Abstract
BACKGROUND
Helicobacter pylori infection is a risk factor for the development of gastric cancer, and the bacterial oncoprotein CagA contributes to gastric carcinogenesis.
METHODS
We analyzed H. pylori isolates from persons in Colombia and observed that there was marked variation among strains in levels of CagA expression. To elucidate the basis for this variation, we analyzed sequences upstream from the CagA translational initiation site in each strain.
RESULTS
A DNA motif (AATAAGATA) upstream of the translational initiation site of CagA was associated with high levels of CagA expression. Experimental studies showed that this motif was necessary but not sufficient for high-level CagA expression. H. pylori strains from a region of Colombia with high gastric cancer rates expressed higher levels of CagA than did strains from a region with lower gastric cancer rates, and Colombian strains of European phylogeographic origin expressed higher levels of CagA than did strains of African origin. Histopathological analysis of gastric biopsy specimens revealed that strains expressing high levels of CagA or containing the AATAAGATA motif were associated with more advanced precancerous lesions than those found in persons infected with strains expressing low levels of CagA or lacking the AATAAGATA motif.
CONCLUSIONS
CagA expression varies greatly among H. pylori strains. The DNA motif identified in this study is associated with high levels of CagA expression, and may be a useful biomarker to predict gastric cancer risk.
IMPACT
These findings help to explain why some persons infected with cagA-positive H. pylori develop gastric cancer and others do not.
Keywords: Helicobacter pylori, CagA, gastric cancer, Colombia
INTRODUCTION
Helicobacter pylori is a gram negative bacterium that persistently colonizes the human gastric mucosa. Most H. pylori-infected persons remain asymptomatic, but the presence of H. pylori is associated with an increased risk of gastric cancer. The clinical outcome of H. pylori infection is determined by a combination of bacterial, host, and environmental factors (1–6).
An important virulence factor produced by some H. pylori strains but not others is the CagA protein (7, 8). Epidemiological studies have shown that infection with cagA-positive strains is associated with an increased risk of gastric cancer, compared to infection with cagA-negative strains (9–14). CagA is an antigenic protein that is translocated into host cells by a type IV secretion apparatus (15–19). Upon entry into host cells, CagA is tyrosine phosphorylated by Src kinases at conserved EPIYA motifs within the CagA protein. CagA interacts with multiple host cell components and causes numerous alterations in cell signaling and morphology (17, 20–23).
The incidence of gastric cancer varies considerably throughout the world. Within Colombia, two regions located less than 200 km apart in the state of Nariño have a markedly different incidence of gastric cancer. In a study conducted in 1976, the incidence of gastric cancer in the Andean mountainous region was estimated to be 150 cases per 100,000 inhabitants, whereas in the coastal region, gastric cancer rates were estimated to be 6 cases per 100,000 inhabitants (24, 25). The H. pylori infection rates are very similar in the populations inhabiting these regions (26). The prevalence of cagA-positive strains in the low risk region is slightly (~ 9%) lower than that found in the high risk region (27), but this difference is unlikely to account for the 25-fold difference in gastric cancer rates. CagA proteins with a greater number of EPIYA motifs undergo more tyrosine phosphorylation and potentially have increased intracellular activity compared to proteins with fewer EPIYA motifs (28–30), but the number of CagA EPIYA motifs is not significantly different in strains from the low risk and high risk regions of Colombia (31). Recently, it was noted that cagA-positive strains found in the low and high risk regions of Colombia differ in phylogeographic origin (32). Multilocus sequence typing (MLST) analysis indicated that all of the tested H. pylori strains from the high-risk region were of European origin, whereas the majority of strains from the low-risk region were of African origin (32).
In this study, we investigated CagA expression in H. pylori strains isolated from Colombian patients, and observed that there is considerable variation in the levels of CagA expressed by different strains. We identified a motif upstream of the CagA ATG initiation site that is present more frequently in strains expressing high levels of CagA than in strains expressing low levels of CagA, and we demonstrate experimentally that this motif is a determinant of CagA expression levels. We report that H. pylori strains of European and African phylogeographic origin differ in the levels of CagA expressed. Finally, we report that, in comparison to H. pylori strains expressing low levels of CagA, strains expressing higher levels of CagA are associated with more advanced precancerous lesions in gastric biopsy samples.
MATERIALS and METHODS
Bacterial strains
H. pylori strains used in this study were isolated from antral mucosa biopsy samples from subjects in the state of Nariño, Colombia, and H. pylori strain 26695 (33) was used as a reference strain. The strains analyzed in this study were randomly selected from a previously characterized set of Colombian strains (32) using a blinded code. All strains were cagA-positive and contain type s1m1 vacA (32). Several strains were excluded from analysis due to contamination or failure to grow in liquid broth culture. The 36 strains analyzed in this study (17 from subjects in a region of low cancer risk and 19 from subjects in a region of high cancer risk) are shown in Table 1. H. pylori strains were grown in room air supplemented with 5% CO2 at 37° C. For routine growth, H. pylori strains were maintained on trypticase soy agar plates supplemented with sheep blood. In addition, H. pylori strains were grown in modified Brucella broth containing 5% fetal bovine serum (FBS) [BB-FBS] or on BB-FBS agar plates. When necessary, BB-FBS agar plates were supplemented with metronidazole (7.5 µg/ml) or chloramphenicol (5 µg/ml). Escherichia coli strains were grown on Luria Bertani medium. When necessary, the E. coli culture medium was supplemented with ampicillin (50 µg/ml) or chloramphenicol (25 µg/ml).
Table 1.
Expression of CagA in Colombian H. pylori strains
| strain | CagA expressiona |
Number of AATAAGATA motifsb |
Regionc | MLST typed | Histopathology scoree |
|---|---|---|---|---|---|
| 5001 | 1.0 | 0 | low | hpAfrica1 | 2.3 |
| 5004 | 1.4 ± 0.2 | 0 | low | hpAfrica1 | 2.3 |
| 5005 | 0.4 ± 0.2 | 0 | low | hpAfrica1 | 1.7 |
| 5007 | 1.4 ± 0.3 | 0 | low | hpAfrica1 | 2.0 |
| 5009 | 1.4 ± 0.2 | 1 | low | hpEurope | 1.7 |
| 5010 | 0.5 ± 0.3 | 0 | low | hpAfrica1 | 2.0 |
| 5016 | 1.8 ± 0.2 | 0 | low | hpAfrica1 | 3.5 |
| 5017 | 0.9 ± 0.3 | 0 | low | hpAfrica1 | 2.3 |
| 5018 | 1.6 ± 0.5 | 1 | low | hpAfrica1 | 3.5 |
| 5022 | 0.9 ± 0.3 | 0 | low | European | 4.5 |
| 5024 | 1.9 ± 0.1 | 0 | low | hpAfrica1 | 2.0 |
| 5028 | 1.1 ± 0.4 | 1 | low | hpEurope | 3.8 |
| 5032 | 2.3 ± 0.2 | 1 | low | hpEurope | 3.5 |
| 5038 | 3.2 ± 0.5 | 1 | low | hpEurope | 3.5 |
| 5043 | 3.7 ± 0.7 | 1 | low | hpEurope | 5.3 |
| 5051 | 2.4 ± 0.2 | 1 | low | hpAfrica1 | 2.0 |
| 5054 | 1.6 ± 0.6 | 0 | low | hpAfrica1 | 2.3 |
| 5056 | 4.0 ± 0.8 | 1 | high | hpEurope | 4.5 |
| 5057 | 2.4 ± 0.4 | 1 | high | hpEurope | 3.5 |
| 5068 | 2.4 ± 0.4 | 1 | high | hpEurope | 4.3 |
| 5069 | 3.5 ± 0.6 | 1 | high | hpEurope | 4.3 |
| 5072 | 0.7 ± 0.2 | 1 | high | hpEurope | 3.5 |
| 5074 | 5.1 ± 1.1 | 1 | high | hpEurope | 4.7 |
| 5080 | 0.5 ± 0.3 | 1 | high | hpEurope | 3.5 |
| 5082 | 4.4 ± 1.3 | 1 | high | hpEurope | 4.8 |
| 5086 | 4.8 ± 1.1 | 1 | high | hpEurope | 4.6 |
| 5090 | 1.0 ± 0.2 | 0 | high | hpEurope | 2.0 |
| 5091 | 1.9 ± 0.2 | 1 | high | hpEurope | 4.5 |
| 5093 | 3.5 ± 0.3 | 1 | high | hpEurope | 4.5 |
| 5095 | 2.0 ± 0.2 | 1 | high | hpEurope | 2.0 |
| 5096 | 1.5 ± 0.1 | 0 | high | hpEurope | 4.3 |
| 5097 | 1.0 ± 0.3 | 1 | high | hpEurope | 4.4 |
| 5100 | 2.2 ± 0.1 | 1 | high | hpEurope | 2.3 |
| 5104 | 4.7 ± 0.6 | 1 | high | hpEurope | 3.5 |
| 5112 | 2.2 ± 0.1 | 1 | high | hpEurope | 4.3 |
| 5114 | 0.9 ± 0.3 | 0 | high | hpEurope | 4.4 |
CagA expression was analyzed by immunoblotting, and relative levels of expression were quantified by comparing the CagA signal intensity of each strain with that of a standard (CagA from H. pylori strain 5001). For each strain, the CagA signal was calculated for 3 independent biological samples, and the average of these values is presented. Values represent mean ± SEM.
