Abstract
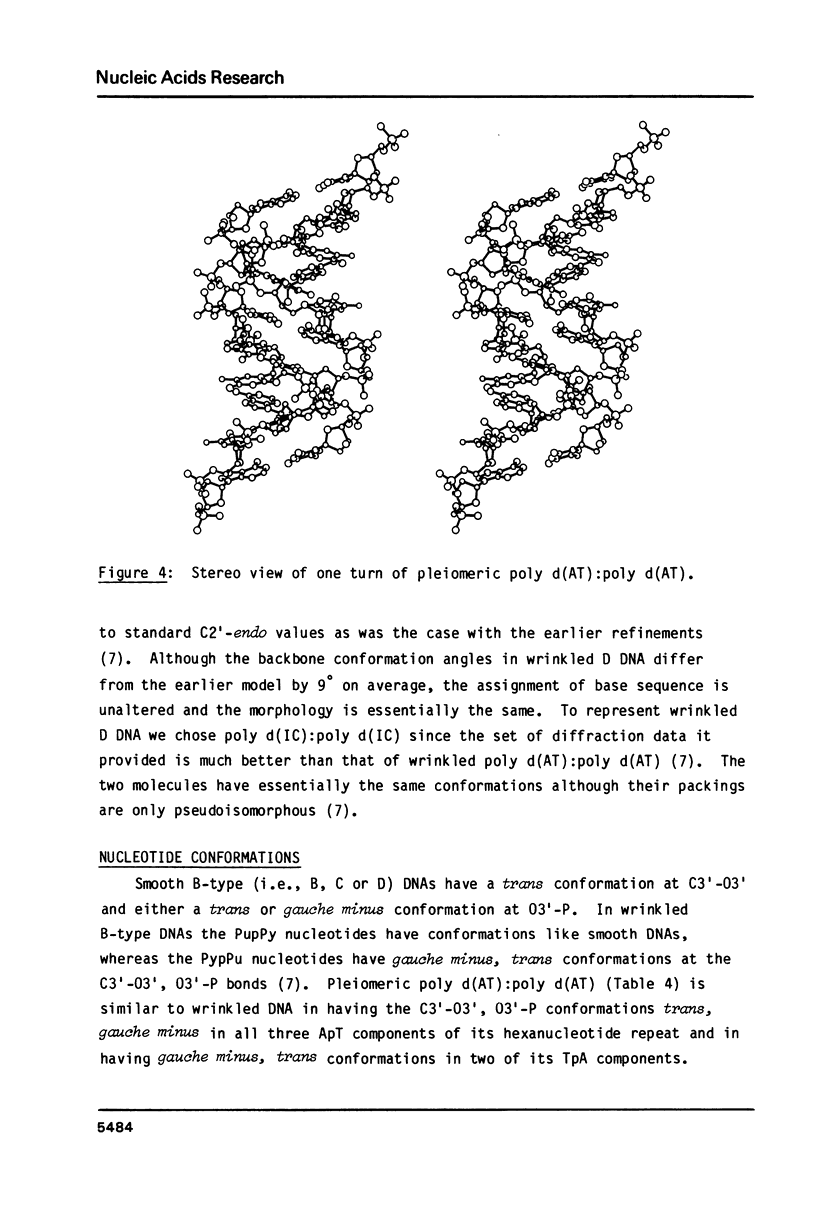
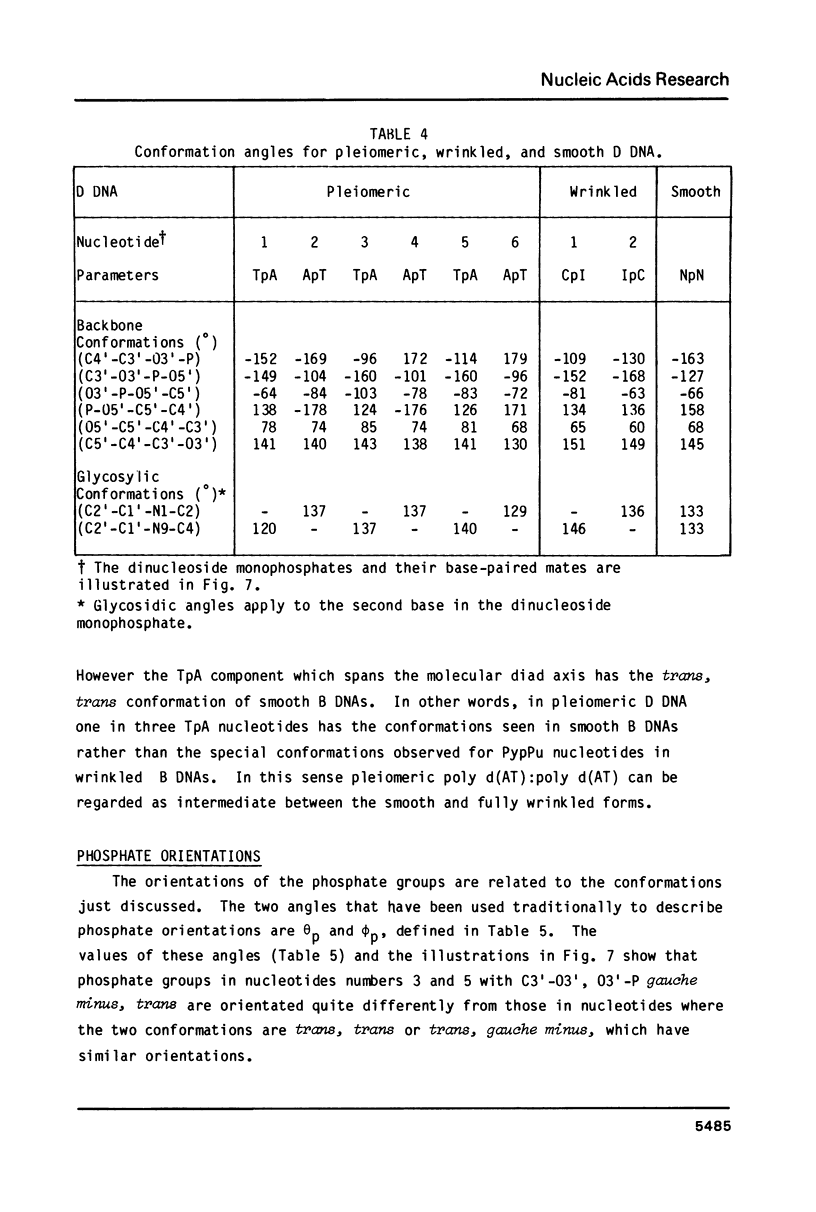
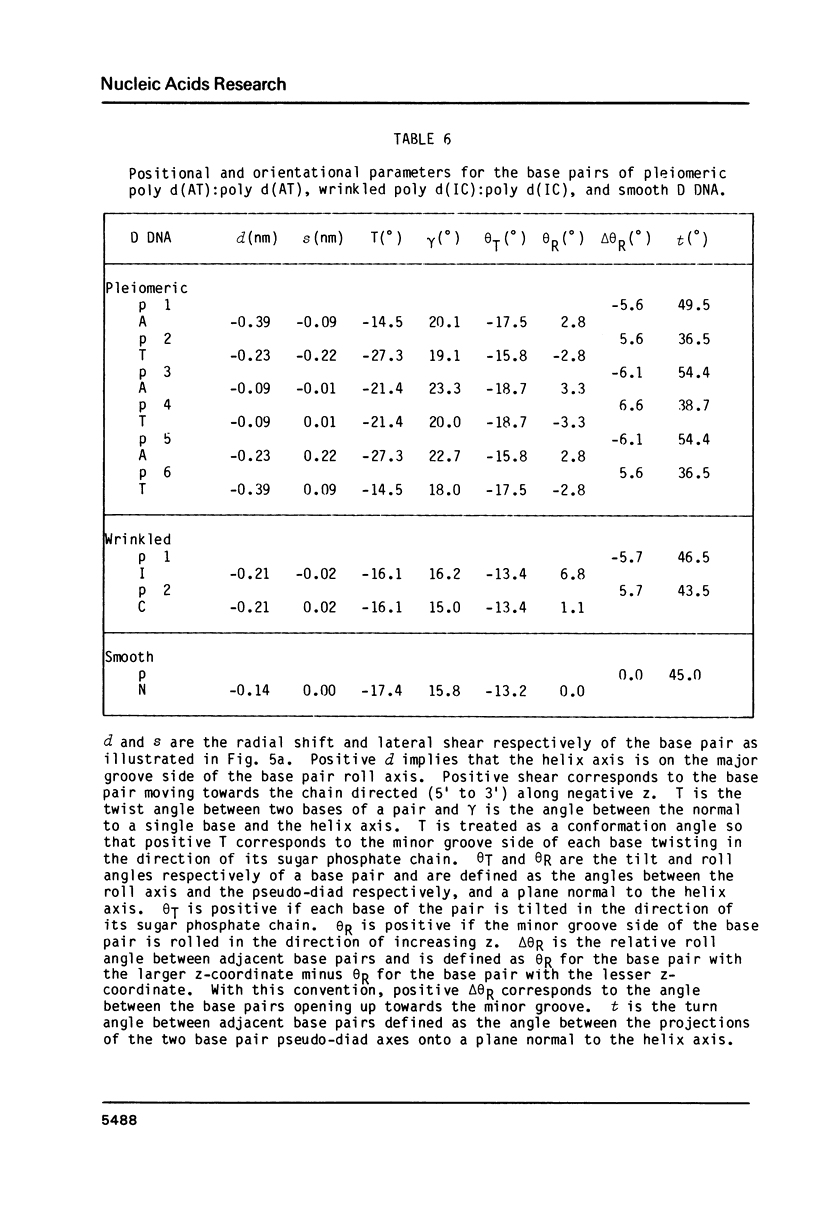
A chemically simple polynucleotide duplex, poly d(AT):poly d(AT), has been trapped in a fibrous form with a complex helical secondary structure with a large (7.4 nm) axial repeat 24 nucleotides long. The motif which is repeated by the symmetry elements is a hexanucleotide in which two residues (both TpA) have the less common gauche minus conformation at C3'-O3' and consequently distinctive phosphate orientations. This reinforces earlier conclusions that PypPu nucleotides tend to have different shapes from PupPy nucleotides and that DNA surfaces may signal what base sequences lie beneath them. The morphological differences between this pleiomeric DNA polymer and closely-related, but more symmetrical allomorphs are just as great as those observed in short DNA fragments in crystals.
