Abstract
Amongst bacterial cell envelopes, the Borrelia burgdorferi outer membrane (OM) is structurally unique in that the identities of many protein complexes remain unknown; however, their characterization is the first step towards our understanding of membrane protein interactions and potential functions. Here, we used two-dimensional blue native/SDS-PAGE/mass spectrometric analysis for a global characterization of protein-protein interactions as well as to identify protein complexes in OM vesicles isolated from multiple infectious sensu stricto isolates of B. burgdorferi. Although we uncovered the existence of at least 10 distinct OM complexes harboring several unique subunits, the complexome is dominated by the frequent occurrence of a limited diversity of membrane proteins, most notably P13, outer surface protein (Osp) A, -B, -C and -D and Lp6.6. The occurrence of these complexes and specificity of subunit interaction were further supported by independent two-dimensional immunoblotting and co-immunoprecipitation assays as well as by mutagenesis studies, where targeted depletion of a subunit member (P66) selectively abolished a specific complex. Although a comparable profile of the OM complexome was detected in two major infectious isolates, such as B31 and 297, certain complexes are likely to occur in an isolate-specific manner. Further assessment of protein complexes in multiple Osp-deficient isolates showed loss of several protein complexes but revealed the existence of additional complex/subunits that are undetectable in wild-type cells. Together, these observations uncovered borrelial antigens involved in membrane protein interactions. The study also suggests that the assembly process of OM complexes is specific and that the core or stabilizing subunits vary between complexes. Further characterization of these protein complexes including elucidation of their biological significance may shed new light on the mechanism of pathogen persistence and the development of preventative measures against the infection.
Keywords: Borrelia burgdorferi, Lyme disease, microbial pathogenesis, outer membrane, protein complex
Introduction
Lyme disease is a common tick-borne illness in the United States and an emerging public health threat in many parts of the world. Borrelia burgdorferi, the spirochetal agent of Lyme disease, is maintained in nature via a complex enzootic life cycle involving wild rodents and Ixodes ticks 1, 2. The spirochete alters its antigenic composition via intragenic recombination and regulation of gene expression when encountering new host or vector environments 3–6 (for details see a recent review 7). This assists the pathogen to navigate between a diverse array of host microenvironments and to establish a persistent infection 8. Although microarray studies have identified a large set of borrelial genes that are highly responsive to environmental cues 9–12, relatively limited information is available on the composition of or potential changes in the borrelial proteome 13–15, especially those contained in the outer membrane (OM). OM proteins are expected to play critical roles in B. burgdorferi persistence through the vector-host infection cycle. Therefore, characterization of OM complexes is important to our understanding of the intriguing biology of spirochetes and to the development of novel preventative and therapeutic measures against Lyme borreliosis.
In Gram-negative bacterial pathogens, OM proteins often contribute to various stages in the infection process including tissue adhesion, colonization, immune cell activation and evasion of the host immune system 16, 17. The B. burgdorferi OM undergoes constant antigenic alterations induced by the surrounding environment. In contrast to other Gram-negative bacteria, the B. burgdorferi OM features an absence of usual lipopolysaccharide elements. Instead, the borrelial OM contains numerous surface lipoproteins that do not have membrane-spanning topology and are anchored to the membrane via amino-terminal lipid motifs 18, 19. It is also likely that many soluble proteins tether to the membrane through non-covalent interactions with the constitutive lipids and proteins. Lastly, although B. burgdorferi OM retains much lower density of membrane-spanning proteins compared to that in other Gram-negative bacteria, it contains more proteins than that found in the OM of the related pathogenic spirochete Treponema pallidum 20, 21.
Generally, proteins often assemble into multi-protein complexes that carry out important biochemical processes 22 including specific roles in membrane biogenesis and function, such as energy generation, protein assembly, lipoprotein trafficking and small molecule transportation 23. These functions, in turn, contribute to the microbial pathogenesis 24. Two-dimensional (2D) blue native (BN)/PAGE technology has been widely applied for the isolation of protein complexes in native conditions and generation of global overviews of protein-protein interactions in biological membranes 23–27. For better understanding of spirochete biology, we sought to use the 2D-BN/SDS-PAGE technology to identify protein complexes in the OM of multiple pathogenic isolates of B. burgdorferi. We also explored potential alterations in the dynamics of protein complexes in isogenic mutants lacking a single or multiple subunit members. Overall, we believe that further characterization of protein complexes will shed new light on the molecular mechanisms of B. burgdorferi enzootic cycle persistence and aid development of novel ways to prevent the infection.
Experimental section
Bacterial strains
The following B. burgdorferi isolates were used in the study: a clonal and low-passage infectious B. burgdorferi B31 isolate A3 28, a non-infectious mutant B314 that lacks many linear endogenous plasmids 29, and an infectious wild-type isolate 297, clone BbAH130 30. All spirochetes were cultured in BSK-H medium supplemented with 6% rabbit serum at 33°C and grown until a density of 5×107 – 108 cells/ml.
Isolation of outer membrane vesicles
Isolation of the outer membrane (OM) vesicles of B. burgdorferi was performed as described 31. Briefly 5 × 1010 – 1011 cells were harvested by centrifugation and the pellets were washed twice with phosphate buffered saline pH 7.4 (PBS) supplemented with 0.1% bovine serum albumin (BSA). The cells were resuspended in ice-cold 25 mM citrate buffer (pH 3.2) containing 0.1% BSA and subsequently incubated on a rocker at room temperature for 2 hours. The OM vesicles were isolated from the protoplasmic cylinder by using sucrose density gradient centrifugation as detailed 32. The OM vesicles were monitored for purity by immunoblotting using antibodies against OspA and FlaB as described 33. Generation of polyclonal rabbit antiserum against isolated OM proteins was performed as described 32.
Generation of recombinant proteins and antisera
Recombinant proteins, BmpA, BB0028, OspA, -B, -C, -D, La7, FlaB and Lp6.6 were produced as described 32, 34–38. Briefly, the coding region of genes without respective signal peptides were amplified as separate DNA inserts using published primers as detailed 32, 34–38, whereas primers specific for bb0028 are indicated in Supplementary Table S6. DNA inserts were directionally cloned into EcoRI and XhoI sites of the expression vector pGEX-6P-1 (GE Healthcare). The expression, purification, and enzymatic cleavage of the glutathione-S-transferase (GST) fusion proteins were carried out as described previously 37. To generate polyclonal antisera, the proteins (without the GST tag) produced in Escherichia coli were emulsified in complete Freund’s adjuvant and injected into groups of 2 rabbits (100 μg/animal) or 5 mice (10 μg/animal). The animals were boosted twice at 3-week intervals with the same dose of antigen in incomplete Freund’s adjuvant, and the serum samples were collected two weeks following the second boost. Development of antibodies was analyzed by enzyme-linked immunosorbent assay (ELISA) and immunoblotting, as described 32.