The number of AATAAGATA motifs upstream of the ATG translation initiation site is shown.
H. pylori strains were isolated from regions of Colombia with either high risk or low risk for the development of gastric cancer.
MLST analysis (31) was previously used to characterize the ancestral origin of the H. pylori strains. hpAfrica1 denotes isolates classified as either West African (hspWAfrica) or South African (hspSAfrica).
Histopathology scores describing the severity of lesions observed in gastric tissue from which the H. pylori strains were isolated.
Human subjects
The H. pylori strains used in this study were obtained from 36 male subjects between the ages of 39 and 60 years with dyspeptic symptoms who underwent gastrointestinal tract endoscopy in two public hospitals in the State of Nariño, Colombia (32). The hospitals are located in two cities with contrasting gastric cancer risks: Túquerres in the Andes Mountains where gastric cancer incidence is high, and Tumaco on the coast where gastric cancer incidence is low. The 36 individuals included in this study were part of a larger series of subjects that has been described previously (32).
Gastric mucosa biopsy samples from the 36 subjects were obtained from the antrum, incisura angularis, and corpus, and embedded in paraffin for histology. One additional antral biopsy from each subject was immediately frozen in glycerol and thioglycolate for H. pylori culture, and kept at −80°C. The samples were shipped on dry ice to Vanderbilt University (Nashville, Tennessee, USA) for analysis. All participants provided informed consent and all samples were sent to Vanderbilt University without identifiers. The protocol was approved by the Committees on Ethics of Universidad del Valle and Hospital Departamental in Nariño, Colombia and by the Institutional Review Board at Vanderbilt University.
Mutagenesis of the cagA promoter region in H. pylori 26695
To facilitate introduction of unmarked mutations into H. pylori, a cat-rdxA cassette was synthesized and cloned into pUC57 vector (Genscript). This cassette confers resistance to chloramphenicol mediated by the chloramphenicol acetyl-transferase (cat) gene from Campylobacter coli, and susceptibility to metronidazole is mediated by an intact rdxA gene (HP0954) from H. pylori 26695 (34). To generate unmarked mutant strains, we used a method that is a variant of counterselection methods used previously in H. pylori (35, 36). As a first step, the rdxA gene, which confers resistance to metronidazole, as well as approximately 0.5 kb of flanking DNA on each side, was PCR-amplified from H. pylori 26695 and cloned into pGEM-T Easy to yield pMM670. By using pMM670 as the template for inverse PCR and then ligating the PCR product, we generated a modified plasmid (pMM672) in which the coding region of rdxA was deleted. H. pylori 26695 was transformed with pMM672 (which is unable to replicate in H. pylori) and metronidazole-resistant colonies were selected (34). PCR analysis confirmed that the rdxA locus was deleted from a mutant strain designated H. pylori 26695ΔrdxA.
We next modified H. pylori ΔrdxA to generate a strain (26695 cagA::catrdx-9) containing a cat-rdxA cassette immediately downstream of the ATG intiation site of the cagA gene. To generate this mutant, primers 5’-GTCGTTGTAGAATTGTAGATCTTTAGG-3' and 5’-TATCCAACCAATCCCCACCAG-3' were utilized to PCR amplify a DNA fragment from H. pylori 26695 comprising approximately 650 bp upstream and 650 bp downstream of the cagA translational start site. This PCR product was cloned into pGEMT-Easy (Promega), and the resultant plasmid was used as a template for inverse PCR with primers that introduced a BamHI site immediately following the cagA translation start site. The resultant plasmid was digested with BamHI and a cassette encoding the cat-rdxA cassette (described above) was ligated into the restriction site. The cagA::cat-rdxA plasmid, which is unable to replicate in H. pylori, was transformed into H. pylori 26695 ΔrdxA, and single colonies resistant to chloramphenicol (5 µg/ml) but sensitive to metronidazole (7.5 µg/mL) were selected. Transformant 26695 cagA::catrdx-9 was selected for further studies. Immunoblot analysis revealed the loss of CagA expression in this strain, and the insertion of the cat-rdxA cassette into the cagA gene was confirmed by PCR amplification and DNA sequencing. To introduce alterations into the cagA promoter region, 26695 cagA::catrdx-9 was transformed with various plasmids described below, and transformants resistant to metronidazole were selected.
cagA promoter swapping experiments
DNA fragments encompassing approximately 0.5 kb upstream and 0.7 kb downstream from the cagA transcription start site (0.6 kb upstream and 0.6 kb downstream of the ATG initation site) were PCR amplified from Colombian strains using primers 5’-GCAAAAACAAACCCAAGCTGA-3’ and 5’-ACCAGTAGGCCCTCCATTTTTTTC-3’, and the amplified products were cloned into the pGEMT-Easy (Promega) vector. The resultant plasmids, which are unable to replicate in H. pylori, were then used to transform H. pylori strain 26695 cagA::catrdx-9 (described above). Transformations were plated onto metronidazole containing BB-FBS agar plates, and metronidazole resistant clones were isolated. These metronidazole resistant colonies represent recombination events resulting in the removal of the cat-rdxA cassette from the cagA ORF of H. pylori strain 26695 cagA::catrdx-9. As the removal of the cat-rdxA insertion should result in the restoration of an intact cagA gene, all H. pylori transformants were subsequently screened for CagA expression. To ensure that the 26695 cagA promoter region had been replaced with the corresponding wild-type Colombian sequences, the cagA promoter regions of transformants expressing CagA were sequenced.
