Abstract
Correlative microscopy is a sophisticated approach that combines the capabilities of typically separate, but powerful microscopy platforms: often including, but not limited, to conventional light, confocal and super-resolution microscopy, atomic force microscopy, transmission and scanning electron microscopy, magnetic resonance imaging and micro/nanoCT (computed tomography). When targeting rare or specific events within large populations or tissues, correlative microscopy is increasingly being recognized as the method of choice. Furthermore, this multi-modal assimilation of technologies provides complementary and often unique information, such as internal and external spatial, structural, biochemical and biophysical details from the same targeted sample. The development of a continuous stream of cutting-edge applications, probes, preparation methodologies, hardware and software developments will enable realization of the full potential of correlative microscopy.
INTRODUCTION
In this era of genetic manipulation, the widespread adoption of fluorescent protein chimeras has a prominent role in enhancing our understanding of cellular and molecular phenomena. As new molecules of interest are discovered and known molecules reassessed, fluorescent protein fusions have become a critical tool to analyze their functions from the whole organism to sub-cellular machinery to single molecules. However, it is clear that no single imaging technology can reveal all the details of a biological sample. This readily can be appreciated when one considers while transmission electron microscopy has the best resolution, live cell imaging is not possible and has restricted field-of-view and severe limitations in three-dimensionality. Conversely, photon-based optical microscopy, enables live-cell, large field-of-view and is far more amenable to extended 3D imaging, but spatial resolution often prevents definitive localization. It is thus a natural extension of the various imaging tools, each with inherent capabilities and limitations, to expand the overall information content by correlative microscopy.
The concept of correlative microscopy is not new. Virtually all classically-trained electron microscopists screen semi-thick resin sections from a microtome and by light microscopy (LM) to locate a desired region of the sample for ultrathin sections and transmission electron microscopy (TEM). Although the same exact region is not viewed by both LM and TEM, this simple approach exploits four fundamental features of correlative microscopy: 1) identifying a specific target (structure, region, cell type, tissue or biological phenomenon), 2) putting the target into context of a much larger region, 3) unique information content from different probes, stains or contrasting reagents, and 4) resolution improvements by EM that provide ultrastructural detail. Any one of these features justifies the appropriate use of correlative microscopy. Indeed, this basic approach has naturally evolved into to the most commonly applied correlative combination referred to as Correlative Light and Electron Microscopy (CLEM).
While the merits of basic CLEM are well documented, there are numerous other correlative combinations that can broaden the range of data content from individual targets. However, more sophisticated correlative methods require the precise relocation of the same target using very different sample preparation protocols, support structures, contrasting mechanisms and physical sample size. Here, we describe major advances the key areas of sample preparation, correlative probes [1,2], automation and image alignment that will have the biggest impact on driving the development and mainstream application of correlative microscopy. Detailed protocols are beyond the scope of this opinion, however references to excellent reviews [3] and methods are provided to guide those interested in exploring recent advanced adaptations of correlative microscopy.
Sample Preparation
Careful consideration of fixation is paramount to minimize artifacts that lead to data misinterpretation. While conventional fixation has its place (and in some cases is the only way to preserve samples) physical fixation using cryogenic methods is the gold-standard for ultrastructural preservation. This is attributed to the rapid cessation of cellular activity within milliseconds as opposed to minutes with conventional chemical fixation. Cryo-fixation technologies such as high-pressure freezing (HPF) are invaluable for following highly dynamic events such as vesicular trafficking [4•] or nuclear division in C. elegans [5] and Drosophila embryos [6]. Placement of the Leica EM PACT2 Rapid Transfer System HPF adjacent to a light microscope, allows extended live-cell time-lapse imaging of a dynamic cellular phenomenon and then rapid freezing during an event of interest in less than 5 seconds. While not instantaneous, many biological phenomena work well in this kinetic time-frame. Speigelhalter et al. [7] used this elegant approach with confocal microscopy to follow effects of hyper-osmotic treatment on COS-1 cells transfected with several fluorescent protein fusions, subsequently high-pressure frozen, resin embedded and immunogold labeled for serial section EM tomography (Fig 1). Current HFP designs require sample transfer from a separate microscope to a freezing apparatus. It is tempting to consider the possibility of a high-end optical microscope as an integrated as part of a high-pressure freezer. Without the need for transferring the sample, freezing could occur in less than one second. If digital images are acquired, pre-defined pattern or intensity based recognition of an event could trigger freezing without user intervention.