Blue native-PAGE, two-dimensional SDS-PAGE and Western blotting
Analysis of OM complexes was performed under native conditions by BN/PAGE, as described 32. The isolated OM vesicles were solubilized with n-dodecyl β-D-maltoside (DDM) (DDM/protein = 40/1 w/w) and subjected to first dimensional BN/PAGE at 4 °C in a 5–14% polyacrylamide gradient gel. Native protein markers (Invitrogen) were used to estimate the approximate size of protein complexes in the gel. The intact individual lanes from the BN/PAGE gel were excised and transferred onto a nitrocellulose membrane for immunodetection with anti-OM, anti-P66 or anti-OspA antibodies. Parallel BN/PAGE gels were incubated for 30 min in 25mM Tris/HCl, pH 7.5, containing 1% SDS (w/v) and 1% dithiothreitol (w/v), and subjected to a second dimensional SDS-PAGE for determination of subunit compositions of the protein complexes using 12–20% linear gradient polyacrylamide gels containing 7 M urea. Pre-stained protein markers (New England Biolabs) were used for estimation of molecular masses of proteins in SDS-PAGE gels. The proteins separated in 2D-BN/SDS-PAGE gel were either stained by SYPRO Ruby (Invitrogen) or transferred onto a nitrocellulose membrane. The membrane was blocked with 3% BSA-PBS overnight and incubated with subunit-specific primary antibodies. The signals were developed by the addition of horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit IgG at a 1:10,000 dilution. Protein signals were visualized using chemiluminescence Western Blotting Detection Reagent (GE Healthcare).
Liquid chromatography-mass spectrometry and data analysis
Samples for tandem mass spectrometry were prepared by tryptic in-gel digestion of excised protein bands as described 32. Briefly, protein complexes from gels were excised, chopped into pieces and transferred to tubes that were previously rinsed with 0.1% trifluoroacetic acid (TFA) and 60% acetonitrile (ACN). The gels were then serially washed with 50% ACN, a mixture of 50% ACN, 50 mM NH4HCO3, and 10 mM NH4HCO3. The gel pieces were dried, digested with trypsin (0.1 mg/ml) (Promega Corporation, Madison) and further extracted with 100 μl of 50% ACN and 5% TFA and concentrated to a final volume of 40 μl. Tryptic digests were injected onto a Zorbax SB300 C18 column (1.0 × 100 mm) (Agilent Technologies, Santa Clara, CA) connected to an Accela HPLC system (Thermo Electron, San Jose, CA) and interfaced to a Thermo Finnigan LTQOrbitrap XL mass spectrometer equipped with an Ion Max electrospray source. Separation of peptides was achieved by a linear gradient of 5–35% solvent B at 50 μl/min for 45 min (solvent A: 95% water, 5% ACN, 0.1% formic acid, solvent B: 5% water, 95% can, 0.1% formic acid). The LTQ Orbitrap XL mass spectrometer (Thermo Electron) was operated in positive ion mode with data dependent MS/MS acquisition. The instrument was set to complete a full mass scan from mass-to-charge ratio (m/z) 350–2000 with resolution 60,000 (m/z 400) in the Orbitrap followed by data dependent MS/MS analysis of up to five of the most intense ions with CID in the linear ion trap at unit mass resolution. Dynamic exclusion was turned on to exclude ions from being selected again for MS/MS analysis for 30 sec with a window of [−0.5 – +1.5] m/z. LC-MS/MS data files were searched using Sequest search engine through Bioworks (Thermo Electron) and Mascot search engine through the in-house Mascot Server (Matrix Science, London, U.K.) against a borrelial protein databases extracted from the UniProt database. Results were combined using Scaffold Distiller (Proteome Software, Portland, OR) for identification of proteins. Predicted localization of B. burgdorferi proteins to the spirochete membrane was determined according to the database annotation and PSORT Prediction, as detailed 39.
Protein cross-linking and co-immunoprecipitation assay
The chemical cross-linking using dithiobis(succinimidyl) propionate (DSP) was performed as described previously 32. Briefly, 1010 B. burgdorferi cells were collected, washed with PBS, and further incubated in PBS containing 1 mM DSP and 4% dimethyl sufoxide for 2 hours on the ice. The reaction was terminated using Tris-HCl (pH 7.5) with a final concentration of 10 mM and centrifuged. The pellet was resuspended in PBS, sonicated and equally split into three tubes and incubated with mouse monoclonal antibodies against OspA, P66 and GST for 4 hours at 4°C. The reaction mixtures were further incubated with protein A-sepharose beads (Pierce Biotechnology Inc., Rockford, IL) overnight at 4°C. The protein bound antibody complexes were then washed five times with immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 1% NP-40 substitute, 150 mM NaCl, 1 mM EDTA) and dissolved in Laemmli sample buffer for SDS-PAGE and immunoblot analysis.
PCR
The oligonucleotide primers used for the PCR or RT-PCR analysis of B. burgdorferi genes are listed in Supplementary Table S6. Total RNA was extracted from B. burgdorferi using Trizol reagent (Invitrogen), further digested with RNase-free DNaseI (New England Biolabs, Ipswich, MA), purified using the RNeasy kit (Qiagen, Valencia, CA) and then converted to cDNA using the AffinityScript cDNA synthesis kit (Agilent Technologies). The gene transcripts were normalized to the B. burgdorferi flaB levels, as detailed 39.
Generation of p66-deficient B. burgdorferi B31
The infectious B. burgdorferi isolate A3 was used to create an isogenic bb0603 (p66) deficient B. burgdorferi by an exchange of the p66 open reading frame with a kanamycin-resistance cassette via homologous recombination. DNA fragments flanking up- and downstream regions of the p66 gene were PCR-amplified using primers P1-P4 and inserted into two multiple-cloning sites flanking the kanAn cassette in plasmid pXLF10601, as detailed 39. The recombinant plasmid was sequenced to confirm the identity of the insert and electroporated into B. burgdorferi. Transformants were then selected on kanamycin (350 μg/ml). Of the antibiotic resistant clones, one harboring the intended recombination and carrying the endogenous set of plasmids present in the wild-type isolate was selected by PCR 39 and used for additional experiments.
Generation of OspA/B/C-deficient B. burgdorferi 297
Inactivation of ospAB and ospC individually has been described previously 30, 38. To construct an ospAB and ospC double mutant, an ospAB mutant was first re-constructed with an erythromycin-resistant (ErmR) marker instead of a streptomycin resistant (StrepR) marker that was reported previously. To do so, the previously described suicide plasmid pXT-OspAB that harbors 4 kb DNA fragment with ospAB and flanking sequence 30 was digested with BstAPI and AccI and replaced with a 1016bp ermR cassette 40. The resulting suicide plasmid DNA was transformed into an infectious clone of B. burgdorferi strain 297, BbAH130 30, to produce the ospAB mutant BbXY326. To inactivate ospC in BbXY326, the previously described suicide vector pOspC-Strep used for disrupting ospC 38, was transformed into BbXY326. ErmR- and StrepR-positive clones were selected and the absence of OspA, OspB and OspC in the mutant clones was confirmed by immunoblot analysis (data not shown).
