [40].
Figure 1. Central metabolic pathways of T. tenax.
Pathways for carbon metabolism (glucose uptake, glucose metabolism, carbon dioxide fixation, amino acid biosynthesis) and energy metabolism (electron transfer chains, A1A0-ATP synthase, PPiase, sulfate reduction) are depicted. The amino acid pathways are indicated in blue, dashed lines in order to better distinguish from the other carbon and energy metabolic pathways. Assumed electron transport is given in dotted lines and trehalose as well as glycogen metabolism are implied in black, dashed lines. For abbreviations and gene IDs see respective text sections.
Table 2. T. tenax candidate genes coding for the key enzymes involved in the dicarboxylate/4-hydroxbutyrate cycle.
| Enzyme | T. tenax ORF ID | Gene ID best hit aa identity and e-value |
| PEP-carboxylase (pepC) | TTX_1107 | Tneu_0418 (100%, 0.0) |
| Igni_0341 (91%, 6e-60) | ||
| Succinyl-CoA reductase | TTX_1101 | Tneu_0421 (99%, 0.0) |
| Succinic semialdehyde reductase | TTX_1106 | Tneu_0419 (100%, 5e-138) |
| 4-Hydroxybutyrate-CoA ligase (AMP-forming) | TTX_1100 | Tneu_0420 (82%, 0.0) |
| 4-Hydroxybutyryl-CoA dehydratase | TTX_1102 | Tneu_0422 (100%, 0.0) |
| Crotonyl-CoA hydratase/(S)-3-Hydroxybutyryl-CoA DH | TTX_1028 | Tneu_0541 (99%, 0.0) |
| Igni_1058 (99%, 1e-160) | ||
| Acetoacetyl-CoA β-ketothiolase | TTX_0886 | Tneu_0249 (99%, 1e-166) |
| Igni_1401 (99%, 2e-104) |
For the biosynthesis of the aromatic amino acids erythrose 4-phosphate (E4P) is required as precursor, which is formed from F6P and glyceraldehyde 3-phosphate via transketolase (Figure 1). In T. tenax two ORFs encoding the N- and the C-terminus of transketolase (tktA, tktB; TTX_1754, TTX_1753) have been identified, which cluster with genes involved in the synthesis of the aromatic amino acids.
Amino acid biosynthesis
From the genome data it can be assumed that T. tenax possesses pathways for the biosynthesis of all 20 proteinogenic amino acids (Figure 1; Table S3). Most of the genes involved in amino acid biosynthesis are organized in large gene clusters, e.g. genes involved in histidine, aromatic and branched chain amino acid synthesis (Table S3). Interestingly, most of the reconstructed pathways resemble the common pathways of the Bacteria (e.g. Escherichia coli, Bacillus subtilis) and the Eucarya (e.g. yeast). For example, all genes encoding enzymes required for the complex biosynthesis of tryptophane, tyrosine and phenylalanine from phosphoenolpyruvate and E4P via shikimate and chorismate, could be identified in the T. tenax genome (Table S3). There is no evidence for the existence of the recently described archaeal aspartate-semialdehyde pathway [41].
Also, all genes encoding the enzymes for the conventional synthesis of the branched chain amino acids valine, leucine and isoleucine, could be found in T. tenax (Table S3). Interestingly, this is contrast to the closely related T. neutrophilus [42] as well as other Archaea [43], [44], which use the citramalate cycle. In many Archaea, most of the genes encoding enzymes of the conventional pathway for proline synthesis from glutamate (e.g. glutamate 5-kinase; EC 2.7.2.11) are absent, and it has previously been shown that varying pathways for the synthesis of proline are used [45], [46], [47]. From the genome data two possible routes for the synthesis of proline could be proposed in T. tenax: (i) Biosynthesis from glutamate via 1-pyrroline-5-carboxylate dehydrogenase (putA, EC1.5.1.12; TTX_1787), which catalyzes the formation of γ-glutamic semialdehyde leading to pyrroline-5-carboxylate, and pyrroline-5-carboxylate reductase (proC, EC1.5.1.2; TTX_1730) converting pyrroline-5-carboxylate into proline. (ii) Cyclization of the non-proteinogenic amino acid L-ornithine catalyzed by the ornithine cyclodeaminase (OCD, arcB, EC 4.3.1.12; TTX_2070, TTX_0618), like it is described for M. jannaschii [45].
The genome data further indicate that arginine is most likely not synthesized from glutamate via the conventional route due to the lack of a gene encoding acetylglutamate synthase (EC 2.3.1.1) catalyzing the first step of arginine synthesis. However, alternative pathways for arginine synthesis either from carbamoyl-phosphate (via ornithine carbamoyltransferase (argF, EC 2.1.3.3; TTX_0091), argininosuccinate synthase (argG, EC 6.3.4.5; TTX_0123) and argininosuccinate lyase (argH, EC 4.3.2.1; TTX_0467) in the urea cycle, or from aspartate (via argininosuccinate synthase (argG, EC 6.3.4.5; TTX_0123) and argininosuccinate lyase (argH, EC 4.3.2.1; TTX_0467) could be identified in T. tenax.
For methionine synthesis, T. tenax most likely uses a pathway starting from homocysteine via methionine synthase (metE, EC 2.1.1.14; TTX_1021), like it is also described for M. thermautotrophicus [48] and supposed for M. jannaschii [49].
Like in many other Archaea, e.g. T. neutrophilus [42], P. aerophilum [50], I. hospitalis [44], and lower Eucarya [51], lysine synthesis in T. tenax proceeds via the aminoadipate pathway from 2-oxoglutarate and acetyl-CoA. The complete set of genes has been identified in the T. tenax genome (Table S3), whereas four of nine genes encoding enzymes required for the alternative synthesis of lysine via the widespread diaminopimelate pathway [52] are missing.
All but one gene (hisB) encoding enzymes for histidine synthesis from phosphoribosyl pyrophosphate (PRPP) have been identified in the T. tenax genome (Table S3). However, the lack of histidinol phosphatase (hisB, EC 3.1.3.15) has also previously been reported in other Archaea (e.g. Archaeoglobus fulgidus, M. thermautotrophicus [53], [47], [54] and a substitution of histidinol phosphatase by HD superfamily phosphohydrolases [55] has been suggested [47]. The T. tenax ORF TTX_1708 is coding for a phosphohydrolase of the HD superfamily (COG1078).
Energy metabolism
Chemolithoautotrophic growth
Like other obligatory sulfur reducers (e.g. Acidianus ambivalens, Pyrodictium occultum, Pyrodictium abyssi and T. neutrophilus), T. tenax gains energy from anaerobic H2 oxidation with sulfur as terminal electron acceptor (hydrogen-sulfur autotrophy).
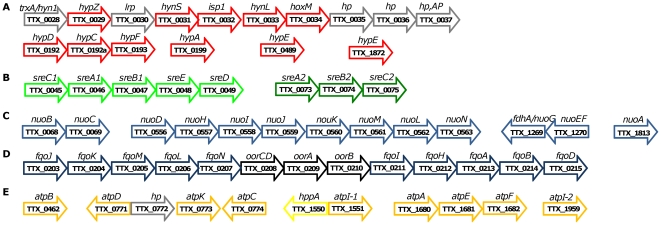
Hydrogen oxidation and sulfur reduction require the presence of a hydrogenase and a sulfur reductase. The T. tenax genome contains a single set of genes encoding the Iron-Nickel hydrogenase subunits including the large NiFe subunit HynL (TTX_0033), the smaller FeS subunit HynS (TTX_0031), and the membrane anchor protein Isp1 (hemeB; TTX_0032). The corresponding accessory genes required for the maturation of HynL (hypACDEF TTX_0192, TTX_192a, TTX_0193, TTX_0199; TTX_0489; TTX_1872; Fig. 2A) and two maturation proteases hybD/hoxM (TTX_0029, TTX_0034) are scattered over the genome. The presence of a single set of hydrogenase genes suggests that the gene products are responsible for hydrogen uptake during chemolithoautotrophic growth (Figure 1).
Figure 2. Gene organization.
