Abstract
Signaling mucins are large transmembrane glycoproteins that regulate signal transduction pathways. Recent advances have shown that two major types of post-translational modifications, protein glycosylation and proteolytic processing, play important and unexpected roles in regulating signaling mucin function. New O-glycosyltransferases and proteases have been identified, and the structure of the domain that undergoes autoproteolysis has been solved. A picture is beginning to emerge where specific glycosyl modifications and regulated processing control the signaling and adherence properties of signaling glycoproteins and contribute to the routing of signals to specific pathways.
Keywords: signal transduction, protein glycosylation, protein processing, protease, glycoprotein
Introduction
Signaling mucins are high molecular weight transmembrane glycoproteins that regulate signal transduction pathways. These molecules have gained widespread popularity as markers for many different types of cancers and because of their prominent roles in regulating a wide variety of signal transduction pathways [1,2]. Signaling mucins are regulated by two major types of post-translational modifications, protein glycosylation and proteolytic processing. This review highlights what the precise modifications are, the enzymes that carry out those modifications, and the consequences of protein modification on signaling mucin function and regulation. Other signaling glycoproteins are discussed in the context of signaling mucin regulation.
Although generally variable in primary amino acid sequence, signaling mucins have a number of common features and the same overall topology (Figure 1). Signaling mucins are single-pass cell-surface glycoproteins with a rod-like extracellular domain that is connected to a cytosolic C-terminal domain by a transmembrane α helix. The cytosolic signaling domain distinguishes signaling-type mucins from non-signaling mucins (like MUC2), which do not contain cytosolic domains. A defining feature of mucin glycoproteins are tandem repeats rich in proline, threonine, and serine residues (PTS domain) in the extracellular domain (Figure 1).
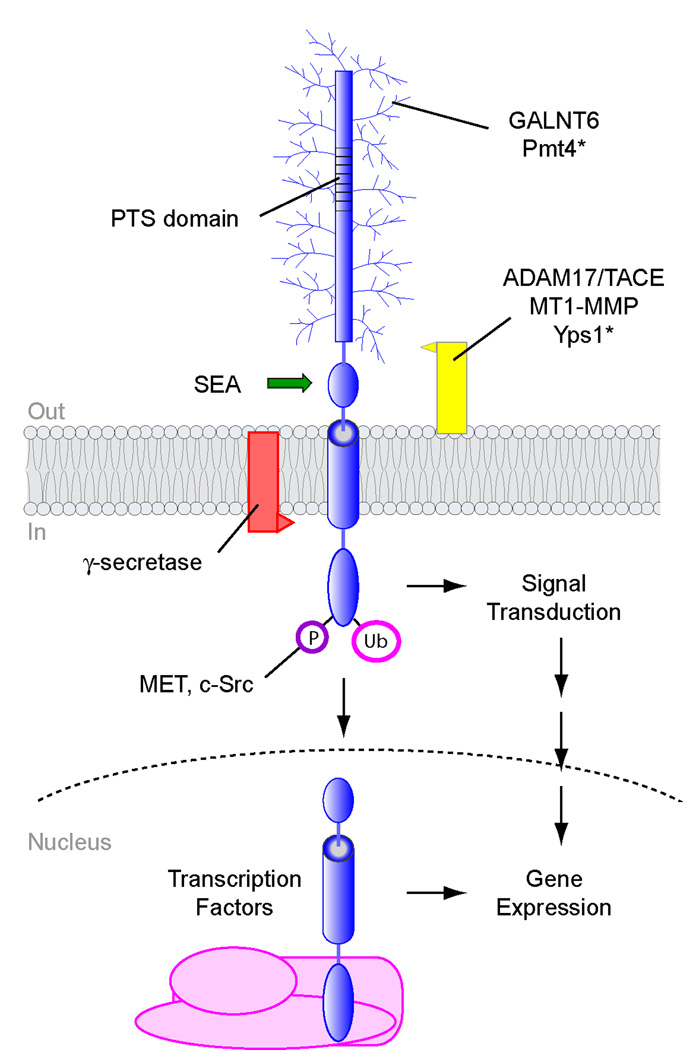
Figure 1. Post-translational modifications of signaling mucins.
A schematic representation of a signaling mucin is shown. Proteins that modify signaling mucins include O-glycosyltransferases, like GALNT6 and Pmt4, and proteases γ-secretase, Yps1, ADAM17/TACE, and MT1-MMP. Autocatalytic processing by the SEA domain is shown by a green arrow. The cytosolic domain is modified by ubiquitin (Ub), and phosphate (P) moieties. The cytosolic and transmembrane domains of MUC1 can translocate to the nucleus and associate with transcription factors to regulate gene expression. Modifiers of yeast mucin-like proteins are marked with an asterisk. Not all signaling mucins undergo all the modifications shown. The extracellular domain is not to scale and can be much larger than shown.