Results
Identification of B. burgdorferi outer membrane protein complexes and their subunit compositions
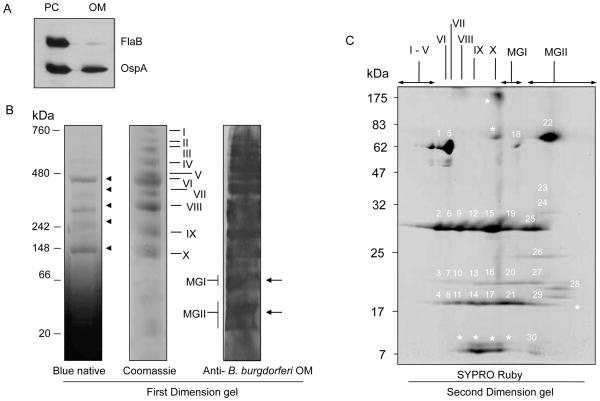
To characterize protein complexes in the OM of B. burgdorferi, we purified the OM vesicles from an infectious B31 isolate (Figure 1A) and performed BN/PAGE analysis. Assessment of BN gels reflected the presence of five major bands that migrated with approximate molecular masses between 148–480 kDa (Figure 1B, left panel, arrowheads). Further staining of BN/PAGE gels with Coomassie brilliant blue (Figure 1B, middle panel) identified additional less conspicuous high molecular weight complexes thereby revealing a total of 10 discrete complexes (complex I-X, middle panel, Figure 1B). Although immunoblotting of one-dimensional BN/PAGE gels typically yields poorly resolved protein bands 41, these efforts using anti-B. burgdorferi OM antibodies detected less abundant and lower molecular-weight complexes (Figure 1B, right panel, arrows). This likely represents protein monomers as revealed by mass spectrometry (MS) assays (detailed below) and thus were labeled as monomeric group (MG) I and II. To ascertain the subunit identity of the protein complexes, complexes I-X and MGI-II were separately excised from one-dimensional BN/PAGE gel and digested with trypsin. The enzymatic digests were subjected to MS-based protein identification using liquid chromatography (LC)-MS/MS analysis. The MS analyses were repeated in an independent experiment consistently identifying 20 unique proteins as participating members of 10 B. burgdorferi OM protein complexes whereas MGI and II reflect the existence of 27 proteins (Table S1). Notably, a few of the previously known abundant membrane proteins, OspA, -B and -C occurred in all protein complexes (Table S1). P13, OspD and Lp 6.6 were detectable in all complexes except for one, whereas other proteins, such as P66, BB0405, BB0543, BB0553, BBA03, La7, ErpA, BBH37, BBI39 and BBK45 displayed intermittent distributions between multiple complexes or existed as monomers. At least 27 proteins including BmpA were identified exclusively in MGI-II, indicating their occurrence as free monomers or unstable subunits of protein complexes.
Figure 1. Identification of B. burgdorferi outer membrane (OM) complexes and constituent subunit members.
(A) Assessment of the purity of isolated OM fraction of B. burgdorferi. The OM vesicles were isolated, and the purity of solubilized proteins was tested by immunoblotting using antibodies against known OM (OspA) and periplasmic cylinder (PC) protein (FlaB). (B) Separation of OM protein complexes by blue native/polyacrylamide gel electrophoresis (BN/PAGE). A 5–14% gradient BN/PAGE gel was used for the separation of protein complexes. The first dimensional gel (left panel), which detected five major bands (arrowheads) was either stained with Coomassie brilliant blue (middle panel) or transferred onto a nitrocellulose membrane and probed with antibodies against wild-type B. burgdorferi OM proteins (right panel). The identified complexes were labeled numerically from I to X while monomeric protein groups were indicated as MGI and MGII (arrows). (C) SYPRO Ruby staining of OM complexes resolved by second dimensional SDS-PAGE. The OM protein complexes and monomeric protein groups, as separated by 5–14% gradient BN/PAGE, were resolved by denaturing second dimension 12–20% gradient SDS-PAGE followed by staining with SYPRO Ruby. Protein markers are shown to the left. Thirty gel spots (indicated as white numbers) were excised for mass spectrometry and the proteins identified are tabulated in Table S2. Asterisks indicate gel spots where a precise identification was not possible.
To corroborate subunit identification of OM protein complexes from one-dimensional BN/PAGE gels, individual proteins in each complex were further separated in second dimension SDS-PAGE and then identified by LC-MS/MS analysis. As complexes greater than 500 kDa (complex I-V, Figure 1B) were overlapping and difficult to resolve by second-dimensional gel (data not shown), we focused on characterizing the remaining complexes (complex VI-X) and monomeric groups (MGI-II, Figure 1B). The OM protein complexes were first resolved by one-dimensional BN/PAGE and then the excised gel strips were subjected to a second dimension SDS-PAGE and stained with SYPRO Ruby enabling visualization of individual subunits (Figure 1C). To identify the subunit composition of individual protein complexes visualized in 2D-BN/SDS-PAGE, the gel spots from each complex were excised (numbered 1–30, Figure 1C), and digested with trypsin, and proteins were identified by LC-MS/MS analysis (Table S2). Note that gel spot 22 contained a relatively high molecular weight (67-kDda) protein without identifiable membrane localization signal and thus likely represent a contaminating protein (Table S2). Both series of MS data, the one-dimensional BN/PAGE (Table S1) and the 2D-BN/SDS-PAGE (Table S2) were compared, and common subunits of complexes as well monomers were further tabulated (Table S3). We concluded that complexes VI-X contained at least 10 unique subunits. Among them, five membrane-anchored proteins, OspA, -B, -C, -D and Lp6.6, and three transmembrane proteins, P66, P13 and BB0405 were found to be the most common members of OM complexes. Note that the OspB protein detected in the current B. burgdorferi isolate migrates as a ~19-kDa protein, which represents a truncated version of the larger OspB protein, as reported earlier 28.
Specificity of protein-protein interactions amongst the selected members of outer membrane protein complexes
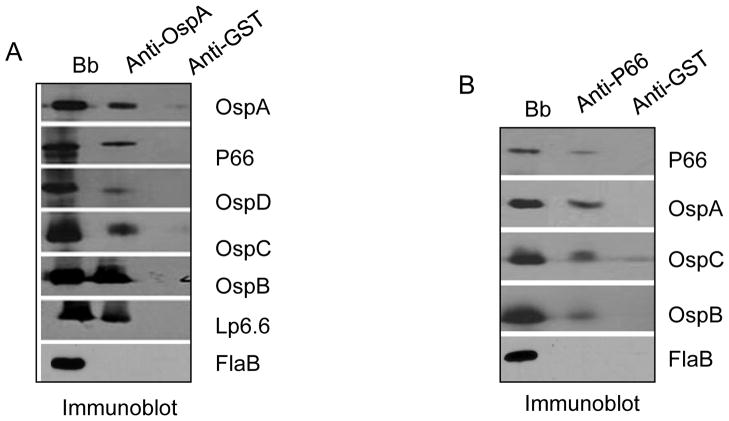
As MS-based identity of complex subunits revealed ubiquitous occurrence of a few Osps that are known to be abundant in the OM, we sought to obtain additional experimental data to corroborate the MS results. To achieve this, a series of immunoblot analyses were performed using antibodies available against selected subunits that were detected in one or multiple complexes. As presented in Figure 2, the results supported the MS-based subunit identity of protein complexes showing a common distribution of OspA, -B, -C, -D and Lp6.6 in most complexes. Similarly, a selective distribution was observed for other proteins, such as P66 (complex VI) or BB0405 (complex X).
Figure 2. Assessment of OM protein complexes by immunoblot analysis using antibodies specific to selected subunits.
(A) Detection of OM protein complexes containing OspA or P66. Protein complexes were separated by first dimensional BN/PAGE, transferred to a nitrocellulose membrane and blotted with anti-P66 (left panel) and anti-OspA (right panel). P66 antibody recognized a major band (arrowhead) that corresponds to complex VI (Figure 1B), while the OspA antibody detected multiple bands (arrows) comparable to the migration of complexes I-X and monomeric protein groups MGI-II. (B) Detection of subunit members of OM protein complexes in second dimensional SDS-PAGE by immunoblot analysis. Protein complexes, as separated by BN/PAGE, were further resolved into subunits by second dimensional SDS-PAGE, transferred to a nitrocellulose membrane and blotted with specific antibodies against P66, BmpA, OspA, -D, BB0405, OspC, La7, OspB and Lp6.6.
