Abstract
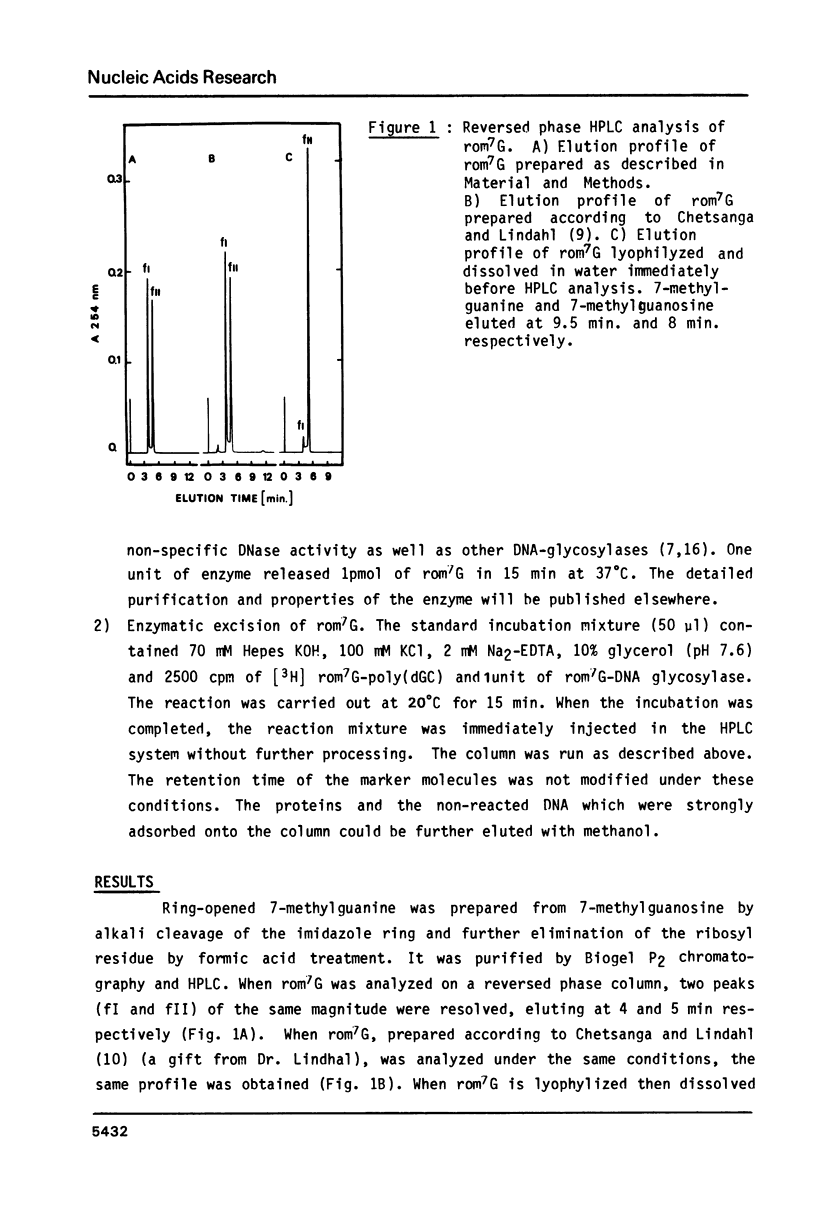
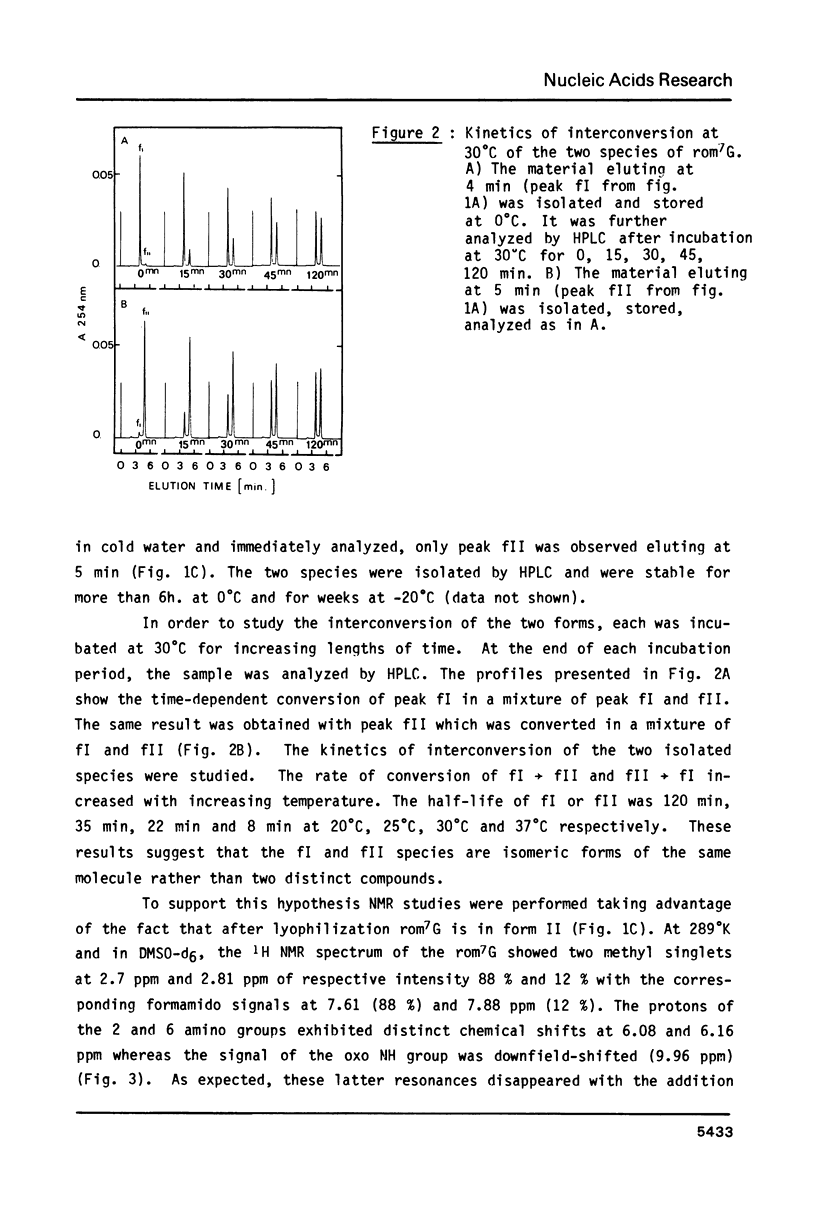
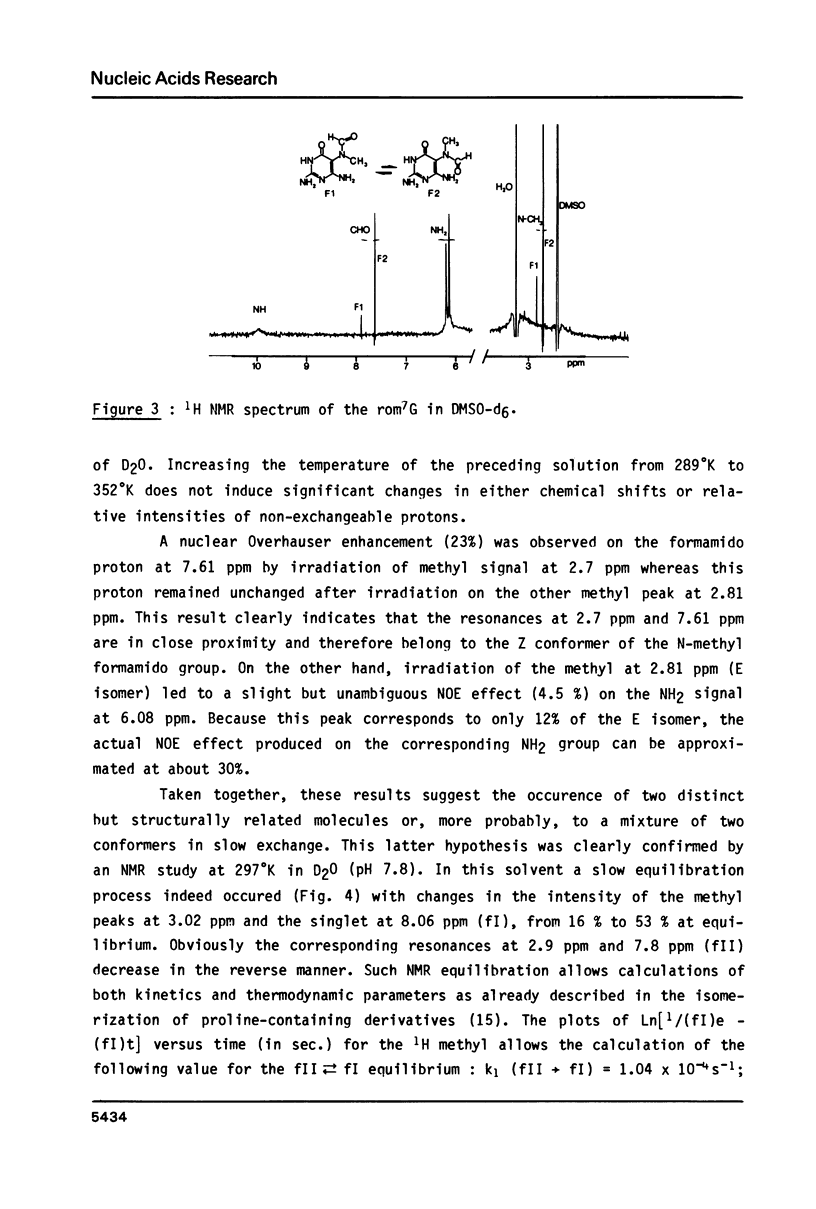
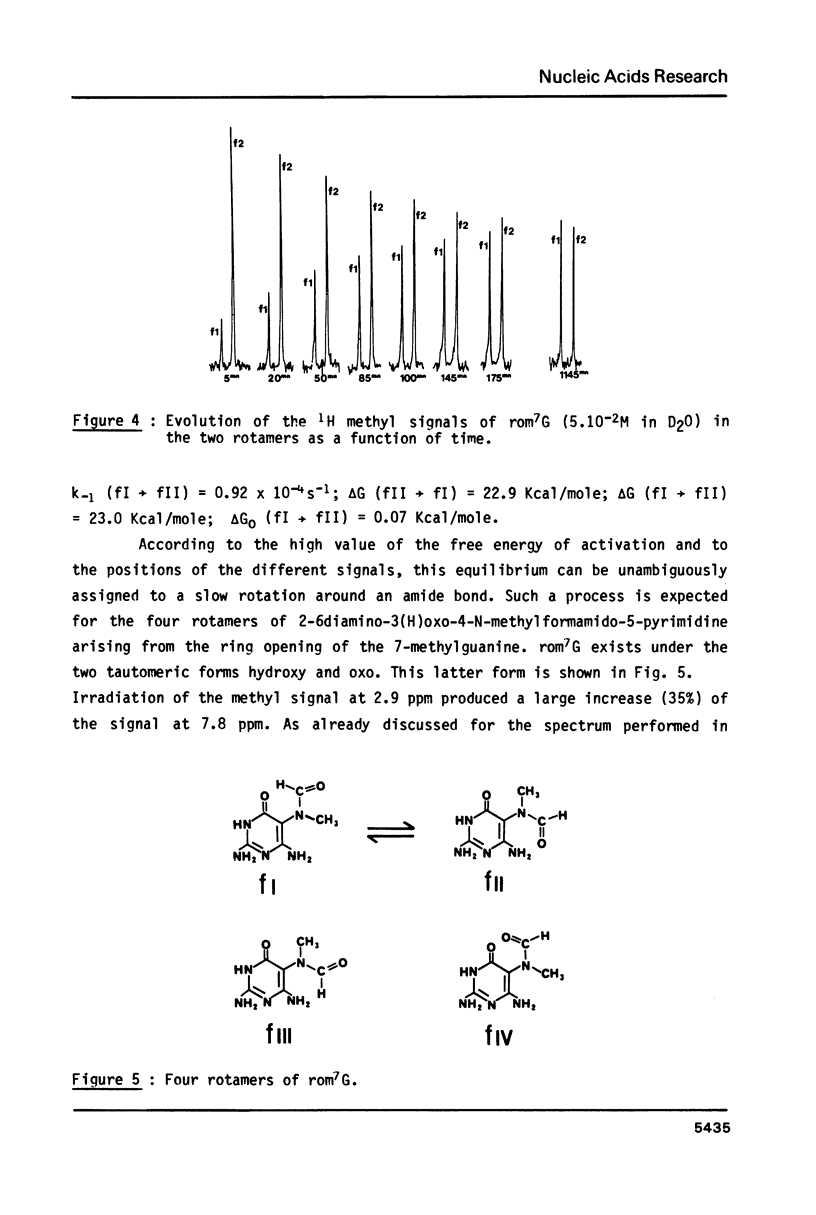
High performance liquid chromatography analysis of imidazole open ring 7-methylguanine, 2-6 diamino-4-hydroxy-5N-methyl-formamidopyrimidine (rom7G), showed two well-separated peaks (fI and fII) of the same magnitude. Rechromatography of each isolated component indicated that they are slowly interconverted to give a 1:1 mixture. NMR analysis demonstrated that the two species observed on reversed phase HPLC are rotational isomers. Thermodynamic measurements strongly suggested that the equilibrium can be assigned to rotation