Abstract
Coordinated replication of eukaryotic nuclear genomes is asymmetric, with copying of a leading strand template preceding discontinuous copying of the lagging strand template. Replication is catalyzed by DNA polymerases α, δ and ε, enzymes that are related yet differ in physical and biochemical properties, including fidelity. Recent studies suggest that Pol ε is normally the primary leading strand replicase, whereas most synthesis by Pol δ occurs during lagging strand replication. New studies show that replication asymmetry can generate strand-specific genome instability resulting from biased deoxynucleotide pools and unrepaired ribonucleotides incorporated into DNA during replication, and that the eukaryotic replication machinery has evolved to most efficiently correct those replication errors that are made at the highest rates.
Introduction
Three main processes contribute to replication fidelity [1]. (i) Replicative DNA polymerases almost always select correct deoxynucleoside triphosphates (dNTPs) for incorporation onto correctly aligned primer-templates. (ii) Rare mismatches made during replication can be excised by the 3´ to 5´ exonucleases associated with certain replicases. Nucleotide selectivity and proofreading depend on the relative and absolute concentrations, respectively, of the dNTPs. (iii) Rare errors that escape proofreading can be corrected by mismatch repair (MMR). Operating in series, these processes generate very few replication errors, such that DNA based organisms generate much less than one spontaneous mutation per genome per generation [2]. This is truly amazing accuracy in light of the enormous complexity of replication. This complexity partly derives from the fact that the two DNA strands are anti-parallel yet DNA polymerases only synthesize DNA in the 3´–5´ direction. Thus coordinated replication of the two strands is intrinsically asymmetric, with copying of a leading strand template preceding discontinuous copying of the complementary lagging strand template [3,4]. During replication of the eukaryotic nuclear genome, which is the focus of this review, the lagging strand is synthesized as Okazaki fragments of about 200–300 nucleotides, requiring an enormous amount of end processing to complete replication. This review considers studies conducted in the last few years on the roles of Pols δ and ε in copying the leading and lagging strand templates, on strand-specific genome instability resulting from asymmetric replication, and on how the balance between generating and correcting errors achieves high fidelity replication of both strands.
Eukaryotic replication requires three very different polymerases
Replication of nuclear genomes is initiated at replication origins spaced at about 30–100 kilobase intervals. Two replication forks assemble at origins and travel in opposite directions until they merge with forks migrating from adjacent origins. Three eukaryotic DNA polymerases are required for replication, Pol α, Pol δ and Pol ε [4,5]. Their catalytic subunits share the sequence homology of B family polymerases, they contain the fingers, palm and thumb subdomains typical of most polymerases, and they are thought to share a common two-metal-ion catalytic mechanism for polymerization. Nonetheless, Pols α, δ and ε differ substantially in subunit composition and biochemical properties. For example, Pol α is the least processive, and also the least accurate [1] because it lacks an intrinsic proofreading exonuclease. Both properties are consistent with an essential but limited role in initiating replication at origins and of Okazaki fragments. Pol α extends RNA primers synthesized by its associated primase activity until sufficient duplex DNA is created to allow a switch to Pol δ and/or Pol ε. Pols δ and ε are better suited for extensive DNA synthesis because they are highly processive when operating with their accessory proteins. Their polymerase catalytic subunits possess 3´ exonucleolytic proofreading activity, such that they are more accurate than Pol α [1]. Studies in vitro indicate that yeast Pol ε is more accurate than yeast Pol δ with regard to single base-base [6] and single base deletion mismatches [7]. Because Pol δ and Pol ε have similar nucleotide selectivity [8], the higher fidelity of Pol ε likely reflects more efficient proofreading.