To obtain further evidence of the specificity of protein-protein interaction between subunit members, we sought to perform co-immunoprecipitation analyses using selected antibodies. In order to stabilize the protein complexes, cultured B. burgdorferi cells were treated with DSP, a membrane-permeable, amine-reactive homobifunctional crosslinker with a spacer arm of 12Å. Subsequently, cells were subjected to co-immunoprecipitation analyses. As OspA and P66 are subunit members of multiple protein complexes, antibodies specific to these proteins were used to immunoprecipitate other associated subunits (Table S3 and Figure 2) and later identified by immunoblot analysis. OspA antibodies were able to pull-down OspA and associated subunits, such as P66, OspB, -C, -D and Lp6.6 (Figure 3A). Similarly, P66 antibodies were able to pull down P66, OspA, -B and -C (Figure 3B). The OspA or P66 antibodies did not immunoprecipitate other proteins, such as FlaB, that were not expected to be present in the protein complex (Figure 3A and 3B). Overall, as suggested by BN/MS-based analysis, these results support the possible interaction of selected proteins showing their occurrence in common OM protein complexes.
Figure 3. Interaction of subunits assessed by protein cross-linking/co-immunoprecipitation.
B. burgdorferi (Bb) lysates were treated with a protein cross-linker (DSP) and immunoprecipitated either with OspA (panel A) or P66 (panel B) antibodies. Antibodies against glutathione-S-transferase (GST) were used as a control. Left lane (Bb) donates antibody recognition of target proteins in B. burgdorferi lysates without the immunoprecipitation step. Immunoprecipitated proteins (middle and right lanes) were identified by antibodies against OspA, P66, OspD, -C, -B and Lp6.6. As an additional control, immunoprecipitates were also probed with antibodies against the B. burgdorferi protein (FlaB), which was not expected to be present in the complex.
P66 is essential for the occurrence of complex VI
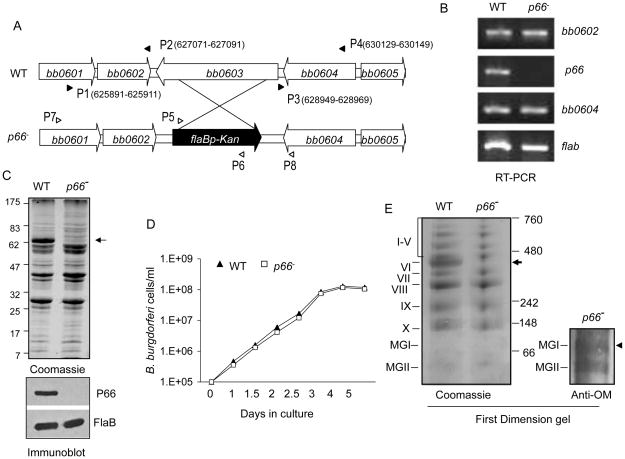
Since certain integral membrane proteins are common members of OM complexes, we next assessed the occurrence of protein complexes using spirochetes deficient in a selected integral membrane protein. Based on the molecular weight, occurrence of P66 in MGI is likely monomeric, however, the protein is detectable in a high-molecular weight complex (complex VI, Table S3 and Figure 2B). We therefore, sought to generate an isogenic p66 mutant using the infectious parental isolate and then assess the formation of OM protein complexes. As shown in Figure 4A, a mutant was created by replacing the entire open reading frame of bb0603 (p66) with an antibiotic resistance gene via allelic exchange. The selected mutant clone retained the same set of plasmids as the wild-type spirochetes (data not shown) and failed to produce p66 mRNA (Figure 4B) or protein (Figure 4C). The genetic manipulation did not introduce apparent polar effects, as the mutant expressed neighboring genes bb0602 and bb0604 at levels similar to the wild-type isolate (Figure 4B). Additionally, deletion of p66 did not alter B. burgdorferi growth kinetics in vitro (Figure 4D). To assess if P66 deficiency affects the occurrence of B. burgdorferi OM protein complexes, wild-type spirochetes and p66 mutants were grown to stationary phase, OM vesicles were isolated, and protein complexes were analyzed using BN/PAGE. Results showed that while p66 mutants maintained a similar profile of protein complexes as parental spirochetes, complex VI was conspicuously absent (Figure 4E, left panel, arrow) where P66 either exists as homo-oligomer or alternatively as an essential member of a heteromeric complex. As expected, P66 is non-essential for the occurrence of MGI, which existed in the p66 mutant (Figure 4E, right panel, arrowhead).
Figure 4. Generation of isogenic B. burgdorferi p66 mutants and assessment of OM protein complexes.
(A) Schematic representation of wild type (WT) and p66 mutant (p66−) B. burgdorferi at the bb0603 (p66) locus. Genes bb0601- bb0605 (white box arrows) and the kanamycin-resistance cassette driven by the B. burgdorferi flaB promoter (flaBp-Kan, black box arrow) are indicated. The regions up- and down-stream of the p66 locus were amplified using primers P1-P4 (black arrow-heads) and cloned to BamHI-SacII and XhoI-KpnI sites flanking the flaBp-Kan cassette. (B) RT-PCR analysis for assessment of p66 transcripts and polar effects of mutagenesis. Total RNA was isolated from wild type (WT) and p66 mutant (p66−) B. burgdorferi, converted to cDNA for detection of p66, flaB, bb0602 and bb0604 transcripts. (C) Protein analysis of wild type (WT) and p66 mutant (p66−). Equal amounts of protein were separated on SDS-PAGE gels and either stained with Coomassie blue (upper panel) or transferred onto a nitrocellulose membrane and probed with P66 and FlaB antibodies (lower panels). Protein standards are shown to the left in kDa. Arrow indicates the missing P66 band in mutant lysates. (D) p66 mutant lacks detectable growth defects in vitro. Wild type (WT) and p66 mutant (p66−) spirochetes were diluted to a density of 105cells/ml, grown at 33°C in BSK-H medium and counted under a dark-field microscope. (E) Analysis of OM protein complexes in p66 mutants. The OM fraction from wild type or p66 mutant B. burgdorferi was isolated, and protein complexes were separated using first dimensional BN/PAGE (left panel). A parallel first dimensional BN/PAGE gel containing protein complexes of the p66 mutants was transferred onto a nitrocellulose membrane and immunoblotted with anti-OM antibodies (right panel). Complex VI was absent in the p66 mutant (arrow), while monomeric protein group MGI was present (arrowhead).
