Abstract
The Crk and Crk-like (CrkL) adaptor proteins play important roles in numerous signaling pathways, bridging tyrosine kinase substrates to downstream signaling effectors by virtue of their phosphotyrosine-binding SH2 domains and their effector-binding SH3 domains. Critical to understanding the diverse roles of Crk/CrkL is the identification of tissue- and signal-specific tyrosine phosphorylated substrates to which they are recruited and the tissue-specific effector proteins they chaperone into signaling complexes. Crk and CrkL are known biochemically and genetically to be essential mediators of Reelin/Disabled-1 (Dab1) signaling which governs proper mammalian brain development. Multimeric Reelin clusters its receptors as well as the receptor-bound intracellular scaffolding protein Dab1. Clustering induces Fyn/Src-dependent Dab1 tyrosine phosphorylation which recruits Crk/CrkL and SH3-bound effectors. Previously 21 Crk/CrkL-SH3 binding proteins were identified from diverse cell types. We present here the proteomic identification of 101 CrkL-SH3 binding proteins from embryonic murine brain. The identified proteins are enriched in the Crk/CrkL-SH3 binding motif and signaling activities regulating cell adhesion and motility. These results suggest Reelin-induced Dab1 tyrosine phosphorylation may generate a multi-faceted signaling scaffold containing a rich array of Crk/CrkL-SH3 binding effectors and may explain a growing diversity of cellular activities suggested to be influenced by Reelin/Dab1 signaling.
Keywords: Crk-like (CrkL), Crk, Src Homology 2 (SH2), Src Homology 3 (SH3), Reelin, Disabled-1 (Dab1), Proteomics, mass spectrometry, neuronal migration, brain development
Introduction
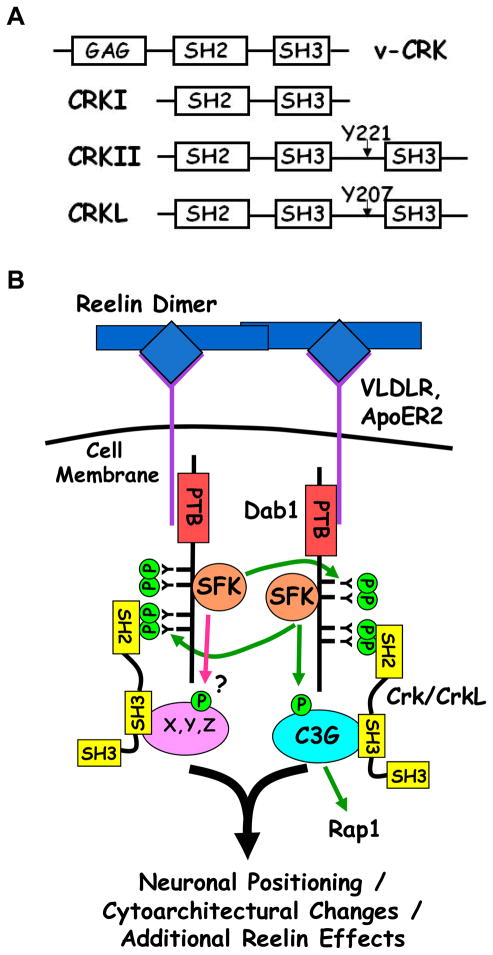
The emergence of the field of signal transduction was spurred along by discoveries identifying that the transforming agents within a number of tumor-causing retroviruses were oncogenes encoding under-regulated, hyperactive or otherwise altered proteins with diverse signaling activities. While some viral oncogenes were found to encode signaling proteins with enzymatic activities such as kinases and G-proteins1 the protein encoded by the oncogene within the CT10 avian sarcoma virus curiously neither had enzymatic activity, nor was it a growth or transcription factor. Its discoverers, Mayer et al., found instead that the oncogene product was a fusion between the major Group specific antigen (Gag) protein of the virus and a host-derived sequence containing protein-protein interaction domains highly conserved in many signaling proteins. They found this aberrant fusion led to an increase in tyrosine phosphorylated proteins within infected cells and named the oncogene protein product v-Crk for CT10 regulator of kinase2. Since its original discovery in 1988, two cellular homologs of v-Crk were identified: c-Crk (Crk) and Crk-Like (CrkL) encoded by separate genes3. In mammals Crk is primarily found in two forms (Crk I and CrkII) due to alternative splicing3. A schematic of the domain structures of Crk homologs is shown in Figure 1A and a multiple sequence alignment of v-Crk and avian, murine and human Crk protein isoforms is shown in Supporting Information Figure 1.
Figure 1. A. Schematic of the domain structures of Crk/CrkL isoforms. (B) Model of Reelin/Dab-1 Signaling involving Crk/CrkL.
Dimeric Reelin clusters its receptors ApoER2 and/or VLDLR leading to tyrosine phosphorylation of Dab1 by SFKs at 4 critical residues (Y185, Y198, Y220 and Y232). Dab1 phosphorylated at Y220 and Y232 binds to Crk and CrkL via their SH2 domains. Crk/CrkL-SH3 binding partners such as C3G or unknown partners (X, Y, Z) are thereby recruited to the emerging signaling complex where they could be locally regulated. See text for details as well as Supporting Information Figure 2 for a more elaborate model of Reelin/Dab1 signaling.
The roles of Crk and CrkL are best understood in the context of growth and differentiation factor signaling3, but recently we have shown biochemically, and others genetically that Crk and CrkL play essential roles in signaling cascades downstream of Reelin4–7, a secreted ligand critical for proper development of the vertebrate central nervous system8–12. Persistent effort from a number of laboratories has led to a model for early signaling events in typical Reelin signaling (Figure 1B and in greater detail in Supporting Information Figure 2). Reelin acting as a dimer or higher order multimer13 clusters its transmembrane receptors ApoER2 and VLDLR14, 15 thereby clustering the Reelin receptor-associated scaffolding protein Dab115, 16. This results in increased Dab1 tyrosine phosphorylation by Src family kinases (SFKs)17–19 at four important residues (Y185, Y198, Y220 and Y232)20, 21. Crk/CrkL bind to Dab1 phosphorylated at Y220 and Y232 which contain the preferred Crk-SH2 binding motif (pY(D/K/N/Q)(V/H/F)P)22–24, and mice homozygous for a dab1Y220F/Y232F allele show dramatic brain defects intermediate between wildtype and complete loss of Dab121. These results are in agreement with, previous binding studies in cell lines4, dominant negative effects of Dab1 Y220F/Y232F injected in the developing murine neocortex25 and conditional ablation of Crk and CrkL during murine brain development7.
As adaptor proteins the assumed role of Crk and CrkL in Reelin signaling is the recruitment of SH3-binding effectors to the pY-Dab1 scaffold (Fig. 1B). Furthermore Crk/CrkL-SH3 binding effectors may bring functionality that can be locally regulated by or act in concert with Reelin-induced SFK, PI3K or Akt kinase activities18, 26–28 (see Supporting Information Figure 2). Indeed the Crk/CrkL-SH3 binding protein, and Rap1 GEF, C3G becomes tyrosine phosphorylated and activated following Reelin stimulation4. Furthermore, Reelin-induced C3G activation is dependent upon Dab1 tyrosine phosphorylation and the presence of Crk/CrkL in the pY-Dab1 signaling complex4, 7, 21. Given Reelin can generate receptor clusters13 we hypothesize Crk/CrkL recruit a diversity of proteins to the assemblage of pY-Dab1 scaffolds, resulting in localized regulatory events that vary depending on the composition and relative abundance of the Crk/CrkL-SH3 binding effectors in different tissues and/or at different developmental stages. As a first step toward testing this hypothesis we endeavored to identify the repertoire of CrkL-SH3 binding partners present specifically in embryonic brain. We report here a large-scale proteomic and subsequent bioinformatic analysis of CrkL-SH3 binding proteins from embryonic murine brain extracts. We discuss the identified proteins primarily in the context of their potential roles in Reelin signaling. However, this dataset should also provide insight into the variety of signaling pathways in which Crk and CrkL have important roles, particularly in embryonic brain.