Genes encoding Iron-Nickel hydrogenase (including accessory genes) (A), sulfur/polysulfide reductases (B), and gene clusters of the two sets of complex I (C and D) as well as A1A0-ATP synthase (E) from T. tenax are shown. The annotated ORFs and their orientation is indicated by arrows (not to scale), the gene names and the respective gene IDs are given. A: trxA/hyn1 – Thioredoxin/Rieske ferrdoxin; hypZ – [Ni,Fe]-hydrogenase maturation factor; lrp – Transcriptional regulator, Lrp/AsnC family; hynS – [Ni,Fe]-hydrogenase I, small (41 kDa) subunit; isp1- [Ni,Fe]-hydrogenase, cytochrome b subunit (29 kDa); hynL - [Ni,Fe]-hydrogenase I, large (66 kDa) subunit; hoxM - [Ni,Fe]- hydrogenase maturation factor for HynL; hp – hypothetical protein; hp,AP - phosphodiesterase/nucleotide pyrophosphatase, AP (anchored protein) superfamily; hypDCFAE, hydrogenase expression/formation proteins. B: sreA1/2, sulfur reductase large 100 kDa subunit (Mo-FeS protein); sreB1/2, sulfur reductase FeS subunit; sreC1/2, membrane protein; sreD, FeS electron transfer protein; sreE, reductase assembly protein. C and D: nuoABCDEFGHIJKLMN, subunits of the NADH:quinone oxidoreductase (complex I); fdhA/nuoG, fdhA/NADH-oxidizing subunit; fqoAB- and fqoHIJM-N subunits of the second set of NADH:quinone oxidoreductase (complex I); oorA-B-CD, 2-oxoacid oxidoreductase subunits. E: atpABCDEI-1I-2 subunits of the membrane-bound A1A0- ATP synthase; hp hypothetical protein; hppA, membrane-bound proton-translocating pyrophosphatase (vacuolar-type H+-pyrophosphatase).
Similar to the Archaea A. ambivalens, P. abyssi and the bacterium Wolinella succinogenes [56], [57], [58], the hydrogenases form short and rather simple electron transport chains with sulfur or polysulfide reductases (SR/PSR) in T. tenax (Figure 1). One pentacistronic operon in T. tenax shows exactly the same gene composition as the A. ambivalens SR operon with a 30-50% amino acid identity of the reading frames. Both operons comprise genes encoding the MoPterin (sreA1) and the FeS subunits (sreB1), a membrane anchor protein (sreC1), a polyferredoxin of unknown function (sreD), and a system-specific chaperone (sreE) similar to nitrate reductase maturation proteins NirD (Figure 2B; TTX_0045-0049). The presence of a TAT motif in the large MoPterin and the FeS subunit suggests the export of these subunits across the membrane. In addition to the pentacistronic SR operon, a second, tricistronic operon with SR homologs (sreA2-sreB2-sreC2; TTX_0073-0075; Figure 2B) could be identified. In contrast to the previously described operon, TAT motifs in sreA2 and sreB2 are absent, suggesting a cellular orientation of these subunits (Figure 1).
Electron transfer between hydrogenase and SR is most probably mediated by quinones (Figure 1), since no indication for the presence of c-type cytochromes was found. The TAT motif in the hydrogenase FeS and the SR MoPterin proteins suggests that the catalytic subunits are oriented outwardly, extending into the “quasi-periplasmic space” (Figure 1) [59], [60]. Therefore, the question arises, how a proton motive force is generated during hydrogen oxidation and sulfur reduction. We assume that a Q cycle is in operation facilitating the uptake of protons in the cytoplasm during quinone reduction by the hydrogenase and the release into the quasi-periplasm upon re-oxidation by the SR.
Chemoorganoheterotrophic growth. T. tenax completely oxidizes organic compounds to CO2 via the oxidative TCA cycle [8], [33]. Energy is conserved by a membrane-bound electron transport chain and sulfur has been suggested as final electron acceptor (sulfur respiration). The NADH:quinone oxidoreductase (complex I) is encoded by minimum of 14 genes in aerobic microorganisms (nuoA-N or nqo1-14). [61]. Complex I genes were found in many genomes of anaerobic Archaea including Archaeoglobus and also in several methanogens. Only 11 of the 14 subunits are conserved in most of the anaerobes, while the others are replaced by non-homologous ferredoxin or F420-oxidizing subunits. The three other subunits (Nqo1-3 or NuoEFG) catalyze NADH binding and oxidation in complex I of aerobes.
Surprisingly, T. tenax has two presumably complete sets of complex I genes (Figure 1), one of which seems to include the NADH-binding subunits (nuoA-N). The nuo genes are spread over four operons across the genome (TTX_1813; TTX_0068-0069; TTX_0556-0563; TTX_1269-1270; Figure 2). 13 out of 14 of these genes can be assigned unambiguously, only one of the NADH-oxidizing subunits (NuoG) gives ambiguous results. The second set of complex I genes is located at a single site in the genome (fqo/oor; TTX_0203-0215), however, it includes uncommon subunits. The fqo genes are strikingly similar to the F420H2:quinone oxidoreductase known from several methanogens and from Archaeoglobus fulgidus. 10 out of 12 A. fulgidus fqo genes are conserved in T. tenax (Figure 2), while the F420-oxidizing fqoF subunit is missing in accordance with the fact that the organism does not use this cofactor. In the middle of this region three 2-oxoacid oxidoreductase genes are found (oorA-D, TTX_0208-0210; Figure 2). This unprecedented observation raises the question, whether these oor genes encode a separate soluble enzyme with OOR activity or, whether the protein replaces the substrate-oxidizing subunits in the membrane-bound complex I to funnel electrons directly from the oxidative 2-oxoacid decarboxylation into the quinone pool.
Succinate dehydrogenase (complex II; Figure 1) provides another electron entry point in the respiratory chain. One complete set of sdh genes, including membrane anchor proteins, is present (TTX_0861-0864) as well as additional genes encoding a second flavoprotein and FeS subunit, respectively (TTX_1104-1105). It cannot be convincingly decided, without biochemical analyses, which of the genes encodes SDH, present in the TCA cycle, and whether, some of these genes might also encode the fumarate reductase [8].
In addition, genes encoding an analog of the bc1 complex (complex III; Figure 1) are also present. The genes are arranged in the same order as in C. maquilingensis: One bicistronic operon encodes a Rieske protein (SoxL, TTX_0319, the only Rieske protein or Rieske ferredoxin in the genome) and a b-type cytochrome, respectively (SoxN, TTX_0318), while the other operon is transcribed in the opposite direction from the same promoter region and encodes another b-type cytochrome (CbsA, TTX_0320) and a membrane protein of unknown function (CbsB, TTX_0321). This bc1-analogous complex was previously identified in S. acidocaldarius and in A. ambivalens [62], [63], and supposedly transfers electrons from quinol to an unknown high-potential electron carrier in the Sulfolobales, which finally transfers them to the terminal oxidase. A bona fide terminal oxidase was not identified in the T. tenax genome, however, two paralogous copies of the subunit I of a bd oxidase are present (cydA, TTX_0142 and TTX_0143). Many of the essential residues are conserved in TTX_0142 [64], [65], and therefore, the questions arise, whether this is indeed an oxygen-reducing enzyme and whether it is part of an oxygenic electron transport chain. Many anaerobic Archaea carry either this combination of terminal oxidase genes or alternatively, homologs of subunits I and II of these enzymes [64], [65]. Their in vivo structure and function remains to be elucidated, however, it is tempting to speculate that they might play a role in T. tenax under microaerobic growth conditions, although T. tenax is described as obligate anaerobe. The presence of multiple ferredoxin genes is characteristic for many Archaea. At least six different fdx genes are identified in T. tenax (TTX_0439, TTX_0681, TTX_0731, TTX_0985, TTX_1318, TTX_2019) either encoding 4Fe4S or 7-8Fe ferredoxins. They have been implicated in oxygen protection, in electron transfer between organic substrates and electron transport chains and as general redox carrier in the absence of c-type cytochromes. The involvement in electron transfer is supported by the presence of multiple genes encoding for example 2-oxoacid:ferredoxin oxidoreductases or aldehyde:ferredoxin oxidoreductases [8]. The link to membrane-bound electron transport chains could be provided by two sets of electron transfer flavoprotein complex (etf) genes encoding oxidoreductases that shuffle electrons to or from unknown membrane-bound proteins and the ferredoxin:quinone oxidoreducase subunits of complex I (see above) [66].
