Abstract
Stark contrasts in clade species diversity are reported across the tree of life and are especially conspicuous when observed in closely related lineages. The explanation for such disparity has often been attributed to the evolution of key innovations that facilitate colonization of new ecological niches. The factors underlying diversification in bees remain poorly explored. Bees are thought to have originated from apoid wasps during the Mid-Cretaceous, a period that coincides with the appearance of angiosperm eudicot pollen grains in the fossil record. The reliance of bees on angiosperm pollen and their fundamental role as angiosperm pollinators have contributed to the idea that both groups may have undergone simultaneous radiations. We demonstrate that one key innovation—the inclusion of foreign material in nest construction—underlies both a massive range expansion and a significant increase in the rate of diversification within the second largest bee family, Megachilidae. Basal clades within the family are restricted to deserts and exhibit plesiomorphic features rarely observed among modern bees, but prevalent among apoid wasps. Our results suggest that early bees inherited a suite of behavioural traits that acted as powerful evolutionary constraints. While the transition to pollen as a larval food source opened an enormous ecological niche for the early bees, the exploitation of this niche and the subsequent diversification of bees only became possible after bees had evolved adaptations to overcome these constraints.
Keywords: bees, key innovation, diversification, Megachilidae, nesting biology, bee–flower relationships
1. Introduction
Bees provide a mixture of pollen and nectar as food for their developing larvae. To protect these provisions from microbial infection or liquefaction that may result from exposure to moisture, most bees coat the inside of their brood cells with a hydrophobic lining secreted by Dufour's gland [1,2]. By contrast, megachilid bees use an eclectic array of foreign material to line their cells. The French naturalist Jean-Henri Fabre commented extensively on the nesting habits of megachilids and posed the following question: ‘… the Osmiae make their partitions with mud or with a paste of chewed leaves; the Mason-bees build with cement; … the Megachiles made disks cut from leaves into urns; the Anthidia felt cotton into purses; the Resin-bees cement together little bits of gravel with gum; … Why all these different trades …?’ (p. 333 in [3]).
It has been demonstrated that the foreign material used by megachilid bees is hydrophobic and shows antimicrobial activity [4,5], thus serving a similar function to the secreted cell lining in other bee groups. Not all megachilids, however, use foreign material in nest construction. Bees of the tribe Lithurgini do not line their nest cells at all; instead, they excavate burrows in wood or stems [6,7]. The absence of nest-lining in this group was originally attributed to a behavioural loss associated with above-ground nesting [8], but the phylogenetic position of Lithurgini at the base of Megachilinae [9] suggests that it represents an ancestral trait [10]. Bees of the subfamily Fideliinae build unlined nests that they excavate in sandy soil [11–14]. Two distinct tribes of fideliine bees are recognized: Fideliini and Pararhophitini, which are both entirely restricted to deserts; the absence of cell lining in these bees may be related to the arid conditions of their habitats, which may make nest-lining unnecessary [15]. It remains unclear, however, whether cell-lining behaviour, using either secretions or foreign material, has been secondarily lost in these lineages or whether the absence of cell lining represents an ancestral state. To answer these questions, we present a robust molecular phylogeny of Megachilidae and trace the evolution of nesting biology within the family. We demonstrate that the use of foreign material in nest construction was a key innovation that triggered both range expansion and diversification in megachilid bees, and also propose that the ancestral biology of this family, which is still reflected in several extant megachilid lineages, mirrors the ancestral behaviour of bees in general. Similarities in the biology of the early megachilid lineages pertaining to nesting and foraging behaviour are numerous, conspicuous, and challenge our understanding of the evolution and diversification of bees.
2. Material and methods
(a). Taxon sample
We selected 98 ingroup taxa representing all seven tribes of the family Megachilidae. Our ingroup includes 12 Fideliini, two Pararhophitini, eight Lithurgini, three Dioxyini, 23 Anthidiini, 17 Osmiini and 33 Megachilini. We chose 31 outgroup taxa to represent the diversity of the rest of the bees, comprising one Colletidae, one Halictidae, one Andrenidae, five Melittidae and 23 Apidae. Electronic supplementary material, table S1 lists the DNA voucher numbers and collection localities for each of the specimens used in this study. We sampled more densely in the families Melittidae and Apidae to accommodate the placement of fossil calibration points. Voucher specimens are deposited in the Cornell University Insect Collection.
