Highlights
► We examine structures of viral proteins in complex with sialic acids. ► We focus on human pathogens. ► We compare binding modes and specificities of recognition. ► We identify common parameters of interaction. ► We provide a platform for understanding the specificities of other viral proteins.
Abstract
Viral infections are initiated by specific attachment of a virus particle to receptors at the surface of the host cell. For many viruses, these receptors are glycans that are linked to either a protein or a lipid. Glycans terminating in sialic acid and its derivatives serve as receptors for a large number of viruses, including several human pathogens. In combination with glycan array analyses, structural analyses of complexes of viruses with sialylated oligosaccharides have provided insights into the parameters that underlie each interaction. Here, we compare the currently available structural data on viral attachment proteins in complex with sialic acid and its variants. The objective is to define common parameters of recognition and to provide a platform for understanding the determinants of specificity. This information could be of use for the prediction of the location of sialic acid binding sites in viruses for which structural information is still lacking. An improved understanding of the principles that govern the recognition of sialic acid and sialylated oligosaccharides would also advance efforts to develop efficient antiviral agents.
Introduction
The monosaccharide sialic acid decorates all eukaryotic cell surfaces, capping many different oligosaccharide structures on N-linked and O-linked glycoproteins as well as on glycolipids [1]. Glycans terminating in sialic acid have emerged as a key class of receptors for an impressive number of viruses, many of which are human pathogens. The highly pathogenic Influenza A, B and C viruses as well as the human parainfluenza viruses attach to sialic acids (reviewed in [2, 3, 4]). Coxsackievirus A24 variant and enterovirus 70, which cause Acute Hemorrhagic Conjunctivitis and have pandemic potential, also attach to sialylated oligosaccharides [5, 6]. Epidemic Keratoconjunctivitis (EKC) has been linked to several human D-type adenoviruses, and one of these has recently been shown to attach to the disialylated GD1a motif [7••]. The human JC and BK polyomaviruses (JCV and BKV, respectively) cause a fatal demyelinating disease and kidney graft loss, respectively, in immunocompromised individuals. Both viruses use glycans terminating in sialic acid as their receptors [8••, 9]. Moreover, the recently identified Merkel cell polyomavirus, a human oncovirus, likely uses the trisialylated ganglioside GT1b as a receptor [10•, 11], and other mammalian polyomaviruses such as Simian Virus 40 (SV40) and murine polyomavirus (Polyoma) also bind glycans terminating in sialic acid [12, 13•, 14]. Most human noroviruses, the causative agents of violent gastrointestinal illnesses, attach to non-sialylated histo-blood group antigens, in contrast to murine norovirus, which binds to a sialylated oligosaccharide [15]. However, some strains of human noroviruses were recently shown to bind to the sialyl-Lewis X motif as well [16•]. Rotaviruses cause severe gastroenteritis in children. They have long been classified into strains that can be inhibited by neuraminidase treatment, which cleaves sialic acid from glycan sequences on host cells, and those that are insensitive to it [17, 18]. Neuraminidase-insensitive strains were presumed to use non-sialylated receptors. Interestingly, the ‘neuraminidase-insensitive’ rotavirus strain Wa was recently shown to attach to the ganglioside GM1, which carries a branching sialic acid [19••]. Because of its branched structure, this particular carbohydrate is difficult to cleave with neuraminidases [20, 21]. SV40 had likewise been presumed to attach to a non-sialylated carbohydrate [22, 23, 24] before GM1 was identified as its receptor [12]. Sialic acid, therefore, has to be considered as a possible receptor component even for viruses whose infectivity cannot be modulated by treatment of cells with commonly used neuraminidases.
The structure of the most common sialic acid in humans, α-5-N-acetyl-neuraminic acid (Neu5Ac), features four protruding functional groups (carboxylate, hydroxyl, N-acetyl and glycerol functions). Compared to more simple monosaccharides, the large number of functional groups enables sialic acids to participate in an unparalleled number of hydrogen bonds, salt bridges and non-polar interactions. Since sialic acid is typically located at the terminus of a glycan, its functions are easily accessible for interactions. Perhaps it is not surprising, therefore, that the sialic acid itself serves as the major point of contact with the glycan-binding viral attachment protein in all cases where structural information of sufficient resolution is available [8••, 13•, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35•, 36•, 37, 38, 39, 40, 41•].
In this review, we compare binding modes of sialic acids and sialylated receptors in representative structures of virus-receptor complexes in order to derive parameters that guide the recognition of sialic acid and its derivatives. Such parameters could be useful for the prediction of new sialic acid binding sites in viral proteins, or of altered modes of sialic acid (and its derivatives) binding in different viral serotypes and strains.