A simple model for leading and lagging strand replicases
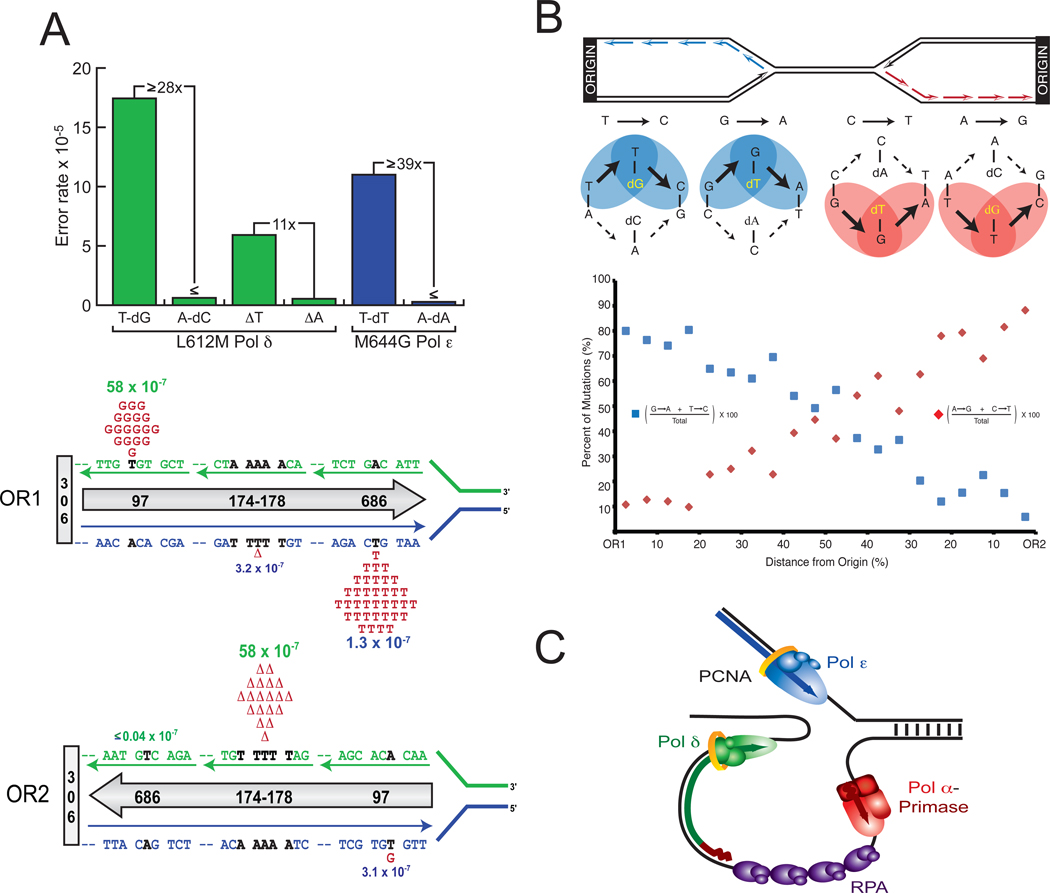
While Pol α clearly initiates DNA synthesis at origins and of Okazaki fragments, the roles of Pols δ and ε in leading and lagging strand replication in vivo have been difficult to discern, in part because mutations in the yeast genes encoding these polymerases (POL3 for Pol δ and POL2 for Pol ε) are lethal or nearly so [9,10]. The situation changed when structure-function studies identified novel variants of yeast Pol α [11], Pol δ [12–14] and Pol ε [15] that were used for tracking where each polymerase synthesizes DNA in a cell. Each variant contains a replacement for a conserved leucine (α/δ) or methionine (ε) at the polymerase active site. These variants have high replication capacity allowing normal cell growth. They also have low fidelity that elevates spontaneous mutation rates in vivo, thus identifying the polymerase responsible for making a replication error. Most importantly, they have error signatures that distinguish whether the error was made during copying the leading or lagging strand template. For example, between two mismatches that could give rise to T-A to C-G transition substitutions, L612M Pol δ generates T-dGTP mismatches at a ≥ 28-fold higher rate than A-dCTP mismatches (Fig. 1A, top) during DNA synthesis in vitro [14]. In a MMR-defective pol3-L612M msh2Δ strain that monitors uncorrected replication errors made by L612M Pol δ in vivo, T to C substitutions at base pair 97 in the URA3 gene were created at a high rate (58 × 10−7) when URA3 was located close to a well characterized replication origin (ARS306), and distant from the next closest origin [16]. Given the direction of fork movement as base pair 97 is being replicated, and inferring that the T to C substitution results from a T-dGTP mismatch, the error would be generated by Pol δ during lagging strand replication (Fig. 1A, OR1). Importantly, T to C substitutions at base pair 97 occurred at a much lower rate (3.1 × 10−7) when the URA3 orientation (OR2) was reversed relative to ARS306. In this orientation, the T-dGTP mismatch would be a leading strand error, so the much lower rate implies that Pol δ has at most a minor role in replicating the leading strand template. When a similar logic was applied to other orientation-dependent mutations across the URA3 open reading frame (e.g., deletion of a T-A base pair (Fig. 1A) and see other events in [16]), the results supported the idea that Pol δ primarily copies the lagging strand template.