(b). Datasets and alignment
We sequenced fragments from four protein-coding genes (CAD, 882 bp; NAK, 1489 bp; EF1-alpha, 1111 bp; LW rhodopsin, 673 bp) and one ribosomal gene (28S, 1306 bp), following the DNA extraction and sequencing protocols outlined by Danforth et al. [16]. All taxa and GenBank accession numbers are listed in electronic supplementary material, table S2. PCR primers and conditions are listed in electronic supplementary material, table S3. The four protein-coding genes were aligned using MAFFT [17] and then adjusted by eye in MacClade [18]; all introns were removed. The ribosomal gene (28S) was aligned via secondary structure according to the method described by Kjer [19]; all unalignable regions were excluded. The secondary structure alignment was based on the 28S map of Apis mellifera [20]. Details regarding data partitioning and model-testing are included in the electronic supplementary material.
(c). Phylogenetic analyses
Phylogenetic analyses were performed using both Bayesian and maximum-likelihood methods. Bayesian analyses were performed using MrBayes v. 3.1.2 [21,22]. A GTR + I + Γ model was used for all partitions except for the stem partition of 28S, which was analysed using the doublet model. All parameters were unlinked between partitions. Preliminary analyses resulted in poor mixing of chains, so the default temperature setting of 0.2 was adjusted to 0.03, which improved mixing and increased the chain swap acceptance rate to within the range recommended by the MrBayes users' manual. We ran six independent analyses, for a total of 180 000 000 generations. Sampling was performed every 2000 generations. An appropriate burn-in was discarded from each analysis using Tracer [23], leaving 96 956 000 post-burn-in generations; these were further sampled using LogCombiner v. 1.6.1 [24] to ensure independent sampling of trees. The final combined posterior distribution of 25 239 trees was used to build a maximum clade credibility tree using TreeAnnotator v. 1.6.1 [24] (electronic supplementary material, figure S1).
Maximum-likelihood analyses were performed using RAxML v. 7.0.4 (sequential version raxmlHPC [25]). We used the rapid bootstrapping algorithm with a GTR + CAT approximation to perform 1000 bootstrap replicates. The maximum-likelihood bootstrap tree is shown in electronic supplementary material, figure S2.
(d). Divergence dating analysis using BEAST
We used BEAST v. 1.6.1 to perform a Bayesian divergence dating analysis [24]. Each partition was analysed using a GTR + I + Γ model; substitution models were unlinked across partitions. We used an uncorrelated lognormal relaxed-clock model with a Yule tree prior. Trees were sampled every 2000 generations. We randomly chose a starting tree from the posterior distribution of trees from the MrBayes analysis; we used TreeEdit v. 1.0 [26] to scale the root height to 130 Myr in order to conform to the constraints imposed by prior distributions on divergence times. Ten independent analyses were run for a total of 300 000 000 generations. An appropriate burn-in was discarded from each analysis using Tracer [23], leaving 217 068 000 total post-burn-in generations. In order to ensure independent sampling of trees, we sampled every third tree from the post-burn-in posterior distribution of trees using LogCombiner v. 1.6.1 [24] and then used TreeAnnotator v. 1.6.1 [24] to build a maximum clade credibility tree from this posterior distribution of trees (electronic supplementary material, figure S3).
(e). Calibration of internal nodes and root node in BEAST
We used fossils to time-calibrate seven internal nodes on our tree. Five of these calibration points were assigned a lognormal prior distribution, while two were assigned a normal prior distribution. We present the details of these calibration points, as well as a discussion of fossils that were unusable for the purposes of calibrating our phylogeny, in the electronic supplementary material.
Bees are thought to be the sister group to the apoid wasps [27]. Apoids first appear in the fossil record during the Cretaceous [28]; Engel [28] proposes that bees originated some time after this and gives an uppermost boundary for their age of 125 Myr. There is no direct fossil evidence to suggest that bees arose at this time, however, and we believe that the age of the bees may be older than previously estimated. The Late Cretaceous (approx. 65 Ma) origin of Cretotrigona prisca, a highly derived eusocial meliponine bee, indicates that a significant amount of bee diversification had already taken place by the Late Cretaceous. Furthermore, it has been widely speculated that the origin of bees happened after the origin of the angiosperms [28–31]; recent molecular evidence [32] places the origin of the angiosperms in the Late Triassic, 30–80 Myr earlier than previously estimated. We find both of these arguments compelling reasons to explore the possibility that bees arose earlier than current estimates suggest.