Experimental Procedures
Mice, plasmids, small molecules and antibodies
Timed pregnant and three week old (P21) CD-1 mice were ordered from Charles River Canada (Saint-Constant, Québec) and housed and treated according to an institutionally-approved IACUC protocol. Dissections were conducted when embryos were at embryonic day 16.5 (E16.5) or just after birth (PO). Plasmids encoding GST and GST-CrkL-SH3 were gifts of Akira Imamoto (University of Chicago). Plasmids encoding GST-Fyn-SH3, GST-RasGap-SH3 and the GST-Grb2 C-terminal SH3 were gifts of Jon Cooper (Fred Hutchinson Cancer Research Center). Plasmids encoding GST-Src-SH3 and GST-NCK1 N-terminal SH3 were gifts of Shawn Shun-Cheng Li (University of Western Ontario). FKBP-Dab1 wildtype and Y5F constructs were gifts of Johannes Nimpf (Medical University of Vienna). The FKBP dimerizing reagent AP20187 was a gift of Ariad Pharmaceuticals Incorporated and was used at 200nM for 20 minutes before lysis. Antibodies were obtained from the following sources: α-SOS1 and α-Tks4 (Upstate Biotechnology/Millipore, Billerica, MA); α-C3G (H-300), α-CIN85 (H-300), α-Dock4 (R6Y), α-DDEF2/ASAP2 (H-300), α-ARAP (N-20), α-Lamellipodin (H-150), α-N-WASP (H-100) and α-LPP (8B3A11) (Santa Cruz Biotechnology, Santa Cruz, CA).
Affinity chromatography and large-scale GST fusion protein pull down assay
E16.5 murine whole brain extracts were generated by dounce homogenization in ice-cold brain complex lysis buffer (BCLB: 25 mM Tris pH 7.2, 137 mM NaCl, 10% glycerol, 1% Igepal, 25 mM NaF, 10 mM Na4P2O7, 1 mM Na3VO4, 1 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml pepstatin A). Insoluble material was cleared by centrifugation at 4°C for 15 minutes at 16,000 × g and the supernatant was collected and used for further study. 20 ml of supernatant, corresponding to 30 mg of protein and generated from 21 E16.5 brains, was pre-cleared by rocking with 100 μl packed glutathione agarose resin (G-Biosciences, Maryland Heights, MO) for one hour at 4°C, centrifuging briefly and collecting the supernatant. The supernatant was then similarly pre-cleared by rocking for 3 hours at 4°C with 100 μl gutathione agarose bound to 100 μg of GST. The pre-cleared supernatant was then split into two equal halves and rocked at 4°C with 50 μl of glutathione agarose beads bound to either 50 μg GST or 50 μg GST-CrkL-SH3 overnight. Resins with bound proteins were collected by centrifugation and washed 3 times with BCLB and drained. Protein sample buffer (125 mM Tris pH 6.8, 2% SDS, 5% β-mercaptoethanol, 7.5% glycerol) was added and samples were heated to 95°C for five minutes and proteins were separated using a 10% (37.5:1 acrylamide:bis-acrylamide) SDS-PAGE gel and then stained with coomassie blue. This initial screen was performed once.
In-gel digestion and liquid chromatography tandem mass spectrometry (LC-MS/MS)
The Coomassie-stained gel from the glutathione-S-transferase (GST) and the GST-CrkL-SH3 lanes were each cut into 24 pieces above the Crk-SH3 band. The gel pieces were sliced into 1 mm cubes, washed with 1 ml HPLC grade water and incubated in 1 ml destain solution (50 mM ammonium bicarbonate and 50 % acetonitrile (MeCN)) for 30 minutes at 37°C. For complete removal of the stain, destaining was repeated once again and then subjected to dehydration by adding 200 μl of 100 % MeCN for 10 minutes. The gel pieces were further dried in a speed vacuum for 15 minutes. Proteins were cut into peptides using sequencing grade modified trypsin (Promega, Madison, WI) at a concentration of 6 ng/μl in 50 mm ammonium bicarbonate at 37°C overnight after an initial re-swelling on ice for 30 minutes at 12.5 ng/μl. The digests were centrifuged for 5 minutes at 13,000 × g and the supernatant was transferred to a 0.6 ml tube. 200 μl of extraction buffer (50% MeCN, 2.5% formic acid (FA)) was then added to the gel pieces and spun again at 13,000 × g for 15 minutes. Extracted peptides from a single band were pooled and dried in a speed vacuum. The peptides were resuspended in 2.5% FA and 2.5% MeCN and loaded using a Micro AS autosampler onto a microcapillary column of 100 μm inner diameter packed with 12 cm of reverse-phase magic C18 packing material (5 μm, 200 Å; Michrom Bioresources, Inc., Auburn, CA). After a 14.5 min isocratic loading in 2.5% MeCN, 0.15% FA (Solvent A) peptides were eluted using a 5%–35% gradient of Solvent B (99% MeCN, 0.15% FA) over 30 min and electrosprayed into a LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). The precursor scan was followed by ten collision-induced dissociation (CID) tandem mass spectra for the top 10 most abundant ions. Tandem mass spectra were searched against a concatenated forward and reverse29 mouse NCI protein database using SEQUEST30 (version 27 revision 12) requiring fully tryptic peptides, allowing a precursor mass tolerance of 2 Da; and a mass addition of 80 Da allowing for phosphorylation on serine, threonine, and tyrosine residues; addition of 16 on methionine for oxidation, and requiring the addition of 71 for an acrylamide adduct on cysetine. SEQUEST matches in the first position were then filtered by Xcorr scores of 1.0, 1.5 and 1.8 for the charge states of plus one, two and three respectively. A ΔCn2 score of 0.2 was also required for each peptide. Protein matches from the GST-CrkL-SH3 pulldowns that were not identified in the GST pulldown and were made with three or more peptides were further considered. When such filters were applied to the data searched against the composite forward and reverse mouse NCI protein databases, no reverse peptide hits remained giving a false discovery rate at the peptide level of less than 0.01%.
Small-scale pull down assays, immunoblotting and co-immunoprecipitation
E16.5, P0 and P21 whole brains were either lysed in brain complex lysis buffer as described above or with Triton Lysis Buffer pH 7.2 (1% Triton X-100, 25mM sodium phosphate made up of a ratio of 1.9:8.1 of NaH2PO4:Na2HPO4, 150mM NaCl, 5mM EDTA, 1 mM Na3VO4, 1 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml pepstatin A) with similar results. Each pulldown used 4 mg of protein extract with 40 μg used in each corresponding whole cell extract blot. After pre-clearing the extracts with 15 μg of glutathione agarose-GST in batch format for two hours, glutathione agarose-bound GST-CrkL-SH3, other indicated GST-SH3 fusions or GST alone (15–20 μg of each fusion protein per pulldown) were permitted to incubate with the pre-cleared extracts overnight. The beads were washed three times with lysis buffer and proteins were eluted with protein sample buffer and separated by SDS-PAGE. Proteins were transferred to nitrocellulose membranes, and blocked for 1 hour in 5% dry Milk, Tris-buffered saline with 0.05% Tween-20 (TBST). Primary antibody incubation was carried out in 1.5% BSA in TBST at 4°C overnight and the blots were then washed in TBST three times for 5–10 minutes each. Blots were incubated with horse radish peroxidase-conjugated secondary antibodies diluted in TBST for 1–2 hours followed by three 5–10 minute washes with TBST and detection by enhanced chemiluminescence and exposure to x-ray film. For co-immunoprecipitation experiments, Human Embryonic Kidney (HEK) 293 cells growing in Dulbecco’s Modified Eagles Medium (Mediatech, Manassas, VA), 5% fetal bovine serum (Hyclone, Logan, UT), 5% Cosmic Calf Serum (Hyclone), 50 units/mL penicillin, and 50 μg/mL streptomycin were transfected with FKBP Dab1 constructs15 using Genejammer (Agilent, Santa Clara, CA). Prior to lysis in brain complex lysis buffer, cells were treated for 15 minutes with 200 nM AP20187. Clarified cell extracts were incubated with anti-LPP antibodies while rocking overnight at 4°C. Protein G agarose (G-Biosciences, Maryland Heights, MO) was added and rocking was continued for four hours. Immune complexes were washed four times with lysis buffer, boiled in denaturing sample buffer and then subjected to SDS-PAGE and immunoblotting. Each experiment presented was performed 2–3 times with similar results.