Figure 1. Assigning yeast Pol δ and Pol ε to specific strands.
(A) Biased error rates in vitro and biased, orientation-specific mutation rates in vivo for two variant yeast replicases. See text for description. Adapted from [9] with permission. (B) Whole genome analysis of the distribution of transition substitutions relative to origins of replication in a pol3-L612M msh2Δ yeast strain. See text for description. Adapted from [17] with permission. (C) Model for division of polymerase labor at the eukaryotic replication fork.
A more recent study [17] extended this approach to the whole genome by sequencing 16 genomes from the pol3-L612M msh2Δ strain. This identified 1206 single base substitutions distributed evenly across all 16 chromosomes. The vast majority of these were consistent with formation of T-dGTP and G-dTTP mismatches (Fig. 1B, top). The distribution of these events was strikingly asymmetric relative to the 274 functional replication origins in yeast (Fig. 1B), and occurred with strand biases that varied as predicted if Pol δ is primarily replicating the lagging strand template.
To examine the role of Pol ε in replication, mutagenesis in URA3 was monitored in pol2-M644G strains proficient in MMR, thereby scoring Pol ε errors that escape MMR [15]. Two hotspots were observed in one URA3 orientation but not the other, both for T-A to A-T substitutions (Fig. 1A shows the hotspot at base pair 686). The results, and the fact that M644G Pol ε generates T-dTTP mismatches at a ≥39-fold higher rate than A-dATP mismatches in vitro (top of Fig. 1A), imply that Pol ε participates in replicating the leading strand template [15]. When combined with the evidence that Pol δ has at most a minor role in leading strand replication, the data further imply that Pol ε may be the major leading strand replicase. This leads to a simple model (Fig. 1C) wherein Pols ε and δ primarily replicate the leading and lagging strands, respectively. This model is consistent with a report that Pol δ and Pol ε proofread errors on opposite DNA strands [18], and with more recent evidence [19] suggesting that lagging strand replication errors generated by L868M Pol α are proofread by the 3´ exonuclease of Pol δ but not by the 3´ exonuclease of Pol ε [19]. Similar studies have not yet been performed in higher eukaryotes, but certain asymmetric error signatures for human Pol ε [20] and variants of human Pol δ [21] may be useful for this purpose.
More complex circumstances
Eukaryotic chromosomes vary widely in sequence composition, they are highly organized with respect to transcription and chromatin content, and they are constantly assaulted by endogenous metabolites and external environmental stresses that result in variety of different lesions in DNA. Each of these variables may influence the composition of replication forks, such that a simple model is unlikely to apply to all circumstances [9,10]. When replication stalls at a lesion or a natural fork barrier, the fork can be re-modeled to deal with the dilemma in several ways. One solution is translesion synthesis by one or more specialized DNA polymerases, which may operate at the fork and/or during filling of gaps left behind when replication restarts downstream of the lesion ([22] and references therein). Restart of leading strand synthesis by Pol α-primase and a switch to Pol δ could allow these enzymes to contribute to leading strand replication.