Surprisingly, a complete set of genes required for dissimilatory sulfate reduction could be identified in the T. tenax genome. This set comprises sat encoding an ATP sulfurylase (TTX_0441), apsAB encoding the APS reductase (TTX_0428-0429), and dsrABCGK (TTX_1185-1188, TTX_1191) encoding the dissimilatory siroheme sulfite reductase including a so far unidentified membrane anchor protein (Figure 1). The functionality of the dissimilatory sulfate reduction has been confirmed by chemoorganotrophic growth of T. tenax on sulfate as electron acceptor (Hensel, unpublished data).
Chemiosmosis
An archaeal, membrane-bound A0A1-ATP synthase is present in T. tenax (for review see [67]). As reported for Crenarchaeota, the subunits are spread in the genome and beside single genes two gene clusters were identified (Figure 2). Interestingly, T. tenax harbors two copies of atpI (atpI-1, atpI-2) coding the subunit “a” of A0, which forms the stator in archaeal ATPases (Figure 1). This is so far unique within the Archaea, but the meaning of this gene duplication is yet unknown. Sequence signatures of the membrane integral A0-subunit c (atpK) suggest that protons, rather than Na+, are translocated over the membrane by the T. tenax ATPase [68]. The presence of a membrane-bound pyrophosphatase (hppA, TTX_1550) indicates that the hydrolysis of PPi contributes, at least partially, to the membrane potential, as shown for the vacuolar-type membrane pyrophosphatase of P. aerophilum (V-PPase, PAE1771) [69], [70]. Interestingly, TTX_1550 encoding the respective T. tenax homolog (78% aa identity, (560/717)), is found in a divergent organization with atpI-1 (TTX_1551; Figure 2), suggesting a regulatory function. Additionally, a soluble, cytoplasmic pyrophosphatase is present in T. tenax (ppA, TTX_0388), which is supposed to have an important function to drive biosynthetic processes such as DNA synthesis.
Protein and ion transport
Protein transport
Next to the essential general secretion system, Sec61αβγ (TTX_1416, TTX_1720, TTX_1808), T. tenax possesses the twin arginine translocation (Tat) system (tatA, TTX_2052, tatC, TTX_1059 and tatD, TTX_0685), which transports proteins in their fully folded state across membranes. As in other Archaea, tatB is not present in the T. tenax genome [71]. Possible Tat substrates were predicted using the TatFind program (Table S4.a) [72]. They include HynS, SreA1, ornithine carbamoyltransferase, a hypothetical protein, as well as SoxL, ABC-type branched-chain amino acid transport system (periplasmic component) and formate dehydrogenase (alpha subunit). Three operons (TTX_0962-0973, TTX_1130-1136, TTX_0887-0898) are identified, which might constitute type IV pili (TP4) assembly systems. Bacterial type IV pili are involved in a variety of functions such as twitching motiliy, cell-cell contacts, adherence and DNA uptake [73]. All three operons contain ATPases, which are known to be essential for the assembly processes of TP4. The TTX_0962-0973 and the TTX_1130-1136 operon contain next to the ATPase pilin like proteins, which might either function in the transport process or might be subunits of a pilus. However, T. tenax does not seem to contain a flagellum operon as typical flagellar accessory proteins as FlaI, FlaH or FlaJ are missing.
To be targeted to one of these systems, precursor proteins are equipped with signal peptides. In T. tenax 70 proteins contain a signal peptide (∼3.4%). The majority (48 signal peptides) are class 1 signal peptides that target the protein to the Sec translocase and they have been identified using SignalP [74]. Whereas seven proteins contain putative Tat dependent signal peptides (mentioned above, Table S4.a), 15 exhibit a type IV pilin like signal peptide predicted by the program FlaFind (Table S4.b) [75] and might be pilin subunits. T. tenax contains a clear leader peptidase homolog (TTX_1710), involved in the processing of sec dependent signal peptides. A possible candidate for a type IV prepilin peptidase was also identified (TTX_0979).
Ion and metabolite transport
A total of 412 proteins, 20.1% of the predicted proteome of T. tenax is localized in the membrane. Of these 412 proteins, 133 proteins (6.5% of the total amount of protein coding ORFs) can be classified as transporters (Table S5.a; for classification see I. Paulsen's transport database, http://www.membranetransport.org/) [76]. No indications for the presence of PEP-dependent phosphotransferase (PTS) systems were observed in the T. tenax genome, which is in accordance with most Archaea investigated so far (with the only exception of Haloarcula marismortui, T. pendens and Haloquadratum walsbyi, the latter only harbors enzyme I and HPr) [77]. The distribution of the different transport classes is comparable to the one from S. solfataricus. Both do have two times more secondary transporters than ATP-dependent transporters. About half of all T. tenax transport proteins (66 of 133) share highest similarity with transporters from P. aerophilum, whereas 32 are closest related to proteins from S. solfataricus. Analysis of the 15 substrate binding proteins of ABC transporters of T. tenax (Table S5.b) showed that only two have a N-terminal “bacterial” like sec-dependent signal peptide and are subsequently anchored by a C-terminal transmembrane domain to the membrane (N-terminus outside, C-terminus inside). The transmembrane domain is preceded by a ST-linker, a stretch of serine or threonine residues [78]. These linker regions are often known to be O-glycosylated at the serine or threonine residues. T. tenax binding proteins are glycosylated as they can be isolated by ConA (lectin) affinity chromatography (four binding proteins were identified by mass spectroscopy; Table S5.b), which is specific for terminal mannose residues. The majority of the T. tenax binding proteins has an N-terminal transmembrane domain followed by the ST (or SQ) linker (resulting in N-terminus inside, C-terminus outside). However, the type IV pilin-like signal peptide, identified in S. solfataricus, as well as the cysteine containing consensus motif implying lipidation in Euryarchaeota is absent [79]. Therefore, in T. tenax, similar to P. aerophilum, it is not clear, whether the binding proteins are N-terminally processed. Most probable the N-terminal transmembrane domain is used to anchor the binding protein to the membrane, which is supported by the position of the ST-linker.
Genetics and Information Processing
Replication
The DNA replication machinery of T. tenax conforms to the archaeal norm by resembling that of Eucarya [80]. T. tenax encodes a single candidate initiator protein (TTX_1848) that is homologous to eucaryal Orc1 and Cdc6. Archaeal Cdc6 has been shown to contact the MCM helicase (TTX_0274). MCM acts to unwind DNA, whereupon the exposed single stranded DNA is coated by a single strand binding protein. Interestingly, neither T. tenax nor the closely related Pyrobaculum ssp. possesses obvious homologs of canonical SSBs. However, a recent study has identified a novel single-stranded binding protein, CC1, in T. tenax (TTX_1853/1420 (two genes with identical gene products) and TTX_0308) [81]. Whether CC1 performs the roles of canonical SSBs in the replication process remains to be determined. Archaeal primase is a heterodimer and both subunits are conserved in T. tenax (TTX_0579 and TTX_1586). Recent work has suggested that archaeal primase may be coupled to the progression of the MCM helicase via the bridging action of the GINS complex [82]. In this light, it may be significant that one of the two T. tenax GINS homologs (TTX_0578, GINS15) is encoded within an operon with the catalytic subunit of primase, which is found in many, but not all, Archaea [82]. Once the primer is synthesized, it is extended by the replicative DNA polymerase. T. tenax encodes three members of the family B DNA polymerases (TTX_0168, TTX_1461 and TTX_1917). In Archaea and Eucarya the attachment of Polymerase to their template is conferred by PCNA. Although Eucarya and Euryarchaeota generally have a single PCNA homolog that forms a homotrimer, the Crenarchaeota encode two or more PCNA subunits. T. tenax, like P. aerophilum, has two PCNA homologs (TTX_0580 and TTX_0869). Whether these form homo- or heteromultimers in T. tenax, is currently unknown. PCNA requires an additional factor, RFC, to load it on DNA. Archaeal RFC is normally a pentamer with one large subunit in complex with a homotetramer of a small subunit. T. tenax possesses homologs of both, the large and small subunit encoded within an operon (TTX_1850-1851) and, moreover, the ORF TTX_1485 is coding for a second homolog of the small subunit in T. tenax. The organism possesses a number of topoisomerases; reverse gyrase TTX_1984, a type 1A topoisomerase III homolog (TTX_1447) and a type 2 topoisomerase TopoVI. The latter typically contains two subunits, A (TTX_0746) and B. In T. tenax, the B subunit appears to be split into two halves (TTX_0744 and TTX_0745).