OM complexes are conserved in diverse B. burgdorferi isolates but dramatically altered in Osp-deficient cells
As certain abundant outer surface proteins (Osps) are common members of protein complexes I-X (Table S3), we next assessed possible alteration of complex profiles in diverse infectious isolates and in mutants where multiple Osps are absent. In addition, we wanted to identify less conspicuous complexes or subunits that are potentially hidden in wild-type B. burgdorferi but are detectable in spirochetes deficient in abundant Osp subunits. Analysis of OM vesicles in a linear plasmid-deficient B. burgdorferi mutant, isolate B314 29 deficient in OspA, -B, and -D by BN/PAGE analysis indicated the loss of multiple complexes that are apparent in wild-type cells, such as complexes I-V, VII, VIII and IX (Figure 5A, left panel). MS analysis of detectable protein complexes in the B314 isolate (Figure 5A, right panel) identified a total of 11 unique proteins (Table S4). Similar to wild-type spirochetes, P13, OspC and P66 were detected in most complexes; BB0418, an OM efflux channel protein (BB0142) and a putative phosphomannomutase (BB0835) were intermittently distributed across the complexes. Immunoblot analysis of 2D-BN/SDS-PAGE gels indicated that while La7 is detectable only in MGII group; BB0405, P66, OspC and BmpA subunits are distributed in most protein complexes of the B314 isolate (Figure 5B. arrows). However, MS analysis identified new subunit members, such as BB0406, P83/100, BB0028 and BB0795 in the B314 isolate (Table S4) that were not detectable in wild-type cells (Table S1-S3).
Figure 5. B. burgdorferi osp mutant isolates displays dramatic alterations of OM protein complexes.
(A) Analysis of OM protein complexes in the osp mutant B. burgdorferi. The OM fractions were isolated from the wild type and the osp mutant isolate B314 and corresponding protein complexes were resolved by first dimensional BN/PAGE. Four protein complexes in osp mutant that likely migrated with similar molecular masses of wild-type complexes VI and X as well as monomeric protein groups MGI and MGII (arrowheads) were excised (B314 panel) and protein identification was performed by mass spectrometry, as presented in Table S4. (B) Subunit detection of OM protein complexes in B314 isolate by immunoblot analysis of gels resolved by second dimensional SDS-PAGE. OM protein complexes were resolved by BN/PAGE and second dimensional SDS-PAGE, transferred to a nitrocellulose membrane and blotted with specific antibodies against P66, BmpA, BB0028, BB0405, OspC and La7. Arrows indicate enhanced distribution of BmpA, BB0405 and OspC towards high-molecular-weight complexes, compared to that in wild-type cells (Figure 2B). (C) Comparison of OM protein complexes in the B. burgdorferi isolates B31, 297 and Osp-deficient 297. The OM fractions were purified from indicated spirochete isolates and corresponding protein complexes were resolved by first dimensional BN/PAGE. A few protein complexes in Osp-deficient 297 that likely migrated with comparable molecular masses of wild-type complexes VI, IX and X (arrows) were excised (Osp- panel) and protein identification was performed by mass spectrometry, as presented in Table S5. (D) Subunit detection of OM protein complexes in Osp-deficient 297 isolate by immunoblot analysis of gels resolved by second dimensional SDS-PAGE. OM protein complexes were resolved by BN/PAGE and second dimensional SDS-PAGE, transferred to a nitrocellulose membrane and blotted with specific antibodies against P66, BmpA, BB0028, BB0405, La7 and Lp6.6.
We next assessed the conservation of protein complexes in other major B. burgdorferi infectious isolates, such as a human isolate 297. We also sought to assess the dynamics of OM complexes when three major Osps (OspA/B/C) were depleted in the wild-type 297 isolate, which is also naturally deficient in another major Osp (OspD). We expect that, similar to B314 isolate, Osp deficiency would allow us to identify less conspicuous complexes/subunits. A mutant with serial deletions of ospA, -B and -C was created as described in the Materials and Methods section. Analysis of OM vesicles by BN/PAGE indicated that with the exception of complex IX, other complexes are conserved in both wild-type B31 and 297 isolates, (Figure 5C, left and central lanes). However, the Osp-deficient 297 isolate reflected an apparent loss of complexes I-V, VII, and VIII (Figure 5A, right panel), but harbors four inconspicuous but detectable complexes (Figure 5C, right panel, arrows) with 24 unique subunit members (Table S5) - of which 12 are non-identifiable in wild-type spirochetes or B314 isolates. However, immunoblot analysis of 2D-BN/SDS-PAGE gels indicated a comparable distribution of common subunits in Osp-deficient 297 (Figure 5D) and B314 (Figure 5B) isolates. Together, while these studies identified low abundance protein complex/subunits in Osp-deficient spirochetes, these data together indicated a high conservation of OM complexes in B. burgdorferi sensu stricto isolates, as well as possible existence of OM complexes that are specific to a strain.
Discussion
The outer membrane (OM) of Gram-negative bacteria houses typical β-barrel integral membrane proteins as well as membrane-anchored proteins, some of which exist as membrane protein complexes, serving vital roles in membrane function and biogenesis 42. The B. burgdorferi genome 43, 44 encodes a large number of potential membrane proteins and our current study identified the existence of multiple OM complexes in infectious B. burgdorferi isolates. Although we do not know the stoichiometry of identified complexes or whether low molecular weight complexes simply represent protein monomers, our study has identified a number of unique subunit proteins that were detected in blue native (BN) analysis of wild type or Osp-deficient cells. These results demonstrate that these protein complexes could be involved in protein-protein interaction relevant to unknown function in spirochete biology. While other pathogenic Gram-negative bacteria, such as Helicobacter pylori, possess fewer OM protein complexes, mostly multimeric enzymes 24, borrelial OM harbors a larger number of complexes dominated by a few membrane-spanning proteins, such as P66, P13, BB0405 and membrane-anchored lipoproteins, such as OspA, -B, -C, -D, Lp6.6, which either have unknown function or are known to be involved in membrane transport, tissue adhesion and immune evasion.
BN/PAGE technology represents a reliable approach for the identification of protein complexes in native conditions as well as the assessment of novel protein-protein interactions 23. We have previously reported the existence of seven protein complexes in B. burgdorferi OM 32. Our current study extends the initial observation by showing the existence of at least 10 OM complexes and determining their subunit identity. The occurrence of a limited diversity of proteins, such as a few Osps as ubiquitous subunits raises the possibility that some of these complexes may be derived from unstable higher order complexes. However, we performed a detergent screen involving digitonin 45, n-octyl-β-D-glucoside 46, Triton-X-100 25, IGEPAL CA-360 or n-dodecyl β-D-maltoside 23, 32 and determined that the latter detergent generates a reproducible profile of OM complexes (data not shown). These results suggest that the complexes I-X are unlikely to be derived from unstable higher order complexes and are further supported by independent LC-MS/MS analyses, BN/PAGE gel immunoblotting and co-immunoprecipitation analysis. Finally, analyses of OM complexes in isogenic mutants also supported the specificity of individual complexes. For example, loss of complex VI in p66 deletion mutants suggests the involvement of P66 as an essential subunit. Although P66 is detected in multiple complexes by second-dimensional immunoblot and MS analysis, its participation is redundant in all complexes expect for the primary complex VI, which is detectable even by immunoblot analysis of one-dimensional BN/PAGE gel (Figure 2A) that has a limited capability of resolution of protein bands 41. On the other hand, analysis of complex formation in a targeted Osp-deficient isolate suggested that the missing subunit(s) OspA -B, -C are integral for the occurrence of several complexes, such as complex I-V, VII and VIII. Although occurrence of these limited Osps in multiple complexes is puzzling, previous studies 47 demonstrated that exogenous addition of certain Osps, such as OspA or -D, results in their insertion into borrelial OM, a process primarily attributed to the amino-terminal lipid moiety but also contributed to by protein-protein interaction. In addition, subunit interactions as observed in our study were also previously documented, such as OspA interaction with OspA 48, P66 association with OspA, -B and -C 49 and co-localization of OspA and -B in the spirochete membrane 50. Overall, these studies suggested the existence of discrete OM complexes that are composed of a limited diversity of membrane proteins and that specific subunits are essential or redundant for OM complex formation.