Targeted mutagenesis of cagA promoter region
To generate mutations in the promoter region of cagA, a 1.2 kb fragment encompassing 0.5 kb of DNA upstream and 0.7 kb downstream of the cagA transcriptional start site was PCR amplified from H. pylori strain 5056 with primers 5’-GCAAAACAAACCCAAGCTGA-3’ and 5’-ACCAGTAGGCCCTCCATTTTTTTC-3’ and cloned into the plasmid vector pGEMT-Easy. Targeted mutagenesis was carried out using the Quick-Change mutagenesis kit (Agilent Technologies). The introduction of mutations into plasmids was confirmed by DNA sequencing. These plasmids were then used to transform H. pylori strain 26695 cagA::catrdx-9 and transformants were selected as described above.
Western blot analysis
To compare the levels of CagA expression in H. pylori Colombian strains, the bacterial strains were inoculated into BB-FBS broth and grown overnight. Cultures were then inoculated into fresh BB-FBS to an initial OD600 of 0.1. Broth cultures were grown for 15 h and harvested; the OD600 of the H. pylori cultures at the time of harvest was ~ 0.5 to 0.6, corresponding to cultures in logarithmic phase. H. pylori cells were lysed using NP-40 lysis buffer (37, 38), and each sample (5 µg of protein) was subjected to SDS-PAGE and western blot analysis. CagA expression was analyzed using a 1:10,000 dilution of a CagA-specific polyclonal antibody generated against amino acids 1–300 (Santa Cruz Biotechnology), or a 1:30,000 dilution of an independent polyclonal anti-CagA antiserum generated against a larger segment of CagA (amino acids 1–880). Goat anti-rabbit horseradish peroxidase (HRP) (1: 6,000) was used as a secondary antibody and immunoreactive bands were visualized using a chemiluminescence kit (GE healthcare). Following CagA analysis, blots were treated with Restore Western Blot Stripping Buffer (Thermo), and re-probed with antiserum generated against soluble proteins of H. pylori [anti-HP, 1:10,000, (39)]. To quantify levels of CagA expression, we compared the CagA signal in each sample to the CagA signal of a reference strain (strain 5001). Densitometry was performed using Image J software (NIH), and background signal levels were subtracted from measurements of CagA-specific signal intensities. For each strain, three independent samples were analyzed, and the average of these values was calculated.
Real-time PCR
Total RNA was isolated from H. pylori using Trizol Reagent (Gibco) according to manufacturer’s instructions. The RNA was digested with RQ1 RNAse free DNAse (Promega), and the RNA samples were then subjected to a clean-up step using RNAeasy columns (Qiagen). cDNA synthesis was performed on 100 ng of purified RNA using the first strand cDNA synthesis kit (Biorad). As controls, first strand cDNA reactions were carried out in parallel without reverse transcriptase. The cDNA and control reactions were diluted 1:20 and used in real-time PCR reactions. Real-time PCR was performed using an ABI Real-Time PCR machine, with SYBR green as the fluorochrome. Abundance of transcript was calculated using the ΔΔCT method, with each transcript signal normalized to the abundance of the 16S rRNA internal control. The normalized transcript signal for each sample was then compared to similarly normalized values obtained with the 26695 wild-type strain. The primers used for real-time analysis are as follows:
16S rRNA: 5’-GGAGTACGGTCGCAAGATTAAA-3’ and 5’-CTAGCGGATTCTCTCAATGTCAA-3’; cagA: 5’-GAGTCATAATGGCATAGAACCTGAA-3’ and 5’-TTGTGCAAGAAATTCCATGAAA-3’; HP1588 (encoding a hypothetical protein) : 5’-GCTGTCATTGTTGCGAATGC-3’ and 5’-TGCAATATCAATCGCTGTCCATA-3’ and ureA (encoding urease subunit A): 5’- GAAGACATCACTATCAACGAAGGCAA-3’ and 5’-GTTACCGCCAATGTCAATCAA-3’.
Analysis of histopathology
For each of the 36 subjects from whom H. pylori strains were isolated, a global gastric histologic diagnosis was determined independently by two pathologists (PC and MBP). This diagnosis was based on analysis of all gastric biopsies from antrum, incisura angularis, and corpus, according to the updated Sydney system for the classification of gastritis (40) and to the Padova International Classification for dysplasia (41). Both pathologists were blinded to the geographic origins of the subjects and data pertaining to H. pylori isolates. Global diagnoses were assigned on an ordinal scale from 1 to 6, as follows: 1, mild to moderate non-atrophic gastritis; 2, severe non-atrophic gastritis; 3, multifocal atrophic gastritis without intestinal metaplasia (MAG); 4, intestinal metaplasia (IM); 5, dysplasia; and 6, adenocarcinoma. A previously validated histopathology scoring system taking into account the extent and degree of atrophic, metaplastic and dysplastic changes was also used to quantify differences in morphological variables within each global diagnosis category. The assignment of these values was accomplished as described previously (32).
RESULTS
Expression of CagA in Colombian H. pylori strains
This study began with an analysis of CagA expression in H. pylori strains isolated from subjects in Colombia. We detected considerable variation in the levels of CagA expressed by these strains. For example, immunoblotting studies revealed that CagA expression in strains 5001, 5004, 5005 and 5007 was lower than CagA expression in strains 5043, 5056, 5074 and 5093 (Figure 1A), and this difference was recapitulated when the samples were immunoblotted with a different CagA antiserum (data not shown). The relative level of CagA expression was quantified by comparing the CagA signal of each H. pylori strain to the CagA signal of a strain selected as a reference (strain 5001). As shown in Table 1, the relative expression levels of CagA expression in the panel of Colombian strains ranged from a minimum of 0.4 to a maximum of 5.1.
Figure 1. Heterogeneity among H. pylori strains in expression of CagA.
A. H. pylori strains were cultured in broth and lysed as described in the Methods. Cell extracts were standardized by protein concentration and immunoblotted to detect CagA. Blots then were stripped and reprobed with an anti-H. pylori antiserum. B. H. pylori 26695 cagA::catrdx-9 was transformed with 1.2 kb DNA fragments derived from the indicated Colombian strains (500 bp upstream and 700 bp downstream of the cagA transcriptional start site). The resulting transformants were cultured as described in the Methods, and were analyzed for CagA expression as described above. C. Analysis of transformants depicted in panel B. Nucleotide numbers are relative to the cagA transcriptional start site, and designate the chromosomal region in H. pylori 26695 that was replaced with sequences derived from Colombian strains. Vertical lines indicate the cagA transcriptional start site and the ATG translation initiation site. D. This schematic illustrates the predicted cagA transcriptional start site (TS), an AT-rich inverse repeat identified in the cagA promoter region of H. pylori 26695, and the ATG translation initiation site.
Strain-specific sequences upstream from cagA influence levels of CagA expression
We next sought to determine if the observed heterogeneity in levels of CagA expression among Colombian strains was attributable to variations in sequences upstream of the cagA ATG translation initiation site. As a first approach, we amplified 1.2 kb sequences (0.5 kb upstream and 0.7 kb downstream of the cagA transcriptional start site) from the 8 Colombian strains described above, and introduced these sequences into H. pylori strain 26695 cagA::catrdx-9 (Figure 1B). These experiments resulted in replacement of the 26695 sequence with a minimum of 146 bp upstream of the cagA transcriptional start site (or 250 bp upstream of the cagA ATG initiation site), derived from Colombian strains. The regions of the H. pylori 26695 genome replaced with sequences derived from Colombian strains are shown in Figure 1C. As shown in Figure 1B, H. pylori 26695 cagA::catrdx-9 transformants containing DNA derived from strains 5001, 5004, 5005 and 5007 expressed relatively low levels of CagA compared to transformants containing DNA derived from strains 5043, 5056, 5074 and 5093. Thus, the levels of CagA expression observed in transformants correlated with the relative levels of CagA expression in the corresponding wild-type strains that were the sources of transformed DNA (Figure 1A, B). These results indicate that variation among H. pylori strains in levels of CagA expression is attributable to variations in the region of DNA incorporated into H. pylori 26695 cagA::catrdx-9. The DNA incorporated into H. pylori 26695 cagA::catrdx-9 from Colombian strains contains the transcriptional initiation site of cagA, the cagA promoter, an AT-rich inverted repeat important for cagA transcription (42), and the ATG translation initiation site (Figure 1D).