Attachment to terminal sialic acid
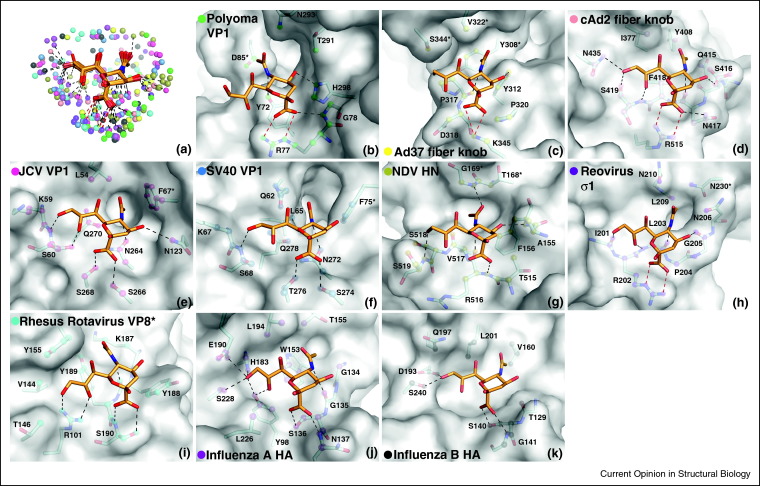
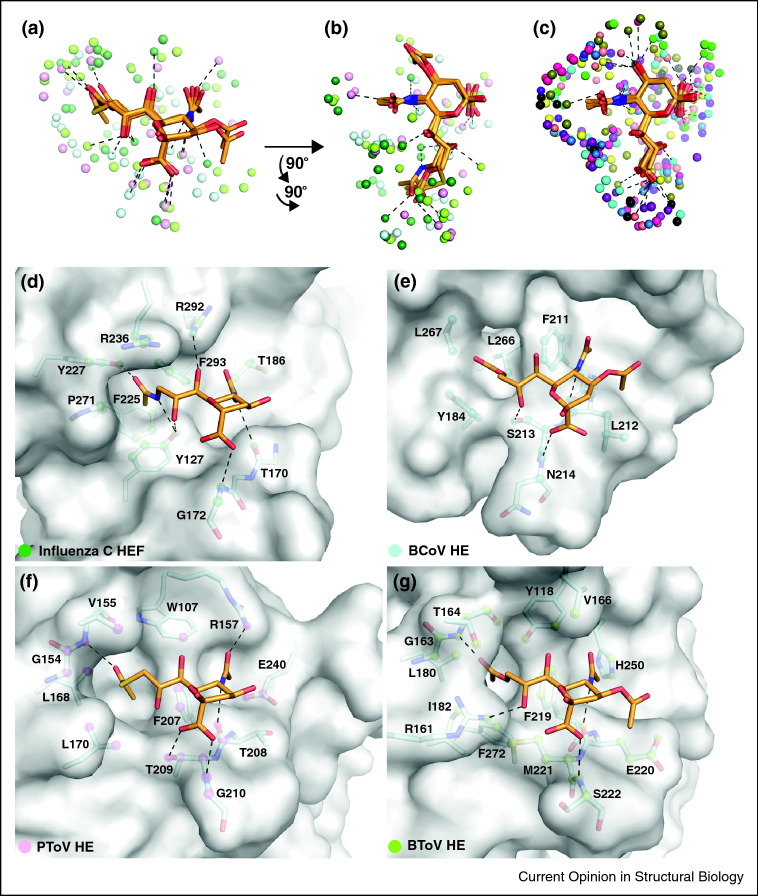
Several structures of viral attachment proteins in complex with sialylated compounds have been determined recently, providing new insights into viral specificity for glycan receptors [8••, 13•, 35•, 36•, 37, 38, 39, 41•]. Taken together, the known structures now form a large database that is suitable for the closer examination of contacts in order to compare the modes of interaction and attempt to define common principles of sialic acid recognition. We have investigated here the mode of Neu5Ac binding for ten different viral attachment proteins (Figure 1 ): the hemagglutinins (HAs) of Influenza A and B viruses [26, 34], the adenovirus serotype 37 (Ad37) fiber knob [7••], the canine adenovirus serotype 2 (cAd2) fiber knob [39], the major capsid proteins VP1 of the polyomaviruses Polyoma [29], SV40 [13•], and JCV [8], the attachment protein σ1 of human type 3 orthoreovirus [41•], the attachment protein VP8* of Rhesus Rotavirus [30], and the hemagglutinin-neuraminidase (HN) of Newcastle Disease Virus (NDV) [31]. As the interactions with terminal sialic acid are very similar among different types of Influenza A HAs, the structure of the H3 type [26] was chosen to represent this group. In addition, we have also analyzed contacts in the hemagglutinin-esterase-fusion (HEF) protein of Influenza C virus [27] and the hemagglutinin-esterase (HE) proteins of Bovine Coronavirus (BCoV) [35], Bovine Torovirus (BToV) [36•] and Porcine Torovirus (PToV) [36•] (Figure 2 ). These four proteins bind to derivatives of Neu5Ac that are O-acetylated at position 9 or at positions 4 and 9. In cases where the attachment protein also has receptor-destroying enzymatic activity, such as in the HN and HE(F) proteins, only the sites that can clearly be attributed to attachment are considered here, thus excluding the dual function neuraminidase site of some HN proteins [32, 42, 43].
Figure 1.