P66 is shown to act as a B. burgdorferi adhesin, which can bind to β3-integrin 51, 52. Additionally, P66 has also been shown to modulate the expression of a specific set of genes in cultured human cells 53. However, similar to other transport proteins, such as P13 54, DipA 55, BB0142 56, MIP family proteins 57, 58 or oligopeptide permeases (Opp) complex 59, 60, P66 is also suggested to be involved in solute transport across the borrelial membrane 61, 62. While we show that some of these proteins involved in formation of OM complexes, the function of most of the unique proteins (Table S1-S3) including their precise participation in a given complex warrants future investigations. The presence of common protein spots in the same vertical line of the 2D-BN/SDS-PAGE (Figure 1C) could be indicative of the protein’s presence in the same complex. However, those proteins could also be part of separate complexes with similar electrophoretic mobility in the BN/PAGE 63. The protein bands that migrated with approximate molecular masses between 20–66 kDa in BN/PAGE gels were labeled as monomeric protein groups (MGI and MGII) as they cannot contain so many full-length subunits, and therefore, largely represent collections of monomeric proteins. Although MGI and MGII could still house complexes of small molecular weight proteins, we speculate that majority of these proteins may exist as such in the membrane or were disrupted from larger unstable complexes during purification. Additionally, we could not exclude the potential occurrence of undetectable low abundance complexes, overlapping monomers or multiple complexes in a single complex. An unstable complex or subunit member could also be lost during membrane extraction procedures. For example, although we have detected one of the members of the Borrelia efflux system, BesC (bb0142), two other members of the Bes protein complex 64, BesB and BesA remained undetectable. Finally, the occurrence of certain abundant subunits, such as Osps, could potentially interfere with the ability of MS to detect relatively scarce but unique peptides. For example, many unique proteins, some possessing identifiable transmembrane motifs (Table S4 and S5), remained detectable only in Osp-deficient isolates. This suggests their likely occurrence as low-abundance proteins yet highlighting their possible involvement in protein-protein interaction. The functions of some of them are either unknown (BB0028, BB0324, BB0406, BppA1, DegP ortholog BB0104), linked as channel forming (BBA01) 65, host-pathogen interaction (OspE paralog BBN38) 66, as a component of β-barrel assembly machine or BAM (BamA or BB0795) 67. In contrast, others are known to be immunogenic and identified or speculated to play an important role in the borrelial enzootic cycle, such as BB0323 68, surface lipoprotein BBA07 69, BB0744 70, MlpJ 71, surface protein RevA1 70, virulent strain-associated repetitive antigen BBI16 72, OM associated protein BBA74 73. Therefore, assessment of additional mutants lacking abundant Osps in BN/PAGE-based assays is likely to be useful to identify unknown subunits of B. burgdorferi OM complexes, such as BAM 67, or other low abundance protein complexes. Adoption of more recently described techniques, such as high-resolution clear native electrophoresis 74, 75 could be attempted for better separation of membrane protein complexes allowing more precise identification of the subunits.
In conclusion, our study uncovered the existence of discrete protein complexes in B. burgdorferi OM vesicles as well as identified a number of unique subunits that are members of the OM complexome. However, the biological significance of the occurrence of a limited diversity of membrane proteins, including few Osps, as members of OM complexes remains to be determined. Whether these enigmatic Osps perform additional redundant functions in pathogen biology, such as stabilization of the OM in addition to their stage-specific expression or function in the B. burgdorferi enzootic infection cycle 76–79, remains to be elucidated. Incidentally, a previous study also suggested redundant and interchangeable roles of at least some of these membrane proteins, such as OspA and -C in the stabilization of borrelial OM and pathogen protection from the hostile host immune environment 80. Further characterization of the membrane protein complexes including their in vivo occurrence, function and the significance of protein-protein interaction will shed new light on understanding how B. burgdorferi evolved to survive in the complex enzootic cycle and in the development of new strategies to interfere with the infection.
Supplementary Material
Acknowledgments
This study was supported by the funding from the NIH/NIAID (AI076684 and AI080615) to UP. We are grateful to Darrin Akins, Aravinda de Silva and Alan Barbour for their help with reagents. We thank Adam Coleman, Alexis Smith, Brian Backstedt, Cara Wilder and Faith Kung for their assistance with the preparation of the manuscript and X. Frank Yang for his invaluable suggestions and critical reading of the manuscript.
Footnotes
Table S1 – S6. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Anderson J. Epizootiology of Lyme borreliosis. Scan J Infect Dis-Suppl. 1991;77:23–24. [PubMed] [Google Scholar]
- 2.Anderson JF, Duray PH, Magnarelli LA. Prevalence of Borrelia burgdorferi in white-footed mice and Ixodes dammini at Fort McCoy, Wis. J Clin Microbiol. 1987;25(8):1495–7. doi: 10.1128/jcm.25.8.1495-1497.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65(5):1193–217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal U, Fikrig E. Adaptation of Borrelia burgdorferi in the vector and vertebrate host. Microbes Infect. 2003;5(7):659–66. doi: 10.1016/s1286-4579(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 5.Tilly K, Rosa PA, Stewart PE. Biology of infection with Borrelia burgdorferi. Infect Dis Clin North Am. 2008;22(2):217–34. v. doi: 10.1016/j.idc.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang JR, Hardham JM, Barbour AG, Norris SJ. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of Vmp-like sequence cassettes. Cell. 1997;89(2):275–85. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 7.Samuels DS. Gene Regulation in Borrelia burgdorferi. Annu Rev Microbiol. 2010 Sep 28; doi: 10.1146/annurev.micro.112408.134040. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Fikrig E, Narasimhan S. Borrelia burgdorferi--traveling incognito? Microbes Infect. 2006;8(5):1390–9. doi: 10.1016/j.micinf.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun. 2003;71(6):3371–83. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, Caimano M, Skare J, Swingle K, Akins D, Schwartz I. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun. 2003;71(4):1689–705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci U S A. 2002;99(3):1562–7. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokarz R, Anderton JM, Katona LI, Benach JL. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun. 2004;72(9):5419–32. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll JA, Garon CF, Schwan TG. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun. 1999;67(7):3181–7. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll JA, Gherardini FC. Membrane protein variations associated with in vitro passage of Borrelia burgdorferi. Infect Immun. 1996;64(2):392–8. doi: 10.1128/iai.64.2.392-398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowalk AJ, Nolder C, Clifton DR, Carroll JA. Comparative proteome analysis of subcellular fractions from Borrelia burgdorferi by NEPHGE and IPG. Proteomics. 2006;6(7):2121–34. doi: 10.1002/pmic.200500187. [DOI] [PubMed] [Google Scholar]