around the N-methyl formamido bond. The two species, fI and fII, separated by HPLC were identified as rotamers E and Z, respectively. The structures of fI and fII were also determined. A polynucleotide containing rom7G was obtained by alkaline treatment of poly (dGC) containing 7-methylguanine. In order to study its structure within the polynucleotide, rom7G was enzymatically excized by E.coli rom7G-DNA glycosylase. The analysis of the products released by the enzyme showed a 1:4 mixture of the two rotamers favoring the Z form (fII).
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
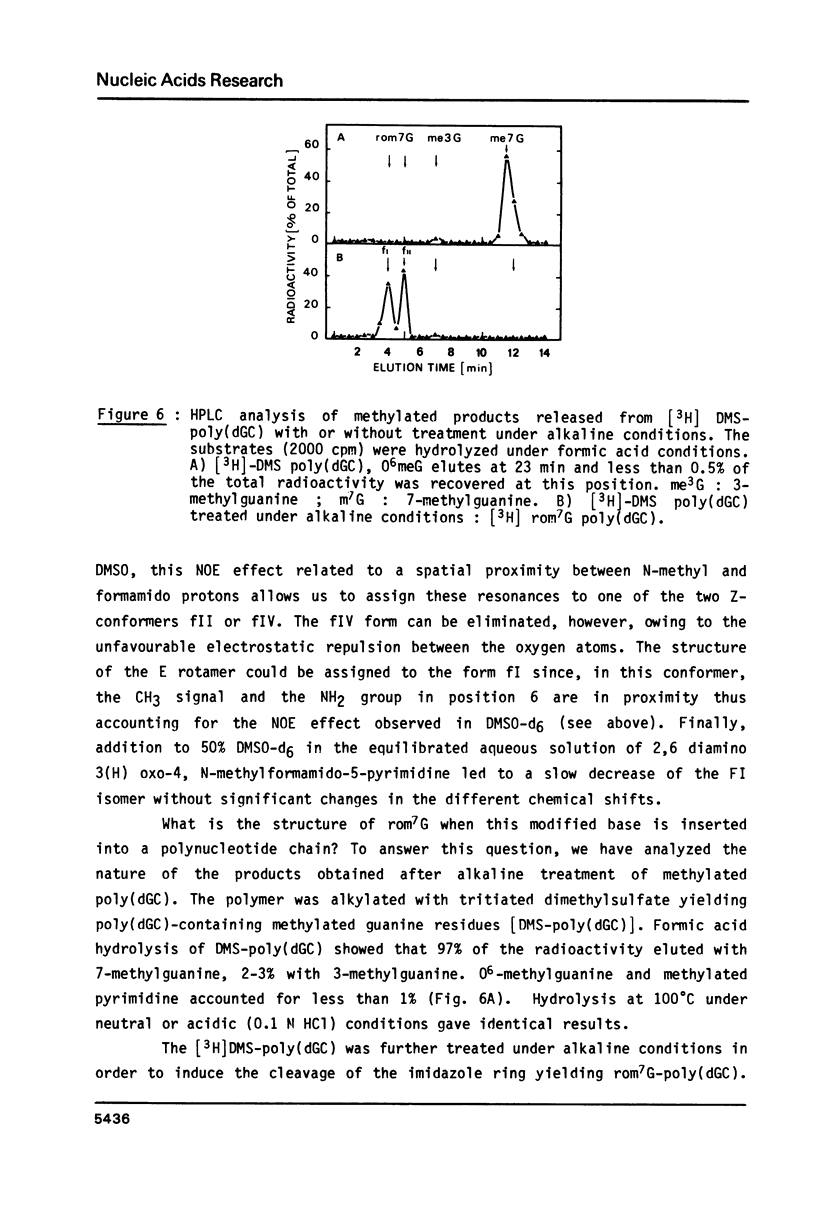