Protein motif analyses
Motif extraction was carried out using the motif-x31 software via its online server: http://motif-x.med.harvard.edu. For the motif analysis of CrkL-SH3 binding proteins, a file containing these protein sequences was uploaded to the motif-x server using the FASTA option with the following parameters: width = 13, occurrences = 20, and significance ≤ 10−6. Proline was designated as the central residue. The background was set as the mouse IPI proteome. Sequences falling in proline-containing motifs were aligned and centered on a proline residue. Logos representing the frequency of residues surrounding the central residue were created using the Weblogo32 software at http://weblogo.berkeley.edu. To determine if an individual protein had a putative Crk/CrkL binding motif, each of the 101 identified CrkL-SH3 binding proteins was subjected to a Scansite23 analysis, scanning for Crk-binding signatures at low, medium or high stringency. Scansite23 was similarly used to identified potential Src target motifs (see Table 2).
Table 2. E16.5 murine brain CrkL-SH3 binding proteins within PANTHER gene ontology categories Cell Adhesion and Cell Communication.
PANTHER gene ontology classification was performed as described in the text and proteins in each category are listed. Additionally, each protein was searched for a possible Src substrate motif as described in the text. Identified Src substrate motifs were predicted by Scansite and their stringency is indicated (high, medium or low). None denotes no Src substrate motif was identified.
| Protein Description | Scansite | |
|---|---|---|
| Protein | Cell Adhesion (PANTHER Category) | Src target |
| Asap1 | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1 | medium |
| Ctnnd1 | Catenin delta-1 | medium |
| Ddef2 (Asap2) | Development and differentiation-enhancing factor 2 | medium |
| Abl1 | Proto-oncogene tyrosine-protein kinase ABL1 | low |
| Abl2 | Tyrosine-protein kinase ABL2 | low |
| ARAP1 | Arf-GAP, Rho-GAP domain, ANK repeat and PH domain-containing protein 1 | low |
| Sema6d | Semaphorin-6D | none |
| Cell Communication (PANTHER Category) | ||
| Dock4 | Dedicator of cytokinesis protein 4 | high |
| Pik3cb | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform | high |
| Pik3r2 | Phosphatidylinositol 3-kinase regulatory subunit beta | high |
| Asap1 | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1 | medium |
| Ctnnd1 | Catenin delta-1 | medium |
| Cyfip2 | Cytoplasmic FMR1-interacting protein 2 | medium |
| Ddef2 (Asap2) | Development and differentiation-enhancing factor 2 | medium |
| Dock3 | Dedicator of cytokinesis protein 3 | medium |
| Dock5 | Dedicator of cytokinesis protein 5 | medium |
| Snx26 (Arhgap33) | TC10/CDC42 GTPase-activating protein | medium |
| Sos1 | Son of sevenless homolog 1 | medium |
| Abl1 | Proto-oncogene tyrosine-protein kinase ABL1 | low |
| Abl2 | Tyrosine-protein kinase ABL2 | low |
| ARAP1 | Arf-GAP, Rho-GAP domain, ANK repeat and PH domain-containing protein 1 | low |
| Caskin1 | Caskin-1 | low |
| Cbl | E3 ubiquitin-protein ligase CBL | low |
| Cblb | E3 ubiquitin-protein ligase CBL-B | low |
| Dock1 | Dedicator of cytokinesis protein 1 | low |
| Eps15 | Epidermal growth factor receptor substrate 15 | low |
| Eps15l1 | Epidermal growth factor receptor substrate 15-like 1 | low |
| Inppl1 | Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2 | low |
| Itsn1 | Intersectin-1 | low |
| Phldb1 | Pleckstrin homology-like domain family B member 1 | low |
| RAPGEF1 (C3G) | Rap guanine nucleotide exchange factor 1 | low |
| Rics (Arhgap32) | Rho/Cdc42/Rac GTPase-activating protein RICS | low |
| Sec23a | Protein transport protein Sec23A | low |
| Sh3bp1 | SH3 domain-binding protein 1 | low |
| Sh3pxd2b | SH3 and PX domain-containing protein 2B | low |
| Sos2 | Son of sevenless homolog 2 | low |
| Trim67 | Tripartite motif-containing protein 67 | low |
| Brsk2 | BR serine/threonine-protein kinase 2 | none |
| EEF1A1 | Elongation factor 1-alpha 1 | none |
| Lats1 | Serine/threonine-protein kinase LATS1 | none |
| Lpp | Lipoma-preferred partner homolog | none |
| RAPH1 | Ras-associated and pleckstrin homology domains-containing protein 1 | none |
| Sema6d | Semaphorin-6D | none |
| Sh3d19 | SH3 domain-containing protein 19 | none |
| Synj1 | Synaptojanin-1 | none |
Gene ontology classifications and analysis
Gene ontology classifications were made using the PANTHER33 classification system and the definitions of these categories can be found in the Supporting Information Experimental Procedures section of this manuscript. Of the 101 identified CrkL-SH3 binding partners, PANTHER had curated gene ontology information for 99. For comparison, the top 181 identified proteins from total E16.5 brain extract, as used and as described previously34, were also subjected to PANTHER analysis and 176 of these proteins had curated gene ontology information in the PANTHER system. The gene symbols of each set were loaded and searched using the online server http://www.pantherdb.org/. The result pages were kept as “gene” and all the databases belonging to Celera, NCBI and Flybase were selected in order to maximize PANTHER output. A PANTHER Biological Processes analysis was conducted and is presented here. Information regarding cellular compartmentalization of proteins used data at the Mouse Genome Informatics resource35, or when not available the LOCATE resource36 (see text and Supporting Information Table 2 for more details).