- 16.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5(6):580–92. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Lin J, Huang S, Zhang Q. Outer membrane proteins: key players for bacterial adaptation in host niches. Microbes Infect. 2002;4(3):325–31. doi: 10.1016/s1286-4579(02)01545-9. [DOI] [PubMed] [Google Scholar]
- 18.Radolf JD, Goldberg MS, Bourell K, Baker SI, Jones JD, Norgard MV. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect Immun. 1995;63(6):2154–63. doi: 10.1128/iai.63.6.2154-2163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radolf JD, Norgard MV, Brandt ME, Isaacs RD, Thompson PA, Beutler B. Lipoproteins of Borrelia burgdorferi and Treponema pallidum activate cachectin/tumor necrosis factor synthesis. Analysis using a CAT reporter construct. J Immunol. 1991;147(6):1968–74. [PubMed] [Google Scholar]
- 20.Radolf JD, Bourell KW, Akins DR, Brusca JS, Norgard MV. Analysis of Borrelia burgdorferi membrane architecture by freeze-fracture electron microscopy. J Bacteriol. 1994;176(1):21–31. doi: 10.1128/jb.176.1.21-31.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker EM, Borenstein LA, Blanco DR, Miller JN, Lovett MA. Analysis of outer membrane ultrastructure of pathogenic Treponema and Borrelia species by freeze-fracture electron microscopy. J Bacteriol. 1991;173(17):5585–8. doi: 10.1128/jb.173.17.5585-5588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92(3):291–4. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 23.Stenberg F, Chovanec P, Maslen SL, Robinson CV, Ilag LL, von Heijne G, Daley DO. Protein complexes of the Escherichia coli cell envelope. J Biol Chem. 2005;280(41):34409–19. doi: 10.1074/jbc.M506479200. [DOI] [PubMed] [Google Scholar]
- 24.Pyndiah S, Lasserre JP, Menard A, Claverol S, Prouzet-Mauleon V, Megraud F, Zerbib F, Bonneu M. Two-dimensional blue native/SDS gel electrophoresis of multiprotein complexes from Helicobacter pylori. Mol Cell Proteomics. 2007;6(2):193–206. doi: 10.1074/mcp.M600363-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Farhoud MH, Wessels HJ, Steenbakkers PJ, Mattijssen S, Wevers RA, van Engelen BG, Jetten MS, Smeitink JA, van den Heuvel LP, Keltjens JT. Protein complexes in the archaeon Methanothermobacter thermautotrophicus analyzed by blue native/SDS-PAGE and mass spectrometry. Mol Cell Proteomics. 2005;4(11):1653–63. doi: 10.1074/mcp.M500171-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Liu K, Qian L, Wang J, Li W, Deng X, Chen X, Sun W, Wei H, Qian X, Jiang Y, He F. Two-dimensional blue native/SDS-PAGE analysis reveals heat shock protein chaperone machinery involved in hepatitis B virus production in HepG2.2.15 cells. Mol Cell Proteomics. 2009;8(3):495–505. doi: 10.1074/mcp.M800250-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stroh A, Anderka O, Pfeiffer K, Yagi T, Finel M, Ludwig B, Schagger H. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J Biol Chem. 2004;279(6):5000–7. doi: 10.1074/jbc.M309505200. [DOI] [PubMed] [Google Scholar]
- 28.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Bono JL, Akins DR, Radolf JD, Schwan TG, Rosa P. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70(4):2139–50. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadziene A, Wilske B, Ferdows MS, Barbour AG. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun. 1993;61(5):2192–5. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med. 2004;199(5):641–8. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skare JT, Shang ES, Foley DM, Blanco DR, Champion CI, Mirzabekov T, Sokolov Y, Kagan BL, Miller JN, Lovett MA. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J Clin Invest. 1995;96(5):2380–92. doi: 10.1172/JCI118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Promnares K, Kumar M, Shroder DY, Zhang X, Anderson JF, Pal U. Borrelia burgdorferi small lipoprotein Lp6.6 is a member of multiple protein complexes in the outer membrane and facilitates pathogen transmission from ticks to mice. Mol Microbiol. 2009;74(1):112–25. doi: 10.1111/j.1365-2958.2009.06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coleman AS, Yang X, Kumar M, Zhang X, Promnares K, Shroder D, Kenedy MR, Anderson JF, Akins DR, Pal U. Borrelia burgdorferi complement regulator-acquiring surface protein 2 does not contribute to complement resistance or host infectivity. PLoS ONE. 2008;3(8):3010e. doi: 10.1371/journal.pone.0003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fikrig E, Pal U, Chen M, Anderson JF, Flavell RA. OspB antibody prevents Borrelia burgdorferi colonization of Ixodes scapularis. Infect Immun. 2004;72(3):1755–9. doi: 10.1128/IAI.72.3.1755-1759.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Neelakanta G, Liu X, Beck DS, Kantor FS, Fish D, Anderson JF, Fikrig E. Role of outer surface protein D in the Borrelia burgdorferi life cycle. Infect Immun. 2007;75(9):4237–44. doi: 10.1128/IAI.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pal U, Dai J, Li X, Neelakanta G, Luo P, Kumar M, Wang P, Yang X, Anderson JF, Fikrig E. A Differential Role for BB0365 in the Persistence of Borrelia burgdorferi in Mice and Ticks. J Infect Dis. 2008;197(1):148–155. doi: 10.1086/523764. [DOI] [PubMed] [Google Scholar]
- 37.Pal U, Wang P, Bao F, Yang X, Samanta S, Schoen R, Wormser GP, Schwartz I, Fikrig E. Borrelia burgdorferi basic membrane proteins A and B participate in the genesis of Lyme arthritis. J Exp Med. 2008;205(1):133–41. doi: 10.1084/jem.20070962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, Norgard MV, Fikrig E. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113(2):220–30. doi: 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Coleman AS, Anguita J, Pal U. A chromosomally encoded virulence factor protects the Lyme disease pathogen against host-adaptive immunity. PLoS Pathog. 2009;5(3):e1000326. doi: 10.1371/journal.ppat.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 2001;98(22):12724–9. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nijtmans LG, Henderson NS, Holt IJ. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26(4):327–34. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- 42.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121(2):235–45. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35(3):490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 44.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Venter JC, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390(6660):580–6. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 45.Oates J, Barrett CM, Barnett JP, Byrne KG, Bolhuis A, Robinson C. The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J Mol Biol. 2005;346(1):295–305. doi: 10.1016/j.jmb.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 46.Tziatzios C, Schubert D, Lotz M, Gundogan D, Betz H, Schagger H, Haase W, Duong F, Collinson I. The bacterial protein-translocation complex: SecYEG dimers associate with one or two SecA molecules. J Mol Biol. 2004;340(3):513–24. doi: 10.1016/j.jmb.2004.04.076. [DOI] [PubMed] [Google Scholar]
- 47.Bunikis J, Mirian H, Bunikiene E, Barbour AG. Non-heritable change of a spirochaete’s phenotype by decoration of the cell surface with exogenous lipoproteins. Mol Microbiol. 2001;40(2):387–96. doi: 10.1046/j.1365-2958.2001.02382.x. [DOI] [PubMed] [Google Scholar]