Strand-specific instability due to imbalanced dNTP pools
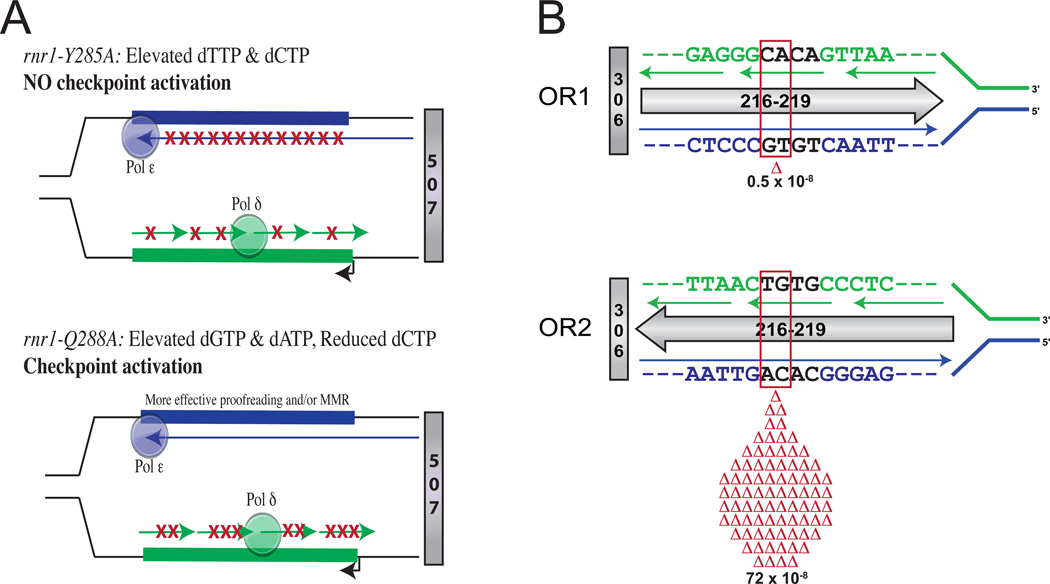
The concentrations of the dNTPs required for eukaryotic replication are not equal even in normal cells [23,24], and this natural pool imbalance can be exacerbated by mutations in the RNR1 gene encoding a subunit of ribonucleotide reductase (RNR), an enzyme involved in the production and regulation of dNTPs. These imbalances can be mutagenic by promoting polymerase misinsertion, and increased concentrations of one or more dNTPs can also reduce proofreading efficiency by increasing mismatch extension. For these reasons, mutagenic specificity can be predicted by the nature of the dNTP pool changes and the sequence of the template being replicated. Recently, two studies [25,26] used this predictability to infer strand-specific mutagenesis driven by two different pool imbalances in yeast strains with amino acid changes in the allosteric specificity site of RNR. In one strain (rnr1-Y285A), high dTTP and dCTP concentrations did not reduce proliferation or initiate an S-phase checkpoint, but strongly increased mutation rates in specific sequence contexts. The mismatches inferred to be responsible for the mutations occurred during both leading and lagging strand replication (Fig. 2A, top), and largely with the specificity predicted by the nature of the dNTP pool changes. In contrast, a strain (rnr1-Q288A) with a different dNTP pool imbalance progressed more slowly through S phase, likely due to the low dCTP concentration, and in this strain the S-phase checkpoint was activated. Surprisingly, the mutagenesis in this strain was inferred to only result from lagging strand replication (Fig. 2A, bottom). These results correlate with a suggested role for Pol ε in S-phase checkpoint control [27], again implying that Pol ε is the leading strand replicase. The checkpoint may selectively reduce leading strand replication errors by affecting one or both replication error correction steps, proofreading and MMR.
Figure 2. Strand-specific genome instability due to replication asymmetry.
(A) Strand-specific distribution of mutational hotspots in yeast strains with rnr1 mutations that differentially affect dNTP pools and checkpoint activation. See text for description. Adapted from [26] with permission. (B) Orientation-dependent CA dinucleotide deletion rates in pol2-M644G rnh201 yeast strains [30]. See text for description.
Strand-specific instability via rNTP incorporation
rNTPs in DNA present a potential risk to genome stability because the reactive 2´ hydroxyl would sensitize the DNA backbone to strand cleavage. This is interesting because rNTP concentrations in eukaryotes are much higher than dNTP concentrations [23,28]. Although most DNA polymerases discriminate well against rNTP insertion into DNA [29], a recent study showed that in reactions containing cellular rNTP and dNTP concentrations, yeast Pols α, δ and ε incorporate substantial numbers of ribonucleotides into DNA during synthesis in vitro [28]. Soon thereafter [30], rNTPs were shown to be incorporated during replication by yeast Pol ε in vivo, and also that they are normally removed by RNase H2-dependent repair. However, failure to repair these ribonucleotides in RNase H2-defective strains slightly increased replicative stress and strongly increased the rate of 2–5 base pair deletions in repetitive sequences. In some cases (e.g., deletion of a CA dinucleotide, Fig. 2B), the deletion rate was higher for one orientation of URA3 as compared to the other. These data suggest strand-specific rNTP incorporation by Pol ε, once again underscoring the asymmetry of replication. Interestingly, the rate of 2–5 base pair deletions in the pol2-M644G rnh201 strain was not strongly increased by loss of mismatch repair [31]. This suggests a model wherein, when rNMPs incorporated by M644G Pol ε during leading strand replication are not repaired by RNase H2, they are processed outside the context of replication in a manner that gives rise to misaligned intermediates that result in short deletions. In support of this hypothesis, a recent study [32] showed that topoisomerase 1 is required for formation of 2–5 base pair deletions in rnh201Δ strains. Topoisomerase 1 normally relieves DNA supercoils formed during transcription, and it can also cleave the backbone of DNA containing a single ribonucleotide. This cleavage creates DNA ends that must be processed to allow ligation, and it is this processing that is proposed to provide the opportunity for strand misalignments in repetitive sequences that ultimately yield the deletions.