The most extensively studied signaling mucin is MUC1, which regulates the RAS-MEK-ERK mitogen-activated protein kinase (MAPK) pathway [3] and many other signaling pathways [1,2]. Other signaling mucins include MUC4 and MUC12 and have also been characterized. Mucin-like glycoproteins also exist in other organisms including genetically tractable model systems like the baker’s yeast Saccharomyces cerevisiae. In yeast, two signaling glycoproteins have been characterized, Msb2 [4] and Hkr1 [5], which like their mammalian counterparts contain PTS domains and regulate MAPK pathways. Msb2 regulates MAPK signaling by associating with the ubiquitous polarity Rho (Ras homology) GTPase Cdc42 [6] and might represent a functional homolog of human MUC12 [7]. Non-signaling glycoproteins proteins have also been studied in yeast such as the adhesion flocculin Flo11 [8].
Glycosyl Modifications in the Extracellular Domain
Signaling mucins are rich in serine and threonine residues (>40% of the total residues). Many of these residues are thought to be modified by O-linked glycosylation, and the PTS domain has been shown to be extensively O-glycosylated [9]. As a result, oligosaccharides can more than double the molecular mass of the proteins. Signaling mucins are modified by different sugars including galactose, N-acetylgalactoosamine (GalNAc), fucose, and/or sialic acid, which can influence the size and overall charge of the protein. MUC1 for example is heavily sialylated, and this modification becomes more extensive after the protein has been internalized from the cell surface and is recycled back to the plasma membrane through the Golgi apparatus [10]. Glycosylation can stabilize mucins at the cell surface, by limiting their endocytosis [11,12] and by protecting the polypeptide chain from degradation by extracellular proteases [13].
What enzymes glycosylate signaling mucins? Mucin-type O-glycosylation is initiated by members of the polypeptide N-acetylglucosaminyltransferase family (gene name GALNT for mammals, PGANT for Drosophila). There are at least 15 GALNT genes in the human genome [14], and several of these enzymes likely participate in O-GalNAc addition to MUC1 depending on the cell and tissue type. Other glycosyltransferases may add various sugars subsequent to O-GalNAc addition [14].
Recently, a specific GALNT, GALNT6, was found to modify MUC1 in breast cancer cells [15]. The expression of the GALNT6 gene was elevated in cancer cells, which also express high levels of MUC1. Down-regulation of GALNT6 expression by small interfering RNAs suppressed the growth of breast cancer cells [15]. The idea that different GALNTs specifically glycosylate mucins in different contexts is supported by studies in model organisms. A specific yeast O-mannosyltransferase, Pmt4, among a large family of Pmt proteins, plays the major role in Msb2 glycosylation [16]. Similarly, in the fruit fly, Drosophila melanogaster, loss of different PGANT genes results in different phenotypes, which suggests that different PGANTs glycosylate non-overlapping targets [17]. An implication of these findings is that individual glycosyltransferases selectively modify signaling mucins to precisely regulate their function and activity.
What role does protein glycosylation play in regulating signaling mucin function? An exciting possibility is to regulate signaling pathway specificity. Signaling pathways typically function in web-like networks composed of many different proteins. In these networks, signals are routed through common protein modules to induce selective responses. How signals are faithfully transmitted along one of many possible paths is not clear. A role for glycosylation in influencing signaling specificity was demonstrated by studies of Notch, a transmembrane glycoprotein which is similar to signaling mucins and which regulates many developmental processes [18]. Notch is activated by binding to proteins expressed on adjacent cells, Delta and Serrate/Jagged. Fringe is a glycosyltransferase (specifically an N-acetylglucosaminyltransferase), that modifies Notch [19] by elongating O-fucosyl residues through the addition of N-acetylglucosamine. The glycosyl modification by Fringe has the effect of potentiating signaling from Delta while inhibiting signaling from the Serrate/Jagged family of ligands (Figure 2A). Thus, the selective glycosylation of Notch by Fringe can evoke a pathway-specific response.