Comparative analysis of sequences upstream from cagA
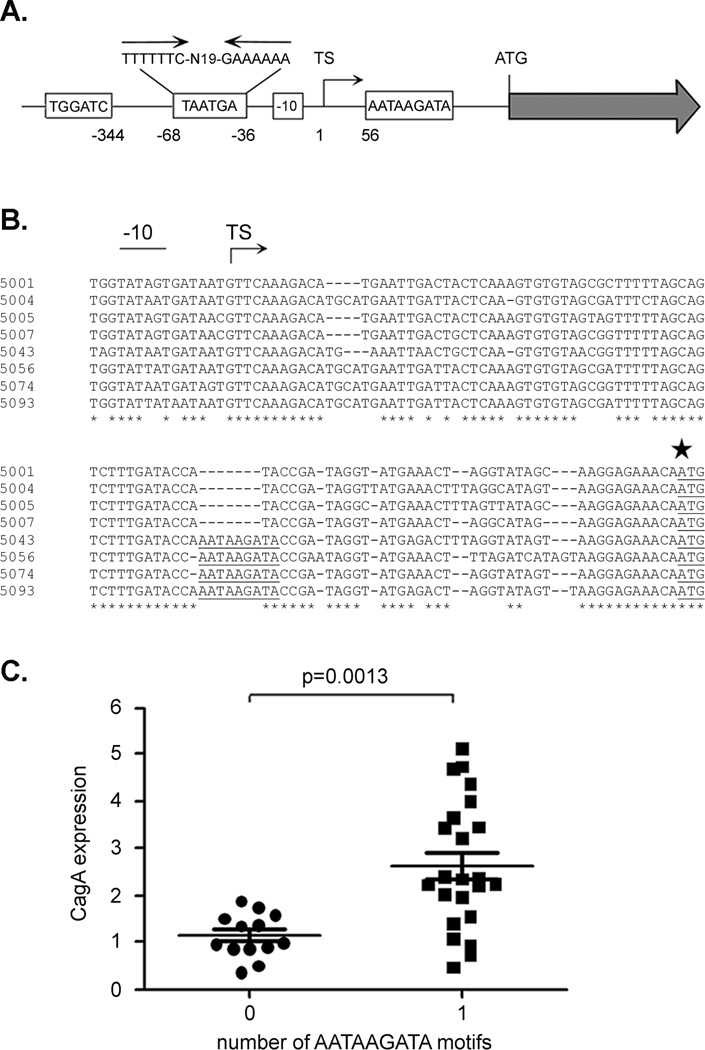
Next, we analyzed the nucleotide sequences upstream from cagA in strains expressing high levels of CagA and strains expressing low levels of CagA, in an effort to identify sequence differences that might account for the differences in CagA expression. Specifically, we sequenced and analyzed a ~ 1.2 kb region (0.5 kb upstream and 0.7 kb downstream of the transcriptional start site) from the 8 strains described above (4 strains expressing relatively high levels of CagA and 4 strains expressing relatively low levels of CagA). Several differences among strains were identified upstream of the cagA transcriptional start site (Figure 2A). These differences include variations in the number of copies of a TGGATC motif (1 or 2 copies) located 344 bp upstream of the transcriptional start site, and variation in the number of copies of a TAATGA motif (0, 1, 2 copies) located within an AT-rich region of the cagA promoter that contains an inverted repeat (Figure 2A). The predicted -10 promoter sequences also differed among the strains; the observed -10 sequences included TATAATGA, TATAGTGA, TATTATGA and TATTATAA (Figure 2B). In addition, there was variation among strains in the presence of aTGn motif (43, 44) located immediately upstream from the -10 promoter sequence. Finally, an examination of a region located between the transcriptional start site and the ATG initiation site revealed a region of nucleotide sequence heterogeneity that correlated with CagA expression levels. Specifically, an AATAAGATA motif located approximately 56 nucleotides downstream of the transcription start site (40 nucleotides upstream of the cagA ATG initiation codon) (Figure 2A) was present in the strains expressing high levels of CagA (5043, 5056, 5074 and 5093), but was absent in the strains expressing low levels of CagA (5001, 5004, 5005 and 5007) (Figure 2B).
Figure 2. Analysis of an AATAAGATA motif.
A. Regions upstream from cagA that exhibit substantial sequence heterogeneity among Colombian H. pylori strains are illustrated. The Colombian strains vary in the number of copies of TGGATC, TAATGA, and AATAAGATA motifs. The TAATGA motifs are located within an AT-rich region of the cagA promoter that contains an inverted repeat. Variations in the cagA -10 sequence are also observed among the Colombian strains. The location of each motif relative to the cagA transcriptional start site is shown. TS denotes the position of the transcriptional start site. B. Sequences immediately upstream of the cagA ATG initiation site (indicated by a large star) in 8 H. pylori strains were aligned. Four strains express relatively low levels of CagA (5001, 5004, 5005, 5007) and four express relatively high levels of CagA (5043, 5056, 5074, 5093). The AATAAGATA motif is underlined and is present in four strains that express high levels of CagA. The -10 sequences and transcriptional start site (TS) are also shown. C. CagA expression in each strain was analyzed by immunoblotting, and levels of CagA expression were quantified by comparing the CagA signal intensity of each strain to that of a standard (CagA in strain 5001). The relationship between number of AATAAGATA motifs and level of CagA expression is shown. The p-value was determined by t-test.
We sequenced the same regions from 28 additional H. pylori strains, scoring the strains for the presence or absence of the AATAAGATA motif. Of the 36 sequenced regions, 23 were found to contain the AATAAGATA motif, while the remaining 13 did not contain the motif (Table 1). A comparison of the level of CagA expression with the presence or absence of the AATAAGATA motif is shown in Figure 2C and Table 1. The presence of the AATAAGATA motif is associated with higher levels of CagA expression, and the absence of the motif is associated with lower levels of CagA expression [Figure 2C, p= 0.0013 (t-test)]. Among 23 strains possessing this motif, 16 expressed CagA at a level that was at least 2-fold greater than the level of CagA expressed by strain 5001 (Table 1). In contrast, all 13 strains lacking this motif expressed relatively low levels of CagA (less than 2-fold greater than the level expressed by strain 5001) (p=0.0006, Fisher’s exact test). No significant association was observed between levels of CagA expression and other sites of heterogeneity illustrated in Figure 2A, including the type of -10 sequence (p=0.15, ANOVA), the presence of a TGn motif corresponding to an extended -10 promoter sequence (p = 0.16, t-test), the number of copies of TGGATC motifs (p=0.68, t-test), or the number of copies of TAATGA motifs (p=0.96, t-test; 0, 1 versus 2).