Overall, these studies highlight three generally understudied subjects, discrimination against an incorrect sugar during DNA replication, the roles of RNases H in repair of ribonucleotides in DNA [33], and mutagenesis due to aberrant processing of unrepaired rNMPs in the genome. Among many interesting issues remaining to be addressed, one is the intriguing speculation that the transient presence of rNMPs in the genome may have important signaling functions (several possibilities discussed in [28]). This seems possible because rNMPs in DNA affect helix parameters (see references in [30]), which may affect protein binding. The cost of rNMPs as signals may be low because large numbers of unrepaired rNMPs can be tolerated in the yeast nuclear genome with only a mild effect on growth [30,34].
A reciprocal relationship between generating and correcting replication errors
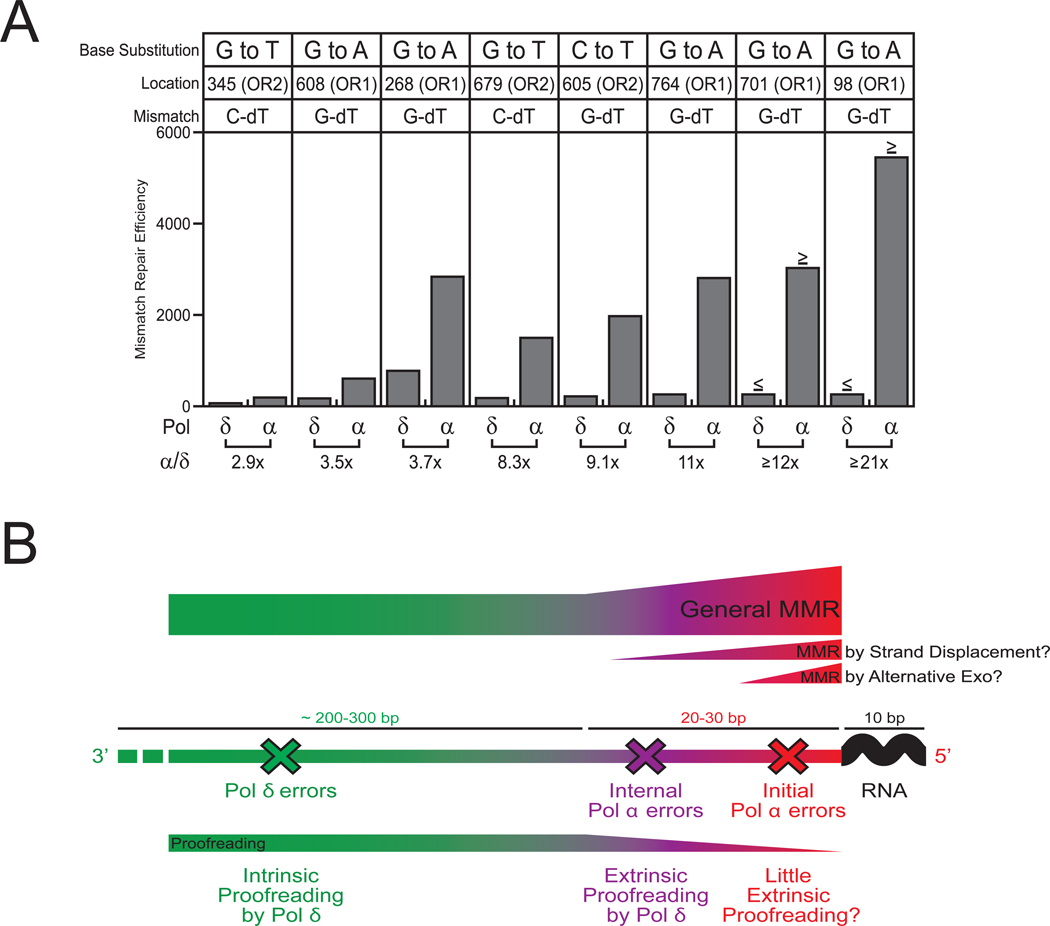
Early genetic studies in E. coli (e.g., see [35] and references therein) revealed that mismatch repair most efficiently corrects mismatches that are most commonly made by its major replicase, DNA polymerase III. This type of ‘reciprocity’ is also seen for replication errors generated by yeast Pol δ. Pol δ most frequently creates mismatches that could result in transitions and single base deletions in mononucleotide runs, and it is these errors that are corrected most efficiently by MMR [16]. Does reciprocity extend even further? Might MMR correct lagging strand replication errors made by less accurate Pol α more efficiently than those made by more accurate Pol δ? One reason to consider this possibility is that errors made by Pol α are closer to the 5´ ends of Okazaki fragments than are errors made by Pol δ. This proximity may be relevant because the 5´ ends of DNA can serve as signals for MMR in vitro [36], and because when working in conjunction with the asymmetric PCNA sliding clamp [37], DNA 5´ ends have been suggested to be strand discrimination signals in vivo [38]. When this type of reciprocity was tested in yeast using the Leu to Met mutator alleles of Pol δ and Pol α, the results (Fig. 3A and [39]) suggest that apparent MMR efficiencies may indeed be higher for base-base mismatches made by Pol α as compared to the same mismatches made by Pol δ. This supports a model (Fig. 3B) wherein the 5′ ends of Okazaki fragments serve as strand discrimination signals. The proximity of a Pol α error to a 5´ DNA end may result in higher efficiency for the general MMR machinery, just as the efficiency of E. coli MMR is proportional to the distance between the mismatch and the strand discrimination signal [36]. It is also possible that mismatches near the 5′ ends of Okazaki fragments are repaired by a MMR pathway specialized to protect the genome against particularly abundant replication errors made by Pol α (Fig. 3B). This could involve mismatch removal via strand displacement, as recently observed in vitro [40], or MMR involving nucleases such as Exo1 or the exonuclease activity of yeast FEN1, a nuclease involved in Okazaki fragment maturation that has also been suggested to participate in Msh2-Msh6-dependent MMR [41].