Transcription
The basic transcription apparatus of Archaea resembles the basal eucaryal RNA polymerase II system including homologs to the general transcription factors TATA-box binding protein (TBP), transcription initiation factor IIB (TFB in Archaea), and the alpha-subunit of transcription initiation factor IIE (TFE) [84], [85].
Clustering of genes encoding proteins of the basal transcription and translation machineries is a general feature of ‘prokaryotic’ genomes. Two separate gene clusters coding for RNA polymerase subunits and ribosomal proteins are conserved within the archaeal domain. The first one encompasses genes coding for the catalytic subunits (rpoB, rpoA1, and rpoA2) as well as subunit H, the second one encompasses the two assembly subunits (rpoD and rpoN) [86]. In T. tenax, P. aerophilum, and T. pendens the two gene clusters are fused and this organization might facilitate assembly of the RNA polymerase. Multiplicity of general transcription factors is commonly found in Archaea. Studies in Halobacterium NRC-1 revealed specific regulons for different TFB paralogs [87]. For TFB3 of S. solfataricus an activation of transcription by interaction with the ternary complex (DNA, TFB1, TBP) has been demonstrated [88]. The genome of T. tenax harbours single homologs for tbp (TTX_0178) and tfe (TTX_1936), but four tfb homologs (tfb1, TTX_1484, tfb2, TTX_2085, tfb3, TTX_1929, and tfb4, TTX_1732). TFB1 exhibits highest overall sequence similarity to the characterized TFB homologs of Sulfolobus shibatae (AAA81380) and S. acidocaldarius (AAF18139) [89], [90]. Like TFB3 of S. solfataricus the T. tenax TFB3 lacks one cyclin fold. Next to a classical homolog of transcription elongation factor S (TTX_0581), an additional paralog to transcription elongation factor S, lacking the conserved C-terminus required for stimulation of the intrinsic endonuclease activity of RNAP, was found in all genomes of the Thermoproteaceae: T. tenax (TTX_0711), P. aerophilum (PAE3480), and C. maquilingensis (Cmaq_0787).
Translation
T. tenax was found to contain one RNA operon comprising 16S and 23S RNA as well as a separate 5S RNA gene (Table S6.a). The T. tenax genome contains a full complement of 46 tRNA predictions, plus one apparent tRNA pseudogene [91], [92]. A total of 28 genes possess introns at non-canonical positions (10 tRNAs have two introns and one has three introns; http://gtrnadb.ucsc.edu/Ther_tena/Ther_tena-summary.html) Table S6.a).
With the only exception of L41e, which shows some uncertain distribution in Archaea, all conserved ribosomal proteins in the hitherto known archaeal genomes are present in T. tenax (Table S6.b). Interestingly, unique to T. tenax within the Archaea, an exact duplication of the ribosomal gene (S30e) is found (TTX_0151 and TTX_0161).
In contrast to Bacteria and most Archaea, which harbor large clusters of genes coding for ribosomal proteins (e.g. str locus containing the S10 - spc – alpha operons; [93]), the genome of T. tenax is characterized by rather short clusters with only up to five genes, a feature which seems to be typical for the members of the Thermoproteaceae (T. tenax, P. aerophilum, P. islandicum, C. maquilingensis). Aminoacyl tRNA synthetase genes for 20 amino acids were found in the genome (Table S6.c). No indications could be observed for the cotranslational incorporation of selenocysteine or pyrrolysine. Thus, all amino acids including asparagine and glutamine seem to be incorporated by direct acylation of the tRNA.
Comparative genomics and phylogenetic position of T. tenax within the Crenarchaeota
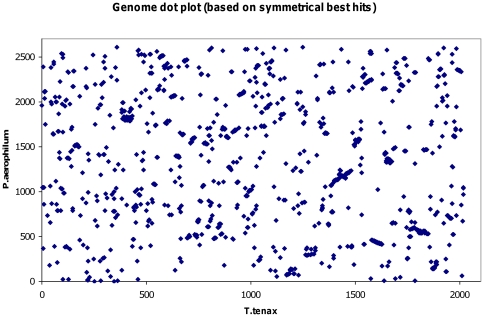
The phylogenetic position of T. tenax within the Crenarchaeota as a sister group of ‘Pyrobacula’, the group that contains all known Pyrobaculum species and T. neutrophilus, is confirmed by the 16S rRNA sequences (ARB-Living-Tree project) [94] and the phylogenetic tree based on three subunits of RNA polymerase (Figure S2). Although, thus, T. tenax is phylogenetically clearly separated from the genus Pyrobaculum, as a group, T. tenax and the Pyrobacula, can be clearly separated from the deeper branching Thermoproteaceae with C. maquilingensis and Thermofiliaceae with T. pendens. Despite the closer relationships between T. tenax and P. aerophilum within the Thermoproteaceae, the synteny in these genomes is minimal, suggesting extensive gene rearrangement, which has occurred after their divergence from the common ancestor (Figure 3).
Figure 3. Genome dot plot comparison of T. tenax and P. aerophilum.
CDSs in genomic order were tested for colinearity between the two genomes of T. tenax and P. aerophilum. Each point represents a matching pair of orthologs with an e-value of <1×e−15 (for approach see material and methods). The calculations yielded a value for C (colinearity factor) of 280. The comparison of bacterial genomes of similar size yielded values in the range of 18 (Helicobacter pylori J99 vs H. pylori 26695) to 238 (Helicobacter acinonychis vs Wolinella succinogenes) [119]. The very limited synteny between the genomes of T. tenax and P. aerophilum that keeps only the gene order of local islands intact, suggests major genomic rearrangements after their divergence from the common ancestor.
We compared the protein complement of T. tenax to the database of clusters of orthologous groups developed specifically for archaeal genomes, arCOGs [16] (database update in preparation). We assigned 1,953 (95%) of the proteins to 1,604 arCOGs; the coverage is comparable to that observed for closely related Pyrobaculum species [16]. Overall, the gene content of T. tenax is typical of Crenarchaeaota. It preserved the intact 157 gene core shared by all archaeal genomes and additional 42 gene families that are missing only in the smallest Archaeum, i.e. N. equitans. T. tenax has not lost any of the 234 arCOGs present in all Crenarchaeota, including nine that are not present in any euryarchaeal genomes. The latter set includes five genes that are shared with Eucarya: the recently described small RPB8 subunit of DNA-directed RNA polymerase (TTX_1930) [95], a Zn-finger containing protein, an apparent transcription elongation factor 1 ortholog (TTX_1715), and ribosomal proteins S25e, S26e and S30e (TTX_0177, TTX_0164, TTX_0151 and TTX_0161 (Table S7). The conserved gene core of the Thermoproteales lineage consists of 607 arCOGs and representatives of only five, i.e. arCOGs 1304, 975, 5463, 921, 5461, are missing specifically in T. tenax (Table S7). Among the core gene families, 19 are not present in other crenarchaeal genomes, including five that are shared only with the deep-branching Archaeum K. cryptofilum and nine, which are so far unique for Thermoproteales and must be implicated in important house-keeping functions. The majority of these 19 families are uncharacterized or only with general function predicted. Interestingly enough, among those there are two genes that could be potentially involved in cell division of Thermoproteales, the only group of Archaea for which the cell division mechanism is not known yet [12]. One of these proteins is a ParA family ATPase (TTX_1301) involved in chromosome and plasmid partitioning [96] and the other is an actin-like protein (TTX_0752), the closest homolog of the major component of the cytoskeleton in Eucarya [97]. Another uncharacterized protein among these 19 is encoded in the same operon with actin, suggesting their functional relationships. These three proteins can be considered as prime candidates for a role in cell division of Thermoproteales (Table S7).