Results and Discussion
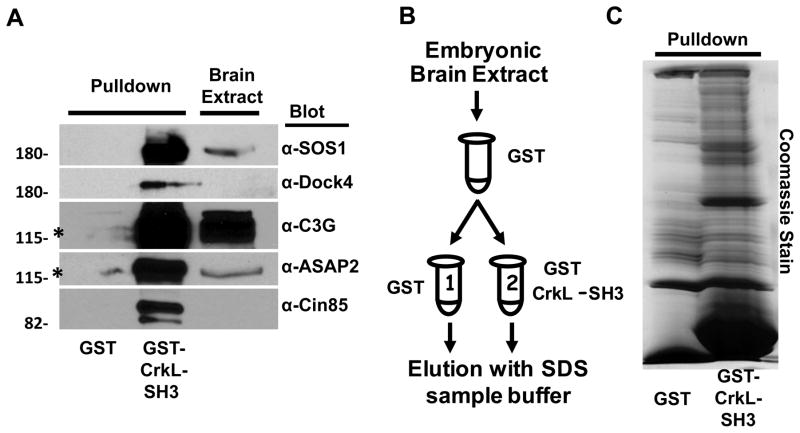
To identify CrkL-SH3 binding proteins with potential relevance to Reelin/Dab1-signaling, we examined E16.5 murine brain extracts for CrkL-SH3 binding partners. Our approach was based on affinity chromatography using a fusion protein of GST and the amino-terminal SH3 domain of CrkL. We developed a standard pulldown assay and tested if our conditions were achieving success by performing immunoblots for known CrkL-SH3 binding partners using pulldowns of either GST-CrkL-SH3 or GST alone. We immunoblotted for five known CrkL-SH3 binding proteins, the Ras GEF SOS1, the Rap GEFs C3G and DOCK4, the Arf GAP Ddef2/ASAP2 and the adaptor protein Cin85. Each of these proteins was found bound to the GST-CrkL-SH3 resin but not to the GST resin (Fig. 2A). To achieve levels of bound protein sufficient for mass spectrometry analysis, we scaled-up our experiment and used the strategy outlined in Fig. 2B. Briefly, E16.5 murine brain extracts were pre-cleared with glutathione agarose and then with agarose bound to GST. The pre-cleared extract was then divided in half. One half was subjected to a pulldown using GST while the other was subjected to a pulldown using GST-CrkL-SH3. After washing, bound proteins were eluted using SDS protein sample buffer and eluates were subjected to SDS-PAGE. The eluted proteins were visualized by staining the gel with coomassie blue which indicated multiple proteins were bound to the GST-CrkL-SH3 resin which were absent from the GST resin (Fig. 2C). Each gel lane was sectioned into 24 regions above the position of the GST-CrkL-SH3 fusion. Care was taken so as not to use the section of the gel where the GST and GST-CrkL-SH3 lanes meet. Each region was diced into 1 mm cubes, digested in-gel with trypsin, and extracted peptides were subjected to liquid chromatography tandem mass spectrometry in a linear ion trap mass spectrometer. Mass spectra were analyzed using SEQUEST30 and a concatenated forward and reverse mouse NCI protein database approach29 that facilitated filtering of top SEQUEST peptide matches to a less than 0.01% false discovery rate as described in the Experimental Procedures section for proteins identified by three or more peptides and which were not identified in GST alone analyses. These requirements eliminated all peptides whose top SEQUEST matches were to the decoy database. This resulted in the identification of 101unique proteins (Supporting Information Table 1). All identified peptides for each of these proteins are listed in Supporting Information Table 2. As Crk/CrkL function within the cytosol we conducted a bioinformatic analysis to determine the cellular compartment of the identified CrkL-SH3 binding proteins using MGI35 as way to determine if certain substrates were more or less likely to interact with CrkL in vivo. When compartmental information was not available in the MGI resource, we used the program LOCATE36, and if neither of these programs contained information for a given protein we examined individual articles. We found no compartmental information for only four proteins (see Supporting Information Table 2). These analyses revealed six proteins that were not reported to have at least some contact with the cytosol: (1) SAM and SH3 domain containing 1, serine/arginine repetitive matrix 1; (2) euchromatic histone methyltransferase 1; (3) X-ray repair complementing defective repair in Chinese hamster cells 1; (4) NCK interacting protein with SH3 domain; (5) PHD finger protein 8, protein phosphatase 1; (6) regulatory subunit 10, splicing factor 3b, subunit 2. These six proteins were deemed exclusively nuclear. However, it is worth noting that some details regarding cellular compartments may not be complete, as the protein NCK interacting protein with SH3 domain is so named given its ability to interact with NCK, a cytosolic protein that is part of the greater Crk/CrkL adapter protein family. Thus >91% of the CkrL-SH3 binding proteins identified in our study are not excluded from the cytosol and therefore and interaction with Crk/CrkL.
Figure 2. Purification of CrkL-SH3 binding partners from embryonic murine brain extract.
(A) Small-scale GST or GST-CrkL-SH3 pulldown analyses were performed on five known Crk/CrkL-SH3 binding proteins and subjected to immunoblotting with the indicated antibodies. Brain extract denotes extract prior to pulldowns. Purification procedure for large-scale pulldown (B): E16.5 brain extract after being pre-cleared with glutathione agarose was pre-cleared with GST-agarose. The supernatant was split equally and incubated with either GST-agarose or GST-CrkL-SH3 agarose. Bound proteins were eluted with denaturing SDS sample buffer and (C) subjected to SDS-PAGE and staining with coomassie blue prior to in-gel tryptic digestion and LC-MS/MS analysis.
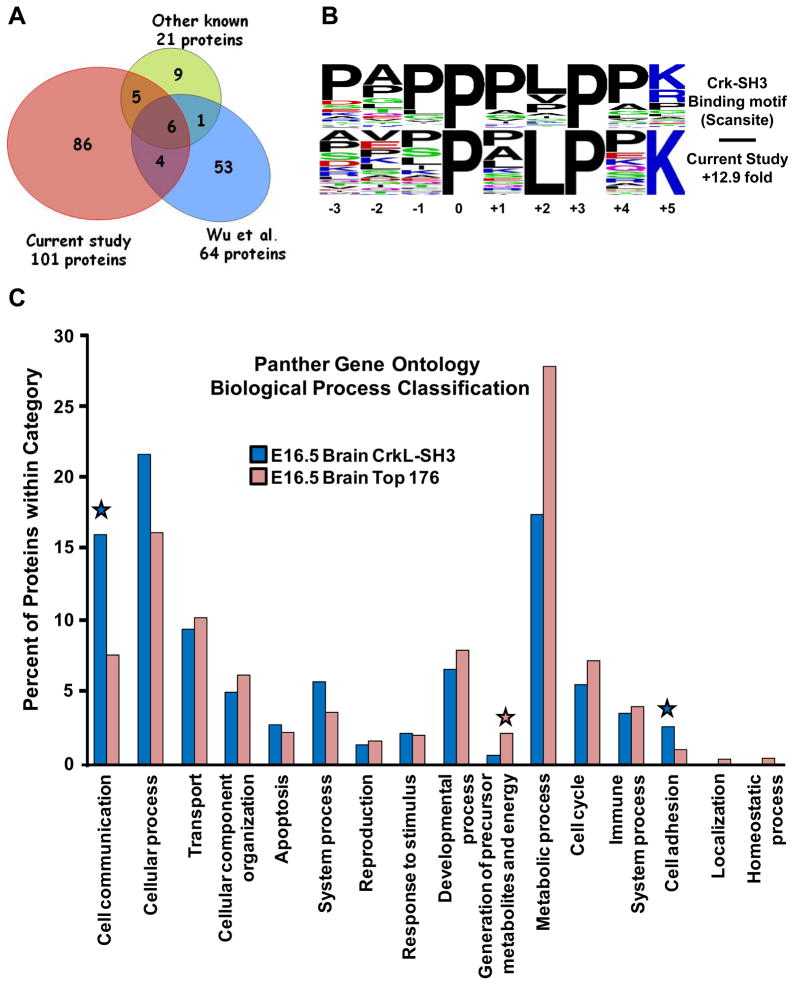
A Venn diagram is shown in Figure 3A comparing the overlap of our list of CrkL-SH3 binding proteins from embryonic brain to the 21 previously known Crk/CrkL-SH3 binding proteins whose interactions were shown biochemically using extracts from various cells or tissues (Supporting Information Table 3). We identified in our analysis 11/21 known Crk/CrkL-SH3 binding partners. The reasons for not identifying eight of the other ten can largely be explained: three of the binding partners are predominantly or exclusively expressed in hematopoietic cells (Map4k1, Dock2 and Cd34), two were identified but eliminated from our list as they were also identified in our GST alone control (Eef1a2 and Map4k5), one (Rac1) was not identified due to its size being less than the GST-CrkL-SH3 domain, below which we did not analyze, one (NS1) was from the Spanish Influenza A virus, and one (Kalrn) was likely too large (340kDa) to be identified in our gel-based analysis. The reasons for not identifying the other two (Kidins220 and Mapk8) cannot be fully explained. Also included in the Venn diagram are 64 proteins that were singled out as potential Crk-SH3 interacting proteins following a binding reaction of His6-Crk-SH3 to an array of 1,536 putative SH3 binding peptides37 (Supporting Information Table 4). Eleven (52%) of the known Crk/CrkL-SH3 binding proteins, and ten (16%) of the peptide array-based Crk-SH3-binding proteins overlapped with our dataset. However, eighty six (86%) of the proteins in our dataset have not previously been reported to interact with Crk/CrkL. This could be explained as our study is the first large-scale analysis of CrkL-SH3 binding proteins from tissue, the fact that we are using a unique embryonic tissue type, as well as the increased sensitivity of today’s mass spectrometers. Supporting Information Table 6 provides a list of the overlapping proteins in the Venn diagram.
Figure 3. Bioinformatic Characterizaton of Crk/CrkL-SH3 binding proteins.
(A) Venn diagram depicting the overlap of the CrkL-SH3 binding proteins identified in this study with the known Crk/CrkL-SH3 binding proteins and potential Crk-SH3 binding proteins as determined by peptide array (Wu et al). (B) Weblogos of a Crk-SH3-like binding motif (lower panel) found enriched in our CrkL-SH3 dataset from embryonic brain. For reference the upper panel shows the Crk-SH3 binding motif extracted from Scansite. (C) PANTHER gene ontology analysis performed on the CrkL-SH3 identified in this study compared to the same performed on the top 176 proteins identified from embryonic murine brain extract. The CrkL-SH3 binding proteins were found to be more than two-fold enriched in cell communication and cell adhesion categories (indicated by blue stars) and more than two-fold less enriched in Generation of precursor metabolites and energy (pink star).