- Beranek D. T., Weis C. C., Evans F. E., Chetsanga C. J., Kadlubar F. F. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem Biophys Res Commun. 1983 Jan 27;110(2):625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
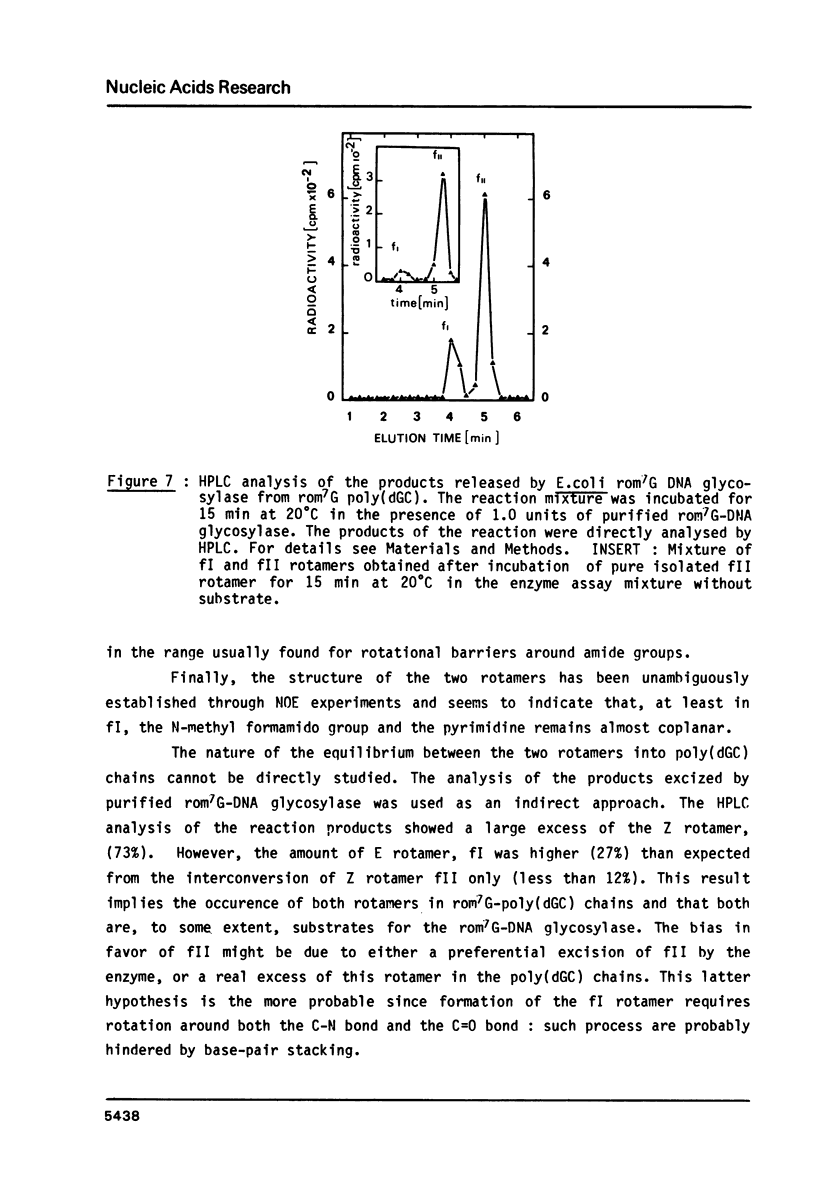
- Boiteux S., Laval J. Imidazole open ring 7-methylguanine: an inhibitor of DNA synthesis. Biochem Biophys Res Commun. 1983 Jan 27;110(2):552–558. doi: 10.1016/0006-291x(83)91185-3. [DOI] [PubMed] [Google Scholar]
- Chetsanga C. J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979 Aug 10;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetsanga C. J., Lozon M., Makaroff C., Savage L. Purification and characterization of Escherichia coli formamidopyrimidine-DNA glycosylase that excises damaged 7-methylguanine from deoxyribonucleic acid. Biochemistry. 