We assign a uniform prior distribution to the root node. While other studies have favoured more informative root priors, such as the lognormal [33] or the normal [34], we feel that the only way to obtain an objective estimate for the origin of Megachilidae is to impose a relatively uninformative prior on the root. The lower bound of the root prior is assigned a value of 100 Myr and is based on an extremely conservative estimate for the origin of bees based on the fossil record [28]. The upper bound is assigned a value of 217 Myr and is based on a recent molecular estimate for the age of crown angiosperms [32]. Our use of a fairly broad uniform prior causes the 95 per cent HPD for divergence date estimates to be larger than those associated with other types of prior distributions. Our dating analyses, however, were run to stationarity, and age estimates from multiple independent runs converged to a single stable value; we accept the broad 95 per cent HPD as a necessary consequence of using a uniform prior distribution.
(f). Biogeographic reconstruction
Biogeographic reconstructions were performed using both S-Diva [35] and Lagrange [36]. Most of our terminal taxa represent genera; for this reason, the most plausible ancestral range for each terminal was coded based on the current distribution of the species represented by the terminal (based on [37]). In both S-Diva and Lagrange analyses, the following areas were considered: Afrotropic, Palaearctic, Southeast Asia, Australia, Nearctic and Neotropic; in case of ambiguity, polymorphism was allowed. Given our near-complete sampling of the basal-most branches, such polymorphisms only concerned the higher megachilid tribes Anthidiini, Osmiini and Megachilini, and did not affect inference at the base of the family. We present the details of both biogeographic analyses in the electronic supplementary material.
(g). Ancestral state reconstruction
We used BayesTraits [38] to reconstruct the ancestral nesting biology of Megachilidae. Cell-lining behaviour was coded for each terminal (including the outgroup) as: totally unlined (0), in Dasypoda, fideliine and lithurgine bees; lined with glandular secretion (1), in all members of the families Andrenidae, Halictidae and Colletidae, as well as in several lineages of Apidae and in the genus Melitta; lined with foreign material (2), in the oil-collecting bees, some Apidae and all higher Megachilidae; or as cleptoparasitic (3). We coded the corbiculate apidae, as well as all lineages for which no information was available, as (012). Meganomia was coded (02), as Rozen (p. 4 in [39]) states that cells of Meganomia contained ‘no built-in lining, i.e. consisting of soil mixed with secretions’, but have a waterproof lining, possibly consisting of nectar. Information on nesting biology was found in [37] and references therein. We present the details of our Bayesian ancestral state reconstructions in the electronic supplementary material.
(h). Correlated trait evolution
We used BayesTraits [38] to test for correlated evolution between the total geographical area occupied by a taxonomic group and diversification rate. We calculated diversification rate using the function lambda.stem.ms01 in the Laser package in R [40,41] and the total geographical range for each terminal taxon using the area calculator provided by the Free Map Tools website [42]. We present the details of this analysis, as well as specific information regarding species distribution, in the electronic supplementary material.
(i). Diversification rate analysis
We used MEDUSA (modelling evolutionary diversification using stepwise Akaike information criterion [43]) to test for changes in the tempo of diversification among the branches of the megachilid phylogeny. We used the final consensus tree from our BEAST analysis and removed the outgroup using Mesquite [44]. We collapsed several taxa into single terminals and calculated the total number of species represented by each terminal; terminals were collapsed in order to more easily quantify the number of species represented. The resulting phylogeny contained 82 taxa. We chose to use corrected Akaike information criterion (AICc) scores instead of AIC scores in order to account for the small sample size of our phylogeny. We used MEDUSA to fit a series of 20 models and used a strict cut-off value of 10 as our ΔAICc threshold. A model with two rate shifts (three sets of birth and death rates) was chosen as the best-fit model.