Given that GST binding proteins were eliminated from the dataset, the CrkL-SH3 binding proteins reported here are arguably of only two types: proteins showing novel and direct binding to the CrkL-SH3 domain or proteins that bind indirectly to the CrkL-SH3 domain. An indication that the proteins in our dataset contain a number of proteins directly binding to the CrkL-SH3 domain would be if they are enriched in the target domain known to bind to Crk-SH3. We assessed this in two ways. We first conducted a motif-x analysis31. Motif-x facilitates the identification of motifs enriched within a defined set of proteins relative to the abundance of the same motif in the proteome of the organism under investigation. Among other proline-containing motifs, we found in our CrkL-SH3 binding protein dataset a greater than 12-fold enrichment of a motif similar to the optimal Crk-SH3 binding motif extracted from the motif scanning tool at Scansite23 (Fig. 3B). Second, we subjected each individual protein sequence in the three Crk/L-SH3 datasets from Fig. 3A to a Scansite23 motif analysis to determine the number and percent of proteins containing a potential Crk/CrkL-SH3 binding motif. Table 1 summarizes the results and each dataset shows a similar profile in number and percent of each stringency type of Scansite-predicted Crk-SH3 binding motifs. Indeed the three datasets show 92%, 94% and 95% of proteins from the dataset presented here, the dataset from the PATS analysis and the dataset from the known Crk/CrkL binding proteins have at least one predicted Crk-SH3 binding domain respectively. 20/21 of the known proteins contained a Scansite-identifiable Crk-SH3 binding motif. The one protein, Rac1, in which a Crk-SH3 binding domain was not identified by Scansite was shown to bind to the Crk-SH3 domain by virtue of a PPPvkkRKRk motif in the C-terminal region of Rac138, with the PPP and RKR sequences shown to be essential, at least as one set or another by mutating each set to a series of alanines. Thus, in this case, it appears that the first proline and the last lysine (underline above) in the PXXPXK motif proved sufficient for binding to the Crk-SH3 domain. Given that the proteins identified in this study have enrichments in Crk/CrkL-SH3 binding motifs and given 92% of the identified proteins have at least one Crk/CrkL-SH3 binding motif as predicted by Scansite, this suggests the embryonic brain CrkL-SH3 binding proteins are enriched in proteins that bind directly to the CrkL-SH3 domain.
Table 1. Summary of Proline-Based Motifs Identified from Crk/CrkL-SH3 binding proteins data sets.
The three protein datasets compared in this table are (A) the CrkL-SH3 binding proteins identified in this study from embryonic murine brain (E16.5 brain), (B) the proteins harboring sequences used for peptide array-based identification of Crk-SH3 binding partners (PATS), (C) the previously identified Crk/CrkL-SH3 binding proteins (Known). Data in column I are the number and percentage of proteins in the given dataset which contain a proline-based motif as determined by motif-x and which were used to generate the Weblogos in Fig. 3B. Column II gives the number and percent of proteins with a Scansite-predicted Crk-SH3 binding motif in each dataset. Column III gives the number and percent of proteins in the given dataset with Scansite-predicted Crk-SH3 binding motifs by Scansite stringency level: high (H), medium (M) and low (L).
| Crk/CrkL-SH3Binding Protein Set | I. # of proteins in dataset | II. # and % of proteins with at least one Scansite-predicted Crk-SH3 binding motif | III. # and % of proteins with top Scansite Crk-SH3 binding motif in stringency category: (H)igh, (M)edium, (L)ow, (N)one |
|---|---|---|---|
| (A) E16.5 Brain | 101 | 93, 92% | 51H (51%), 26M (26%), 16L (16%), 8N (8%) |
| (B) PATS | 64 | 60, 94% | 25H (39%), 20M (31%), 15L (23%), 4N (6.3%) |
| (C) Known | 21 | 20, 95% | 14H (67%), 3M (14%), 3L (14%), 1N (5%) |
To determine if the CrkL-SH3 binding proteins we identified were enriched for biological processes consistent with potential functions of Reelin signaling we used the gene ontology analysis program Panther33. For the purpose of generating a basis of comparison we also performed the Panther analysis on the top 176 proteins identified in a large-scale proteomic analysis of a whole cell extract of E16.5 murine brain34. The Panther program calculates the percentage of the annotated proteins in the dataset falling into one of 16 biological process categories with some proteins conceivably being part of more than one category. The complete Panther analysis we performed for each dataset is presented in Supporting Information Table 5. In Fig. 3C we graphically portray the 16 categories and show the percent of proteins from each of the datasets falling into each category. When we compared the CrkL-SH3 proteins identified in this study to the top 180 most identified proteins from the embryonic brain extract to we found that the CrkL-SH3 binding proteins had two times the representation in the cell communication and cell adhesion PANTHER categories. Conversely, the CrkL-SH3 binding proteins were more than two-fold less enriched in the category “Generation of precursor metabolites and energy.” A similar reduction was also observed in “Metabolic processes.” Supporting Information Table 5 lists the CrkL-SH3 binding proteins in each category. Intriguingly, an enrichment of CrkL-SH3 binding proteins in cellular communication and the regulation of cell adhesion is consistent with cellular mechanisms at play when cells are responding to signaling cues regulating cellular migration or positioning. Given that one of these proteins, C3G, was previously shown to be phosphorylated on tyrosine in a Reelin and Crk/CrkL-dependent manner, likely due to activated SFKs, we asked if other proteins in these categories might have predicted Src target motifs using Scansite23. Intriguingly, 80% of the proteins in these categories had putative Src phosphorylation sites as opposed to 69% of the entire dataset.
It is important to note that proteins in categories where enrichments were not observed may still be important to Reelin signaling. For example, proteins associated with the Golgi may be classified in the transport category, and while this may not seem immediately relevant to Reelin signaling, Reelin was recently reported to dramatically alter Golgi polarity39. However, while regulated intracellular trafficking may be initiated by Crk/CrkL-SH3 binding proteins, downstream trafficking effector proteins may not necessarily be found bound directly to the Crk/CrkL-SH3 domain. An example of Reelin regulated vesicular trafficking was recently reported describing Reelin-induced Cdc42 activation of N-WASP and the Arp2/3 complex40. Of note, not only was N-WASP previously identified as a pY-Dab1 binding protein41, we also identified N-WASP to be a novel CrkL-SH3 binding protein.