1981 Sep 1;20(18):5201–5207. doi: 10.1021/bi00521a016. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Laval J., Pierre J., Laval F. Release of 7-methylguanine residues from alkylated DNA by extracts of Micrococcus luteus and Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):852–855. doi: 10.1073/pnas.78.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval J. Two enzymes are required from strand incision in repair of alkylated DNA. Nature. 1977 Oct 27;269(5631):829–832. doi: 10.1038/269829a0. [DOI] [PubMed] [Google Scholar]
- Lawley P. D., Orr D. J. Specific excision of methylation products from DNA of Escherichia coli treated with N-methyl-N'-nitro-N-nitrosoguanidine. Chem Biol Interact. 1970 Aug;2(2):154–157. doi: 10.1016/0009-2797(70)90047-5. [DOI] [PubMed] [Google Scholar]
- Lawley P. D., Shah S. A. Methylation of ribonucleic acid by the carcinogens dimethyl sulphate, N-methyl-N-nitrosourea and N-methyl-N'-nitro-N-nitrosoguanidine. Comparisons of chemical analyses at the nucleoside and base levels. Biochem J. 1972 Jun;128(1):117–132. doi: 10.1042/bj1280117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Loveless A. Possible relevance of O-6 alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature. 1969 Jul 12;223(5202):206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- Margison G. P., Pegg A. E. Enzymatic release of 7-methylguanine from methylated DNA by rodent liver extracts. Proc Natl Acad Sci U S A. 1981 Feb;78(2):861–865. doi: 10.1073/pnas.78.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roques B. P., Garbay-Jaureguiberry C., Combrisson S., Oberlin R. Determination of the rates and barriers to conformational isomerization in the dipeptide L-Pro-L-4Hyp by direct 13C NMR thermal equilibration. Biopolymers. 1977 Apr;16(4):937–944. doi: 10.1002/bip.1977.360160415. [DOI] [PubMed] [Google Scholar]
- Singer B. The chemical effects of nucleic acid alkylation and their relation to mutagenesis and carcinogenesis. Prog Nucleic Acid Res Mol Biol. 1975;15(0):219–284. [PubMed] [Google Scholar]