3. Results and discussion
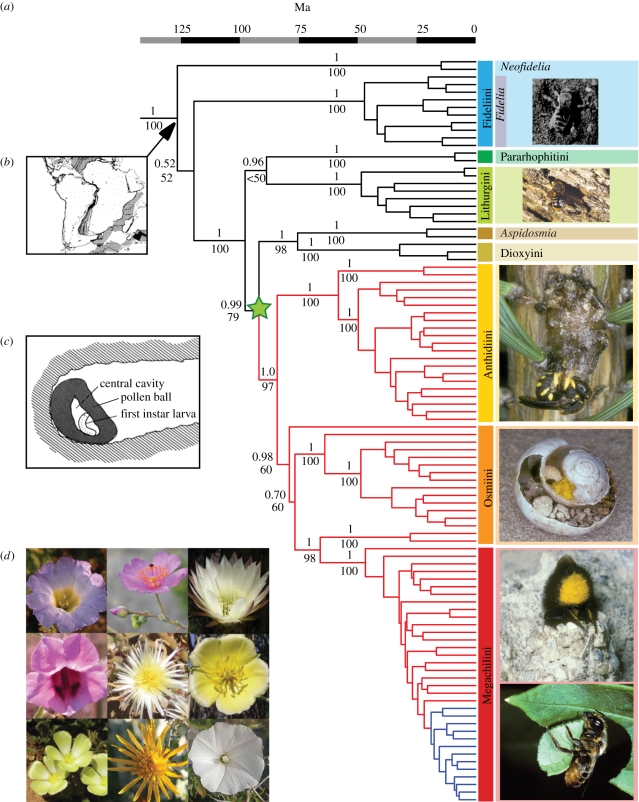
The results of both maximum-likelihood and Bayesian analyses support a non-traditional interpretation of early megachilid phylogeny (figure 1a). According to our phylogenetic hypothesis, the small Palaearctic tribe Pararhophitini is not closely related to the largely austral tribe Fideliini, but appears more closely related to the subfamily Megachilinae; this result is strongly supported in all analyses (figure 1a; electronic supplementary material, figures S1, S2 and S3). Furthermore, the two lineages of Fideliini (the genera Fidelia and Neofidelia) constitute a weakly supported grade at the base of Megachilidae. Further tests using Bayes factors [46] strongly support the non-monophyly of both the subfamily Fideliinae (Bayes factor: hereafter BF = 260.36) and the tribe Fideliini (BF = 33.68).
Figure 1.
(Opposite.) Fossil-calibrated maximum clade credibility tree for bee family Megachilidae. (a) Bayesian posterior probabilities and maximum-likelihood bootstrap values shown above and below nodes, respectively, for all clades older than 50 Myr. Terminals are labelled to tribe according to present taxonomic assignment, even if determined to be paraphyletic in the current analysis. Branch colours correspond to significant changes in diversification rate (black: diversification rate = 0.0164, relative extinction = 0.885; red: diversification rate = 0.0867, relative extinction = 0.848; blue: diversification rate = 0.315, relative extinction = 0.518). The node marked with a green star corresponds to the transition between building unlined nests and building nests using foreign material. There is no reversion to building unlined nests after this point. Photographs to the right of phylogeny from top to bottom: (1) Tribe Fideliini: Fidelia villosa using hind legs to excavate sand from a burrow (photo: Jerome G. Rozen [14], courtesy of the American Museum of Natural History); (2) Tribe Lithurgini: Lithurgus chrysurus entering nest in dead tree trunk (photo: Andreas Müller); (3) Tribe Anthidiini: Anthidium strigatum closing a nest cell of resin (photo: Albert Krebs); (4) Tribe Osmiini: nest of Osmia bicolor built in an abandoned snail shell (photo: Albert Krebs); (5) Tribe Megachilini: (top) Megachile parietina entering her nest made of mud (photo: Albert Krebs); (bottom) Megachile ligniseca using her mandibles to cut a leaf disc (photo: Felix Amiet). (b) Biogeographic reconstructions indicate a Gondwanan origin for Megachilidae, approximately 126 Ma (figure reprinted from [45], copyright 1988, with permission from Elsevier). (c) The ancestor of all Megachilidae built unlined nests in sandy soil, much like extant lineages Fidelia, Neofidelia and Pararhophites (nest of Fidelia villosa shown; picture: Jerome G. Rozen [14], courtesy of the American Museum of Natural History). (d) Host plants of Fideliini (electronic supplementary material, table S4). Top row (left to right): Nolana sp. (Solanaceae; host of Neofidelia longirostris; photo: Michael O. Dillon), Calandrinia sp., Trichocereus sp. (Portulacaceae and Cactaceae, respectively; hosts of N. profuga; photos: Joshua R. McDill, Scott Zona); centre row (left to right): Sesamum sp. (Pedaliaceae; host of Fidelia friesei; photo: Jessica Litman), Psilocaulon sp. (Aizoaceae; host of F. villosa, F. kobrowi, F. paradoxa; photo: Jessica Litman), Sisyndite spartea (Zygophyllaceae; host of F. pallidula; photo: Tomas Hajek); bottom row (left to right): Grielum sp. (Neuradaceae; host of F. hessei, F. major, F. fasciata; photo: Serban Proches), Berkheya fruticosa (Asteraceae; host of F. braunsiana; photo: Henry Brisse), Convolvulus trabutianus (Convolvulaceae; host of F. ulrikei; photo: Pierre-Marie Roux). Not shown: Tribulocarpus dimorphanthus (Aizoaceae; host of F. ornata). Note that all flowers are characterized by radial symmetry and exposed anthers.