Interactions of viral attachment proteins with terminal Neu5Ac. (a) Contacts of viral proteins with terminal Neu5Ac. The complex structures shown in panels (b–k) were superposed using the terminal sialic acid residues. The Neu5Ac residues are shown in stick representation, with carbons colored orange, oxygens red, and nitrogens blue. All protein atoms within a 4.0 Å radius of Neu5Ac atoms are displayed as spheres and colored according to the color code depicted in panels (b–k). Hydrogen bonds and salt bridges are represented with black and red dashed lines, respectively. (b–k) Neu5Ac binding sites of Polyoma VP1 (b, pdb 1VPS [29]), Ad37 fiber knob (c, pdb 3N0I [7]), cAd2 fiber knob (d, pdb 2WBV [39]), JCV VP1 (e, pdb 3NXD [8]), SV40 VP1 (f, pdb 3BWR [13]), NDV HN (g, pdb 1USR [31]), Reovirus σ1 (h, pdb 3S6X [41]), Rhesus Rotavirus VP8* (i, pdb 1KQR [30]), Influenza A HA (j, 1HGG [26]), and Influenza B HA (k, pdb 2RFT [34]). In all cases, Neu5Ac is shown in stick representation and colored as in panel (a). The protein surfaces are colored gray, with residues interacting with Neu5Ac shown in stick representation and colored by element. Protein atoms within a 4.0 Å radius around Neu5Ac are highlighted with colored spheres. In cases where the Neu5Ac binding site is formed by two protein chains, one of the chains is denoted with an asterisk.
Figure 2.
Interactions of viral attachment proteins with O-acetylated Neu5Ac. (a) Contacts of viral proteins with terminal O-acetylated Neu5Ac. Complex structures were superposed on the terminal substituted Neu5Ac. Interacting protein atoms as well as hydrogen bonds and salt bridges are shown as in Figure 1a. Top view of O-acetylated (b) and unsubstituted (c) Neu5Ac, with contacting protein atoms shown as colored spheres. The views are rotated from those in Figure 1 and (a) by two 90° rotations, one around a horizontal and one around a vertical axis. (d–g) Binding sites for O-acetylated Neu5Ac in Influenza C HEF (d) [27], BCoV HE (e, pdb 3CL5 [35]), PToV HE (f, pdb 3I27 [36]) and BToV HE (g, pdb 3I1L [36]). The structures are displayed as in Figure 1b–k.
The investigated viral attachment proteins are, for the most part, not homologous to one another and belong to unrelated viruses that differ in envelope structure and genome type. Nevertheless, their interactions with sialic acid display striking similarities (Figure 1, Figure 2). In all complexes, the sialic acid adopts essentially the same conformation, namely a trans conformation of the 5-N-acetyl group and an α-conformation at the anomeric carbon, which is dominant in biological oligosaccharides. Interestingly, all attachment proteins, including the ones that bind to O-acetylated compounds, make extensive contacts with one face of the sialic acid ring, while the other face is engaged by only few contacts (Figure 1, Figure 2). A likely reason for this preference is the formation of two key contacts that are formed in all complexes in a similar manner. One of these contacts involves the negatively charged carboxylate group, which is most often recognized by two parallel hydrogen bonds or a salt bridge. Each of the analyzed proteins donates at least one hydrogen bond to a carboxylate oxygen atom. The second contact involves the nitrogen atom in the N-acetyl group. With only one exception, all proteins receive a hydrogen bond from this nitrogen atom. The spatial arrangement of carboxyl group and the N-acetyl nitrogen thus helps distinguish sialic acids from other monosaccharides. Both groups project from the same face of the sialic acid ring, accounting for the preferential binding of this face of the monosaccharide. Apart from these two key interactions, the proteins engage in different hydrogen bonding patterns to various hydroxyl groups of the glycerol chain or the ring, or to additional acetyl substituents.
Examination of van der Waals interactions between the viral attachment proteins and Neu5Ac reveals that only about 50% of the contact surface of Neu5Ac participates in such contacts (Figure 1, Figure 2). The resulting shape resembles a rimmed imprint of the binding face of Neu5Ac on the protein surface (Figure 1, Figure 2). In all complexes, van der Waals interactions are formed with the methyl group of the N-acetyl chain. However, the different proteins interacting with Neu5Ac sample different epitopes on the Neu5Ac contact surface. For example, JCV and SV40 VP1, Rhesus Rotavirus VP8*, and Influenza A HA all center their van der Waals contacts on the glycerol and N-acetyl chains (Figure 1e,f,i,j). The surfaces of all four viruses feature subtle protrusions that separate the recessed areas in which the glycerol and N-acetyl chains are bound. Polyoma VP1, on the other hand, mainly contacts Neu5Ac from the other side, and does not interact with the glycerol chain at all (Figure 1b). Examination of the binding surfaces demonstrates that shape complementarity is an important factor in the engagement of sialic acid. As the contact areas are quite small and the sialic acids are partially exposed to solvent, adding or removing a single contact can thus have significant effects on the affinity of a given virus for sialic acid or its variants.