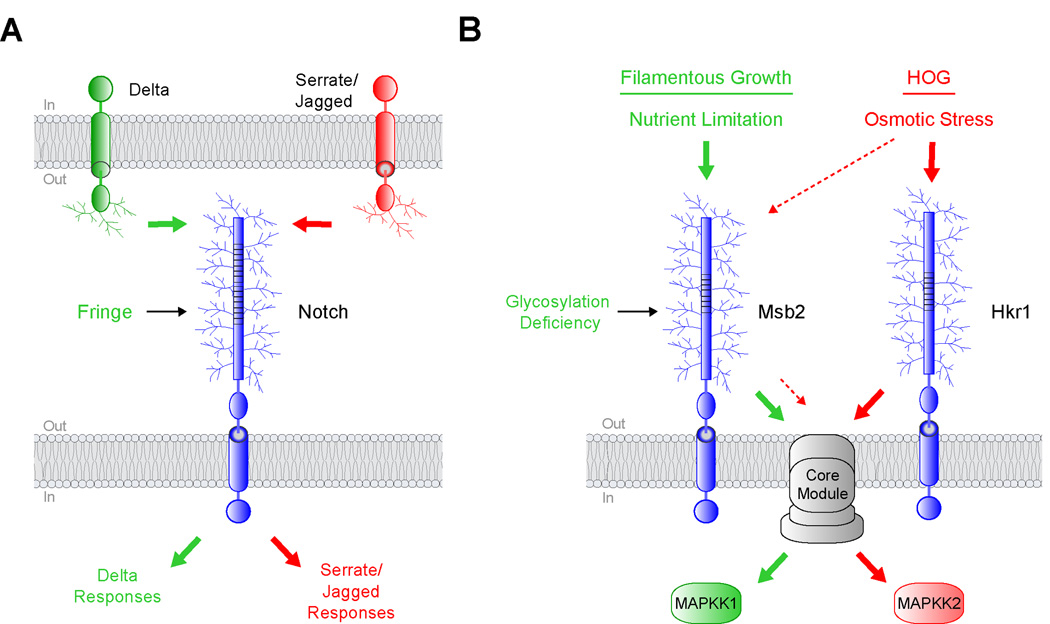
Figure 2. Protein glycosylation can regulate signaling specificity.
A) Notch can bind to transmembrane ligands of the Delta and Serrate/Jagged families. Fringe is a glycosyltransferase that modifies Notch and enhances a Delta-specific response (green arrows) while inhibiting Serrate/Jagged-specific responses (red arrows). B) Two yeast MAPK pathways that share components. Nutrient limitation activates the filamentous growth pathway through Msb2 (green arrows). Osmotic stress activates the HOG pathway through Msb2 and Hkr1 (red arrows). Reduced glycosylation of Msb2 specifically triggers the filamentous growth pathway through a core module composed of proteins that are shared between the two pathways, shown in grey. The lighter dashed red arrows designate a function for Msb2 in the HOG pathway.
A loose parallel can be drawn for yeast mucin-like proteins, in that glycosylation can differentially influence the activity of one MAPK pathway over another. Two yeast glycoproteins, Msb2 and Hkr1, regulate two MAPK pathways (Figure 2B, [20]). The pathways sense different stimuli and induce non-overlapping responses but share a common protein module (Figure 2B, [21]). In fact, one of the mucin-like glycoproteins, Msb2, can itself function in both pathways (Figure 2B, [5]). One difference between the pathways is that they require different downstream kinases (Figure 2B, MAPKK). In a recent landmark study, glycosylation deficiency was shown to activate Msb2 in one of the pathways but not the other (Figure 2B, [16]). This unexpected discovery, together with other evidence [22], suggests that Msb2 and Hkr1 function preferentially in different pathways. It remains unclear how the two proteins activate different MAPKKs through a shared module. Possibly, pathway-specific proteins that remain to be identified differentially associate with the C-termini of the two mucins, which are dissimilar. Therefore, the selective glycosylation of transmembrane glycoproteins is becoming an emerging paradigm for how pathway specificity is achieved.
Processing and Release of the Extracellular Domain
A second major post-translational modification of signaling mucins is proteolytic processing. Signaling mucins can be processed outside the cell in their extracellular domains and inside the cell in their cytosolic domains. These processing events have important consequences on mucin function and regulation. MUC1 undergoes autoproteolytic cleavage [23], which maps between the glycine and serine residues (G↑SVVV) at position 1097 in the extracellular domain of the protein [24,25]. The cleavage site and surrounding region is also found (almost exclusively) in other O-glycosylated proteins and is referred to as the SEA (sea-urchin sperm protein, enterokinase, and agrin) module [26]. The structure of the SEA module has been determined by NMR spectroscopy, which sheds light on the mechanism of cleavage (Figure 3, [27]). Structural analysis of the SEA domain has also provided detailed energetic and thermodynamic information about self-cleavage reaction [28,29].
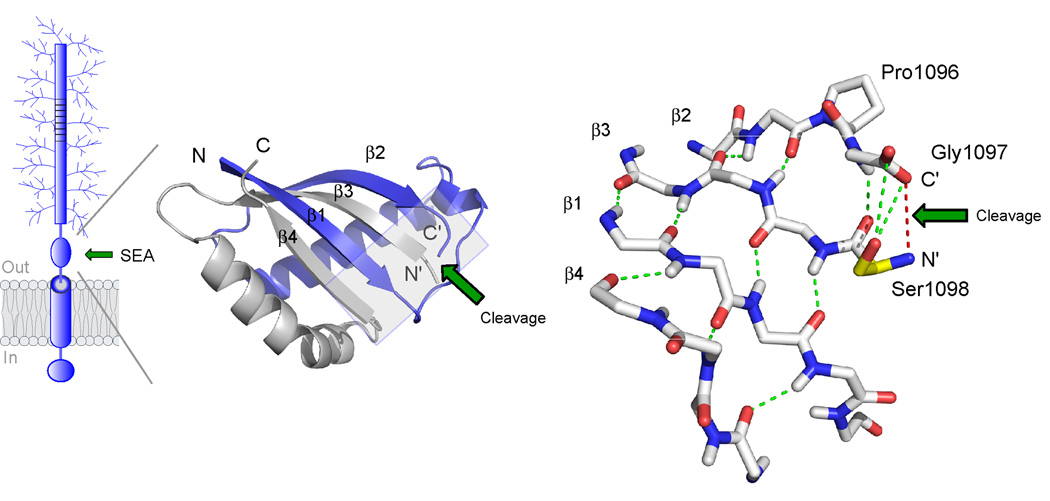
Figure 3. Structure of the self-cleaving SEA domain of MUC1.