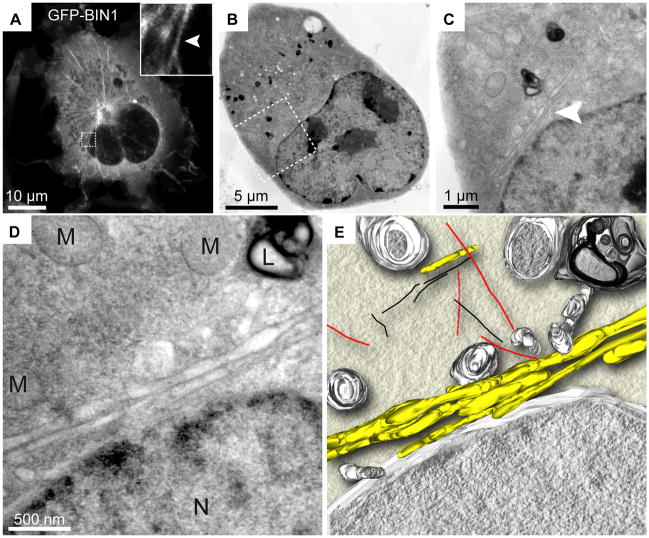
Figure 1. Correlative light microscopy with TEM tomography.
GFP-BIN1 expression in COS-1 cells. Bridging of specific GFP-BIN1 localization from confocal microscopy to high resolution TEM is illustrated by the correlation of a fluorescent image (A) with sequentially higher resolution EM micrographs (B–D). GFP-BIN1 localizes to tubules (arrow heads) and TEM places the localization into the subcellular context of mitochondria (M), lysosome (L) and nucleus (N) (D). An EM tomogram generated from 139 tilted images provides additional 3D information of the relation of tubules (yellow) to mitochondria, nuclear envelope, endosomal vesicles and lysosomes (grey), microtubules (red) and actin (black). Figure adapted from Speigelhalter et al. [8].
Correlative Probes
A current limitation of correlative microscopy is lack of compatibility between probes used in light microscopy and the standard preparation protocols that typically employed on another imaging platform. Many organic dyes and FPs quench when moved from an aqueous to a solvent or dehydrated environment. For FPs, addition of 5% water in solvent or resin steps enables preservation en bloc [8]. Fortunately, even if fluorescence cannot be maintained en bloc, we can garner much information by overlaying images from two different microscopes [9].
The most precise localization is often required for gleaning insights to proteins with unknown function. High-resolution correlative approaches that utilize nanoparticles such as gold [7], FluoroNanogold [10], quantum dots [1,11,12] and DAB photooxidation [13] have proven valuable for permitting contrast in optical as well as electron microscopy. See detailed reviews on correlative probes [1,14•,15]. Ultimately, immune-based probe size considerations, functionalization strategies and requirements for antibodies or other affinity chemistries, require permeablization, whether in the form of extracting detergents, solvents or sectioning methodologies. Furthermore, the precision of localization is reduced to ~15nm for indirect labeling, which is sufficient for localizing many molecules, but spatially, we frequently need better. Furthermore, the suitability for a given project can be impeded by low labeling density and steric hindrance.
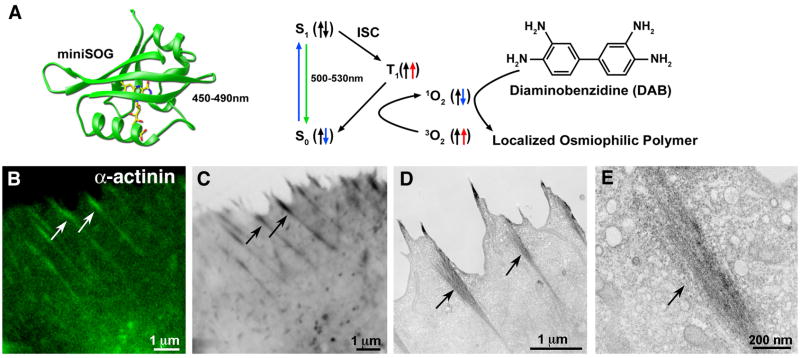
Photooxidation holds great promise to circumvent issues of penetration and resolution found with immune-based nanoparticles. With organic dyes or fluorescent protein fusions, the production of singlet oxygen while photobleaching in the presence of diaminobenzadine (DAB) results in the formation of highly localized and osmiophilic electron dense precipitate at the sight of the fluorophore [13]. Shu and colleagues ([16• •] recently reported the creation of a mini-superoxide generating (miniSOG) tag derived from a plant flavoprotein that fluoresces with blue light excitation (Fig 2A) and has been optimized to generate singlet oxygen more than two orders of magnitude better than GFP, thus dramatically improving the sensitivity of DAB for genetically encoded fusion proteins(Fig 2B–2E).