Recognition of sialic acid variants
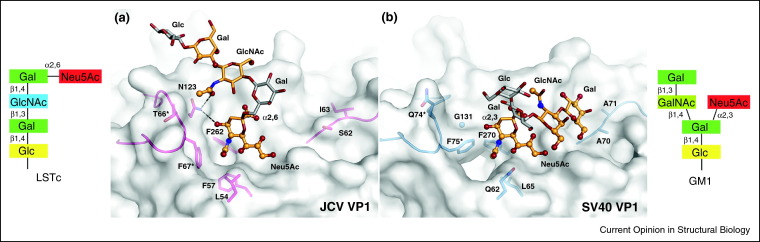
The parent compound of Neu5Ac, neuraminic acid, can feature numerous modifications that give rise to over 40 different known sialic acid variants [44, 45]. Several of these modifications are predominantly found on specific cell types and tissues, or in selected species. It is perhaps not surprising, therefore, that some viruses exploit this divergence and preferentially recognize sialic acids other than Neu5Ac. The database of viral protein structures contains few examples of viruses attaching to O-acetylated Neu5Ac [27, 35•, 36•], but their analysis is nevertheless informative (Figure 2). While the key hydrogen bonds to one face of the sialic acid are the same as described for Neu5Ac, the distribution of van der Waals contacts is somewhat altered. In the four complexes, the majority of van der Waals contacts are centered around the unique 9-O-acetyl groups as well as the adjacent side of the N-acetyl group, while the opposite side of the ring does not engage in as many interactions (Figure 2b). The 9-O-acetyl group inserts deeply into tight-fitting protein cavities, providing selectivity for sialic acids modified in this manner. Recognition of different sialic acids is also a likely cause of changes in tropism and host range. The interactions of SV40 with GM1 ganglioside containing α-N-5-glycolyl neuraminic acid (Neu5Gc), a sialic acid present in simians but not humans, illustrate this point [46]. SV40 VP1 features a large pocket near the Neu5Ac N-acetyl group (Figure 1, Figure 3), and it is tempting to speculate that this pocket serves to accommodate the additional hydroxyl group of Neu5Gc [13•]. VP1 of the human JCV (Figure 1, Figure 3), whose sialic acid binding site is largely similar to that of SV40 VP1, features a much smaller pocket that likely prefers the smaller human Neu5Ac over the simian Neu5Gc.
Figure 3.
Different sialic acid and context specificity of SV40 and JCV VP1. (a) JCV VP1 in complex with LSTc oligosaccharide. (b) SV40 VP1 in complex with GM1 oligosaccharide. The oligosaccharides are shown in stick representation and colored by element, with oxygens in red and nitrogens in blue. Monosaccharides that approach the protein closer than 4.0 Å are colored in bright orange, while those not contacting the protein are colored gray. Oligosaccharide atoms within a 4.0 Å radius around the proteins are highlighted as spheres. The protein surface is shown in gray. Residues that define the different oligosaccharide specificities of the two proteins are shown as sticks and colored blue and pink for SV40 and JCV VP1, respectively. Residues from a different polypeptide chain are denoted with an asterisk.
Conservation of sialic acid binding sites
Sialic acid binding sites are often highly conserved in homologous viruses. This is evident when comparing different HA types of Influenza A, the capsid proteins of JCV and SV40 (Figure 3a,b), or the HE proteins of PToV and BToV (Figure 2f,g). In all three cases, the sialic acid engages the two homologous proteins using similar contacts and is therefore bound in the same orientation and position. However, at least one example exists where homologous proteins, the Ad37 and cAd2 fiber knobs, bind sialic acid at different locations (Figure 1c,d). Interestingly, there are several examples of highly homologous proteins that bind sialic acid at the same site but in different orientations. The VP1 proteins of Polyoma and SV40, for example, feature a very high level of sequence identity, and they bind sialic acid in generally similar areas on the protein surface. However, the orientations of the bound sialic acids differ markedly (Figure 1b,f) [13•]. Similarly, Influenza C HEF and BCoV HE bind sialic acid at the same position, but again in different orientations with respect to the proteins and with contacts provided by different structural elements (Figure 2d,e) [35]. These two examples demonstrate the need for caution when modeling interactions with sialic acid based on a homologous structure.
Specificity for sialic acid in different contexts
As sialic acid is ubiquitous at the cell surface, interactions with subsequent carbohydrates are typically employed to define specificity and tropism. Glycan microarrays have been highly useful in revealing the determinants of such interactions [47]. The critical role of the context of the sialic acid — linkage type, as well as length, sequence and conformational preferences of the remaining oligosaccharide chain — is perhaps best illustrated by its influence on the host range of influenza viruses. Briefly, human Influenza A viruses engage long glycans terminating in α2,6-linked sialic acid that preferentially adopt a bent conformation and that are expressed extensively in the upper airway epithelia of humans. Avian strains predominantly recognize shorter glycans that adopt a linear conformation and that often contain α2,3-linked linked sialic acid [48•]. The vast database on Influenza A HA structures in complex with sialylated ligands, and concurrent glycan array analyses, has been the subject of several excellent recent reviews [2, 3], and will therefore not be discussed in detail here. However, glycan array screening has recently helped to unravel the identities of sialylated glycan receptors for two pathogenic human viruses, and structural biology has defined the nature of interaction in both cases [8]. We review below each of these two examples, which illuminate the importance of the context in which the terminal sialic acid is placed.