There are only six arCOGs that are present in T. tenax but not in other Crenarchaeota. One of them is DNA uptake protein DprA (TTX_0242), which is encoded in a predicted operon with another functionally related protein ComF (TTX_0243), which was never before detected in archaeal genomes and apparently has been transferred horizontally from Bacteria. Another example of potential horizontal gene transfer from Bacteria to T. tenax is the cytochrome b subunit of Ni,Fe-hydrogenase (Isp1, TTX_0032), which is also absent in other archaeal genomes.
We employed the tree representing the consensus view on archaeal taxonomy [98], [12], [99] and arCOG patterns (Figure S1) to reconstruct the gene repertoire of the common ancestor of the T. tenax and Pyrobaculum group and gene loss and gain events during the evolution of T. tenax lineage using the maximum-likelihood approach developed recently by Csurös and Miklos [100]. The estimated gene repertoire of the common ancestor of the T. tenax and Pyrobaculum group consists of 1,619 gene families. Proteins encoded in the T. tenax genome are assigned to 1,604 different arCOGs, whereas gene complement of the common ancestor of Pyrobaculum group is estimated as 1,768 families with the net gain of 149 genes. The estimated gene family drift for T. tenax is not very high, only 62 families were gained and 77 were lost, implying that the T. tenax genome shares 92% of gene families with the ancestor. For comparison, the Pyrobaculum group ancestor shares only 82% with the Pyrobaculum/Thermoproteus ancestor with further erosion of similarity within the Pyrobaculum clade. Similar estimates, restricted to metabolic genes, show that 34% T. tenax gene families are directly inherited from the ancestral metabolic repertoire, whereas Pyrobaculum group species only of 28% on average. The examples of functions (Table S7) include the exchange of genes with analogous functions, like the substitution of a number of amino acid ABC family transporters by PotE-like amino acid transporters (belonging to the amino acid/polyamine/organocation (APC) superfamily); and minor shifts in metabolic preferences, like acquisition of sugar transporters, which suggests an increasing role of sugar utilization for T. tenax. A few other gene families that are lost, have functional substitutes encoded in the genome. For example, the loss of ABC family phosphate transporter can be compensated by inorganic phosphate transporter. All the above indicates that T. tenax largely preserved the functional repertoire of the ancestor.
Materials and Methods
Strain and DNA preparation
T. tenax strain Kra 1 (DSM 2078T; NCBI Taxonomy ID 2271) cells were grown under autotrophic conditions as described previously [1]. Genomic DNA (gDNA) was prepared as described previously [101].
Isolation of membranes and isolation of glycosylated proteins
Cells (400 ml) were spun down and membranes isolation and the purification of glycosylated membrane proteins were performed as described previously [79].
Genome Sequencing
The genome was sequenced with a random (‘shotgun’) sequencing and assembly strategy [102] with gaps closed via primer walking on bridging plasmid clones, and by direct sequencing with chromosomal DNA as a template. Controlled fragmentation of the gDNA for cloning into plasmid vectors was done with a HydroShear (Gene Machines, San Carlos, CA). Fragments of 2.5 and 5 kbp average lengths, repectively, were cloned into TOPO subcloning vectors (Invitrogen, Carlsbad, CA) and sequenced from both ends after plasmid purification with QIAquick (Qiagen, Hilden, Germany) on Qiagen liquid handling stations according to the manufacturer̀s instructions. Sanger-type sequence reactions [103] were analyzed on ABI Prism 377 and 3700 systems (PE Biosystems) and processed for sequence quality (base calling) and assembly with the Phred/Phrap/Consed software package [104]–[106]. The final genome sequence was assembled from 17,638 sequence reads (including 1,244 primer walking sequences) with a mean trimmed read length of 616.2 nt, resulting in an 8-fold sequence coverage with an estimated error rate of less than 0.4×10−4.
Sequence analysis and annotation
Analysis and sequence annotations have been performed as described previously (Table S8) [107]. GCskew analysis and localization of the putative start of the chromosomal replication was performed with the program GenSkew (http://mips.gsf.de/services/analysis/genskew). Repeats were identified and analyzed with REPuter [108]. High and low GC regions were identified by EMBOSS [109]. tRNA genes were located with tRNAscan-SE and GtRNAdb [91], [92]. ORFs were predicted by the expert program REGANOR [110], which is integrated into the GenDB package [111] and combines the gene finding programs GLIMMER [112] and CRITICA [113]. Curation and annotation of the genome were done with the help of the GENDB annotation package [111]. Curation by hand was performed in order to identify and remove false positive ORFs found by GLIMMER and CRITICA.
Annotation of the identified ORFs was accomplished on the basis of sequence similarity searches against a selection of sequence databases followed by manual expert curation. Similarity searches were performed by using blastx [114] against the NCBI nonredundant database on protein level [115], the Swissprot [116], KEGG [117], Clusters of Orthologous Groups (COG) [15] and the archaeal COG [16] database. Genes with a sufficient degree of similarity (cut-off 10E−15) were finally assigned to orthologous groups in COGs. ORFs shorter than 150 bp with best BLAST scores (E-values) higher than 10−15 were deleted from the final reported set of genes.
Gene order and colinearity in T. tenax and P. aerophilum (indicated in Figure 3)
A quantitative co-linearity factor was calculated from the genomic positions (x and y coordinates) of each ortholog pair relative to O, the number of CDSs in the target genome, as follows: For each pair of neighbouring ORFs on the genome of T. tenax (xi, xi+1), the position of the corresponding orthologs on the genome of P. aerophilum (yi, yi+1) was used to calculate D = Min (|yi+1–yi|, O – |yi+1–yi|). The colinearity factor C is defined as C = ΣD/O.
Comparative genomics and reconstruction of gene gain and loss events during the evolution of the Thermoproteales branch
Comparative genomic analysis of T. tenax proteome was done using the archaeal Clusters of Orthologous Groups database (arCOGs) [16], which was recently updated and contains 60 archaeal genomes (available at ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/arCOG/) and the Integrated Microbial Genomes (img) suite at JGI [118]. Proteins of T. tenax were assigned to arCOGs using PSI-BLAST program [114] and arCOGs profiles. T. tenax representation in arCOGs was not included for delineation of archaeal (59 genomes, and 58 genomes with N. equitans excluded), crenarchaeal (18 genomes) and Thermoproteales (7 genomes) core arCOGs (families that are present in all genomes of the respective group).
Count software (http://www.iro.umontreal.ca/~csuros/gene_content/count.html) [100] was used to infer gene gain, loss and duplication rates on the branches of the species tree from the 59x8890 (N. equitans was excluded from consideration) matrix of phyletic patterns (containing a number of proteins in each genome assigned to a corresponding arCOG) by the likelihood maximization method based on a phylogenetic birth-and-death model. The tree representing the consensus view of archaeal phylogeny [98], [99], [16] was used as the guide topology (Figure S1). The model estimates probabilities for each arCOG to be present in each of the ancestral nodes and the rates of evolutionary events. For list of arCOGs present, lost or gained at the branches of interest, we used probability cutoff >0.5.
Nucleotide sequence accession number
The genome sequence has been deposited in the EMBL Nucleotide Sequence Database under the accession number FN869859 and the MIGS compliant metadata in the Genomes Online Database (GOLD, www.genomesonline.org) [7] under the accession number GOLD Gc01285.
Supporting Information
Guide tree topology used for reconstruction of evolutionary events for the Thermoproteales lineage. The tree represents the consensus view of archaeal phylogeny based on recent publications [16], [99], [98]. The Thermoproteales branch is shaded.
(PDF)
Phylogeny of Archaea based on analysis of RNA polymerase subunits. Maximum likelihood tree made from aligned sequences of the three largest RNA polymerase subunits: a, a′, and b as described previously [99]. Bootstrap support numbers are given at the nodes as a percentage (n = 10,000). Scale bars represent the average number of substitutions per residue.
(PDF)
Low G+C regions in the T. tenax genome. Location in the genome, length and G+C content of the three identified regions are given.
(DOCX)
Clusters of regularly interspaced short palindromic repeats (CRISPR).
(DOC)
Amino acid biosynthesis pathways. The identified genes (sorted by respective biosynthesis pathways), their ID as well as gene name and annotation are given.
(DOCX)
TatFind (a) and FlaFind (b) positive ORFs in the T. tenax genome. The tools TatFind (http://signalfind.org/tatfind.html) [75] and FlaFind (http://signalfind.org/flafind.html) [75] have been applied.