Figure 2. MiniSOG: a genetically encoded tag for correlative light and EM microscopy by photooxidation.
(A) Structural model of miniSOG (left) and a diagram showing how blue-light excitation leads to photooxidation of diaminobenzidine (DAB) (right). (B–E) minSOG fused to α-actinin and expressed in HeLa cells. Confocal fluorescence (B) correlates with dark precipitation visible by transmitted light (C) and electron micrographs (D). Higher resolution EM shows discrete localization to a putative focal adhesion site (E). Arrows indicate correlated structures. Figure adapted from Shu and colleagues ([16].
Hybrid Versus Automated Approach
The single most significant bottleneck in correlative microscopy is the slow and laborious process of sample relocation between imaging platforms, hence the appeal for hybrid systems that do not rely on separate microscopes. The most widely used hybrid system is an AFM microscope integrated with an inverted light microscope. In this case, the light microscope serves as an excellent tool for identifying cells or other structures using transmitted light and fluorescence microscopy combined with a host of biophysical measurements derived from AFM [17][19] (Fig 3A). High-speed AFMs [20], molecule dynamics with TIRF [21] and high-resolution force volume mapping to reveal surface ligand/receptor interactions [18] [22], are especially promising for live cell work. Similarly, a number of other novel hybrid integrations with exciting potential have been reported including LM/SEM [23], AFM/SEM [24] , AFM/microtome [25] and LM/TEM [26]. Imaging samples at cryogenic temperatures for both fluorescence and TEM tomography is now also technically possible [27,28]. In virtually all hybrid adaptations, compromises must be made due to the unique physics and space constraints of each modality (i.e. resolution, sample size and/or sensitivity) and their utility will be application dependent.
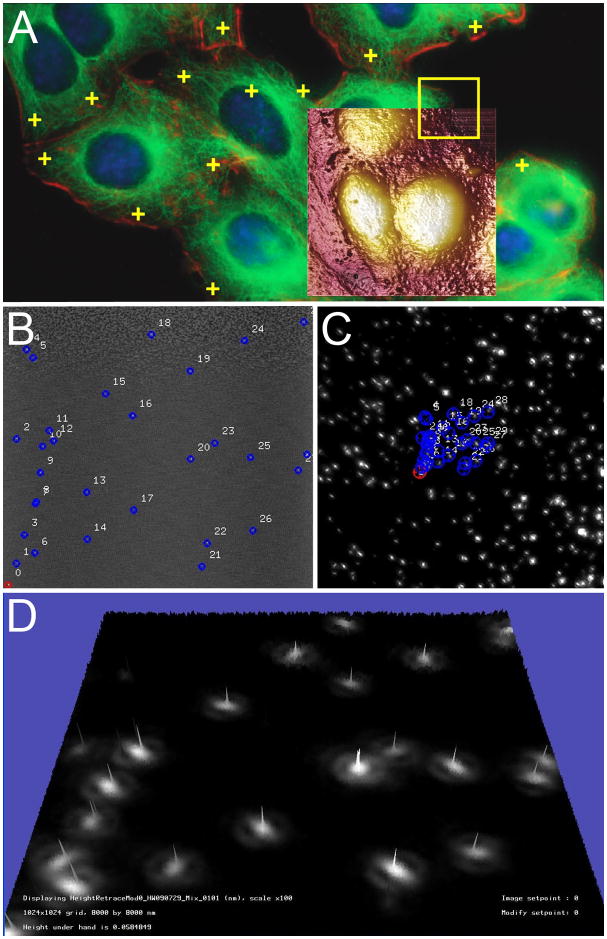
Figure 3. Correlative AFM and fluorescence microcopy automated image alignment.
Correlative microscopy using the Bruker BioScope Catalyst AFM/LM hybrid system with overlay alignment made in real time, pixel by pixel (A). User-defined locations can be targeted for single point force curves for measuring unbinding events (yellow crosses), AFM height image (brown ,35 × 35 μm overlay), or force volume mapping (yellow frame)and with the MIRO canvas fluorescence image can be directly chosen and navigated to using the software. Image provided courtesy of Alexandre Berquand (Bruker), Andreas Holloschi and Petra Kioschis (University of Applied Science). Marker (blue) previously found in an automatic finding stage (B). During registration, these AFM markers are mapped to the corresponding locations in the fluorescence image (C). The two images can then be automatically overlaid with high accuracy. The topography was generated from the AFM images and the colormap originates from the fluorescence image; the numerous bright “hills” indicate that the automatic alignment was successful (D). Panels B–D were provided courtesy of Serdar Cakici (University of North Carolina, Chapel Hill).