The first two branches in our phylogeny are thus the South American genus Neofidelia and the primarily southern African genus Fidelia. The geographical distribution and phylogenetic placement of these lineages reveal an Austral disjunction between the Old and the New World, suggestive of a Gondwanan origin. We find the age of Megachilidae, and thus of the divergence between the South American and African fideliine bees, to be 126 Ma (95% HPD 100–154), pre-dating the separation of the African and South American continental plates (figure 1b). Our estimate of the age of Megachilidae is older than anticipated, given that bees are generally thought to have originated around 125 Ma [28]. Our results indicate an origin for the bees (the root height of our tree) of 149 Ma (95% HPD 119–182). We ran another analysis where the root was constrained to 120 Ma; even under this conservative estimate for the age of the bees [28], the age of Megachilidae is 104 Ma (95% HPD 95–113), which is still consistent with a Gondwanan origin, as the last connections between Africa and South America are thought to have disappeared around 100–110 Ma [47]. Both analyses indicate that the Megachilidae arose relatively rapidly after the origin of the bees.
A Gondwanan origin for Megachilidae is further supported by biogeographic reconstructions. S-Diva results favour a South American/African vicariance (75% of reconstructions) over scenarios involving either African (12.6%) or African/Palaearctic (12.4%) origins and subsequent dispersal to South America. Similarly, in biogeographic inferences using Lagrange [36], analyses where Africa and South America were allowed to be adjacent strongly supported Gondwanan vicariance at the root node (global maximum likelihood −250.4; electronic supplementary material). Analyses where Africa and South America were not adjacent (thus precluding vicariance as a possible outcome and implying Northern Hemisphere migrations) had significantly worse overall likelihood scores (global maximum likelihood −252.3). Dispersal from Africa to South America via Australia and Antarctica (achieved by allowing dispersal between Australia and South America) was even less likely (global maximum likelihood −295.9). However, we agree with Rozen [11] that the most convincing support for vicariance over migration comes from biological evidence. The brood cells of fideliine bees consist of unlined cavities in the sand (figure 1c); for this reason, these bees are entirely restricted to strongly seasonal deserts where annual rainfall is not only low but also extremely unlikely during their nesting season [11]. Alternative biogeographic scenarios to explain their present-day distribution necessarily involve migrations through the Northern Hemisphere or via Antarctica; both of these scenarios imply adaptations to temperate habitats, which we consider extremely unlikely. Indeed, ancestral state reconstructions performed using BayesTraits [38] reveal that the ancestor of Megachilidae built nests that were neither lined with foreign material nor with glandular secretions (average maximum-likelihood probability 0.99, average difference in likelihood 3.6 and 5.4, respectively; posterior probability 0.98, BF 6.0 and 14.4, respectively). All species using foreign material in nest construction form a monophyletic group. The use of foreign material in nest construction has a single origin at the base of the tribes Anthidiini, Dioxyini, Osmiini and Megachilini (average maximum-likelihood probability 0.99, average difference in likelihood 2.5 and 7.3; posterior probability 0.99, BF 4.4 and 10.3).
The use of foreign material in nest construction underlies the ability of megachilid bees to colonize temperate regions and appears to be associated with a dramatic increase in clade species diversity. The lineages Fidelia, Neofidelia and Pararhophites together number 17 species, while the tribes Anthidiini, Osmiini and Megachilini collectively include over 3900 species and exhibit a worldwide distribution. MEDUSA [43] results provide evidence for two significant increases in diversification rate in our phylogeny, the first at the base of the higher megachilids and the second nested within the genus Megachile (figure 1).
The larger of the two rate shifts increases from 0.0164 to 0.0867 and occurs approximately 7 Myr after the advent of nest construction using foreign material, a behaviour that is first observed in the enigmatic genus Aspidosmia [37], the first branch within the subfamily Megachilinae. The increase in diversification rate that occurs after the divergence between Aspidosmia and the rest of Megachilinae suggests that the use of foreign material in nesting may have driven diversification but was not the only factor underlying it.