The structure is based on NMR spectroscopy data ([27], PDB number 2ACM). At left is shown the folding topology and secondary structure of the cleaved SEA heterodimer with the two intertwined subunits colored in blue and grey, respectively. Autoproteolytic cleavage occurs at the edge of a four-stranded β-sheet to generate novel N’ and C’ peptide termini, as indicated. The structure on the right shows a detailed view of the peptide backbone at the cleavage site (boxed area in figure to the left). Autoproteolytic cleavage has occurred between glycine and serine in the tight turn Pro1096-Gly1097-Ser1098 as a result of conformational strain generated by the β-sheet structure (hydrogen bonds in green) and the nucleophilic action of the serine hydroxyl (Ser1098 side chain in yellow). The location of the former glycine-serine peptide bond is illustrated by a red line. The site of cleavage is denoted by a green arrow.
Signaling mucins are also processed by other proteases. The protease that processes MUC4 remains to be identified [30]. The yeast glycoprotein Msb2 is processed by Yps1 [31], a member of an evolutionary conserved family of glycophosphatidylinositol (GPI)-anchored aspartyl proteases [32]. The secreted rat mucin, Muc2, and the yeast flocculin Flo11 [33] are processed by furin [34] (Kex2 in yeast), an evolutionarily conserved pro-protein convertase [35].
Release of MUC1 from the cell surface requires additional processing, because the two fragments generated by auto-proteolysis remain associated by non-covalent interactions. Two “sheddases” are required for the release of MUC1 from cells. One is tumor necrosis factor-alpha converting enzyme (TACE), also called A Disintegrin And Metalloprotease domain-containing protein 17 (ADAM17) [36], and the other is a membrane-type matrix metalloprotease (MT1-MMP) [37]. ADAM-type sheddases are themselves highly regulated at the transcriptional and post-transcriptional levels and regulate the shedding of many different types of glycoproteins (Figure 4 [38]).
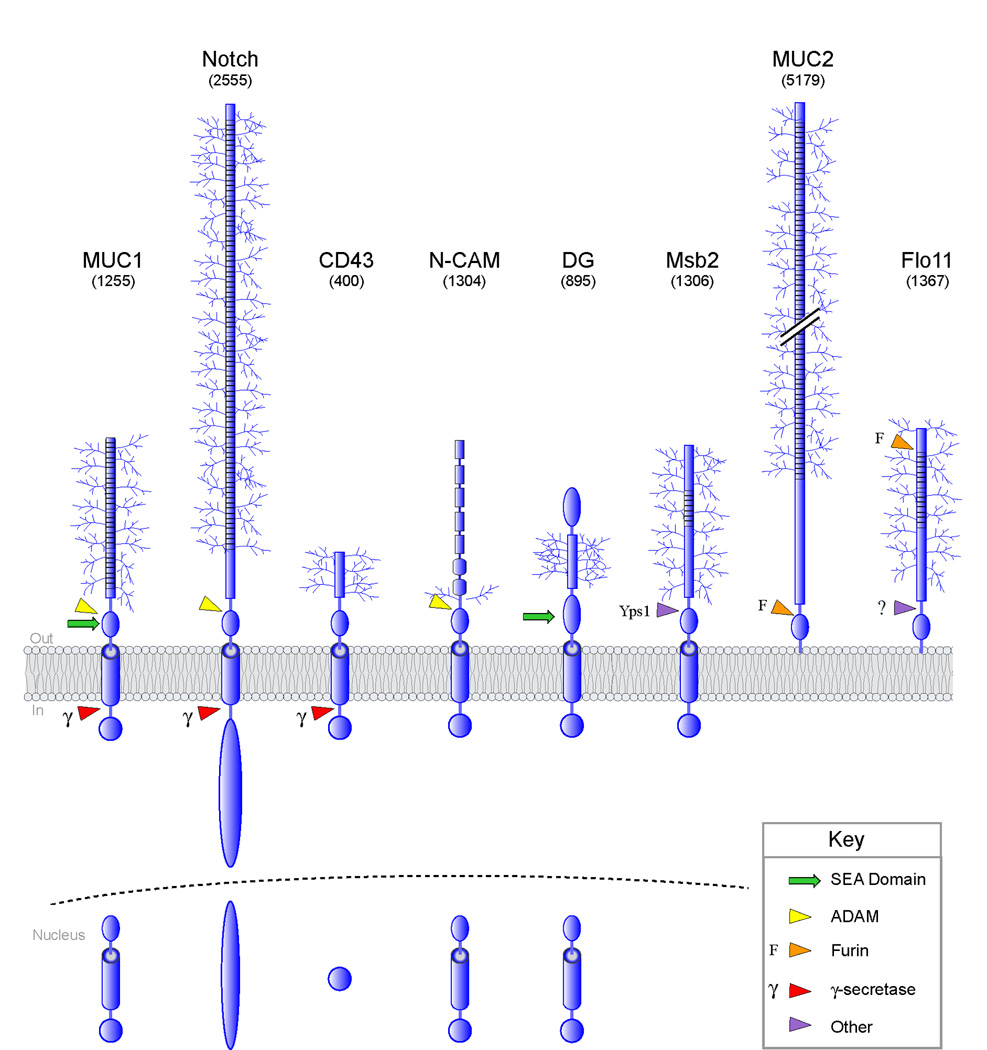
Figure 4. Post-translational processing of cell-surface O-glycosylated proteins.