Where hybrid systems are unfeasible or unavailable, most laboratories currently use a combination of masks, finder grids or mapping and highly skilled operators to facilitate the relocation of samples [9,29]. Relocating a sample on different imaging platforms is non-trivial, often requiring a precision down to a few tens of nanometers. Under these circumstances, automating relocation can substantially increase throughput and reduce technical proficiency required for correlative experiments. However, the specimen preparation requirements for biological microscopy are complex, especially when live-cell imaging is employed, making automation challenging. For example, visualization of live-cells requires working in aqueous solutions to preserve viability of the organism, maintaining fluorescence with many probes, and taking full advantage of the best optical conditions for 3D imaging. In principle, automation (or manual relocation for that matter) allows samples to be processed in an optimal way for each specific platform. Recently, automated solutions have been introduced for correlative microscopy utilizing a coordinate based relocation system between calibrated conventional LM and SEM motorized stages [30]. This simple, yet powerful solution allows quick identification and image documentation in all LM modes (i.e, fluorescence, phase contrast, DIC) and rapid location via SEM, automatically matching field-of-view and a basic overlay routine of corresponding images, shaving hours off of the manual process. Throughput is dramatically increased bringing the efficiency and productivity of correlative microscopy to a level where statistics can be employed.
Image Alignment
Image alignment is a significant challenge for correlative microscopy and this is compounded by the fact that not only can non-linear distortions occur due to inherent differences between various scanning and imaging systems, but also numerous processing steps leave them prone to physical distortions (i.e, shrinkage, sectioning artifacts). Furthermore, because data is often near or beyond the boundaries of what is measurable, each type of microscopy introduces artifacts: AFMs dilate the specimen shape by the shape of their probe, LM convolves fluorescence by point-spread-function, and SEMs produces edge effects. Simple alignment measures are precluded because intensity patterns are often not easily compared and resolution differences between images are common. All of these issues pose challenges with finding and accurately aligning corresponding regions in the images. Possible solutions include: 1) image similarity measures or markers not based on direct intensity comparisons, 2) surrogate images to perform landmark-based alignment manually [31,32] or automatically [33•] (Fig 3B–D), 4) a model-based approach, or 5) a combination of these methods. Alignment can be greatly simplified by adding fiducial markers (gold or beads) which can be easily identified in the images to be aligned [34• •] [35]. Fiducial markers can provide candidate feature points and correspondence can be established through random sampling [33]. Alignment accuracy is not always limited by the lower-resolution image, since model-based fitting approaches such as FIONA [36] can obtain sub-pixel accurate estimations of features. Because model-based optimizations typically require many iterations of forward simulation, the associated microscope simulators must be fast. GPUs have been employed to accelerate AFM simulations for the combined fluorescence/AFM simulator and [37] enabled even more rapid simulation of fluorescence images for the same tool
The Future of Correlative Microscopy
The most significant near term impact on correlative microscopy will come from the rapid development and availability of suitable commercial solutions putting this approach at the fingertips of many more scientists. Furthermore, it must be stressed that the scope of correlative microscopy must be considerably broadened to include any combinations of the numerous more advanced tools that exist such as; Förster Resonance Energy Transfer (FRET) [38,39], Fluorescence Correlation Spectroscopy(FCS) [40•], Raster-scan Image Correlation Spectroscopy (RICS)[40•], Total Internal Reflection Fluorescence [40•], super-resolution [41,42], single molecule fluorescence [43,44] and various other optical techniques [45• •]. The prevalence of correlative approaches in super-resolution (SR) microscopy [46,47] suggests the importance of the technique in bridging the LM-to-EM resolution gap and the need to identify and confirm the labeling of targeted structures and validate SR approaches. This supports the notion that any single microscope technology is unable to provide a comprehensive understanding of the inner workings of a cell.
SEM will also make a huge contribution to correlative efforts with TEM-like images using backscattered electrons and large area mapping capabilities especially suitable for tissues. SEM in combination with serial block face imaging [16,48], serial sectioning [49] and array tomography [8], or focused ion beam [50,51] enables 3D imaging of much larger areas than possible with TEM. When one considers all the possible combinations, an increase in use of more than just two modalities for single targeted structures for ex ample AFM/LM/FIB (Fig 4) is expected. Furthermore whole animal approaches such as magnetic resonance imaging [52], x-ray based computed tomography [53] and bioluminescence [54] will play an important role as we target pathologies down to an individual cell [54] for subsequent high-resolution analysis. To reach its full potential, efforts must include precise alignment (potentially single molecule), segmentation and visualization tools to create an integrated 3D cellular/organism map which ultimately allows coordinate or feature based extraction of data (Fig 4). This could be further extended with automatic macromolecular molecular identification and include embedding 3D molecular structures into high-resolution EM tomograms of cells [55].