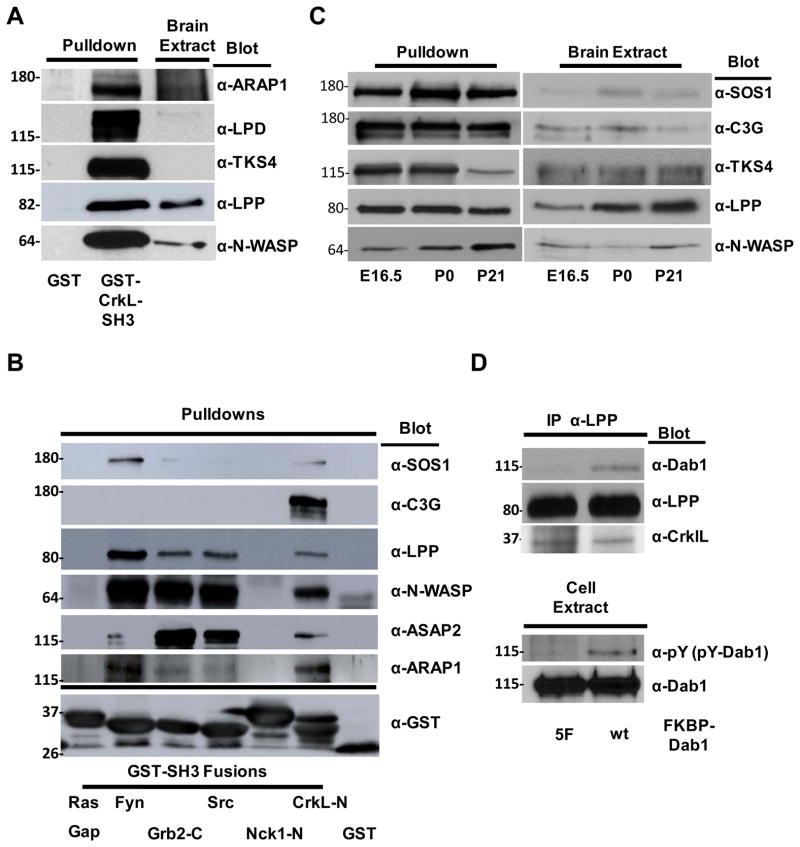
Given that we identified a number of novel CrkL-SH3 binding proteins we conducted again a small-scale GST-CrkL-SH3 pulldown followed by immunoblotting for five of the novel proteins that may have particular relevance to Reelin signaling, ARAP1, LPD, TKS4, LPP and N-WASP. The results are shown in Figure 4A and provide confirmation that these proteins bind to GST-CrkL-SH3 and not to GST alone. ARAP1 is a protein with Arf GAP, Rho GAP, Ankyrin repeat, Ras-associating (RA), and Plekstrin homology (PH) domains and is thought to function at the interface of various signaling activities. Indeed ARAP1 is known to have PI3,4,5P3-dependent Arf-Gap activity and plays roles in regulating receptor recycling and Golgi structure42, 43. Given Reelin is also thought to be involved in regulating actin and golgi dynamics and Reelin-induced PI3K activity appears critical for multiple aspects of Reelin signaling28, 44, 45 ARAP1 is an intriguing, albeit complex candidate in Reelin signaling. LPD (Lamellipodin) is an Ena/VASP binding protein that regulates actin dynamics and induces the formation of lamellipodia requiring the PI3K product PI3,4P2 and is therefore another intriguing potential effector of Reelin signaling46, 47. TKS4 (Tyrosine Kinase Substrate with 4 SH3 domains) is important in the formation of actin rich podosome structures that recruit matrix metalloproteases48, 49. Best understood in cancer paradigms, podosomes are also being studied in the context of non-cancerous migratory cell types50, 51. Intriguingly TKS4 requires phosphorylation by SFKs for its activity and binds the PI3K lipid products PI3P and PI3,4P2 via its Phox homology (PX) domain49, 52. LPP (lipoma preferred (translocation) partner) is a member of the zyxin family of Ena/VASP binding proteins. LPP localizes to sites of cell adhesion and is thought to recruit Ena/VASP proteins for localized actin polymerization and cell protrusion53, 54.
Figure 4. Biochemical Characterization of CrkL-SH3 binding proteins.
(A) Confirmation of five novel CrkL-SH3 binding proteins. Small-scale pulldown analyses were performed as described in Fig. 2A. Following pulldowns and SDS-PAGE, transferred proteins were immunoblotted using the indicated antibodies. (B) Comparison of selected CrkL-SH3 binding proteins with a panel of SH3 domains. Indicated GST-SH3 domains were purified in bacteria and used in pulldown assays as described in Fig. 2A. (C) Comparison of relative binding of selected CrkL-SH3 binding partners in pulldown assays (left panels) from murine brain extracts (right panels) at different stages of development (E16.5, P0 and P21). Following SDS-PAGE, pulldowns and brain extracts were subjected to immunoblotting as indicated. (D) Immunoprecipitation of a trimeric complex between phosphotyrosyl-Dab1, CrkL and LPP. Inducibly-dimerized FKBP-Dab1 (wt) and FKBP-Dab1 (Y5F) were transfected into HEK 293 cells and cells were treated with the bivalent FKBP dimerization agent prior to lysis. Clarified whole cell extracts were immunoprecipitated with anti-LPP antibodies and immune complexes were boiled and subjected to SDS-PAGE and immunoblotting as indicated.
In order to determine if the CrkL-SH3 binding partners we identified might also interact with other SH3 domains, we directly compared the binding of three of the known and three of the novel CrkL-SH3 binding proteins in pulldown assays using the CrkL-SH3 (N-terminal) domain and SH3 domains from five other proteins. Strikingly, only one substrate showed a dominant preference for only one SH3 domain, that being C3G for the CrkL-SH3 domain (Fig. 4B). While highly specific binding preferences can greatly facilitate our understanding of specificity in signaling, we argue that even if some substrates do not bind exclusively to the CrkL-SH3 domain this does not exclude them from contributing to Reelin or other CrkL-dependent signaling pathways. However, our data suggest that one should not assume that the CrkL-SH3 binding proteins that we have identified bind exclusively to CrkL’s N-terminal SH3 domain.
One intriguing possibility is that Reelin-Dab1 signaling can differ at different stages of development or in different tissues depending on the abundance and availability of the various CrkL-SH3 binding proteins. We tested age-specific binding differences for five proteins to the CrkL-SH3 domain from extracts of E16.5, P0 and P21 murine brains. While the majority of differences in binding appear to be due to differences in the individual protein levels in the various brain extracts, this was not true for LPP, TKS4 and N-WASP (Fig. 4C). While LPP increased in protein expression from E16.5 to P21 its binding to the CrkL-SH3 domain at P21 was much reduced compared to the binding at E16.5. TKS4 also showed reduced binding from P21 extracts even though the levels of TKS4 didn’t change. Conversely, while N-WASP levels changed little across the three stages, its binding to the CrkL-SH3 domain increased at older stages. These data may be explained by a regulated interaction of some CrkL-SH3 binding partners. Regulated binding to SH3 domains has precedence, with an excellent example being ERK1/2-dependent phosphorylation of SOS1 leading to a disruption of the binding of SOS1 to the SH3 domain of Grb255. Alternatively, the observed differences could be due to CrkL-SH3 binding partners in some brain extracts being stably associated with either endogenous Crk/L or other proteins such that they are inaccessible during the pulldowns. In addition, should the level of total CrkL-SH3 binding partners at one stage of development exceed the number of GST-CrkL-SH3 in the pulldown this could also give the perception of stage-specific differential binding. Given ~20 μg of GST-CrkL-SH3 fusion protein and 4 mg of extract was used in the small-scale pulldowns the GST-CrkL-SH3 fusion protein would be in excess until the sum of the CrkL-SH3 binding partners in the extract achieved 0.5% of the total protein. Therefore this issue is not likely to be an important factor in our analyses.
While significant work remains to fully characterize each of the identified CrkL-SH3 binding proteins, particularly their possible roles in Reelin signaling, we have further characterized one of the interacting proteins, LPP, by examining complex formation using co-immunopreciptiation. HEK 293 cells were transfected with either a Dab1 wildtype construct or a mutant Dab1 construct (Y5F). The Y5F construct has five tyrosine-to-phenylalanine mutations such that Dab1 cannot be tyrosine phosphorylated. Additionally, each of the Dab1 constructs is fused to an FKBP dimerization domain that can be used to induce dimerization with the bivalent compound AP20187. Importantly, this leads to tyrosine phosphorylation in the case of the wildtype Dab1 construct and not the mutant15. After treating both sets of transfected cells with AP20187, LPP was immunoprecipitated and a trimeric complex of LPP, CrkL and phosphotyrosyl-Dab1 was observed, whereas in the case of the Y5F construct only the LPP-CrkL dimeric complex was observed, showing a trimeric complex dependent on Dab1 tyrosine phosphorylation (Fig. 4D).
Conclusion
Given the potential for multivalent Crk/CrkL signaling complexes generated following Reelin’s clustering of its receptors, a full understanding of Reelin/Dab1 signaling requires knowledge of the SH3 binding partners of Crk/CrkL in the various tissue types and at the developmental stages where Reelin is functioning. This may be particularly true as Reelin/Dab1 signaling has increasingly been suggested to play roles that may be independent of its recognized function in the development of the central nervous system. These include roles in synaptic transmission, learning and cognition56; protection mechanisms against neurodegeneration, schizophrenia and other psychiatric disorders 57–60; and finally adult roles for Reelin in non-neuronal cells including cells of the mammary glands61, liver and the lymphatics62. We present here the first large-scale analysis of CrkL-SH3 binding partners from embryonic murine brain and have identified 101 proteins using affinity chromatography and mass spectrometry analysis. 86 of the identified proteins were entirely novel CrkL-SH3 interacting proteins. Given the important role of Crk/CrkL in murine brain development, particularly as related to Reelin signaling, and given that a number of the identified Crk/CrkL-SH3 partners have roles in motility, adhesion and signaling acitivites, these results will serve to generate a number of targeted studies seeking to delineate Reelin signaling further along its branching path into the cells it modulates.