Processing and nuclear entry of proteins are based on reports for MUC1 [54], Notch [55], dystroglycan (DG) [45,56], N-CAM [57], and CD43 [58]. Notch is processed by ADAM10 and ADAM17. Arrows refer to sites of cleavage: red, γsecretase (γ); green, SEA domain; yellow, ADAM; orange, furin (F); and purple, processing by other protease(s). For some proteins, the exact site of cleavage is not known. In parentheses is the predicted molecular weight of the proteins shown. Only O-glycan (not N-glycan) modifications are depicted.
What effect does processing have on signaling mucin function? A possible insight has emerged from studies of Msb2, through observations that were at first perplexing. Initial observations showed that a version of Msb2 lacking the PTS domain hyper-activates the MAPK pathway, which suggested an inhibitory function for that domain [4]. Subsequent experiments demonstrated that the extracellular inhibitory domain is released from cells as a result of processing [31]. Together these findings suggest a cleavage-dependent activation mechanism [31]. Notch similarly has a large inhibitory domain that is processed and released from cells as part of its activation mechanism [39]. Cleavage-dependent activation may underlie the regulation of other signaling glycoproteins, such as MUC1, although this possibility has not been explicitly tested.
A second function for glycoprotein shedding may be to regulate adhesion. Many cell-surface glycoproteins have adhesive (or anti-adhesive) functions, by associating with proteins on the surface of other cells or in the extracellular matrix. Studies on the yeast flocculin Flo11, a large glycoprotein that is attached to cells by a GPI anchor [8], has shown that the protein is released from cells. Release of Flo11 from cells provides a mechanism for attenuating adherence [33]. The regulated processing of O-glycoproteins therefore represents a means to regulate the signaling and adherence properties of these proteins.
Processing and Modification of the Cytoplasmic Domain
Many transmembrane O-glycosylated proteins including MUC1 [40], Notch [41,42], and other proteins (Figure 4) are processed in their cytosolic domains. For most proteins that have been examined, the membrane-embedded protease complex γ-secretase [43] is responsible for processing at the cytosol-transmembrane boundary. The cytoplasmic domain of MUC1 can enter the nucleus [44] and associate with transcription factors to directly modulate gene expression. At first glance, one might expect that processing of MUC1 by γ-secretase results in nuclear entry. This may be an oversimplification, however, because nuclear forms of MUC1 include the transmembrane domain [45]. How versions of MUC1 that contain the transmembrane domain are liberated from the plasma membrane and trafficked to the nucleus is not clear. Nevertheless, this unconventional trafficking route appears to underlie the nuclear localization other cell-surface glycoproteins such as β-dystroglycan and N-CAM (Figure 4). The cytoplasmic domain of MUC1 may function in other organelles such as the mitochondria. The localization of MUC1 to the mitochondria occurs in an HSP90-dependent manner [46].
The cytosolic domain of MUC1 associates with many different signaling proteins [1,2] and is extensively regulated by multiple post-translational modifications. For example, the C-terminus of MUC1 is phosphorylated by protein kinases including MET [47] and SRC [48]. Phosphorylation of MUC1 by MET induces interaction with p53 [47], and phosphorylation by SRC induces interaction with HSP90 [46]. The cytosolic domain can also be ubiquitinated. Ubiquitination of MUC4 targets the protein for turnover [49].