Figure 3. Reciprocal relationship between generating and correcting replication errors.
(A) Preferential MMR of replication errors generated by Pol α. See text for description. (B) Ideas for reciprocity between generating and correcting errors made by Pols α and δ during lagging strand replication. See text for description. Adapted from [39] with permission.
These studies support an important idea that emerged early in studies of MMR of replication errors, namely that mismatch repair may have “a special relation to the replication complex” [42]. Replication and MMR use many of the same proteins and may be physically coupled [37]. They may also be carefully coordinated in order to deal with nucleosomes, which could potentially influence MMR efficiency and are loaded onto DNA behind the replication fork. Several recent studies reveal a growing interest in this topic [43–46].
Concluding remarks
The studies reviewed here illustrate a growing appreciation of the intricate enzymology required to asymmetrically replicate large and complex eukaryotic nuclear genomes with the accuracy needed to preserve the information content of both strands. The ultimate outcome reflects not just the polymerases discussed here, but also their highly differentiated accessory proteins [5] and the coordination of replication with repair, recombination, transcription [47], nucleosome loading, and chromatin status. This complexity makes it improbable that genome stability can be generally increased [48]. This complexity also provides many opportunities for destabilizing the genome, some undoubtedly yet to be discovered. Achieving a detailed understanding of these opportunities is motivated by the beneficial consequences of mutations to organisms, like evolution, phase variation at contingency loci in microbes and somatic hypermutation of immunoglobulin genes, as well as from the many associations between mutations and human diseases [49,50]. As just one example that may be related to asymmetric relication of the leading and lagging strands, defects in the 3´ exonuclease activities of Pols δ and ε both confer cancer susceptibility in mice, but do so at different ages of onset and with different tissue specificity [51]. Could this be related to differences in strand-specific mutagenesis and/or tissue-specific differences in replication origin firing during development?
Research Highlights.
Pols δ and ε are likely to replicate the leading and lagging strand templates, respectively.
The intrinsic asymmetry of replication generates strand-specific genome instability.