Supplementary Material
Acknowledgments
This work was supported by NSF grant IOS 1021795 (to B.A.B.), the Vermont Genetics Network through NIH Grant P20 RR16462 (support given to B.A.B and M.C.) from the INBRE Program of the National Center for Research Resources (NCRR), and NIH Grant 5 P20 RR016435 from the COBRE Program in Neuroscience funded by the NCRR (support given to B.A.B. and M.C.). We thank A. Imamoto, J. Cooper, J. Nimpf, S. Li and Ariad Pharmaceuticals Inc. for critical reagents.
Abbreviations used
- SH2
Src Homology 2
- SH3
Src Homology 3
- GEF
guanine nucleotide exchange factor
- SFK
Src Family Kinase
- Dab1
Disabled-1
- Crk
CT10 regulator of kinase
- CrkL
Crk-Like
- C3G
Crk SH3-binding GEF
- PI3K
Phosphatidylinositol-3 Kinase
- Gag
Group Specific Antigen
- GAP
GTPase-activating protein
- SOS
Son of Sevenless
- Cin85
Cbl-interacting protein of 85 kDa
- Rap1
Ras-proximate-1
- GST
Glutathione S-transferase
- DOCK4
Dedicator of Cytokinesis 4
- N-WASP
Neural Wiskott-Aldrich syndrome protein
- Ena/VASP
Enabled/Vasodilator-stimulated phosphoprotein
- Cdc42
cell division cycle 42
- Arp2/3
Actin related protein 2/3
- ARAP1
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
- LPD
Lamellipodin
- TKS4
Tyrosine Kinase Substrate with 4 SH3 domains
- LPP
lipoma preferred partner
- GAP
GTP-ase activating protein
- PH
Plextrin Homology
- RA
Ras-associating
- PX
Phox homology
- Lis1
Lissencephaly-1
- NCKβ
Noncatalytic Region of Tyrosine Kinase, Beta
- NMDAR
N-Methyl-D-aspartic acid Receptor
- GSK-3
Glycogen synthase kinase 3
- LimK
Lin-1, Isl-1, and Mec-3 (LIM) kinase
- TSC1/2
Tuberous Sclerosis Complex 1/2 proteins
- SOCS
suppressor of cytokine signaling
- Cul5
E3 ubiquitin ligase component Cullin 5
- MGI
Mouse Genome Informatics
- IACUC
Institutional Animal Care and Use Committee
Footnotes
Supporting Information Available: Two figures, six tables and one text file. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bishop JM. Cellular oncogenes and retroviruses. Annu Rev Biochem. 1983;52:301–54. doi: 10.1146/annurev.bi.52.070183.001505. [DOI] [PubMed] [Google Scholar]
- 2.Mayer BJ, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988;332(6161):272–5. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- 3.Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20(44):6348–71. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- 4.Ballif BA, Arnaud L, Arthur WT, Guris D, Imamoto A, Cooper JA. Activation of a Dab1/CrkL/C3G/Rap1 pathway in Reelin-stimulated neurons. Curr Biol. 2004;14(7):606–10. doi: 10.1016/j.cub.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Magdaleno S, Hopkins R, Slaughter C, Curran T, Keshvara L. Tyrosine phosphorylated Disabled 1 recruits Crk family adapter proteins. Biochem Biophys Res Commun. 2004;318(1):204–12. doi: 10.1016/j.bbrc.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Chen K, Ochalski PG, Tran TS, Sahir N, Schubert M, Pramatarova A, Howell BW. Interaction between Dab1 and CrkII is promoted by Reelin signaling. J Cell Sci. 2004;117(Pt 19):4527–36. doi: 10.1242/jcs.01320. [DOI] [PubMed] [Google Scholar]
- 7.Park TJ, Curran T. Crk and Crk-like play essential overlapping roles downstream of disabled-1 in the Reelin pathway. J Neurosci. 2008;28(50):13551–62. doi: 10.1523/JNEUROSCI.4323-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CC, D’Arcangelo G. The Reelin Gene and Its Functions in Brain Development. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 1–13. [Google Scholar]
- 9.Meyer G. Comparative Anatomy and Evolutionary Roles of Reelin. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 69–87. [Google Scholar]
- 10.D’Arcangelo G. The reeler mouse: anatomy of a mutant. Int Rev Neurobiol. 2005;71:383–417. doi: 10.1016/s0074-7742(05)71016-3. [DOI] [PubMed] [Google Scholar]
- 11.Olson EC, Walsh CA. Reelin/Dab1 Signaling in the Developing Cerebral Cortex. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 89–105. [Google Scholar]
- 12.Hevner RE. Reelin and the Cerebellum. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 141–158. [Google Scholar]
- 13.Takagi J. Crystal Structure of Reelin Repeats. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 57–67. [Google Scholar]
- 14.Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97(6):689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 15.Strasser V, Fasching D, Hauser C, Mayer H, Bock HH, Hiesberger T, Herz J, Weeber EJ, Sweatt JD, Pramatarova A, Howell B, Schneider WJ, Nimpf J. Receptor clustering is involved in Reelin signaling. Mol Cell Biol. 2004;24(3):1378–86. doi: 10.1128/MCB.24.3.1378-1386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrick TM, Cooper JA. High affinity binding of Dab1 to Reelin receptors promotes normal positioning of upper layer cortical plate neurons. Brain Res Mol Brain Res. 2004;126(2):121–8. doi: 10.1016/j.molbrainres.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Arnaud L, Ballif BA, Forster E, Cooper JA. Fyn tyrosine kinase is a critical regulator of disabled-1 during brain development. Curr Biol. 2003;13(1):9–17. doi: 10.1016/s0960-9822(02)01397-0. [DOI] [PubMed] [Google Scholar]
- 18.Bock HH, Herz J. Reelin activates SRC family tyrosine kinases in neurons. Curr Biol. 2003;13(1):18–26. doi: 10.1016/s0960-9822(02)01403-3. [DOI] [PubMed] [Google Scholar]
- 19.Kuo G, Arnaud L, Kronstad-O’Brien P, Cooper JA. Absence of Fyn and Src causes a reeler-like phenotype. J Neurosci. 2005;25(37):8578–86. doi: 10.1523/JNEUROSCI.1656-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell BW, Herrick TM, Hildebrand JD, Zhang Y, Cooper JA. Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr Biol. 2000;10(15):877–85. doi: 10.1016/s0960-9822(00)00608-4. [DOI] [PubMed] [Google Scholar]
- 21.Feng L, Cooper JA. Dual functions of Dab1 during brain development. Mol Cell Biol. 2009;29(2):324–32. doi: 10.1128/MCB.00663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72(5):767–78. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 23.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–41. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ballif BA, Carey GR, Sunyaev SR, Gygi SP. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J Proteome Res. 2008;7(1):311–8. doi: 10.1021/pr0701254. [DOI] [PubMed] [Google Scholar]
- 25.Sanada K, Gupta A, Tsai LH. Disabled-1-regulated adhesion of migrating neurons to radial glial fiber contributes to neuronal positioning during early corticogenesis. Neuron. 2004;42(2):197–211. doi: 10.1016/s0896-6273(04)00222-3. [DOI] [PubMed] [Google Scholar]
- 26.Ballif BA, Arnaud L, Cooper JA. Tyrosine phosphorylation of Disabled-1 is essential for Reelin-stimulated activation of Akt and Src family kinases. Brain Res Mol Brain Res. 2003;117(2):152–9. doi: 10.1016/s0169-328x(03)00295-x. [DOI] [PubMed] [Google Scholar]
- 27.Beffert U, Morfini G, Bock HH, Reyna H, Brady ST, Herz J. Reelin-mediated signaling locally regulates protein kinase B/Akt and glycogen synthase kinase 3beta. J Biol Chem. 2002;277(51):49958–64. doi: 10.1074/jbc.M209205200. [DOI] [PubMed] [Google Scholar]
- 28.Bock HH, Jossin Y, Liu P, Forster E, May P, Goffinet AM, Herz J. Phosphatidylinositol 3-kinase interacts with the adaptor protein Dab1 in response to Reelin signaling and is required for normal cortical lamination. J Biol Chem. 2003;278(40):38772–9. doi: 10.1074/jbc.M306416200. [DOI] [PubMed] [Google Scholar]