Conclusions and Future Directions
Signaling mucins are regulated by many different post-translational modifications. These modifications modulate the charge, stability, and activity of signaling mucins and consequently regulate the strength, duration, and specificity of the signals generated. A future challenge will be to understand which precise glycosyl modifications regulate signaling mucin regulation. The extensive glycosylation of mucin proteins complicates structural and compositional analysis. However, recent advances in the synthesis of glycosylated peptides [50,51], the construction of glycopeptide arrays [52], and the utilization of genomic approaches [53] may facilitate future studies. Future studies on signaling mucins and other glycoproteins will continue to deepen our appreciation of these interesting molecules and assist in the design of effective therapies in the treatment of human disease.
Highlights.
-
>
Signaling mucins are cell-surface glycoproteins that regulate signal transduction pathways
-
>
O-glycosyltransferases modify signaling mucins, including GALNT6 modification of MUC1 in breast cancer cells
-
>
Glycosylation influences the routing of signals to specific pathways
-
>
MUC1 is processed outside the cell by autocleavage and sheddases and inside the cell by γ-secretase
-
>
The structure of the SEA domain by NMR illustrates the mechanism of autoproteolytic cleavage
ACKNOWLEDGEMENTS
Thanks to Dr. Torleif Härd for providing images and the legend for Figure 2, and Drs. Stefan Ruhl, Daniel Carson, and Kermit Carraway for reading the manuscript and providing suggestions. Thanks to Nadia Vadaie for assistance with early drafts and laboratory members Sheelarani Karunanithi, Colin Chavel, and Hema Adhikari for comments. P.J.C. is supported from grants from the National Institutes of Health (1R03DE018425-01) and the American Cancer Society (TBE-114083).
Abbreviations
- ADAM
a disintegrin and metalloprotease
- MT1-MMP
membrane-type 1-matrix metalloprotease
- GalNAc
N-acetylgalactosamine
- GALNT
polypeptide N-acetylglucosaminyltransferase
- MAPK
mitogen-activated protein kinase
- GPI
glycosylphosphatidylinositol
- PAK
p21-activated kinase
- PTS
proline, threonine, and serine
- Rho
Ras homology
- SEA
sea-urchin sperm protein, enterokinase, and agrin
- TACE
tumor necrosis factor alpha-converting enzyme
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
• References of Special Interest:
** References of Outstanding Interest:
- 1.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29:2893–2904. doi: 10.1038/onc.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16:467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Cullen PJ, Sabbagh W, Jr, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF., Jr A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatebayashi K, Tanaka K, Yang HY, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. Embo J. 2007;26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandyopadhyay S, Chiang CY, Srivastava J, Gersten M, White S, Bell R, Kurschner C, Martin CH, Smoot M, Sahasrabudhe S, et al. A human MAP kinase interactome. Nat Methods. 2010;7:801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman HS, Sutton-Smith M, McDermott K, Heal P, Leir SH, Morris HR, Hollingsworth MA, Dell A, Harris A. The contribution of tandem repeat number to the O-glycosylation of mucins. Glycobiology. 2003;13:265–277. doi: 10.1093/glycob/cwg028. [DOI] [PubMed] [Google Scholar]
- 10.Hull SR, Sugarman ED, Spielman J, Carraway KL. Biosynthetic maturation of an ascites tumor cell surface sialomucin. Evidence for O-glycosylation of cell surface glycoprotein by the addition of new oligosaccharides during recycling. J Biol Chem. 1991;266:13580–13586. [PubMed] [Google Scholar]
- 11.Altschuler Y, Kinlough CL, Poland PA, Bruns JB, Apodaca G, Weisz OA, Hughey RP. Clathrin-mediated endocytosis of MUC1 is modulated by its glycosylation state. Mol Biol Cell. 2000;11:819–831. doi: 10.1091/mbc.11.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelmann K, Kinlough CL, Muller S, Razawi H, Baldus SE, Hughey RP, Hanisch FG. Transmembrane and secreted MUC1 probes show trafficking-dependent changes in O-glycan core profiles. Glycobiology. 2005;15:1111–1124. doi: 10.1093/glycob/cwi099. [DOI] [PubMed] [Google Scholar]
- 13.Loomes KM, Senior HE, West PM, Roberton AM. Functional protective role for mucin glycosylated repetitive domains. Eur J Biochem. 1999;266:105–111. doi: 10.1046/j.1432-1327.1999.00824.x. [DOI] [PubMed] [Google Scholar]
- 14.Tian E, Ten Hagen KG. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj J. 2009;26:325–334. doi: 10.1007/s10719-008-9162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park JH, Nishidate T, Kijima K, Ohashi T, Takegawa K, Fujikane T, Hirata K, Nakamura Y, Katagiri T. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70:2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. • This article identifies GALNT6 as one of the polypeptide O-GalNAc transferases responsible for modification of MUC1 in breast cancer cells.
- 16. Yang HY, Tatebayashi K, Yamamoto K, Saito H. Glycosylation defects activate filamentous growth Kss1 MAPK and inhibit osmoregulatory Hog1 MAPK. Embo J. 2009;28:1380–1391. doi: 10.1038/emboj.2009.104. ** This article shows that Msb2 is O-glycosylated by Pmt4. O-glycoslyation of Msb2 regulates activity of one MAPK pathway (Filamentous Growth) but not another (HOG).