(DOCX)
(a) Comparison of transporters in Thermoproteus tenax, Pyrobaculum aerophilum and Sulfolobus solfataricus. Classification according to the Paulsens transport database (http://www.membranetransport.org/) [76]. (b) T. tenax Substrate binding proteins.
(DOCX)
(a) Annotated T. tenax tRNA genes. (b) Annotated genes encoding ribosomal proteins. Rps, ribosomal proteins. (c) Identified T. tenax tRNA synthetase genes.
(DOCX)
Comparative genomic analysis using arCOGs. Core genes: The number of organisms in the corresponding archaeal lineages and number of proteins in individual genomes for respective arCOGs are given. Gain-loss-expansion: Results of the analysis of gene loss, gain and family expansion using arCOG data and COUNT software (http://www.iro.umontreal.ca/~csuros/gene_content/count.html) are given. Abbreviations: CREN - Crenarchaeota; EURY - Euryarchaeota; Tauma - Taumarchaeota; Korar - Korarchaeota; Nano - Nanoarchaeota; Thete - Thermoproteus tenax; All other organism abbreviations are explained in Figure S1.
(XLSX)
Annotated T. tenax genes. The respective ORF ID, location and arCOG annotations as well as COG assignment are given. The last column provides GenBank GI accession numbers for the BLASTP best hits against NCBI Non-redundant database (e-value cutoff 0.001).
(XLSX)
Acknowledgments
The authors thank A. Pühler for providing GenDB (CeBiTec, University of Bielefeld, Germany), the Computing-Centre of the Max-Planck-Society in Garching (RZG, Germany) for providing bioinformatic infrastructure, Krishna Palaniappan for support with img database (Biological Data Management and Technology Center, Berkeley, USA), and V. Müller (Goethe University of Frankfurt) for discussion on A1A0-ATP synthases.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the University of Duisburg-Essen (Germany) and the Deutsche Forschungsgemeinschaft (DFG; SPP1112) by grant He1238/16-2, 3 (Dr. Hensel). Dr. Albers was supported by a VIDI grant of the Dutch Science Organization (NWO) and intramural funds of the Max Planck society. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zillig W, Stetter KO, Schaefer W, Janekovic D, Wunderl S, et al. Thermoproteales: a novel type of extremely thermoacidophilic anaerobic archaebacteria isolated from icelandic solfataras. Zentralbl Mikrobiol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1981;C2:205–227. [Google Scholar]
- 2.Fischer F, Zillig W, Stetter KO, Schreiber G. Chemolithoautotrophic metabolism of anaerobic extremly thermophilic archaebacteria. Nature. 1983;301:511–513. doi: 10.1038/301511a0. [DOI] [PubMed] [Google Scholar]
- 3.Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–23. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci. 2003;100:12984–8. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci 105:8102-7. Comment in: Proc Natl Acad Sci (2008) 1; 2008;105(26):8805–6. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides NC. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. NAR. 2006;34:D332–334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siebers B, Tjaden B, Michalke K, Dörr C, Ahmed H, et al. Reconstruction of the central carbohydrate metabolism of Thermoproteus tenax by use of genomic and biochemical data. J Bacteriol. 2004;186:2179–2194. doi: 10.1128/JB.186.7.2179-2194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaparty M, Zaigler A, Stamme C, Soppa J, Hensel R, et al. DNA microarray analysis of the central carbohydrate metabolism: glycolytic/gluconeogenic carbon switch in the hyperthermophilic Crenarchaeum Thermoproteus tenax. J Bacteriol. 2008;190(6):2231–2238. doi: 10.1128/JB.01524-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber H, Gallenberger M, Jahn U, Eylert E, Berg I, et al. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci USA. 2008;105(22):7851–7856. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baliga NS, Goo YA, Ng WV, Hood L, Daniels Ch J, et al. Is gene expression in Halobacterium NRC-1 regulated by multiple TFB and TFB transcription factors? Mol Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- 12.Samson RY, Bell SD. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 2009;17(11):507–13. doi: 10.1016/j.tim.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, et al. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 14.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, et al. Rfam: updates to the RNA families database. Nucleic Acids Research. 2009;37:136. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 16.Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of Archaea. Biol Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, et al. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4(3):R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naether DJ, Rachel R. The outer membrane of the hyperthermophilic archaeon Ignicoccus: dynamics, ultrastructure and composition. Biochem. Soc. Transact. 2004;32:199–203. doi: 10.1042/bst0320199. [DOI] [PubMed] [Google Scholar]
- 19.Sumper M, Berg E, Mengele R, Strobel I. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J Bacteriol. 1990;172:7111–7118. doi: 10.1128/jb.172.12.7111-7118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veith A, Klingl A, Zolghadr B, Lauber K, Mentele R, et al. Acidianus, Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol Microbiol. 2009;73:58–72. doi: 10.1111/j.1365-2958.2009.06746.x. [DOI] [PubMed] [Google Scholar]
- 21.Julenius K, Mølgaard A, Gupta R, Brunak R. Prediction, conservation analysis and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 22.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in procaryotic genomes. PloS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 24.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-inter-ference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eucaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrangou R, Fermaux C, Deveau H, Richards M, Boyaval P, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 26.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lillestol RK, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann H, Zillig W. Structural variability in the genome of the Thermoproteus tenax virus TTV1. Mol Gen Genet. 1990;222(2-3):435–437. doi: 10.1007/BF00633851. [DOI] [PubMed] [Google Scholar]
- 30.Ahn DG, Kim SI, Rhee JK, Kim KP, Pan JG, et al. TTSV1, a new virus-like particle isolated from the hyperthermophilic crenarchaeote Thermoproteus tenax. Virology. 2006;351:280–290. doi: 10.1016/j.virol.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 31.Haering M, Peng X, Brügger K, Rachel R, Stetter KO, et al. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: a novel virus family, the globuloviridae. Virology. 2004;323:233–242. doi: 10.1016/j.virol.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Zaparty M, Tjaden B, Hensel R, Siebers B. The central carbohydrate metabolism of the hyperthermophilic crenarchaeote Thermoproteus tenax: pathways and insights into their regulation. Arch Microbiol. 2008;190:231–245. doi: 10.1007/s00203-008-0375-5. [DOI] [PubMed] [Google Scholar]
- 33.Selig M, Schoenheit P. Oxidation of organic-compounds to CO2 with sulfur or thiosulfate as electron-acceptor in the anaerobic hyperthermophilic Archaea Thermoproteus tenax and Pyrobaculum islandicum proceeds via the citric-acid cycle. Arch Microbiol. 1994;162:286–294. [Google Scholar]
- 34.Ramos-Vera WH, Berg IA, Fuchs G. Autotrophic carbon dioxide assimilation in Thermoproteales revisited. J Bacteriol. 2009;191(13):4286–97. doi: 10.1128/JB.00145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramos-Vera WH, Weiss M, Strittmatter E, Kockelkorn D, Huchs G. Identification of missing genes and enzymes for autotrophic carbon fixation in crenarchaeota. J Bacteriol. 2011;193(5):1201–1211. doi: 10.1128/JB.01156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soderberg T. Biosythesis of ribose-5-phosphate and erythrose-4-phosphate in Archaea: a phylogenetic analysis of archaeal genomes. Archaea. 2005;1:347–352. doi: 10.1155/2005/314760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verhees CH, Huynen M, Ward D, Schiltz E, de Vos WM, et al. The Phosphoglucose Isomerase from the hyperthermophilic Archaeon pyrococcus furiosus Is a unique enzyme that belongs to the Cupin Superfamily. J Biol Chem. 2001;276:44. doi: 10.1074/jbc.M104603200. [DOI] [PubMed] [Google Scholar]