Targeted mutagenesis of the AATAATAGA motif
Next we sought to experimentally test whether the AATAAGATA motif influenced the level of CagA expression. To do this, we started with a cloned 1.2 kb DNA segment (0.5 kb upstream and 0.7 kb downstream of the cagA transcriptional start site) derived from strain 5056, which, when introduced into strain 26695 cagA::catrdx-9, resulted in higher levels of CagA expression than what was observed in wild-type H. pylori 26695 (Figure 1B). Two separate site-specific mutations were introduced into the AATAAGATA motif of the cloned 5056 DNA. These nucleotide changes involved changing the AATAAGATA motif to either AATAAATAG (designated Mut1) or AAATTGATA (designated Mut2) (Figure 3A). The 5056 sequences (wild-type and mutant derivatives) were introduced into H. pylori strain 26695 cagA::catrdx-9 by natural transformation. A minimum of 355 bp upstream of the transcriptional start site (459 bp upstream of the ATG initiation site) was introduced into H. pylori 26695 cagA::catrdx-9, resulting in the introduction of the 5056 cagA promoter plus wild-type or mutant variants of the AATAAGATA motif (Figure 3B). Consistent with previous results (Figure 1), the introduction of wild-type 5056 DNA into H. pylori strain 26695 cagA::catrdx-9 resulted in a marked increase in CagA expression compared to what was observed in wild-type H. pylori 26695 (Figure 3C, D). In contrast, transformants harboring 5056 DNA containing mutations Mut1 and Mut2 expressed significantly lower levels of CagA (Figure 3C). This provided evidence that the AATAAGATA motif influences levels of CagA expression.
Figure 3. Analysis of CagA expression in 26695 transformants harboring cagA promoter region sequences derived from Colombian strains.
A. A cloned DNA segment containing 0.5 kb of DNA upstream and 0.7 kb downstream of the cagA transcriptional start site from strain 5056 was mutated so that changes (designated Mut1 and Mut2) were introduced into either the AATAAGATA motif (underlined in the 5056 WT sequence) or nucleotides downstream of this motif (Mut3). The mutated nucleotides in Mut1, Mut2 and Mut3 are underlined. Mutated plasmid DNA was transformed into H. pylori 26695 cagA::catrdx-9. B. This panel illustrates the segment of transformed DNA (wild-type, Mut1, Mut2, or Mut3) that was incorporated into the chromosome of H. pylori strain 26695 cagA::catrdx-9. The locations of the cagA promoter region, transcriptional start (TS), and an AATAAGATA motif relative to the cagA translation initiation site are shown. C. Western blot analysis of CagA expression in wild-type H. pylori 26695, 26695 cagA::catrdx-9 transformants harboring a DNA sequence derived from wild-type strain 5056, or transformants harboring 5056 sequences with Mut1 or Mut2 changes. D. Western blot analysis of H. pylori 26695 cagA::catrdx-9 transformants harboring a DNA sequence derived from wild-type strain 5056, or transformants harboring a 5056 sequence with the Mut3 change. E. Real-time PCR analysis of transcript levels for cagA and two controls (HP1588 and ureA) in 26695 cagA::catrdx-9 transformants harboring sequences derived from strain 5056 (wild-type or mutated sequences containing Mut1, Mut2, or Mut3 changes). Real-time PCR was performed as described in the Methods. Mean ± standard deviation from 3 independent samples is shown.
To verify that alterations in the AATAAGATA motif resulted in alterations of cagA transcription, real-time PCR was performed to measure cagA transcript levels in the aforementioned H. pylori 26695 transformants. Consistent with results obtained at the protein level, H. pylori strains transformed with DNA from wild-type strain 5056 showed a significant increase in cagA mRNA levels compared to that of wild-type strain 26695 (Figure 3E). In contrast, little or no increase in cagA transcript levels was observed in transformants harboring 5056 sequences that contain mutations to either the TAA or GATA portions of the AATAAGATA motif. This provided further evidence that the AATAAGATA motif influences levels of CagA transcription.
Targeted mutagenesis of additional nucleotides surrounding the AATAAGATA motif
H. pylori strain 26695 consistently expresses relatively low levels of CagA (Figure 1B), but an analysis of nucleotide sequences upstream of the 26695 cagA ATG initiation site revealed the presence of the AATAAGATA motif that is associated with high CagA expression (Figure 3A). Similarly, we found that some Colombian strains failed to produce high levels of CagA, despite harboring the AATAAGATA motif (Table 1). We sought to elucidate the reason for this apparent discrepancy by further studying strain 26695. Careful inspection of the 26695 sequence revealed that even though it contained the AATAAGATA motif, it differed from strains expressing high levels of CagA in nucleotides just downstream of the AATAAGATA motif. For example, a nucleotide sequence CCGAATAGGTAT immediately follows the AATAAGATA motif in strain 5056. In contrast, in H. pylori 26695, the AATAAGATA motif is followed by the nucleotide sequence CCGATAGTAT (Figure 3A, compare 5056 wild-type with 26695 wild-type). We evaluated the importance of these underlined nucleotides by altering the CCGAATAGGTAT nucleotide sequence of 5056 to CCGATAGTAT (Mut3), thus mimicking the nucleotide sequence surrounding the AATAAGATA motif in strain 26695. As shown in Figure 3D, introduction of the altered sequence (Mut3) into strain 26695 cagA::catrdx-9 resulted in a lower level of CagA expression than was observed following introduction of the wild-type sequence from strain 5056 into strain 26695 cagA::catrdx-9. Consistent with the results of immunoblotting studies, cagA transcript levels were significantly decreased in H. pylori transformants harboring the Mut3 mutation (Figure 3E). These results indicate the importance of not only the AATAAGATA motif, but also additional nucleotides located downstream of this motif in determining the levels of CagA expressed.
Analysis of CagA expression and the AATAAGATA motif in strains from two regions of Colombia
The H. pylori strains used in these studies originated from two regions in Colombia (Túquerres and Tumaco), characterized by a high risk or low risk for the development of gastric cancer, respectively. Therefore, we sought to determine whether there were any detectable differences in levels of CagA expression in strains from the two regions. As shown in Figure 4A, strains isolated from patients in the high risk region expressed higher levels of CagA than did strains isolated from the low risk region (p=0.028). We also detected differences in the presence of the AATAAGATA motif in strains from the two regions (Fig. 4B). The AATAAGATA motif was detected more commonly in H. pylori strains from the high risk region (where strains typically express high levels of CagA), than in strains from the low risk region. Among 19 strains from the high risk region, 16 possessed this motif. In contrast, fewer than half of the strains from the low risk region (7 of 17) possessed the AATAAGATA motif (p=0.014) (Table 1, Figure 4B).
Figure 4. Relationships between H. pylori site of isolation, MLST type, and level of CagA expression.
Levels of CagA expression in individual strains were analyzed by immunoblotting, and CagA expression in each strain is expressed as signal intensity relative to a standard (CagA in strain 5001). The cagA promoter region of H. pylori Colombian strains was sequenced and the presence of an AATAAGATA motif located 40 nucleotides upstream of the ATG initiation site was scored for each strain. The H. pylori strains used in the study were isolated from patients living in regions of Colombia with either a low or high risk for gastric cancer, and were previously analyzed by MLST (32). Panels A and B depict the level of CagA expression and presence or absence of the AATAAGATA motif in strains from the two regions of Colombia (high risk or low risk for gastric cancer), while panels C and D depict the level of CagA expression and presence or absence of the AATAAGATA motif in strains of either European or African ancestral origins, based on MLST analysis. p-values (panels A and C) were determined by using Student’s t-test.