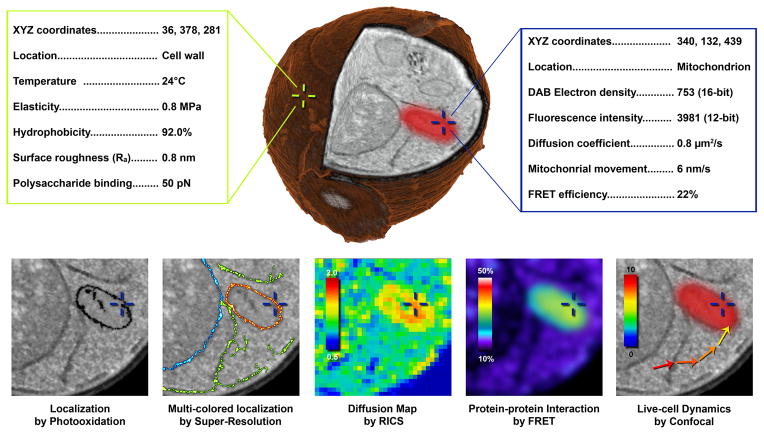
Figure 4. Hypothetical correlative approach to create an interactive and integrated 3D cellular map with biophysical, structural and chemical measurements acquired from a single yeast cell.
Future prospects will likely include correlation of high resolution surface properties collected from different AFM modes (green text box) with characteristics obtained my multiple, advanced light microscopy or spectroscopy techniques (blue text box). This sample could then be rapidly frozen and processed to create high resolution 3D reconstructions from serial block-face imaging with focused ion beam SEM (center yeast cell). We envision this data set may be navigated in an interactive interface (similar to Google Earth), where specific points (cross hairs) can be chosen and a list of characteristics would be displayed (hypothetical values provided). The bottom row illustrates possibilities with such a muti-modal correlative dataset consisting of high resolution localization by DAB photooxidation, 3-color localization by STORM super resolution, diffusion coefficient map by raster image correlation spectroscopy, protein-protein interaction by FRET and live cell dynamics by confocal microscopy.
Conclusions
Recent correlative efforts have placed emphasis on new probes, compatible sample preparation methods, automation, standardized cell and tissue support and transfer devices, fiducial markers and data alignment/integration. The clear need for monitoring multi-parameters for individual events maximizing information content cannot be overstated. However, in order to meet the demands of rigorous scientific protocol, the power of statistics must be a driving factor in this endeavor. Thus, a concerted effort must be placed on automation of the process whenever possible to dramatically increase throughput, ensuring both statistical significance of observations and allowing correlative microscopy to be more broadly adopted. By its very nature, life science-based correlative microscopy is a collaborative approach often minimally requiring expertise in genetic manipulation, cell-culture, immuno-labeling, cryo-preparation, and all other technical skills that accompany light and electron microscopy, not to mention the numerous other imaging modalities described herein. However, many new tools are now available that facilitate a correlative project via novel probe techniques and dramatic reduction in the time from start to finish. Correlative microscopy has arrived and we strongly encourage researchers to take full advantage this directed, multi-faceted and rapidly evolving approach when a single imaging technology is just not enough.
Highlights.
Correlative microscopy combines multiple imaging platforms for targeted analysis.
Sample preparation is critical for successful correlation.
Probes which contrast at light and electron level are discussed.
Improved software and hardware tools for automation facilitate throughput.
Value of correlative microscopy enhanced by broader application and data integration.
Acknowledgments
The work presented here was partly supported by NIH 5-P41-RR02170.
Footnotes
Disclosure Statement
Russell Taylor is the president of and a stockholder in nanoManipulator, Incorporated, which is a company that sells a commercial version of the software used to develop the AFM and fluorescence overlay technology. Kirk Czymmek occasionally serves as a paid consultant for Carl Zeiss to train new users and sales and applications representatives on the basics of confocal or correlative microscopy. He also provides Zeiss feedback for enhancing existing and future correlative products as it relates to biological correlative microscopy and has been invited by Zeiss to speak at national meetings on correlative microscopy but receives no personal compensation for either of these activities - only reimbursement for travel expenses.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jeffrey Caplan, Email: jcaplan@udel.edu.
Marc Niethammer, Email: marc.niethammer@gmail.com.
Russell M. Taylor, II, Email: taylorr@cs.unc.edu.