- 38.Orita I, Sato T, Yurimoto H, Kato N, Atomi H, et al. The Ribulose Monophosphate Pathway Substitutes for the Missing Pentose Phosphate Pathway in the Archaeon Thermococcus kodakaraensis. . J Bacteriol. 2006;188(13):4698–4704. doi: 10.1128/JB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orita I, Yurimoto H, Kawarabayasi Y, Sakai Y, Kato N. The archaeon Pyrococcus horikoshii possesses a bifunctional enzyme for formaldehyde fixation via the ribulose monophosphate pathway. J Bacteriol. 2005;187(11):3636–42. doi: 10.1128/JB.187.11.3636-3642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van der Oost J, Siebers B. The glycolytic pathways of Archaea: evolution by tinkering. editors. Blackwell Publishing. Garrett RA, Klenk HP, editors. Archaea: evolution, physiology and molecular biology. 1st edition. 2007;22:247–260. [Google Scholar]
- 41.White RH. L-Aspartate Semialdehyde and a 6-Deoxy-5-ketohexose 1-Phosphate Are the Precursors to the Aromatic Amino Acids in Methanocaldococcus jannaschii. Biochemistry. 2004;43:7618–7627. doi: 10.1021/bi0495127. [DOI] [PubMed] [Google Scholar]
- 42.Schaefer S, Paalme T, Vilu R, Fuchs G. 13C-NMR study of acetate assimilation in Thermoproteus neutrophilus. Eur J Biochem. 1989;186:695–700. doi: 10.1111/j.1432-1033.1989.tb15262.x. [DOI] [PubMed] [Google Scholar]
- 43.Eikmanns B, Linder D, Thauer RK. Unusual pathway of isoleucine biosynthesis in Methanobacterium thermoautotrophicum. Arch Microbiol. 1983;136:111–113. [Google Scholar]
- 44.Jahn U, Huber H, Eisenreich W, Huegler M, Fuchs G. Insights into the Autotrophic CO2 Fixation Pathway of the Archaeon Ignicoccus hospitalis: Comprehensive Analysis of the Central Carbon Metabolism. J Bacteriol. 2007;189(11):4108–4119. doi: 10.1128/JB.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graupner M, White RH. Methanococcus jannaschii Generates L-Proline by Cyclization of L-Ornithine. J Bacteriol. 2001;183(17):5203–5205. doi: 10.1128/JB.183.17.5203-5205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Higuchi S, Kawashima T, Suzuki M. Comparison of pathways for amino acid biosynthesis in archaebacteria using their genomic DNA sequences. Proc Jpn Acad Ser B. 1999;75:241–245. [Google Scholar]
- 47.Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, et al. Comparative Genomics of the Archaea (Euryarchaeota): Evolution of Conserved Protein Families, the Stable Core and the Variable Shell. Genome Research. 1999;9:608–628. [PubMed] [Google Scholar]
- 48.Vaupel M, Dietz H, Linder D, Thauer RK. Primary structure of cyclohydrolase (Mch) from Methanobacterium thermoautotrophicum (strain Marburg) and functional expression of the mch gene in Escherichia coli. Eur J Biochem. 1996;236(1):294–300. doi: 10.1111/j.1432-1033.1996.00294.x. [DOI] [PubMed] [Google Scholar]
- 49.Tsoka S, Simon D, Ouzounis CA. Automated metabolic reconstruction for Methanococcus jannaschii. Archaea. 2003;1:223–229. doi: 10.1155/2004/324925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fitz-Gibbon ST, Ladner H, Kim UJ, Stetter KO, Simon MI, et al. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc Natl Acad Sci USA. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zabriskie TM, Jackson MD. Lysine biosynthesis and metabolism in fungi. Natural Products Rep. 2000;17(3):85–97. doi: 10.1039/a801345d. [DOI] [PubMed] [Google Scholar]
- 52.Born TL, Blanchard JS. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Curr Opin Chem Biol. 1999;3:607–613. doi: 10.1016/s1367-5931(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 53.Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, et al. The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. 1997 doi: 10.1038/37052. Nature 27, 390(6658):364-70; Erratum in: Nature 2, 394 (6688):101. [DOI] [PubMed] [Google Scholar]
- 54.Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 56.Dirmeier R, Keller M, Frey G, Huber H, Stetter KO. Purification and properties of an extremely thermostable membrane-bound sulfur-reducing complex from the hyperthermophilic Pyrodictium abyssi. Eur J Biochem. 1998;252:486–491. doi: 10.1046/j.1432-1327.1998.2520486.x. [DOI] [PubMed] [Google Scholar]
- 57.Hedderich R, Klimmek O, Kroeger A, Dirmeier R, Keller M, et al. Anaerobic respiration with elemental sulfur and with sulfides. FEMS Microbiol Rev. 1999;22:353–381. [Google Scholar]
- 58.Laska S, Lottspeich F, Kletzin A. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology. 2003;149:2357–2371. doi: 10.1099/mic.0.26455-0. [DOI] [PubMed] [Google Scholar]
- 59.Baumeister W, Lembcke G. Structural Features of Archaebacterial Cell Envelopes. J Bioenerg Biomembr. 1992;24:567–575. doi: 10.1007/BF00762349. [DOI] [PubMed] [Google Scholar]
- 60.Wildhaber W, Baumeister W. The cell envelope of Thermoproteus tenax: three-dimensional structure of the surface layer and its role in shape maintenance. EMBO J. 1987;6:1475–1480. doi: 10.1002/j.1460-2075.1987.tb02389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 62.Hiller A, Henninger T, Schafer G, Schmidt CL. New genes encoding subunits of a cytochrome bc1-analogous complex in the respiratory chain of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J Bioenerg Biomembr. 2003;35:121–131. doi: 10.1023/a:1023742002493. [DOI] [PubMed] [Google Scholar]
- 63.Bandeiras TM, Refojo PN, Todorovic S, Murgida DH, Hildebrandt P, et al. The cytochrome ba complex from the thermoacidophilic crenarchaeote Acidianus ambivalens is an analog of bc(1) complexes. Biochim Biophys Acta. 2009;1787:37–45. doi: 10.1016/j.bbabio.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 64.Tsubaki M, Hori H, Mogi T. Probing molecular structure of dioxygen reduction site of bacterial quinol oxidases through ligand binding to the redox metal centers. J Inorg Biochem. 2000;82:19–25. doi: 10.1016/s0162-0134(00)00140-9. [DOI] [PubMed] [Google Scholar]
- 65.Mogi T. Probing the haem d-binding site in cytochrome bd quinol oxidase by site-directed mutagenesis. J Biochem. 2009;145:763–770. doi: 10.1093/jb/mvp033. [DOI] [PubMed] [Google Scholar]
- 66.Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, et al. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lewalter K, Müller V. Bioenergetics of Archaea: Ancient energy conserving mechanisms developed in the early history of life. Biochim Biophys Acta. 2006;1757:437–445. doi: 10.1016/j.bbabio.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 68.Müller V. An exceptional variability in the motor of archael A1A0 ATPases: from multimeric to monomeric rotors comprising 6-13 ion binding sites. . J Bioenerg Biomembr. 2004;36:115–125. doi: 10.1023/b:jobb.0000019603.68282.04. [DOI] [PubMed] [Google Scholar]
- 69.Drozdowicz YM, Lu YP, Patel V, Fitz-Gibbon S, Miller JH, et al. A thermostable vacuolar-type membrane pyrophosphatase from the archaeon Pyrobaculum aerophilum: implication for the origins of pyrophosphate-energized pumps. FEBS Lett. 1999;460:505–512. doi: 10.1016/s0014-5793(99)01404-0. [DOI] [PubMed] [Google Scholar]
- 70.Drozdowicz YM, Rea PA. Vacuolar H+ pyrophosphatases: from the evolutionary backwaters into the mainstream. Trends Plant Sci. 2001;6:206–211. doi: 10.1016/s1360-1385(01)01923-9. [DOI] [PubMed] [Google Scholar]
- 71.Albers SV, Szabó Z, Driessen AJM. Protein secretion in Archaea: Multiple Paths towards a Unique Cell Surface. Nature Reviews Microbiology, 2006;4:537–47. doi: 10.1038/nrmicro1440. [DOI] [PubMed] [Google Scholar]
- 72.Rose RW, Brueser T, Kissinger JC, Pohlschröder M. Adaptation of protein secretion to extremely high salt concentrations by extensive use of the twin arginine translocation pathway. Mol Microbiol. 2002;5:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 73.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2(5):363–78. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 74.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nature Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 75.Szabó Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJM, et al. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol. 2007;189(3):772–8. doi: 10.1128/JB.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;3:D274–9. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anderson I, Rodriguez J, Susanti D, Porat I, Reich C, et al. Genome sequence of Thermofilum pendens reveals an exceptional loss of biosynthetic pathways without genome reduction. J Bacteriol. 2008;190(8):2957–2965. doi: 10.1128/JB.01949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elferink MGL, Albers SV, Konings WN, Driessen AJM. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein dependent ABC transporters. Mol Microbiol. 2001;39:1494–1503. doi: 10.1046/j.1365-2958.2001.02336.x. [DOI] [PubMed] [Google Scholar]