Analysis of CagA expression and the AATAAGATA motif in strains of European or African phylogeographic origins
MLST analysis was previously used to determine the phylogeographic origin of Colombian H. pylori strains, and all of the strains analyzed were determined to be either European or African in origin (32). As shown in Figure 4C, strains expressing the highest levels of CagA were European in origin, and the strains of African origin expressed relatively low levels of CagA (p=0.0187). The AATAAGATA motif was present more commonly in European strains (21 out of 25 strains) than in African strains (2 of 11 strains) (p=0.0003) (Figure 4D). When comparing African strains (all from the low risk region) with European strains from the same low risk region, the AATAAGATA motif was again found to be present more commonly in European strains (5 of 6 strains), compared to African strains (2 of 11 strains) (p=0.03). Thus, in comparison to strains of African origin, strains of European origin are more likely to contain the AATAAGATA cagA motif and express high levels of CagA.
Correlation of CagA expression and the AATAAGATA motifs with histopathology scores
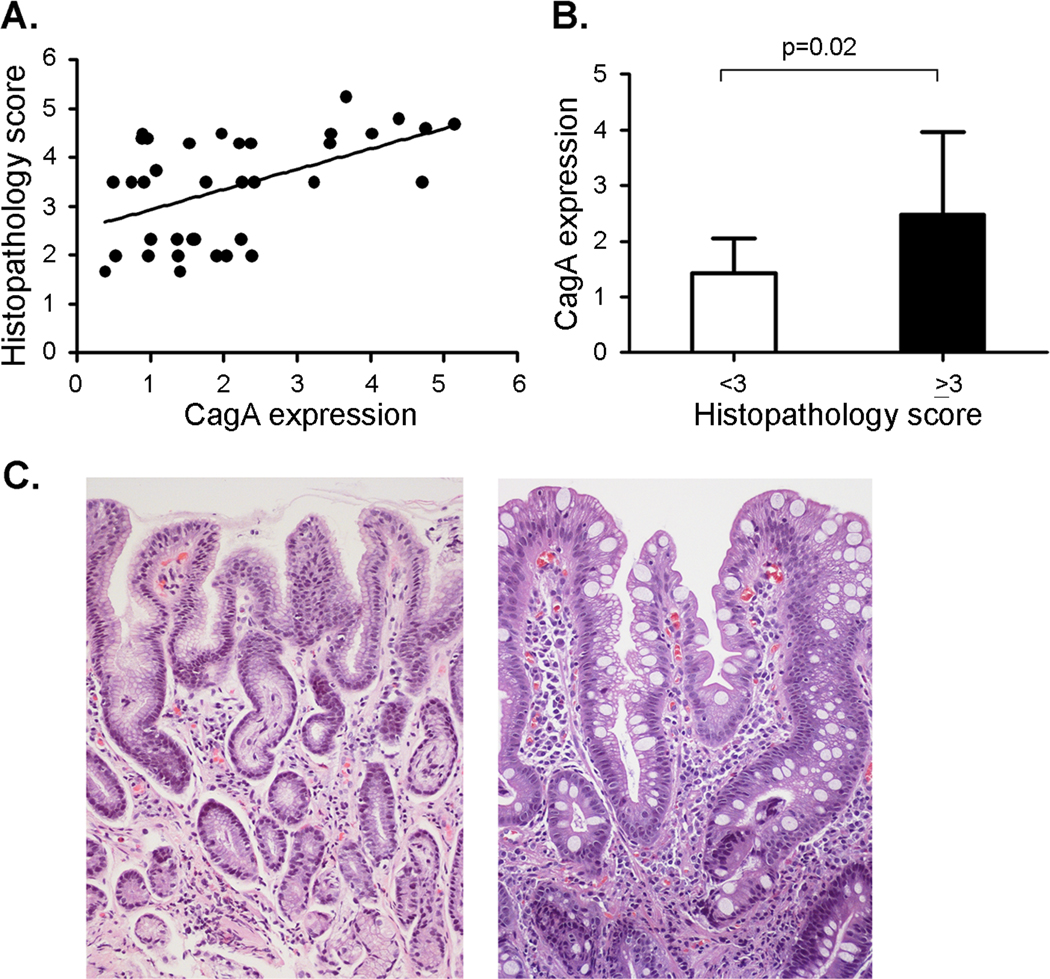
All of the Colombian H. pylori strains analyzed in the current study were isolated from gastric biopsies, and the severity of gastric pathology (focusing on premalignant lesions) was scored as described in the Methods. To determine if levels of CagA expression are associated with severity of gastric disease, we compared the histopathology score of each patient with the levels of CagA expressed by the corresponding H. pylori strain. As shown in Figure 5A, there was a significant correlation between these two parameters (Rs = 0.453, p = 0.0057). H. pylori strains expressing high levels of CagA were consistently associated with high histology scores, and the lowest histology scores were associated with strains expressing low levels of CagA (Figure 5A). Figure 5B analyzes levels of CagA expression in strains from patients with gastric histologic diagnoses that are considered either precancerous (histology score ≥ 3, indicative of atrophic gastritis with and without intestinal metaplasia or dysplasia) or non-precancerous (histology score <3, indicative of non-atrophic gastritis). The level of CagA expression in H. pylori strains from patients with precancerous lesions was significantly higher than the level of CagA expression in H. pylori strains from patients with non-precancerous lesions (p=0.02, Figure 5B). Representative images illustrating gastric lesions of varying severity are shown in Figure 5C.
Figure 5. Relationship between levels of CagA expression and gastric histopathology score.
A. Levels of CagA expression in individual strains were analyzed by immunoblotting, and CagA expression in each strain is expressed as signal intensity relative to a standard (strain 5001). Spearman rank order correlation analysis (Sigma stat) indicated that there was a significant correlation between CagA expression levels and the corresponding gastric histopathology scores (Rs = 0.453, p = 0.0057). B. The average CagA expression was calculated for H. pylori strains associated with histology scores < 3 (non-atrophic gastritis, n= 13 strains) or ≥ 3 (precancerous lesions, n=23 strains). p-values were calculated using t-test. Mean ± standard deviation is shown. C. Representative micrographs of H & E-stained gastric tissues. Left panel is from a subject with non-atrophic gastritis (histopathology score 1.67) infected with strain 5005 (Hp Africa1 MLST type, low CagA expression). Right panel is from a subject with intestinal metaplasia (of mixed complete and incomplete types, and histopathology score 4.8) infected with strain 5082 (European MLST type, high CagA expression). In the center of the micrograph there are 2 glands. One (on the left) displays intestinal metaplasia of the incomplete type, with irregular mucus goblets and absence of a brush border; the other one (on the right) is of the complete type with regular mucus goblets and a well-defined brush border. Magnification 200×.
We next analyzed whether the presence of the AATAAGATA motif was associated with histopathology scores (Figure 6). Strains containing the cagA AATAAGATA motif were associated with higher histopathology scores, compared to strains in which the motif is absent (p=0.025, Figure 6A). We also analyzed whether there was any correlation between histology scores and other sites of heterogeneity upstream from cagA, including predicted -10 sequences and the previously identified TGGATC and TAATGA motifs (Figure 2A). As shown in Figure 6, there was no significant correlation between histopathology scores and the number of TGGATC motifs (Figure 6B), number of TAATGA motifs (Figure 6C), or type of -10 sequence (Figure 6D). Therefore, an increased severity of preneoplastic lesions was correlated with both high levels of CagA expression (Figure 5) and the presence of the AATAAGATA motif (Figure 6A).
Figure 6. Relationship between histopathology scores and cagA upstream sequences.