References
- 1.Giepmans BN. Bridging fluorescence microscopy and electron microscopy. Histochemistry and cell biology. 2008;130:211–217. doi: 10.1007/s00418-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown E, Verkade P. The use of markers for correlative light electron microscopy. Protoplasma. 2010;244:91–97. doi: 10.1007/s00709-010-0165-1. [DOI] [PubMed] [Google Scholar]
- 3.Mironov AA, Beznoussenko GV. Correlative microscopy: a potent tool for the study of rare or unique cellular and tissue events. Journal of microscopy. 2009;235:308–321. doi: 10.1111/j.1365-2818.2009.03222.x. [DOI] [PubMed] [Google Scholar]
- 4*.Brown E, Mantell J, Carter D, Tilly G, Verkade P. Studying intracellular transport using high-pressure freezing and Correlative Light Electron Microscopy. Seminars in cell & developmental biology. 2009;20:910–919. doi: 10.1016/j.semcdb.2009.07.006. With HeLa cells and combining live-cell time-lapse microscopy and high-pressure freezing to follow endocytosis, the authors showed merits of STEM over TEM for visualization of individual quantun dot and gold conjugated probes. [DOI] [PubMed] [Google Scholar]
- 5.Muller-Reichert T, Mantler J, Srayko M, O'Toole E. Electron microscopy of the early Caenorhabditis elegans embryo. Journal of microscopy. 2008;230:297–307. doi: 10.1111/j.1365-2818.2008.01985.x. [DOI] [PubMed] [Google Scholar]
- 6.McDonald KL. A review of high-pressure freezing preparation techniques for correlative light and electron microscopy of the same cells and tissues. Journal of microscopy. 2009;235:273–281. doi: 10.1111/j.1365-2818.2009.03218.x. [DOI] [PubMed] [Google Scholar]
- 7.Spiegelhalter C, Tosch V, Hentsch D, Koch M, Kessler P, Schwab Y, Laporte J. From dynamic live cell imaging to 3D ultrastructure: novel integrated methods for high pressure freezing and correlative light-electron microscopy. PloS one. 2010;5:e9014. doi: 10.1371/journal.pone.0009014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modla S, Mendonca J, Czymmek KJ, Akins RE. Identification of neuromuscular junctions by correlative confocal and transmission electron microscopy. Journal of neuroscience methods. 2010;191:158–165. doi: 10.1016/j.jneumeth.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheutin T, Sauvage C, Tchelidze P, O'Donohue MF, Kaplan H, Beorchia A, Ploton D. Visualizing macromolecules with fluoronanogold: from photon microscopy to electron tomography. Methods in cell biology. 2007;79:559–574. doi: 10.1016/S0091-679X(06)79022-7. [DOI] [PubMed] [Google Scholar]
- 11.Dukes MJ, Peckys DB, de Jonge N. Correlative fluorescence microscopy and scanning transmission electron microscopy of quantum-dot-labeled proteins in whole cells in liquid. ACS nano. 2010;4:4110–4116. doi: 10.1021/nn1010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deerinck TJ, Giepmans BN, Smarr BL, Martone ME, Ellisman MH. Light and electron microscopic localization of multiple proteins using quantum dots. Methods in molecular biology. 2007;374:43–53. doi: 10.1385/1-59745-369-2:43. [DOI] [PubMed] [Google Scholar]
- 13.Meisslitzer-Ruppitsch C, Rohrl C, Neumuller J, Pavelka M, Ellinger A. Photooxidation technology for correlated light and electron microscopy. Journal of microscopy. 2009;235:322–335. doi: 10.1111/j.1365-2818.2009.03220.x. [DOI] [PubMed] [Google Scholar]
- 14*.Weston AE, Armer HE, Collinson LM. Towards native-state imaging in biological context in the electron microscope. Journal of chemical biology. 2009;3:101–112. doi: 10.1007/s12154-009-0033-7. Authors provide helpful technical insights on the methods for succesful DAB photoxidation with various probes for correlative experiments. Authors also provide useful review of recent application of DAB for flourescent probes including GFP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortese K, Diaspro A, Tacchetti C. Advanced correlative light/electron microscopy: current methods and new developments using Tokuyasu cryosections. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2009;57:1103–1112. doi: 10.1369/jhc.2009.954214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS biology. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. This group demonstrated excellent DAB conversion results with a novel fluorescent mutagenized flavoprotein called miniSOG that when expressed as a chimera with a number of organelles and proteins in HeLa cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EH. Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. Journal of cell science. 2009;122:1665–1679. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- 18.Berquand A, Roduit C, Kasas S, Holloschi A, Ponce L, Hafner M. Atomic Force Microscopy Imaging of Living Cells. Microscopy Today. 2010;18:8–14. [Google Scholar]