Achieving specificity through a limited set of additional interactions
Several members of the polyomavirus family use sialylated receptors for cell attachment. Crystal structures of two members of the family in complex with their cognate receptors have been determined recently: SV40 VP1 has been crystallized in complex with the oligosaccharide portion of its ganglioside receptor GM1 [13•], whereas the structure of the VP1 protein of human JCV has been solved with the pentasaccharide receptor fragment LSTc (Lacto-series tetrasaccharide c) [8]. Both receptors feature terminal Neu5Ac, which is α2,3-linked in the branched GM1 molecule and α2,6-linked in the linear LSTc structure (Figure 3). In each case, glycan array screening has unequivocally identified the type of the receptor [8••, 13•]. Moreover, although both GM1 and LSTc were present on the arrays, JCV VP1 failed to interact with GM1, and SV40 VP1 also did not recognize the LSTc compound. Thus, both proteins are highly specific for their cognate receptors. A comparison of the two structures shows that the sialic acid portions of the two receptors form largely equivalent interactions with their respective proteins (Figure 1, Figure 3). The remarkable specificity for each receptor can be attributed to contacts that involve the remaining parts of the oligosaccharides. The LSTc compound assumes a bent conformation, forming additional contacts via the N-acetyl group of its third sugar, GlcNAc, to N123 of JCV (Figure 3a). An α2,3-linked Neu5Ac would not adopt a similarly bent conformation, explaining why sialylparagloboside, which is identical to LSTc except for its α2,3-linked Neu5Ac, does not bind JCV. Modeling LSTc into the SV40 VP1 binding site by superimposing the two sialic acid structures suggests that LSTc could be tolerated by SV40. However, as the residue equivalent to N123 in JCV is a glycine (G131) in SV40, no favorable contacts to the GlcNAc residue can be generated (Figure 3b). The inability to form such an interaction most likely explains why SV40 cannot bind LSTc. It, therefore, appears that the formation of a very small number of contacts is largely responsible for defining the specificity of VP1 for LSTc. The reverse combination, a GM1 ligand bound to JCV, would likely be disfavored due to steric clashes with JCV VP1 residue S62, which is an alanine in SV40.
Achieving specificity through multivalent binding
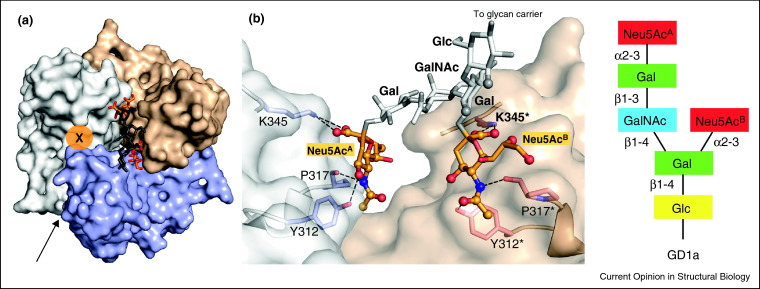
Because of their small contact surfaces and solvent-exposed binding sites, interactions between individual viral attachment proteins and sialylated oligosaccharides are typically of low affinity, with dissociation constants in the millimolar range [13•, 28, 49, 50]. In many cases, high-affinity adherence to the target cell is achieved through the utilization of several low-affinity binding sites. However, receptor clustering is not always necessary to achieve higher-affinity binding. The interaction of Ad37 with its recently identified glycan receptor GD1a illustrates another strategy. It has long been established that Ad37 fiber knobs bind receptors terminating in sialic acid [28, 51], but the nature of the glycan has remained elusive. Glycan array screening has recently revealed that Ad37 fiber knobs specifically recognize the oligosaccharide GD1a, a disialylated compound that features two branches, each terminating in sialic acid [7••]. A structural analysis of the trimeric Ad37 fiber knob in complex with GD1a established that the two terminal sialic acid residues bind to two different Ad37 fiber knob protomers in an identical manner, thus engaging two of the three possible binding sites [7••] (Figure 4 ). This bivalent interaction results in a 250-fold higher affinity (K d = 19 μM) [7••] compared to the monovalent sialyllactose–Ad37 knob interaction (K d = 5 mM) [28]. Thus, although each protomer in an Ad37 fiber knob would be able to bind sialic acid attached to different oligosaccharide structures, specificity for GD1a is generated by a multivalent interaction in which two protomers interact with the same receptor in an identical manner. It is conceivable that trivalent compounds that engage all three binding sites of the Ad37 fiber knobs would have even higher affinity, thus providing a platform for the development of antiviral inhibitors. Using such a strategy, a multivalent inhibitor has been developed that is able to neutralize pentameric shiga-like toxins with very high efficiency [52]. A similar strategy could be useful to develop molecules that inhibit viral attachment proteins, which usually occur as multimers at the viral surface.
Figure 4.
Binding of GD1a glycan to the Ad37 fiber knob protein. (a) Structure of Ad37 fiber knob in complex with the GD1a oligosaccharide [7]. The three different Ad37 fiber knob protomers are shown in surface representation and are colored gray, light red and blue. The GD1a glycan is drawn in stick representation, with both Neu5Ac residues highlighted in color (carbons in orange, oxygens in red and nitrogens in blue). The bridging glycan residues are shown in dark gray. The third binding site (marked by ‘X’) is blocked due to crystal contacts. The arrow indicates the viewing direction shown in panel (b). (b) Interactions between Ad37 fiber knob residues and GD1a. Two different Ad37 fiber knob protomers are shown in transparent surface representation (white and light red). The third protomer is not shown for clarity. Ad37 residues contacting GD1a are shown in stick representation, with oxygens in red and nitrogens in blue. The GD1a glycan is shown in stick representation, with both terminal Neu5Ac residues highlighted in color (carbons in orange, oxygens in red and nitrogens in blue). The bridging glycan residues are shown in gray. Glycan atoms within a distance of 4 Å to Ad37 protein atoms are drawn as spheres. Hydrogen bonds to GD1a glycan are represented with black dashes. Residues from different protomers are denoted with an asterisk.