The second shift in diversification rate occurs within the genus Megachile, from 0.087 to 0.315. The increase in diversification tempo happens approximately 8 Myr after the origin of the true leafcutting Megachile (Michener's group I) from the paraphyletic assemblage of the Chalicodoma group of subgenera (Michener's group II) [37]. Despite their relatively recent origin (22 Ma; 95% HPD 16–27), leafcutting Megachile are extremely diverse and abundant on all continents. The explanation for such species richness may be related to their high reproductive output [48] and their ability to colonize an extremely broad range of habitats, from moist tropics to extreme deserts.
In association with the ancestral state reconstructions of nesting biology, the diversification rate analysis reveals an intimate association between nesting biology, distribution and diversification. The single origin of nest-lining behaviour in Megachilidae makes it difficult to test for correlated evolution between nesting and other traits of interest. In contrast, the total geographical area occupied by the terminal taxa varies from lineage to lineage throughout the phylogeny, allowing us to test for an association between area and diversification rate. The results of BayesTraits analyses [38] indicate strongly correlated evolution between geographical area and diversification rate (BF = 25.8). Consistent with other studies where geographical area has been correlated with diversification [49], we envision a scenario where nest-lining behaviour promoted the widespread colonization of temperate habitats, which in turn drove the diversification seen in the higher megachilids.
Ancestral state reconstructions strongly indicate that the three fideliine lineages are restricted to deserts owing to their plesiomorphic nesting biology, rather than as a secondary adaptation. The use of foreign material in nest construction has a single origin at the base of the tribes Anthidiini, Dioxyini, Osmiini and Megachilini. It has enabled these bees to repeatedly colonize temperate habitats and catalysed a massive shift in diversification rate. Surprisingly, Lithurgini manage to survive in temperate and tropical conditions, although they do not line their brood cells. All Lithurgines dig burrows in wood or stems and their pollen provisions are protected from humidity in these above-ground substrates. In other respects, the pollen provisions and nest architecture of lithurgine bees are very similar to those of fideliine bees. The pollen mass is neither worked nor manipulated by the female; it does not form a spherical mass but rather occupies the entire rear portion of the nest cell. Their burrows are mostly branched and the cells are either not partitioned or partitioned using sawdust or wood particles obtained from the excavation of brood cells. These bees then fill their completed burrows with sawdust, in much the same way that fideliine bees do with sand [6,7].
The identification of nest-lining behaviour as a key innovation also offers an explanation for the behavioural conservatism seen in the early megachilids. The two basal lineages, Fidelia and Neofidelia, which emerged prior to the advent of this innovation, have retained highly similar and comparatively unusual behaviours on two different continents for more than 100 Myr, suggesting powerful evolutionary constraints on these behaviours. A comparison of their nesting biology and host-plant associations provides a unique glimpse into the biology of early megachilids over 120 Ma, early in bee evolution.
(a). Nesting
Unlined nests similar to those observed in fideliine bees are rare among bees. All members of the species-rich short-tongued bee families Andrenidae, Halictidae and Colletidae, which probably form a monophyletic group [9], apply secreted lining to their brood cells [1,2]. Curiously, some desert andrenids apply a secreted lining not to the walls of their nests but to the pollen provisions themselves [50]. In the family Apidae, the evolution of nest-lining behaviour is obscured by three probable origins of oil or resin collection, the unknown phylogenetic positions of lineages that apparently do not line their brood cells (e.g. Eremapis; [51]), four independent origins of cleptoparasitism and the evolution of social behaviour [34]. Lastly, unlined nests are known in several members of the melittid bees [37], a species-poor group that may represent the earliest lineages of extant bees [9]. Many melittids are restricted to xeric areas, especially several species-poor genera for which the nesting biology is not documented (e.g. Eremaphanta, Afrodasypoda, Promelitta). The few genera that are present in temperate regions either collect floral oil (Macropis and Rediviva), have evolved secreted cell lining (Melitta) or shape their pollen balls into peculiar, tripod-like structures that reduce contact between the provisions and the cell wall (Dasypoda). In fact, according to the most comprehensive phylogenetic hypothesis currently available for bees [9], the construction of unlined nests is a behaviour restricted to a few primitive lineages; among all bees, there is not a single documented instance of a reversion to building unlined nests after the evolution of nest-lining behaviour occurs. These observations strongly suggest that the ancestor of bees did not line its nest cells [52] and that cell lining, using either glandular secretions or foreign material, has multiple origins in bees.