- 19.Labernadie A, Thibault C, Vieu C, Maridonneau-Parini I, Charriere GM. Dynamics of podosome stiffness revealed by atomic force microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21016–21021. doi: 10.1073/pnas.1007835107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ando T, Uchihashi T, Kodera N, Yamamoto D, Miyagi A, Taniguchi M, Yamashita H. High-speed AFM and nano-visualization of biomolecular processes. Pflugers Archiv : European journal of physiology. 2008;456:211–225. doi: 10.1007/s00424-007-0406-0. [DOI] [PubMed] [Google Scholar]
- 21.Brown AE, Hategan A, Safer D, Goldman YE, Discher DE. Cross-correlated TIRF/AFM reveals asymmetric distribution of force-generating heads along self-assembled, “synthetic” myosin filaments. Biophysical journal. 2009;96:1952–1960. doi: 10.1016/j.bpj.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller DJ, Helenius J, Alsteens D, Dufrene YF. Force probing surfaces of living cells to molecular resolution. Nature chemical biology. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 23.Kanemaru T, Hirata K, Takasu S, Isobe S, Mizuki K, Mataka S, Nakamura K. A fluorescence scanning electron microscope. Ultramicroscopy. 2009;109:344–349. doi: 10.1016/j.ultramic.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Williams PA, Papadakis SJ, Flavo MR, Patel AM, Sinclair M, Seeger A, Hesler A, Taylor RM, Washburn S, Superfine R. Controlled Placement of an Individual Carbon Nanotube onto a Microelectromechanical Structure. Applied Physics Letters. 2002;80:2574–2576. [Google Scholar]
- 25.Efimov AE, Tonevitsky AG, Dittrich M, Matsko NB. Atomic force microscope (AFM) combined with the ultramicrotome: a novel device for the serial section tomography and AFM/TEM complementary structural analysis of biological and polymer samples. Journal of microscopy. 2007;226:207–217. doi: 10.1111/j.1365-2818.2007.01773.x. [DOI] [PubMed] [Google Scholar]
- 26.Agronskaia AV, Valentijn JA, van Driel LF, Schneijdenberg CT, Humbel BM, van Bergen en Henegouwen PM, Verkleij AJ, Koster AJ, Gerritsen HC. Integrated fluorescence and transmission electron microscopy. Journal of structural biology. 2008;164:183–189. doi: 10.1016/j.jsb.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Plitzko JM, Rigort A, Leis A. Correlative cryo-light microscopy and cryo-electron tomography: from cellular territories to molecular landscapes. Current opinion in biotechnology. 2009;20:83–89. doi: 10.1016/j.copbio.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Steven AC, Baumeister W. The future is hybrid. Journal of structural biology. 2008;163:186–195. doi: 10.1016/j.jsb.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Jahn KA, Barton DA, Su Y, Riches J, Kable EP, Soon LL, Braet F. Correlative fluorescence and transmission electron microscopy: an elegant tool to study the actin cytoskeleton of whole-mount (breast) cancer cells. Journal of microscopy. 2009;235:282–292. doi: 10.1111/j.1365-2818.2009.03223.x. [DOI] [PubMed] [Google Scholar]
- 30.Wepf R. Correlative Microscopy. GIT Imaging and Microscopy. 2010;12:20–22. [Google Scholar]
- 31.Kondra S, Laishram J, Ban J, Migliorini E, Di Foggia V, Lazzarino M, Torre V, Ruaro ME. Integration of confocal and atomic force microscopy images. Journal of neuroscience methods. 2009;177:94–107. doi: 10.1016/j.jneumeth.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 32.Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. The Journal of cell biology. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Preibisch S, Saalfeld S, Schindelin J, Tomancak P. Software for bead-based registration of selective plane illumination microscopy data. Nature methods. 2010;7:418–419. doi: 10.1038/nmeth0610-418. Using rotation and translation invariant descriptors obtained from sets of 4 beads, the authors obtain an automatic alignment procedure for selective plane illumination data. This paper demonstrates a possible way to obtain robust automatic alignment between images using beads with utility for any kind of alignment (which is close to a rigid transformation) based on fiducial markers. [DOI] [PubMed] [Google Scholar]