DNA sequencing revealed 4 regions upstream of the cagA ATG initiation site that varied among the different Colombian strains (see Figure 2A). The strains vary in the number of copies of the indicated DNA sequences (Panels A, B, C) or vary in -10 sequences (Panel D). The histology scores are compared with the number of AATAAGATA motifs (Panel A), TGGATC motifs (Panel B), TAATGA motifs (Panel C), and type of -10 sequence (Panel D). p-values were calculated using t-test (Panels A, B, C) or one-way analysis of variance (Panel D).
DISCUSSION
Gastric cancer develops in a small subset of H. pylori-infected persons, and the remainder remain asymptomatic. The risk of gastric cancer is determined by bacterial factors, host factors, and environmental factors (1–6). Numerous studies have shown that cagA-positive H. pylori strains are associated with an increased risk of gastric cancer compared to strains that lack cagA (9–14). The risk of gastric cancer is also determined by several related bacterial features, including sequence variation within CagA (such as the number and type of EPIYA motifs) (28–30), and the presence or absence of a functional cag type IV secretion system (which translocates CagA into host cells) (15–19). In this study, we demonstrate that there is considerable variation among H. pylori strains in expression of CagA, and we provide evidence that the level of CagA expression may be an additional factor that influences gastric cancer risk.
By comparing DNA sequences upstream of the cagA ATG initiation site, we identified an AATAAGATA DNA motif that was present significantly more frequently in strains expressing high levels of CagA than in strains expressing low levels of CagA. All of the strains expressing high levels of CagA contained this motif, whereas only 35% of strains expressing low levels of CagA contained this motif. Experimental studies revealed that the AATAAGATA motif is required for high levels of CagA expression, and sequences immediately downstream from this motif are also required for high-level CagA expression. Collectively, these results indicate that the AATAAGATA motif is necessary but not sufficient for high levels of CagA expression. In future studies, it will be important to precisely map the boundaries of the region that is required for high-level CagA expression. Nucleotide sequences located near promoters are often sites for binding of regulatory proteins, which can positively or negatively regulate gene transcription. In this case, since this AATAAGATA motif is located downstream of the cagA transcriptional start site within the 5’ untranslated region (5’-UTR) (42), at a site somewhat distant from the promoter, it seems relatively unlikely that it serves as a binding site for a regulatory protein. Instead, we speculate that presence of the AATAAGATA motif may confer increased transcript stability.
One possible mechanism by which RNA stability is regulated is through base pairing with small regulatory RNA. The existence of numerous H. pylori sRNAs potentially involved in gene regulation has recently been described (45). In most cases, binding of sRNA to the target mRNA, typically in the 5’-UTR, results in reduced stability of the target mRNA through occlusion of the ribosome binding site and degradation of the mRNA transcript (46). However, at least one example of increased transcript stability mediated by sRNAs has been described (47). In this case, it is thought that in the absence of the sRNAs, a hairpin structure in the 5’ UTR forms, and that this hairpin structure forms a recognition site for RNA degradation. Base pairing of the sRNAs to binding sites in the 5’ UTR results prevents the stem loop structure, leading to increased stability of the mRNA transcript. Given the location of the AATAAGATA motif on the 5’ UTR of CagA, we speculate that a similar phenomenon might be operative with cagA.
The H. pylori strains used in this study were isolated from patients in two regions of Colombia characterized by either a high risk or low risk for the development of gastric cancer. It was recently demonstrated by MLST analysis that cagA-positive H. pylori strains from the high-risk region are predominantly European in origin, and strains from the low-risk region are either African or European in origin (32). In the present study, we observe that H. pylori isolates from the region with a high risk of gastric cancer express higher levels of CagA than do H. pylori isolates from the region with a low cancer risk, and we find that H. pylori isolates of European origin express CagA at higher levels than do H. pylori strains of African origin. Consistent with these observations, the AATAAGATA motif was found predominantly in H. pylori strains from the high risk region and in strains of European origin. Thus far, there are relatively very few robust genetic markers that allow differentiation of African and European strains (48). It will be important in future studies to test additional H. pylori isolates, including H. pylori strains derived from native European or African subjects, and evaluate whether presence of the AATAAGATA consistently correlates with geographic origin of strains and whether this motif provides a robust marker for CagA expression levels.
In a previous study, it was noted that Colombian H. pylori strains of European origin were associated with increased severity of preneoplastic lesions and increased DNA damage in gastric biopsy samples, compared to Colombian strains of African origin (32). In the current study we observed that H. pylori strains expressing the highest levels of CagA were consistently associated with precancerous lesions; in contrast, only about half of the strains expressing lower levels of CagA were associated with precancerous lesions, and the remainder had non-atrophic gastritis. We propose that the differences in gastric cancer rates between the low and high risk regions may be attributed, at least in part, to differences in levels of CagA produced by the corresponding H. pylori strains. The differences in the gastric cancer rates in these two regions of Colombian are also influenced by many other microbial, host, and environmental factors (49–51). For example, persons in the low-risk region typically consume higher amounts of fruit, fresh vegetables, and seafood compared to persons in the high-risk region. Vitamin C, found in fruits and vegetables, and selenium, found in fish, may provide protection against the development of gastric cancer (52–54). As another example, intestinal parasites are more common in the low-risk region than in the high-risk region, and may also influence gastric cancer rates by diminishing the inflammatory response to H. pylori (51, 55).
In summary, many previous studies have shown the importance of CagA in the development of gastric diseases, and specific polymorphisms in CagA (such as the number and type of EPIYA motifs) have been associated with the severity of disease. In the current study, we provide evidence that variation among strains in levels of CagA expression is also a factor that correlates with the severity of disease.
ACKNOWLEDGEMENTS
Supported by NIH R01 AI068009, P01 CA116087, P01 CA028842, PD30DK058404 (Vanderbilt Digestive Disease Research Center), and the Department of Veterans Affairs.