- 79.Albers SV, Elferink MGL, Charlebois RL, Sensen CW, Driessen AJM, et al. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high affinity membrane-integrated binding protein. J Bacteriol. 1999;181:4285–4291. doi: 10.1128/jb.181.14.4285-4291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barry ER, Bell SD. DNA replication in the Archaea. Micro Mol Biol Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luo X, Schwarz-Linek U, Botting CH, Hensel R, Siebers B, et al. CC1, a novel crenarchaeal DNA binding protein. J Bacteriol. 2006;189(2):403–409. doi: 10.1128/JB.01246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swiatek A, MacNeill S. The archaeo-eukaryotic GINS proteins and the archaeal primase catalytic subunit PriS share a common domain. Biology Direct. 2010;5:17. doi: 10.1186/1745-6150-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, et al. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 2006;7:539–545. doi: 10.1038/sj.embor.7400649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bell SD, Jackson SP. Mechanism and regulation of transcription in Archaea. Curr Opin Microbiol. 2001;4:208–213. doi: 10.1016/s1369-5274(00)00190-9. [DOI] [PubMed] [Google Scholar]
- 85.Reeve JN. Archaeal chromatin and transcription. Mol Microbiol. 2003;48:587–598. doi: 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- 86.Werner F. Structure and function of archaeal RNA polymerases. Mol Micobiol. 2007;65:1395–1404. doi: 10.1111/j.1365-2958.2007.05876.x. [DOI] [PubMed] [Google Scholar]
- 87.Facciotti MT, Reiss DJ, Pan M, Kaur A, Vuthoori M, et al. General transcription factor specified global gene regulation in Archaea. Proc Natl Acad Sci. 2007;104:4630–4635. doi: 10.1073/pnas.0611663104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paytubi S, White MF. The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription. Mol Microbiol. 2009;72(6):1487–99. doi: 10.1111/j.1365-2958.2009.06737.x. [DOI] [PubMed] [Google Scholar]
- 89.Bell SD, Jackson SP. The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J Biol Chem. 2000;275:12934–12940. doi: 10.1074/jbc.275.17.12934. [DOI] [PubMed] [Google Scholar]
- 90.Qureshi SA, Bell SD, Jackson SP. Factor requirements for transcription in the archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lowe T, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Coenye T, Vandamme P. Organisation of the S10, spc and alpha ribosomal protein gene cluster in prokaryontic genomes. FEMS Microbiol Lett. 2005;242:117–126. doi: 10.1016/j.femsle.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 94.Yarza P, Richter M, Peplies J, Euzeby J, Amann R, et al. The All Species Living Tree project: A 16S rRNAbased phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 95.Koonin EV, Makarova KS, Elkins JG. Orthologs of the small RPB8 subunit of the eukaryotic RNA polymerases are conserved in hyperthermophilic Crenarchaeota and "Korarchaeota". Biol Direct. 2007;14(2):38. doi: 10.1186/1745-6150-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Easter J, Jr, Gober JW. ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol Cell. 2002;10(2):427–34. doi: 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- 97.Yutin N, Wolf MY, Wolf YI, Koonin EV. The origins of phagocytosis and eukaryogenesis. Biol Direct. 2009;26(4):9. doi: 10.1186/1745-6150-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6(3):245–52. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 99.Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci 105:8102-7. Comment in: Proc Natl Acad Sci (2008) 1; 2008;105(26):8805–6. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Csurös M, Miklós I. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol. 2009;26(9):2087–95. doi: 10.1093/molbev/msp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schramm A, Siebers B, Tjaden B, Brinkmann H, Hensel R. Pyruvate kinase of the hyperthermophilic crenarchaeote Thermoproteus tenax: physiological role and phylogenetic aspects. J Bacteriol. 2000;182:2001–2009. doi: 10.1128/jb.182.7.2001-2009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae. Rd Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 103.Sanger F, Donelson JE, Coulson AR, Koessel H, Fischer D. Use of DNA polymerase I by a synthetic oligonucleotide to determine a nucleotide sequence in phage fl DNA. Proc Natl Acad Sci USA. 1973;70:1209–13. doi: 10.1073/pnas.70.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 105.Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 107.Baar C, Eppinger M, Raddatz G, Simon J, Lanz C, et al. Complete genome sequence and analysis of Wolinella succinogenes. Proc Natl Acad Sci. 2003;100(20):11690–11695. doi: 10.1073/pnas.1932838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, et al. REPuter: the manifold applications of repeat analysis on a genome scale. Nucleic Acids Res. 2001;29:4633–42. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rice P, Longden I, Bleasby EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 110.Linke B, McHardy AC, Neuweger H, Krause L, Meyer F. REGANOR: a gene prediction server for prokaryotic genomes and a database of high quality gene predictions for prokaryotes. Appl Bioinformatics. 2006;5(3):193–8. doi: 10.2165/00822942-200605030-00008. [DOI] [PubMed] [Google Scholar]
- 111.Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, et al. GenDB-An open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Badger JH, Olsen GJ. CRITICA: Coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 114.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, et al. The Swiss Prot Protein Knowledgebase and its supplement TrEMBL. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Research. 2009;1–9 doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eppinger M, Baar C, Linz B, Raddatz G, Lanz C, et al. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLOS Genetics. 2006;2(7):1097–1110. doi: 10.1371/journal.pgen.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Guide tree topology used for reconstruction of evolutionary events for the Thermoproteales lineage. The tree represents the consensus view of archaeal phylogeny based on recent publications [16], [99], [98]. The Thermoproteales branch is shaded.
(PDF)
Phylogeny of Archaea based on analysis of RNA polymerase subunits. Maximum likelihood tree made from aligned sequences of the three largest RNA polymerase subunits: a, a′, and b as described previously [99]. Bootstrap support numbers are given at the nodes as a percentage (n = 10,000). Scale bars represent the average number of substitutions per residue.
(PDF)
Low G+C regions in the T. tenax genome. Location in the genome, length and G+C content of the three identified regions are given.
(DOCX)
Clusters of regularly interspaced short palindromic repeats (CRISPR).
(DOC)
Amino acid biosynthesis pathways. The identified genes (sorted by respective biosynthesis pathways), their ID as well as gene name and annotation are given.
(DOCX)
TatFind (a) and FlaFind (b) positive ORFs in the T. tenax genome. The tools TatFind (http://signalfind.org/tatfind.html) [75] and FlaFind (http://signalfind.org/flafind.html) [75] have been applied.
(DOCX)
(a) Comparison of transporters in Thermoproteus tenax, Pyrobaculum aerophilum and Sulfolobus solfataricus. Classification according to the Paulsens transport database (http://www.membranetransport.org/) [76]. (b) T. tenax Substrate binding proteins.
(DOCX)
(a) Annotated T. tenax tRNA genes. (b) Annotated genes encoding ribosomal proteins. Rps, ribosomal proteins. (c) Identified T. tenax tRNA synthetase genes.
(DOCX)
Comparative genomic analysis using arCOGs. Core genes: The number of organisms in the corresponding archaeal lineages and number of proteins in individual genomes for respective arCOGs are given. Gain-loss-expansion: Results of the analysis of gene loss, gain and family expansion using arCOG data and COUNT software (http://www.iro.umontreal.ca/~csuros/gene_content/count.html) are given. Abbreviations: CREN - Crenarchaeota; EURY - Euryarchaeota; Tauma - Taumarchaeota; Korar - Korarchaeota; Nano - Nanoarchaeota; Thete - Thermoproteus tenax; All other organism abbreviations are explained in Figure S1.
(XLSX)
Annotated T. tenax genes. The respective ORF ID, location and arCOG annotations as well as COG assignment are given. The last column provides GenBank GI accession numbers for the BLASTP best hits against NCBI Non-redundant database (e-value cutoff 0.001).
(XLSX)