- 34**.Watanabe S, Punge A, Hollopeter G, Willig KI, Hobson RJ, Davis MW, Hell SW, Jorgensen EM. Protein localization in electron micrographs using fluorescence nanoscopy. Nature methods. 2011;8:80–84. doi: 10.1038/nmeth.1537. The authors generate a useful comparison/optimization of various resins in an effort to preserve fluorescence from fluorescence proteins en bloc for subsequent super-resolution, confocal and TIRF micorscopy for correlative EM. Using 100 nm gold particles as fiducial markers, the authors align STED and PALM fluorescence images to electron tomography images to localize proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fronczek DN, Quammen C, Wang H, Kisker C, Superfine R, Taylor R, Erie DA, Tessmer I. High accuracy FIONA-AFM hybrid imaging. Ultramicroscopy. 2011;111:350–355. doi: 10.1016/j.ultramic.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1. 5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 37.Quammen CW, Richardson AC, Haase J, Harrison BD, Taylor RM, Bloom KS. FluoroSim: A Visual Problem-Solving Environment for Fluorescence Microscopy. Eurographics Workshop on Visual Computing for Biomedicine. 2008;2008:151–158. doi: 10.2312/VCBM/VCBM08/151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jares-Erijman EA, Jovin TM. Imaging molecular interactions in living cells by FRET microscopy. Current opinion in chemical biology. 2006;10:409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Current opinion in structural biology. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Digman MA, Brown CM, Horwitz AR, Mantulin WW, Gratton E. Paxillin dynamics measured during adhesion assembly and disassembly by correlation spectroscopy. Biophysical journal. 2008;94:2819–2831. doi: 10.1529/biophysj.107.104984. While not a correlative paper, this manuscript explores in the utility of various single molecule biophysical imaging techniques to determine molecular dynamics within living cells and ultimately compatible with more advanced implementations of correlative microscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patterson GH. Fluorescence microscopy below the diffraction limit. Seminars in cell & developmental biology. 2009;20:886–893. doi: 10.1016/j.semcdb.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toomre D, Bewersdorf J. A new wave of cellular imaging. Annual review of cell and developmental biology. 2010;26:285–314. doi: 10.1146/annurev-cellbio-100109-104048. [DOI] [PubMed] [Google Scholar]
- 43.Agrawal A, Deo R, Wang GD, Wang MD, Nie S. Nanometer-scale mapping and single-molecule detection with color-coded nanoparticle probes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3298–3303. doi: 10.1073/pnas.0712351105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinterdorfer P, Van Oijen A. Handbook of Single-Molecule Biophysics. New York, New York: Springer Science + Business Media; 2009. [Google Scholar]
- 45**.Ji N, Shroff H, Zhong H, Betzig E. Advances in the speed and resolution of light microscopy. Current opinion in neurobiology. 2008;18:605–616. doi: 10.1016/j.conb.2009.03.009. Thie review article provodes a perspective on optical microscopy advances with special emphasis on neurobiological applications. It provides a very useful comparison of super-resolution tachniques including their limitations and includes a discussion/example of super-resolution and correlative EM. [DOI] [PubMed] [Google Scholar]
- 46.Perinetti G, Muller T, Spaar A, Polishchuk R, Luini A, Egner A. Correlation of 4Pi and electron microscopy to study transport through single Golgi stacks in living cells with super resolution. Traffic. 2009;10:379–391. doi: 10.1111/j.1600-0854.2008.00875.x. [DOI] [PubMed] [Google Scholar]
- 47.Lippincott-Schwartz J, Manley S. Putting super-resolution fluorescence microscopy to work. Nature methods. 2009;6:21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS biology. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayworth KJ, Kasthuri N, Schalek R, Lichtman JW. Automating the Collection of Ultrathin Serial Sections for Large Volume TEM Reconstructions. Microscopy and Microanalysis. 2006;12:86–87. [Google Scholar]
- 50.Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merchan-Perez A, Rodriguez JR, Alonso-Nanclares L, Schertel A, Defelipe J. Counting Synapses Using FIB/SEM Microscopy: A True Revolution for Ultrastructural Volume Reconstruction. Frontiers in neuroanatomy. 2009;3:18. doi: 10.3389/neuro.05.018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cleary JO, Modat M, Norris FC, Price AN, Jayakody SA, Martinez-Barbera JP, Greene ND, Hawkes DJ, Ordidge RJ, Scambler PJ, et al. Magnetic resonance virtual histology for embryos: 3D atlases for automated high-throughput phenotyping. NeuroImage. 2011;54:769–778. doi: 10.1016/j.neuroimage.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 53.Metscher BD. MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238:632–640. doi: 10.1002/dvdy.21857. [DOI] [PubMed] [Google Scholar]
- 54.Kim JB, Urban K, Cochran E, Lee S, Ang A, Rice B, Bata A, Campbell K, Coffee R, Gorodinsky A, et al. Non-invasive detection of a small number of bioluminescent cancer cells in vivo. PloS one. 2010;5:e9364. doi: 10.1371/journal.pone.0009364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stahlberg H, Walz T. Molecular electron microscopy: state of the art and current challenges. ACS chemical biology. 2008;3:268–281. doi: 10.1021/cb800037d. [DOI] [PMC free article] [PubMed] [Google Scholar]