Conclusions
A large number of viruses, including many serious human pathogens, use sialylated oligosaccharides for cell attachment. Common principles of interaction can be established by comparing the sialic acid binding modes of different viruses. In most cases, interactions between a viral attachment protein and its glycan receptor involve primarily the sialic acid itself, which is bound with a relatively small contact area in a solvent-exposed region of the protein. Consistent with this, the affinities of such interactions are, at least in cases where they have been measured, very low. Nevertheless, many of the viruses discussed here achieve remarkable specificity for a single type of sialylated oligosaccharide by establishing a small number of auxiliary interactions with functional groups that lie beyond the sialic acid, and by excluding some possible ligands through steric clashes. The auxiliary interactions generally involve fewer hydrogen bonds and bury a smaller amount of surface compared to the interactions that involve the sialic acid itself. It thus appears that many viruses use the unique properties of sialic acid as a ‘hook’ that allows them to adhere to the cell, and modulate binding in different strains or families by subtly altering structural elements in the vicinity of this hook. In a (so far) unique variation of this strategy, the Ad37 knob establishes selectivity for its GD1a glycan receptor by multivalent binding to a single receptor carrying two terminal sialic acid moieties, thus adhering to two identical ‘hooks’ separated by a defined spacer. The prominence of sialic acid in viral attachment may form a basis for new approaches to combat viruses. Compounds that mimic sialic acid have already proved useful as inhibitors of the influenza virus neuraminidase [53] and can also efficiently inhibit the receptor-binding site of the Influenza A virus hemagglutinin [54]. The structural analysis of the Ad37–GD1a interaction has also led to the design and synthesis of a trivalent compound designed to block attachment of adenoviruses that cause EKC [55]. Glycan microarrays have been extraordinarily useful in identifying the correct receptors for many viral proteins [8••, 13•, 47, 56••], which is a prerequisite for structural studies. However, proper interpretation of the information provided by glycan array screening and structural analyses requires affinity data. Such data are often difficult to obtain and compare, and they are currently lacking for many complexes. Being able to correlate affinity measurements with structural data would significantly advance the design of antiviral agents, and, together with oligosaccharide expression data, help to explain viral tropism.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgments
The authors wish to thank Peter Rosenthal for providing the coordinates for Influenza C HEF in complex with 9-N-acetylated Neu5Ac α-methyl sialoside. Because of the large number of available structures of viral proteins in complex with sialylated receptors, we had to select some representative examples for our review. We apologize to all colleagues whose contributions we could not adequately discuss due to space constraints. This work has been supported by grants from the Deutsche Forschungsgemeinschaft (SFB-685), the National Institutes of Health (P01-NS065719) and the Baden-Württemberg Stiftung to TS as well as a student fellowship of the University of Tübingen to JB.
References
- 1.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamblin S.J., Skehel J.J. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010;285:28403–28409. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viswanathan K., Chandrasekaran A., Srinivasan A., Raman R., Sasisekharan V., Sasisekharan R. Glycans as receptors for influenza pathogenesis. Glycoconj J. 2010;27:561–570. doi: 10.1007/s10719-010-9303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villar E., Barroso I.M. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview. Glycoconj J. 2006;23:5–17. doi: 10.1007/s10719-006-5433-0. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson E.C., Jamshidi F., Johansson S.M., Oberste M.S., Arnberg N. Sialic acid is a cellular receptor for coxsackievirus A24 variant, an emerging virus with pandemic potential. J Virol. 2008;82:3061–3068. doi: 10.1128/JVI.02470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander D.A., Dimock K. Sialic acid functions in enterovirus 70 binding and infection. J Virol. 2002;76:11265–11272. doi: 10.1128/JVI.76.22.11265-11272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Nilsson E.C., Storm R.J., Bauer J., Johansson S.M., Lookene A., Angstrom J., Hedenstrom M., Eriksson T.L., Frangsmyr L., Rinaldi S. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat Med. 2011;17:105–109. doi: 10.1038/nm.2267. [DOI] [PubMed] [Google Scholar]; The authors show by glycan array screening that the receptor-recognizing knob domain of the Ad37 fiber protein specifically binds a branched hexasaccharide that is present in the GD1a ganglioside and that features two terminal sialic acids. Structural analysis reveals that the two terminal sialic acids dock into two of the three previously established sialic acid-binding sites in the trimeric Ad37 knob. The findings form a basis for the design and development of sialic acid-containing antiviral drugs for topical treatment of EKC.
- 8••.Neu U., Maginnis M.S., Palma A.S., Ströh L., Feizi T., Atwood W.J., Stehle T. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe. 2010;8:309–319. doi: 10.1016/j.chom.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using glycan array screening, the authors identify the LSTc glycan, a sialylated pentasaccharide present on glycoproteins and glycolipids, as a specific receptor recognition motif for the human JC polyomavirus. The crystal structure of the capsid protein of this virus was solved alone and in complex with LSTc, explaining its distinct receptor specificity.
- 9.Low J.A., Magnuson B., Tsai B., Imperiale M.J. Identification of gangliosides GD1b and GT1b as receptors for BK virus. J Virol. 2006;80:1361–1366. doi: 10.1128/JVI.80.3.1361-1366.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes a new human polyomavirus that is causally linked to an aggressive type of skin cancer, the Merkel cell carcinoma.