- 29.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 30.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67(8):1426–36. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23(11):1391–8. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 32.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31(1):334–41. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zappaterra MD, Lisgo SN, Lindsay S, Gygi SP, Walsh CA, Ballif BA. A comparative proteomic analysis of human and rat embryonic cerebrospinal fluid. J Proteome Res. 2007;6(9):3537–48. doi: 10.1021/pr070247w. [DOI] [PubMed] [Google Scholar]
- 35.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39(Database issue):D842–8. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fink JL, Aturaliya RN, Davis MJ, Zhang F, Hanson K, Teasdale MS, Kai C, Kawai J, Carninci P, Hayashizaki Y, Teasdale RD. LOCATE: a mouse protein subcellular localization database. Nucleic Acids Res. 2006;34(Database issue):D213–7. doi: 10.1093/nar/gkj069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C, Ma MH, Brown KR, Geisler M, Li L, Tzeng E, Jia CY, Jurisica I, Li SS. Systematic identification of SH3 domain-mediated human protein-protein interactions by peptide array target screening. Proteomics. 2007;7(11):1775–85. doi: 10.1002/pmic.200601006. [DOI] [PubMed] [Google Scholar]
- 38.van Hennik PB, ten Klooster JP, Halstead JR, Voermans C, Anthony EC, Divecha N, Hordijk PL. The C-terminal domain of Rac1 contains two motifs that control targeting and signaling specificity. J Biol Chem. 2003;278(40):39166–75. doi: 10.1074/jbc.M307001200. [DOI] [PubMed] [Google Scholar]
- 39.Matsuki T, Matthews RT, Cooper JA, van der Brug MP, Cookson MR, Hardy JA, Olson EC, Howell BW. Reelin and stk25 have opposing roles in neuronal polarization and dendritic Golgi deployment. Cell. 2010;143(5):826–36. doi: 10.1016/j.cell.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leemhuis J, Bouche E, Frotscher M, Henle F, Hein L, Herz J, Meyer DK, Pichler M, Roth G, Schwan C, Bock HH. Reelin signals through apolipoprotein E receptor 2 and Cdc42 to increase growth cone motility and filopodia formation. J Neurosci. 2010;30(44):14759–72. doi: 10.1523/JNEUROSCI.4036-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suetsugu S, Tezuka T, Morimura T, Hattori M, Mikoshiba K, Yamamoto T, Takenawa T. Regulation of actin cytoskeleton by mDab1 through N-WASP and ubiquitination of mDab1. Biochem J. 2004;384(Pt 1):1–8. doi: 10.1042/BJ20041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, Resau J, Zheng Y, Randazzo PA. ARAP1: a point of convergence for Arf and Rho signaling. Mol Cell. 2002;9(1):109–19. doi: 10.1016/s1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- 43.Daniele T, Di Tullio G, Santoro M, Turacchio G, De Matteis MA. ARAP1 regulates EGF receptor trafficking and signalling. Traffic. 2008;9(12):2221–35. doi: 10.1111/j.1600-0854.2008.00823.x. [DOI] [PubMed] [Google Scholar]
- 44.Jossin Y, Goffinet AM. Reelin signals through phosphatidylinositol 3-kinase and Akt to control cortical development and through mTor to regulate dendritic growth. Mol Cell Biol. 2007;27(20):7113–24. doi: 10.1128/MCB.00928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chai X, Forster E, Zhao S, Bock HH, Frotscher M. Reelin stabilizes the actin cytoskeleton of neuronal processes by inducing n-cofilin phosphorylation at serine3. J Neurosci. 2009;29(1):288–99. doi: 10.1523/JNEUROSCI.2934-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H, Way M, Yaffe MB, Boussiotis VA, Gertler FB. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. 2004;7(4):571–83. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 47.Bae YH, Ding Z, Das T, Wells A, Gertler F, Roy P. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1002309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buschman MD, Bromann PA, Cejudo-Martin P, Wen F, Pass I, Courtneidge SA. The novel adaptor protein Tks4 (SH3PXD2B) is required for functional podosome formation. Mol Biol Cell. 2009;20(5):1302–11. doi: 10.1091/mbc.E08-09-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iqbal Z, Cejudo-Martin P, de Brouwer A, van der Zwaag B, Ruiz-Lozano P, Scimia MC, Lindsey JD, Weinreb R, Albrecht B, Megarbane A, Alanay Y, Ben-Neriah Z, Amenduni M, Artuso R, Veltman JA, van Beusekom E, Oudakker A, Millan JL, Hennekam R, Hamel B, Courtneidge SA, van Bokhoven H. Disruption of the podosome adaptor protein TKS4 (SH3PXD2B) causes the skeletal dysplasia, eye, and cardiac abnormalities of Frank-Ter Haar Syndrome. Am J Hum Genet. 2010;86(2):254–61. doi: 10.1016/j.ajhg.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spinardi L, Marchisio PC. Podosomes as smart regulators of cellular adhesion. Eur J Cell Biol. 2006;85(3–4):191–4. doi: 10.1016/j.ejcb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Block MR, Badowski C, Millon-Fremillon A, Bouvard D, Bouin AP, Faurobert E, Gerber-Scokaert D, Planus E, Albiges-Rizo C. Podosome-type adhesions and focal adhesions, so alike yet so different. Eur J Cell Biol. 2008;87(8–9):491–506. doi: 10.1016/j.ejcb.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Gianni D, Taulet N, DerMardirossian C, Bokoch GM. c-Src-mediated phosphorylation of NoxA1 and Tks4 induces the reactive oxygen species (ROS)-dependent formation of functional invadopodia in human colon cancer cells. Mol Biol Cell. 2009;21(23):4287–98. doi: 10.1091/mbc.E10-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petit MM, Fradelizi J, Golsteyn RM, Ayoubi TA, Menichi B, Louvard D, Van de Ven WJ, Friederich E. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol Biol Cell. 2000;11(1):117–29. doi: 10.1091/mbc.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petit MM, Meulemans SM, Van de Ven WJ. The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J Biol Chem. 2003;278(4):2157–68. doi: 10.1074/jbc.M206106200. [DOI] [PubMed] [Google Scholar]
- 55.Corbalan-Garcia S, Yang SS, Degenhardt KR, Bar-Sagi D. Identification of the mitogen-activated protein kinase phosphorylation sites on human Sos1 that regulate interaction with Grb2. Mol Cell Biol. 1996;16(10):5674–82. doi: 10.1128/mcb.16.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu S, Weeber EJ. Reelin and Cognition. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 171–191. [Google Scholar]
- 57.Fatemi SH, Reutiman TJ, Folsom TD. The Role of Reelin in Etiology and Treatment of Psychiatric Disorders. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 317–339. [Google Scholar]
- 58.Abdolmaleky HM, Smith CL, Zhou JR, Thiagalingam S. Epigenetic Modulation of Reelin Function in Schizophrenia and Bipolar Disorder. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 365–384. [Google Scholar]
- 59.Lintas C, Persico AM. Reelin Gene Polymorphisms in Autistic Disorder. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 385–399. [Google Scholar]
- 60.Botella-López A, Sáez-Valero J. Alzheimer’s Diease and Reelin. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 401–409. [Google Scholar]
- 61.Khialeeva E, Lane TF, Carpenter EM. Disruption of reelin signaling alters mammary gland morphogenesis. Development. 138(4):767–76. doi: 10.1242/dev.057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samama B, Boehm N. Reelin, Liver, and Lymphatics. In: Fatemi SH, editor. Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease. Springer; New York, NY: 2008. pp. 251–261. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.