By contrast, unlined nests are prevalent among apoid wasps [53,54], the paraphyletic group from which bees arose. In fact, the nesting biology of fideliine bees is reminiscent of that of many sand-nesting apoids [10] whose nests consist of unlined burrows in the sand. Apoid wasps store paralysed prey that may stay alive for several weeks before being consumed by their larvae. While stored provisions are always susceptible to spoilage [55], the transition from prey-hunting to pollen-collecting in the early bees may have dramatically exacerbated the problems associated with the storage of provisions, given the hygroscopic properties of pollen and its susceptibility to fungal infection, and driven selective pressure to protect provisions from moisture.
(b). Foraging behaviour and host-plant associations
Interactions with angiosperms have often been cited as important driving factors underlying diversification in phytophagous insects [56]. Our results, however, suggest that the shift to pollen collection in early bees did not simply open a vast new ecological niche. First, if the biology of the earliest extant megachilids indeed mirrors the biology of ancestral bees, early bees were constrained to xeric and strongly seasonal habitats, and highly limited in their phenology. Second, another aspect of the behaviour of early bees may have seriously hampered them from fully using all available angiosperm hosts: a pronounced floral specificity (oligolecty). Comparisons of the well-documented foraging behaviour of the basal members of Megachilidae (figure 1d; electronic supplementary material, table S4) provide unique insights into bee–flower relationships prevalent more than 100 Myr ago. Fideliine bees, both in South America and Africa, are notorious oligoleges. Rozen [12] states that on both continents, fideliine bees tend to forage on large flowers with well-exposed anthers (figure 1d); even the narrowly polylectic Neofidelia profuga appears to restrict pollen collection to a few hosts with similar flower architecture, namely large flowers with radial symmetry and well-exposed stamens. The same appears to be true for many lithurgine bees: distantly related species of the genera Lithurgus and Microthurge in Australia, Africa and South America forage exclusively or predominantly on Malvaceae with large flowers, such as Hibiscus, Sida and Turnera (electronic supplementary material, table S4); Asian species appear polylectic but restrict pollen collection to flowers of Malvaceae and Convolvulaceae; and two lineages, the subgenus Lithurgopsis and the genus Trichothurgus, have maintained a close association with the large flowers of Cacteaceae in both South and North America. Lastly, the two species of Pararhophites for which host-plant information is available restrict their foraging to morphologically similar but phylogenetically unrelated flowers that have exposed stamens and five white petals (electronic supplementary material, table S4). In summary, a narrow host range clearly appears to be the plesiomorphic condition in Megachilidae. Moreover, there is a striking lack of bilaterally symmetrical flowers among the hosts of the basal megachilid lineages. By contrast, bilaterally symmetrical flowers, such as Fabaceae and Lamiaceae, which are typical bee-pollinated flowers, are common hosts of a significant proportion of the higher megachilids.
These observations strongly support the view that host choices in bees are evolutionarily constrained [57], as well as the widely discussed assertion that oligolecty is a primitive (rather than a derived) state in bees (reviewed in [57]). Müller [58] suggested that oligolecty might be a behavioural constraint related to flower manipulation, pollen collecting or pollen digestion, rather than a secondary specialization. Interestingly, Müller [58] notes that most apoid wasps are specialized hunters. In fact, the foraging behaviour of apoid wasps is similar in many ways to that of primitive bees. It is evolutionarily conserved, with related species exhibiting similar behaviour on different continents. Most species restrict their host range to distantly related prey belonging to the same order (e.g. grasshoppers, spiders or leafhoppers), which are often similar in size and appearance [53,54], and co-occur in the same habitat. Evans [59] elegantly summarizes the foraging behaviour of the philanthine wasp tribe Cercerini as follows: ‘I suggest that these wasps are not necessarily “good taxonomists”, but that they are programmed to hunt in certain situations and to respond to prey of a certain size and behaviour’ (p. 521). We hypothesize that early bees inherited foraging specificity as a behavioural constraint from their apoid wasp ancestors.
4. Conclusion
Our work reveals that two extant lineages are ‘living fossils’ among the bees. The Mid-Cretaceous origin of Fidelia and Neofidelia, and their bizarre, plesiomorphic biology, strongly support the possibility that these bees reflect the biology of the earliest bees more closely than any other extant lineage. The evolutionary patterns we report in Megachilidae lay the initial framework for understanding patterns of nesting behaviour, distribution, host-plant preference and diversification in all bees.