- 48.Pal U, de Silva AM, Montgomery RR, Fish D, Anguita J, Anderson JF, Lobet Y, Fikrig E. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J Clin Invest. 2000;106(4):561–9. doi: 10.1172/JCI9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bunikis J, Barbour AG. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect Immun. 1999;67(6):2874–83. doi: 10.1128/iai.67.6.2874-2883.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escudero R, Halluska ML, Backenson PB, Coleman JL, Benach JL. Characterization of the physiological requirements for the bactericidal effects of a monoclonal antibody to OspB of Borrelia burgdorferi by confocal microscopy. Infect Immun. 1997;65(5):1908–15. doi: 10.1128/iai.65.5.1908-1915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coburn J, Chege W, Magoun L, Bodary SC, Leong JM. Characterization of a candidate Borrelia burgdorferi beta3-chain integrin ligand identified using a phage display library. Mol Microbiol. 1999;34(5):926–40. doi: 10.1046/j.1365-2958.1999.01654.x. [DOI] [PubMed] [Google Scholar]
- 52.Coburn J, Cugini C. Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin alpha-v-beta-3. Proc Natl Acad Sci U S A. 2003;100(12):7301–7306. doi: 10.1073/pnas.1131117100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lafrance ME, Pierce JV, Antonara S, Coburn J. The Borrelia burgdorferi Integrin Ligand, P66, Affects Gene Expression by Human Cells in Culture. Infect Immun. 2011;79(8):3249–61. doi: 10.1128/IAI.05122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadziene A, Thomas DD, Barbour AG. Borrelia burgdorferi mutant lacking Osp: biological and immunological characterization. Infect Immun. 1995;63(4):1573–80. doi: 10.1128/iai.63.4.1573-1580.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergström S, Zückert WR. Structure, Function and Biogenesis of the Borrelia Cell Envelope. In: Samuels DS, Radolf JD, editors. Borrelia, Molecular Biology, Host Interaction and Pathogenesis. Caister Academic Press; Norfolk, UK: 2010. pp. 139–166. [Google Scholar]
- 56.Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta. 2002;1562(1–2):6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- 57.Froger A, Rolland JP, Bron P, Lagree V, Le Caherec F, Deschamps S, Hubert JF, Pellerin I, Thomas D, Delamarche C. Functional characterization of a microbial aquaglyceroporin. Microbiology. 2001;147(Pt 5):1129–35. doi: 10.1099/00221287-147-5-1129. [DOI] [PubMed] [Google Scholar]
- 58.Schwan TG, Battisti JM, Porcella SF, Raffel SJ, Schrumpf ME, Fischer ER, Carroll JA, Stewart PE, Rosa P, Somerville GA. Glycerol-3-phosphate acquisition in spirochetes: distribution and biological activity of glycerophosphodiester phosphodiesterase (GlpQ) among Borrelia species. J Bacteriol. 2003;185(4):1346–56. doi: 10.1128/JB.185.4.1346-1356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bono JL, Tilly K, Stevenson B, Hogan D, Rosa P. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology. 1998;144(Pt 4):1033–44. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- 60.Lazazzera BA. The intracellular function of extracellular signaling peptides. Peptides. 2001;22(10):1519–27. doi: 10.1016/s0196-9781(01)00488-0. [DOI] [PubMed] [Google Scholar]
- 61.Bunikis J, Noppa L, Bergstrom S. Molecular analysis of a 66-kDa protein associated with the outer membrane of Lyme disease Borrelia. FEMS Microbiol Lett. 1995;131(2):139–45. doi: 10.1111/j.1574-6968.1995.tb07768.x. [DOI] [PubMed] [Google Scholar]
- 62.Skare JT, Mirzabekov TA, Shang ES, Blanco DR, Erdjument-Bromage H, Bunikis J, Bergstrom S, Tempst P, Kagan BL, Miller JN, Lovett MA. The Oms66 (p66) protein is a Borrelia burgdorferi porin. Infect Immun. 1997;65(9):3654–61. doi: 10.1128/iai.65.9.3654-3661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camacho-Carvajal MM, Wollscheid B, Aebersold R, Steimle V, Schamel WW. Two-dimensional Blue native/SDS gel electrophoresis of multi-protein complexes from whole cellular lysates: a proteomics approach. Mol Cell Proteomics. 2004;3(2):176–82. doi: 10.1074/mcp.T300010-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Bunikis I, Denker K, Ostberg Y, Andersen C, Benz R, Bergstrom S. An RND-type efflux system in Borrelia burgdorferi is involved in virulence and resistance to antimicrobial compounds. PLoS Pathog. 2008;4(2):e1000009. doi: 10.1371/journal.ppat.1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pinne M, Denker K, Nilsson E, Benz R, Bergstrom S. The BBA01 protein, a member of paralog family 48 from Borrelia burgdorferi, is potentially interchangeable with the channel-forming protein P13. J Bacteriol. 2006;188(12):4207–17. doi: 10.1128/JB.00302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hovis KM, Tran E, Sundy CM, Buckles E, McDowell JV, Marconi RT. Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect Immun. 2006;74(3):1967–72. doi: 10.1128/IAI.74.3.1967-1972.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lenhart TR, Akins DR. Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol Microbiol. 2009;75:692–709. doi: 10.1111/j.1365-2958.2009.07015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X, Yang X, Kumar M, Pal U. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J Infect Dis. 2009;200(8):1318–30. doi: 10.1086/605846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu H, He M, He JJ, Yang XF. Role of the surface lipoprotein BBA07 in the enzootic cycle of Borrelia burgdorferi. Infect Immun. 78(7):2910–8. doi: 10.1128/IAI.00372-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nowalk AJ, Gilmore RD, Jr, Carroll JA. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect Immun. 2006;74(7):3864–73. doi: 10.1128/IAI.00189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Porcella SF, Fitzpatrick CA, Bono JL. Expression and immunological analysis of the plasmid-borne mlp genes of Borrelia burgdorferi strain B31. Infect Immun. 2000;68(9):4992–5001. doi: 10.1128/iai.68.9.4992-5001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Labandeira-Rey M, Baker EA, Skare JT. VraA (BBI16) protein of Borrelia burgdorferi is a surface-exposed antigen with a repetitive motif that confers partial protection against experimental Lyme borreliosis. Infect Immun. 2001;69(3):1409–19. doi: 10.1128/IAI.69.3.1409-1419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mulay VB, Caimano MJ, Iyer R, Dunham-Ems S, Liveris D, Petzke MM, Schwartz I, Radolf JD. Borrelia burgdorferi bba74 is expressed exclusively during tick feeding and is regulated by both arthropod- and mammalian host-specific signals. J Bacteriol. 2009;191(8):2783–94. doi: 10.1128/JB.01802-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marzoa J, Abel A, Sanchez S, Chan H, Feavers I, Criado MT, Ferreiros CM. Analysis of outer membrane porin complexes of Neisseria meningitidis in wild-type and specific knock-out mutant strains. Proteomics. 2009;9(3):648–56. doi: 10.1002/pmic.200800486. [DOI] [PubMed] [Google Scholar]
- 75.Wittig I, Karas M, Schagger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007;6(7):1215–25. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 76.Coburn J, Fischer JR, Leong JM. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol Microbiol. 2005;57(5):1182–95. doi: 10.1111/j.1365-2958.2005.04759.x. [DOI] [PubMed] [Google Scholar]
- 77.de Silva AM, Fikrig E. Arthropod- and host-specific gene expression by Borrelia burgdorferi. J Clin Invest. 1997;99(3):377–9. doi: 10.1172/JCI119169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosa PA, Tilly K, Stewart PE. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat Rev Microbiol. 2005;3(2):129–43. doi: 10.1038/nrmicro1086. [DOI] [PubMed] [Google Scholar]
- 79.Schwan TG. Temporal regulation of outer surface proteins of the Lyme-disease spirochaete Borrelia burgdorferi. Biochem Soc Trans. 2003;31(Pt 1):108–12. doi: 10.1042/bst0310108. [DOI] [PubMed] [Google Scholar]
- 80.Xu Q, McShan K, Liang FT. Essential protective role attributed to the surface lipoproteins of Borrelia burgdorferi against innate defences. Mol Microbiol. 2008;69(1):15–29. doi: 10.1111/j.1365-2958.2008.06264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.