- 17. Zhang L, Ten Hagen KG. Dissecting the biological role of mucin-type O-glycosylation using RNA interference in Drosophila cell culture. J Biol Chem. 2010;285:34477–34484. doi: 10.1074/jbc.M110.133561. • In this work, different polypeptide O-GalNAc transferases are shown to have different roles in the cell.
- 18.Haltiwanger RS. Regulation of signal transduction pathways in development by glycosylation. Curr Opin Struct Biol. 2002;12:593–598. doi: 10.1016/s0959-440x(02)00371-8. [DOI] [PubMed] [Google Scholar]
- 19.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 20.Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Methods Enzymol. 2007;428:29–45. doi: 10.1016/S0076-6879(07)28002-4. [DOI] [PubMed] [Google Scholar]
- 21.Bardwell L. Mechanisms of MAPK signalling specificity. Biochem Soc Trans. 2006;34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitoniak A, Birkaya B, Dionne HS, Vadiae N, Cullen PJ. The Signaling Mucins Msb2 and Hkr1 Differentially Regulate the Filamentation MAPK Pathway and Contribute to a Multimodal Response. Mol Biol Cell. 2009;20:3101–3114. doi: 10.1091/mbc.E08-07-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levitin F, Stern O, Weiss M, Gil-Henn C, Ziv R, Prokocimer Z, Smorodinsky NI, Rubinstein DB, Wreschner DH. The MUC1 SEA module is a self-cleaving domain. J Biol Chem. 2005;280:33374–33386. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- 24.Lillehoj EP, Han F, Kim KC. Mutagenesis of a Gly-Ser cleavage site in MUC1 inhibits ectodomain shedding. Biochem Biophys Res Commun. 2003;307:743–749. doi: 10.1016/s0006-291x(03)01260-9. [DOI] [PubMed] [Google Scholar]
- 25.Parry S, Silverman HS, McDermott K, Willis A, Hollingsworth MA, Harris A. Identification of MUC1 proteolytic cleavage sites in vivo. Biochem Biophys Res Commun. 2001;283:715–720. doi: 10.1006/bbrc.2001.4775. [DOI] [PubMed] [Google Scholar]
- 26.Bork P, Patthy L. The SEA module: a new extracellular domain associated with O-glycosylation. Protein Sci. 1995;4:1421–1425. doi: 10.1002/pro.5560040716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macao B, Johansson DG, Hansson GC, Hard T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 28.Johansson DG, Macao B, Sandberg A, Hard T. SEA domain autoproteolysis accelerated by conformational strain: mechanistic aspects. J Mol Biol. 2008;377:1130–1143. doi: 10.1016/j.jmb.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 29.Sandberg A, Johansson DG, Macao B, Hard T. SEA domain autoproteolysis accelerated by conformational strain: energetic aspects. J Mol Biol. 2008;377:1117–1129. doi: 10.1016/j.jmb.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 30.Soto P, Zhang J, Carraway KL. Enzymatic cleavage as a processing step in the maturation of Muc4/sialomucin complex. J Cell Biochem. 2006;97:1267–1274. doi: 10.1002/jcb.20718. [DOI] [PubMed] [Google Scholar]
- 31.Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol. 2008;181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1364–1374. doi: 10.1128/EC.4.8.1364-1374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karunanithi S, Vadaie N, Chavel CA, Birkaya B, Joshi J, Grell L, Cullen PJ. Shedding of the Mucin-Like Flocculin Flo11p Reveals a New Aspect of Fungal Adhesion Regulation. Curr Biol. 2010;20:1389–1395. doi: 10.1016/j.cub.2010.06.033. ** This article shows that Flo11 is processed and shed from cells. Processing of Flo11 requires the yeast furin homolog Kex2.