- 11.Erickson K.D., Garcea R.L., Tsai B. Ganglioside GT1b is a putative host cell receptor for the Merkel cell polyomavirus. J Virol. 2009;83:10275–10279. doi: 10.1128/JVI.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai B., Gilbert J.M., Stehle T., Lencer W., Benjamin T.L., Rapoport T.A. Gangliosides are receptors for murine polyoma virus and SV40. Embo J. 2003;22:4346–4355. doi: 10.1093/emboj/cdg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Neu U., Woellner K., Gauglitz G., Stehle T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc Natl Acad Sci U S A. 2008;105:5219–5224. doi: 10.1073/pnas.0710301105. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors use glycan array screening, affinity measurements and structural analysis to define the interactions of Simian Virus 40 with its ganglioside receptor GM1.
- 14.Stehle T., Yan Y., Benjamin T.L., Harrison S.C. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- 15.Taube S., Perry J.W., Yetming K., Patel S.P., Auble H., Shu L., Nawar H.F., Lee C.H., Connell T.D., Shayman J.A. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J Virol. 2009;83:4092–4101. doi: 10.1128/JVI.02245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Rydell G.E., Nilsson J., Rodriguez-Diaz J., Ruvoen-Clouet N., Svensson L., Le Pendu J., Larson G. Human noroviruses recognize sialyl Lewis × neoglycoprotein. Glycobiology. 2009;19:309–320. doi: 10.1093/glycob/cwn139. [DOI] [PubMed] [Google Scholar]; This study shows that some human norovirus strains have at least two binding specificities: one related to α1,2-fucosylated carbohydrates and another related to sialylated carbohydrates.
- 17.Isa P., Arias C.F., Lopez S. Role of sialic acids in rotavirus infection. Glycoconj J. 2006;23:27–37. doi: 10.1007/s10719-006-5435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciarlet M., Ludert J.E., Iturriza-Gomara M., Liprandi F., Gray J.J., Desselberger U., Estes M.K. Initial interaction of rotavirus strains with N-acetylneuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J Virol. 2002;76:4087–4095. doi: 10.1128/JVI.76.8.4087-4095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Haselhorst T., Fleming F.E., Dyason J.C., Hartnell R.D., Yu X., Holloway G., Santegoets K., Kiefel M.J., Blanchard H., Coulson B.S. Sialic acid dependence in rotavirus host cell invasion. Nat Chem Biol. 2009;5:91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]; Using NMR analysis and infectivity assays, the authors present strong evidence that N-acetylneuraminic acid is a key determinant for binding of some rotaviruses, in contrast to the widely accepted paradigm that sialic acids are irrelevant in host cell recognition by sialidase-insensitive rotaviruses.
- 20.Miller-Podraza H., Bradley R.M., Fishman P.H. Biosynthesis and localization of gangliosides in cultured cells. Biochemistry. 1982;21:3260–3265. doi: 10.1021/bi00257a002. [DOI] [PubMed] [Google Scholar]
- 21.Svennerholm L. Chromatographic separation of human brain gangliosides. J Neurochem. 1963;10:613–623. doi: 10.1111/j.1471-4159.1963.tb08933.x. [DOI] [PubMed] [Google Scholar]
- 22.Clayson E.T., Compans R.W. Characterization of simian virus 40 receptor moieties on the surfaces of Vero C1008 cells. J Virol. 1989;63:1095–1100. doi: 10.1128/jvi.63.3.1095-1100.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah K.V., Valis J.D., Daniel R.W. Hemagglutination with simian papovavirus SA12. J Clin Microbiol. 1978;7:396–398. doi: 10.1128/jcm.7.4.396-398.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clayson E.T., Compans R.W. Entry of simian virus 40 is restricted to apical surfaces of polarized epithelial cells. Mol Cell Biol. 1988;8:3391–3396. doi: 10.1128/mcb.8.8.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weis W.I., Brown J.H., Cusack S., Paulson J.C., Skehel J.J., Wiley D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 26.Sauter N.K., Hanson J.E., Glick G.D., Brown J.H., Crowther R.L., Park S.J., Skehel J.J., Wiley D.C. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry. 1992;31:9609–9621. doi: 10.1021/bi00155a013. [DOI] [PubMed] [Google Scholar]
- 27.Rosenthal P.B., Zhang X., Formanowski F., Fitz W., Wong C.H., Meier-Ewert H., Skehel J.J., Wiley D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burmeister W.P., Guilligay D., Cusack S., Wadell G., Arnberg N. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J Virol. 2004;78:7727–7736. doi: 10.1128/JVI.78.14.7727-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stehle T., Harrison S.C. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 1997;16:5139–5148. doi: 10.1093/emboj/16.16.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dormitzer P.R., Sun Z.Y., Wagner G., Harrison S.C. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002;21:885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaitsev V., von Itzstein M., Groves D., Kiefel M., Takimoto T., Portner A., Taylor G. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan P., Thompson T.B., Wurzburg B.A., Paterson R.G., Lamb R.A., Jardetzky T.S. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Blanchard H., Yu X., Coulson B.S., von Itzstein M. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*) J Mol Biol. 2007;367:1215–1226. doi: 10.1016/j.jmb.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q., Tian X., Chen X., Ma J. Structural basis for receptor specificity of influenza B virus hemagglutinin. Proc Natl Acad Sci U S A. 2007;104:16874–16879. doi: 10.1073/pnas.0708363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Zeng Q., Langereis M.A., van Vliet A.L., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci U S A. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]; A structure analysis of the coronavirus hemagglutinin-esterase in complex with its receptor establishes an evolutionary link to the influenza C-like hemagglutinin-esterase fusion protein.