REFERENCES
- 1.Polk DB, Peek RM., Jr Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 3.Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest. 2009;119:2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process-First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 7.Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–2115. [PubMed] [Google Scholar]
- 10.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40:297–301. doi: 10.1136/gut.40.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang JQ, Zheng GF, Sumanac K, Irvine EJ, Hunt RH. Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology. 2003;125:636–644. doi: 10.1053/j.gastro.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 12.Plummer M, van Doorn LJ, Franceschi S, Kleter B, Canzian F, Vivas J, et al. Helicobacter pylori cytotoxin-associated genotype and gastric precancerous lesions. Natl Cancer Inst. 2007;99:1328–1334. doi: 10.1093/jnci/djm120. [DOI] [PubMed] [Google Scholar]
- 13.Figueiredo C, Machado JC, Pharoah P, Seruca R, Sousa S, Carvalho R, et al. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J Natl Cancer Inst. 2002;94:1680–1687. doi: 10.1093/jnci/94.22.1680. [DOI] [PubMed] [Google Scholar]
- 14.Blaser MJ. The biology of cag in the Helicobacter pylori-human interaction. Gastroenterology. 2005;128:1512–1515. doi: 10.1053/j.gastro.2005.03.053. [DOI] [PubMed] [Google Scholar]
- 15.Tegtmeyer N, Wessler S, Backert S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011;278:1190–1202. doi: 10.1111/j.1742-4658.2011.08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourzac KM, Guillemin K. Helicobacter pylori-host cell interactions mediated by type IV secretion. Cell Microbiol. 2005;7:911–919. doi: 10.1111/j.1462-5822.2005.00541.x. [DOI] [PubMed] [Google Scholar]
- 17.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 18.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 19.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci U S A. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatakeyama M. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci. 2011;102:36–43. doi: 10.1111/j.1349-7006.2010.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000407. e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15:163–176. doi: 10.1111/j.1523-5378.2010.00759.x. [DOI] [PubMed] [Google Scholar]
- 23.Muyskens JB, Guillemin K. Helicobacter pylori CagA disrupts epithelial patterning by activating myosin light chain. PLoS One. 2011;6:e17856. doi: 10.1371/journal.pone.0017856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Correa P, Cuello C, Duque E, Burbano LC, Garcia FT, Bolanos O, et al. Gastric cancer in Colombia. III. Natural history of precursor lesions. J Natl Cancer Inst. 1976;57:1027–1035. doi: 10.1093/jnci/57.5.1027. [DOI] [PubMed] [Google Scholar]
- 25.Cuello C, Correa P, Haenszel W, Gordillo G, Brown C, Archer M, et al. Gastric cancer in Colombia. I. Cancer risk and suspect environmental agents. J Natl Cancer Inst. 1976;57:1015–1020. doi: 10.1093/jnci/57.5.1015. [DOI] [PubMed] [Google Scholar]
- 26.Camargo MC, Yepez MC, Ceron C, Guerrero N, Bravo LE, Correa P, et al. Age at acquisition of Helicobacter pylori infection: comparison of two areas with contrasting risk of gastric cancer. Helicobacter. 2004;9:262–270. doi: 10.1111/j.1083-4389.2004.00221.x. [DOI] [PubMed] [Google Scholar]
- 27.Bravo LE, van Doom LJ, Realpe JL, Correa P. Virulence-associated genotypes of Helicobacter pylori: do they explain the African enigma? Am J Gastroenterol. 2002;97:2839–2842. doi: 10.1111/j.1572-0241.2002.07031.x. [DOI] [PubMed] [Google Scholar]
- 28.Naito M, Yamazaki T, Tsutsumi R, Higashi H, Onoe K, Yamazaki S, et al. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology. 2006;130:1181–1190. doi: 10.1053/j.gastro.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 29.Higashi H, Yokoyama K, Fujii Y, Ren S, Yuasa H, Saadat I, et al. EPIYA motif is a membrane-targeting signal of Helicobacter pylori virulence factor CagA in mammalian cells. J Biol Chem. 2005;280:23130–23137. doi: 10.1074/jbc.M503583200. [DOI] [PubMed] [Google Scholar]
- 30.Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, Rhead JL, et al. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135:91–99. doi: 10.1053/j.gastro.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 31.Sicinschi LA, Correa P, Peek RM, Camargo MC, Piazuelo MB, Romero-Gallo J, et al. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2009;16:369–378. doi: 10.1111/j.1469-0691.2009.02811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Sablet T, Piazuelo MB, Shaffer CL, Schneider BG, Asim M, Chaturvedi R, et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut. 2011 doi: 10.1136/gut.2010.234468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 34.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, et al. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 2011 doi: 10.1371/journal.ppat.1002237. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Copass M, Grandi G, Rappuoli R. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect Immun. 1997;65:1949–1952. doi: 10.1128/iai.65.5.1949-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dailidiene D, Dailide G, Kersulyte D, Berg DE. Contraselectable streptomycin susceptibility determinant for genetic manipulation and analysis of Helicobacter pylori. Appl Environ Microbiol. 2006;72:5908–5914. doi: 10.1128/AEM.01135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Busler VJ, Torres VJ, McClain MS, Tirado O, Friedman DB, Cover TL. Protein-protein interactions among Helicobacter pylori cag proteins. J Bacteriol. 2006;188:4787–4800. doi: 10.1128/JB.00066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh JT, Torres VJ, Cover TL. Regulation of Helicobacter pylori cagA expression in response to salt. Cancer Res. 2007;67:4709–4715. doi: 10.1158/0008-5472.CAN-06-4746. [DOI] [PubMed] [Google Scholar]
- 39.Cao P, McClain MS, Forsyth MH, Cover TL. Extracellular release of antigenic proteins by Helicobacter pylori. InfectImmun. 1998;66:2984–2986. doi: 10.1128/iai.66.6.2984-2986.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International workshop on the histopathology of gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 41.Rugge M, Correa P, Dixon MF, Hattori T, Leandro G, Lewin K, et al. Gastric dysplasia: the Padova international classification. Am J Surg Pathol. 2000;24:167–176. doi: 10.1097/00000478-200002000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Spohn G, Beier D, Rappuoli R, Scarlato V. Transcriptional analysis of the divergent cagAB genes encoded by the pathogenicity island of Helicobacter pylori. Mol Microbiol. 1997;26:361–372. doi: 10.1046/j.1365-2958.1997.5831949.x. [DOI] [PubMed] [Google Scholar]
- 43.Sabelnikov AG, Greenberg B, Lacks SA. An extended -10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 44.Forsyth MH, Cover TL. Mutational analysis of the vacA promoter provides insight into gene transcription in Helicobacter pylori. J Bacteriol. 1999;181:2261–2266. doi: 10.1128/jb.181.7.2261-2266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 46.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCullen CA, Benhammou JN, Majdalani N, Gottesman S. Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol. 2010;192:5559–5571. doi: 10.1128/JB.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McNulty SL, Mole BM, Dailidiene D, Segal I, Ally R, Mistry R, et al. Novel 180- and 480-base-pair insertions in African and African-American strains of Helicobacter pylori. J Clin Microbiol. 2004;42:5658–5663. doi: 10.1128/JCM.42.12.5658-5663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camargo MC, Burk RF, Bravo LE, Piazuelo MB, Hill KE, Fontham ET, et al. Plasma selenium measurements in subjects from areas with contrasting gastric cancer risks in Colombia. Arch Med Res. 2008;39:443–451. doi: 10.1016/j.arcmed.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piazuelo MB, Camargo MC, Mera RM, Delgado AG, Peek RM, Jr, Correa H, et al. Eosinophils and mast cells in chronic gastritis: possible implications in carcinogenesis. Hum Pathol. 2008;39:1360–1369. doi: 10.1016/j.humpath.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whary MT, Sundina N, Bravo LE, Correa P, Quinones F, Caro F, et al. Intestinal helminthiasis in Colombian children promotes a Th2 response to Helicobacter pylori: possible implications for gastric carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2005;14:1464–1469. doi: 10.1158/1055-9965.EPI-05-0095. [DOI] [PubMed] [Google Scholar]
- 52.Blot WJ, Li JY, Taylor PR, Guo W, Dawsey S, Wang GQ, et al. Nutrition intervention trials in Linxian, China: supplementation with specific vitamin/mineral combinations, cancer incidence, and disease-specific mortality in the general population. J Natl Cancer Inst. 1993;85:1483–1492. doi: 10.1093/jnci/85.18.1483. [DOI] [PubMed] [Google Scholar]
- 53.Jenab M, Riboli E, Ferrari P, Sabate J, Slimani N, Norat T, et al. Plasma and dietary vitamin C levels and risk of gastric cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) Carcinogenesis. 2006;27:2250–2257. doi: 10.1093/carcin/bgl096. [DOI] [PubMed] [Google Scholar]
- 54.Gonzalez CA, Pera G, Agudo A, Bueno-de-Mesquita HB, Ceroti M, Boeing H, et al. Fruit and vegetable intake and the risk of stomach and oesophagus adenocarcinoma in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) Int J Cancer. 2006;118:2559–2566. doi: 10.1002/ijc.21678. [DOI] [PubMed] [Google Scholar]
- 55.Elliott DE, Summers RW, Weinstock JV. Helminths as governors of immune-mediated inflammation. Int J Parasitol. 2007;37:457–464. doi: 10.1016/j.ijpara.2006.12.009. [DOI] [PubMed] [Google Scholar]