Full text
PDF
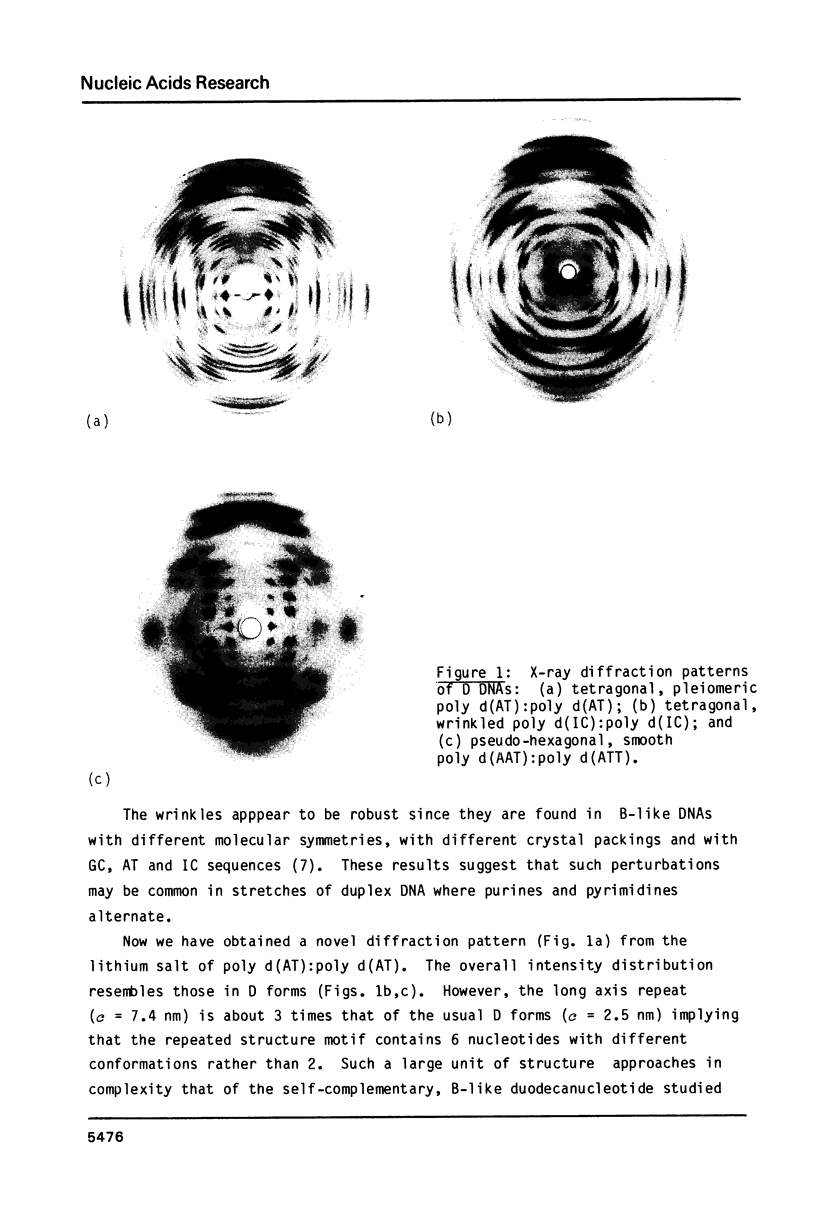



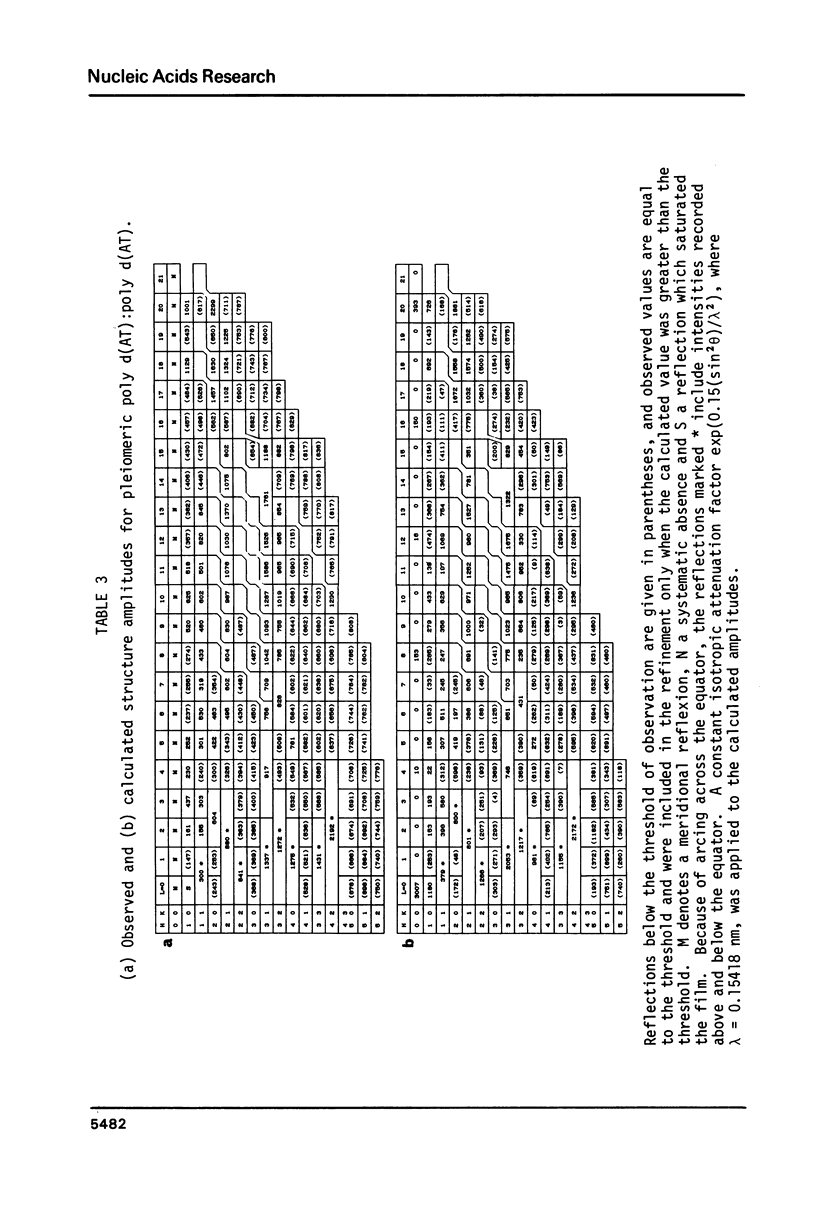








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Banerjee A. K., He R., Walker J. K. New wrinkles on polynucleotide duplexes. J Biomol Struct Dyn. 1983 Oct;1(2):437–452. doi: 10.1080/07391102.1983.10507453. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Puigjaner L. C., Walker J. K., Hall I. H., Birdsall D. L., Ratliff R. L. Wrinkled DNA. Nucleic Acids Res. 1983 Mar 11;11(5):1457–1474. doi: 10.1093/nar/11.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. The dimensions and shapes of the furanose rings in nucleic acids. Biochem J. 1972 Nov;130(2):453–465. doi: 10.1042/bj1300453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Leslie A. G., Arnott S., Chandrasekaran R., Ratliff R. L. Polymorphism of DNA double helices. J Mol Biol. 1980 Oct 15;143(1):49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- Selsing E., Arnott S. Conformations of poly(d(A-T-T))-poly(d(A-A-T)). J Mol Biol. 1975 Oct 15;98(1):243–248. doi: 10.1016/s0022-2836(75)80112-4. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Cruse W. B., Egert E., Kennard O., Sala G., Salisbury S. A., Viswamitra M. A. Crystalline A-dna: the X-ray analysis of the fragment d(G-G-T-A-T-A-C-C). Proc R Soc Lond B Biol Sci. 1981 Nov 24;213(1193):479–487. doi: 10.1098/rspb.1981.0076. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]