- 34.Xu G, Bell SL, McCool D, Forstner JF. The cationic C-terminus of rat Muc2 facilitates dimer formation post translationally and is subsequently removed by furin. Eur J Biochem. 2000;267:2998–3004. doi: 10.1046/j.1432-1033.2000.01319.x. [DOI] [PubMed] [Google Scholar]
- 35.Seidah NG. What lies ahead for the proprotein convertases? Ann N Y Acad Sci. 2011;1220:149–161. doi: 10.1111/j.1749-6632.2010.05883.x. [DOI] [PubMed] [Google Scholar]
- 36.Thathiah A, Blobel CP, Carson DD. Tumor necrosis factor-alpha converting enzyme/ADAM 17 mediates MUC1 shedding. J Biol Chem. 2003;278:3386–3394. doi: 10.1074/jbc.M208326200. [DOI] [PubMed] [Google Scholar]
- 37.Thathiah A, Carson DD. MT1-MMP mediates MUC1 shedding independent of TACE/ADAM17. Biochem J. 2004;382:363–373. doi: 10.1042/BJ20040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy G. Regulation of the proteolytic disintegrin metalloproteinases, the 'Sheddases'. Semin Cell Dev Biol. 2009;20:138–145. doi: 10.1016/j.semcdb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Yu WH, Ren J, Chen W, Huang L, Kharbanda S, Loda M, Kufe D. Heregulin targets gamma-catenin to the nucleolus by a mechanism dependent on the DF3/MUC1 oncoprotein. Mol Cancer Res. 2003;1:765–775. [PubMed] [Google Scholar]
- 41.Fortini ME, Rebay I, Caron LA, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 42.Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 43.Wolfe MS. The gamma-secretase complex: membrane-embedded proteolytic ensemble. Biochemistry. 2006;45:7931–7939. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 44.Wen Y, Caffrey TC, Wheelock MJ, Johnson KR, Hollingsworth MA. Nuclear association of the cytoplasmic tail of MUC1 and beta-catenin. J Biol Chem. 2003;278:38029–38039. doi: 10.1074/jbc.M304333200. [DOI] [PubMed] [Google Scholar]
- 45.Oppizzi ML, Akhavan A, Singh M, Fata JE, Muschler JL. Nuclear translocation of beta-dystroglycan reveals a distinctive trafficking pattern of autoproteolyzed mucins. Traffic. 2008;9:2063–2072. doi: 10.1111/j.1600-0854.2008.00822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren J, Bharti A, Raina D, Chen W, Ahmad R, Kufe D. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- 47.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, Gendler SJ, Bennett EP, Hollingsworth MA. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283:26985–26995. doi: 10.1074/jbc.M805036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Kuwahara H, Ren J, Wen G, Kufe D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3 beta and beta-catenin. J Biol Chem. 2001;276:6061–6064. doi: 10.1074/jbc.C000754200. [DOI] [PubMed] [Google Scholar]
- 49.Lomako WM, Lomako J, Soto P, Carraway CA, Carraway KL. TGFbeta regulation of membrane mucin Muc4 via proteosome degradation. J Cell Biochem. 2009;107:797–802. doi: 10.1002/jcb.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsushita T, Nishimura S. Novel synthesis of functional mucin glycopeptides containing both N- and O-glycans. Methods Enzymol. 2010;478:485–502. doi: 10.1016/S0076-6879(10)78023-X. [DOI] [PubMed] [Google Scholar]
- 51.Matsushita T, Sadamoto R, Ohyabu N, Nakata H, Fumoto M, Fujitani N, Takegawa Y, Sakamoto T, Kurogochi M, Hinou H, et al. Functional neoglycopeptides: synthesis and characterization of a new class of MUC1 glycoprotein models having core 2-based O-glycan and complex-type N-glycan chains. Biochemistry. 2009;48:11117–11133. doi: 10.1021/bi901557a. [DOI] [PubMed] [Google Scholar]
- 52.Ohyabu N, Hinou H, Matsushita T, Izumi R, Shimizu H, Kawamoto K, Numata Y, Togame H, Takemoto H, Kondo H, et al. An essential epitope of anti-MUC1 monoclonal antibody KL-6 revealed by focused glycopeptide library. J Am Chem Soc. 2009;131:17102–17109. doi: 10.1021/ja903361f. [DOI] [PubMed] [Google Scholar]
- 53.Chavel CA, Dionne HM, Birkaya B, Joshi J, Cullen PJ. Multiple signals converge on a differentiation MAPK pathway. PLoS Genet. 2010;6:e1000883. doi: 10.1371/journal.pgen.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Julian J, Dharmaraj N, Carson DD. MUC1 is a substrate for gamma-secretase. J Cell Biochem. 2009;108:802–815. doi: 10.1002/jcb.22292. • This article identifies γ-secretase as the protease complex responsible for processing MUC1 at the cytosol-TM boundary.
- 55.Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Akhavan A, Crivelli SN, Singh M, Lingappa VR, Muschler JL. SEA domain proteolysis determines the functional composition of dystroglycan. Faseb J. 2008;22:612–621. doi: 10.1096/fj.07-8354com. [DOI] [PubMed] [Google Scholar]
- 57.Kleene R, Mzoughi M, Joshi G, Kalus I, Bormann U, Schulze C, Xiao MF, Dityatev A, Schachner M. NCAM-induced neurite outgrowth depends on binding of calmodulin to NCAM and on nuclear import of NCAM and fak fragments. J Neurosci. 2010;30:10784–10798. doi: 10.1523/JNEUROSCI.0297-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersson CX, Fernandez-Rodriguez J, Laos S, Baeckstrom D, Haass C, Hansson GC. Shedding and gamma-secretase-mediated intramembrane proteolysis of the mucin-type molecule CD43. Biochem J. 2005;387:377–384. doi: 10.1042/BJ20041387. [DOI] [PMC free article] [PubMed] [Google Scholar]