Biased dNTP pools and unrepaired rNMPs in DNA result in strand-specific mutagenesis.
The most common replication errors are corrected most efficiently by mismatch repair.
Acknowledgements
To the many scientists whose work contributed to the topics considered here but whose publications were not cited due to space limitations, I thank them for their understanding. Interested readers are encouraged to consult the recent articles that are cited, where citations to many other important studies can be found. I also thank Peter Burgers and Andrei Chabes for thoughtful comments on this article. Research performed in my laboratory is supported by Projects Z01 ES065070 and Z01 ES065089 from the Division of Intramural Research, NIEHS, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake JW. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann N Y Acad Sci. 1999;870:100–107. doi: 10.1111/j.1749-6632.1999.tb08870.x. [DOI] [PubMed] [Google Scholar]
- 3.Yao NY, O'Donnell M. Replisome structure and conformational dynamics underlie fork progression past obstacles. Curr Opin Cell Biol. 2009;21:336–343. doi: 10.1016/j.ceb.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci. 2010;35:339–347. doi: 10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto K, Shimizu K, Nakashima N, Sugino A. Fidelity of DNA polymerase delta holoenzyme from Saccharomyces cerevisiae: the sliding clamp proliferating cell nuclear antigen decreases its fidelity. Biochemistry. 2003;42:14207–14213. doi: 10.1021/bi0348359. [DOI] [PubMed] [Google Scholar]
- 7.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 8.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J Biol Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavlov YI, Shcherbakova PV. DNA polymerases at the eukaryotic fork-20 years later. Mutat Res. 2010;685:45–53. doi: 10.1016/j.mrfmmm.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Murphy KM, Kanevets U, Reha-Krantz LJ. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase delta in Saccharomyces cerevisiae. Genetics. 2005;170:569–580. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase delta. J Biol Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 14.Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larrea AA, Lujan SA, Nick McElhinny SA, Mieczkowski PA, Resnick MA, Gordenin DA, Kunkel TA. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci U S A. 2010;107:17674–17679. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shcherbakova PV, Pavlov YI. 3'-->5' exonucleases of DNA polymerases epsilon and delta correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Korona DA, Lecompte KG, Pursell ZF. The high fidelity and unique error signature of human DNA polymerase {varepsilon} Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 23.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 24.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 25.Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38:3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar D, Abdulovic AL, Viberg J, Nilsson AK, Kunkel TA, Chabes A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pursell ZF, Kunkel TA. DNA polymerase epsilon: a polymerase of unusual size (and complexity) Prog Nucleic Acid Res Mol Biol. 2008;82:101–145. doi: 10.1016/S0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nick McElhinny SA, Watts B, Kumar D, Watt DL, Lundström E-B, Burgers PMJ, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci U S A. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair. 2011 doi: 10.1016/j.dnarep.2011.02.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim N, Huang SY, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watt DL, Arana ME, Johansson E, Burgers PM, Kunkel TA. Replication of ribonucleotide containing DNA templates by yeast replicative polymerases. DNA Repair. 2011 doi: 10.1016/j.dnarep.2011.05.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. Journal of Biological Chemistry. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 36.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 37.Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 38.Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- 39.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci U S A. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci U S A. 2009;106:8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson RE, Kovvali GK, Prakash L, Prakash S. Requirement of the yeast RTH1 5' to 3' exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 42.Wagner R, Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976;73:4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F, Tian L, Gu L, Li GM. Evidence that nucleosomes inhibit mismatch repair in eukaryotic cells. J Biol Chem. 2009;284:33056–33061. doi: 10.1074/jbc.M109.049874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Javaid S, Manohar M, Punja N, Mooney A, Ottesen JJ, Poirier MG, Fishel R. Nucleosome remodeling by hMSH2-hMSH6. Mol Cell. 2009;36:1086–1094. doi: 10.1016/j.molcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadyrova LY, Blanko ER, Kadyrov FA. CAF-I-dependent control of degradation of the discontinuous strands during mismatch repair. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1015914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci. 2010;35:333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Drake JW. General antimutators are improbable. J Mol Biol. 1993;229:8–13. doi: 10.1006/jmbi.1993.1002. [DOI] [PubMed] [Google Scholar]
- 49.Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 50.DePamphilis M. DNA Replication and Human Disease. Cold Spring Harbor Monograph. 2006;46 [Google Scholar]
- 51.Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]