- 36•.Langereis M.A., Zeng Q., Gerwig G.J., Frey B., von Itzstein M., Kamerling J.P., de Groot R.J., Huizinga E.G. Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases. Proc Natl Acad Sci U S A. 2009;106:15897–15902. doi: 10.1073/pnas.0904266106. [DOI] [PMC free article] [PubMed] [Google Scholar]; The structure analysis of two torovirus hemagglutinin-esterases sheds light on the evolution of these proteins and provides insight into mechanisms of substrate binding, substrate recognition, and receptor selection in this important class of viral proteins.
- 37.Liu J., Stevens D.J., Haire L.F., Walker P.A., Coombs P.J., Russell R.J., Gamblin S.J., Skehel J.J. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci U S A. 2009;106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin T., Wang G., Li A., Zhang Q., Wu C., Zhang R., Cai Q., Song W., Yuen K.Y. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology. 2009;392:73–81. doi: 10.1016/j.virol.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 39.Seiradake E., Henaff D., Wodrich H., Billet O., Perreau M., Hippert C., Mennechet F., Schoehn G., Lortat-Jacob H., Dreja H. The cell adhesion molecule “CAR” and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution. PLoS Pathog. 2009;5:e1000277. doi: 10.1371/journal.ppat.1000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H., Chen L.M., Carney P.J., Donis R.O., Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 2010;6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Reiter D.M., Frierson J.M., Halvorson E.E., Kobayashi T., Dermody T.S., Stehle T Crystal structure of reovirus attachment protein σ1 in complex with sialylated oligosaccharides. PLoS Pathog. 2011;7:e1002166. doi: 10.1371/journal.ppat.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first example of a viral protein engaging a carbohydrate receptor through a fibrous, trimeric structure formed from repeating units, the β-spiral. The ligand-binding site is formed by a compact, trimeric module that could potentially be grafted onto other trimeric proteins, endowing them with carbohydrate binding capabilities.
- 42.Crennell S., Takimoto T., Portner A., Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 43.Lawrence M.C., Borg N.A., Streltsov V.A., Pilling P.A., Epa V.C., Varghese J.N., McKimm-Breschkin J.L., Colman P.M. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 44.Schauer R. Achievements and challenges of sialic acid research. Glycoconj J. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varki A., Schauer R. Sialic acids. In: Varki A., Cummings R.D., Esko J.D., editors. Essentials of Glycobiology. Cold Spring Harbor Press; 2009. [PubMed] [Google Scholar]
- 46.Campanero-Rhodes M.A., Smith A., Chai W., Sonnino S., Mauri L., Childs R.A., Zhang Y., Ewers H., Helenius A., Imberty A. N-glycolyl GM1 ganglioside as a receptor for simian virus 40. J Virol. 2007;81:12846–12858. doi: 10.1128/JVI.01311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Palma A.S., Feizi T. Carbohydrate microarrays: key developments in glycobiology. Biol Chem. 2009;390:647–656. doi: 10.1515/BC.2009.071. [DOI] [PubMed] [Google Scholar]
- 48•.Chandrasekaran A., Srinivasan A., Raman R., Viswanathan K., Raguram S., Tumpey T.M., Sasisekharan V., Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]; In this report, the authors show that a characteristic structural topology — not the α2–6 linkage itself — enables specific binding of the influenza virus hemagglutinin to α2–6 sialylated glycans and that recognition of this topology may be critical for adaptation of hemagglutinin to bind glycans in the upper respiratory tract of humans.
- 49.Stehle T., Harrison S.C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure Fold Des. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 50.Sauter N.K., Bednarski M.D., Wurzburg B.A., Hanson J.E., Whitesides G.M., Skehel J.J., Wiley D.C. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry. 1989;28:8388–8396. doi: 10.1021/bi00447a018. [DOI] [PubMed] [Google Scholar]
- 51.Arnberg N., Edlund K., Kidd A.H., Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor. J Virol. 2000;74:42–48. [PMC free article] [PubMed] [Google Scholar]
- 52.Kitov P.I., Sadowska J.M., Mulvey G., Armstrong G.D., Ling H., Pannu N.S., Read R.J., Bundle D.R. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature. 2000;403:669–672. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 53.von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat Rev Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 54.Matsubara T., Onishi A., Saito T., Shimada A., Inoue H., Taki T., Nagata K., Okahata Y., Sato T. Sialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J Med Chem. 2010;53:4441–4449. doi: 10.1021/jm1002183. [DOI] [PubMed] [Google Scholar]
- 55.Spjut S., Qian W., Bauer J., Storm R.J., Frangsmyr L., Stehle T., Arnberg N., Elofsson M A potent trivalent sialic acid inhibitor of adenovirus type 37 infection of human corneal cells. Angewandte Chemie. 2011;50:6519–6521. doi: 10.1002/anie.201101559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56••.Childs R.A., Palma A.S., Wharton S., Matrosovich T., Liu Y., Chai W., Campanero-Rhodes M.A., Zhang Y., Eickmann M., Kiso M. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using glycan array screening, the authors have compared the receptor-binding characteristics of two isolates of the novel pandemic H1N1 influenza virus with those of established influenza H1N1 virus strains. The paper presents an exemplary application of glycan microarray technology.