Acknowledgements
This work was supported by a National Science Foundation grant to B.N.D. and Terry Griswold (DEB-0742998); a National Geographic Society grant to B.N.D. (8448-08); a grant from the Swiss National Science Foundation (PBEZP3-122970) to C.J.P.; grants from the Graduate School and the Mario Einaudi Center at Cornell University to J.R.L.; a grant through the Griswold Endowment (Department of Entomology, Cornell University) to J.R.L.; and a grant from the Linnean Society of London and the Systematics Association to J.R.L. We thank Andreas Müller, Robert Paxton and Antonella Soro for comments on the manuscript; Simon Ho for assistance with BEAST; Rick Ree and Stephen Smith for assistance with Lagrange; Anurag Agrawal for assistance with BayesTraits; Stuart Campbell for assistance with statistical analysis; Werner Arens, Sophie Cardinal, Victor Gonzalez, Terry Griswold, Michael Kuhlmann, Denis Michez, Laurence Packer, Sébastien Patiny, Jerry Rozen, Maximilian Schwarz and Claudio Sedivy for providing specimens; and Philip Perkins at the Harvard University Museum of Comparative Zoology for access to bee fossils.
References
- 1.Cane J. H. 1983. Chemical evolution and chemosystematics of the Dufour's gland secretions of the lactone-producing bees (Hymenoptera: Colletidae, Halictidae, and Oxaeidae). Evolution 37, 657–674 10.2307/2407908 (doi:10.2307/2407908) [DOI] [PubMed] [Google Scholar]
- 2.Hefetz A. 1987. The role of the Dufour's gland secretion in bees. Physiol. Entomol. 12, 243–253 10.1111/j.1365-3032.1987.tb00749.x (doi:10.1111/j.1365-3032.1987.tb00749.x) [DOI] [Google Scholar]
- 3.Fabre J. H. 1920. Bramble-bees and others, 2nd edn. New York, NY: Dodd, Mead, and Company [Google Scholar]
- 4.Messer A. C. 1985. Fresh dipterocarp resins gathered by megachilid bees inhibit growth of pollen-associated fungi. Biotropica 17, 175–176 10.2307/2388512 (doi:10.2307/2388512) [DOI] [Google Scholar]
- 5.Müller A., Topfl W., Amiet F. 1996. Collection of extrafloral trichome secretions for nest wool impregnation in the solitary bee Anthidium manicatum. Naturwissenschaften 83, 230–232 [Google Scholar]
- 6.Garófalo C. A., Camillo E., Campos M. J. O., Zucchi R., Serrano J. C. 1981. Binomial aspects of Lithurgus corumbae (Hymentoptera, Megachilidae), including evolutionary considerations on the nesting behaviour of the genus. Braz. J. Genet. 4, 165–182 [Google Scholar]
- 7.Roberts R. B. 1978. The nesting biology, behaviour and immature stages of Lithurge chrysurus, an adventitious wood-boring bee in New Jersey (Hymenoptera: Megachilidae). J. Kansas Entomol. Soc. 51, 735–745 [Google Scholar]
- 8.Malyshev S. I. 1930. Nistgewohnheiten der Steinbeinen Lithurgus Latr. Z. Morphol. Ökol. Tiere 23, 754–809 10.1007/BF00407240 (doi:10.1007/BF00407240) [DOI] [Google Scholar]
- 9.Danforth B. N., Sipes S., Fang J., Brady S. G. 2006. The history of early bee diversification based on five genes plus morphology. Proc. Natl Acad. Sci. USA 103, 15 118–15 123 10.1073/pnas.0604033103 (doi:10.1073/pnas.0604033103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eickwort G. C., Matthews R. W., Carpenter J. 1981. Observations on the nesting behaviour of Megachile rubi and M. texana with a discussion of the significance of soil nesting in the evolution of megachilid bees (Hymenoptera: Megachilidae). J. Kansas Entomol. Soc. 54, 557–570 [Google Scholar]
- 11.Rozen J. G. 1973. Life history and immature stages of the bee Neofidelia (Hymenoptera, Fideliidae). Am. Mus. Novit. 2519, 1–14 [Google Scholar]
- 12.Rozen J. G. 1977. The ethology and systematic relationships of fideliine bees, including a description of the mature larva of Parafidelia (Hymenoptera, Apoidea). Am. Mus. Novit. 2637, 1–15 [Google Scholar]
- 13.McGinley R. J., Rozen J. G. 1987. Nesting biology, immature stages, and phylogenetic placement of the Palaearctic bee Pararhophites (Hymenoptera: Apoidea). Am. Mus. Novit. 2903, 1–21 [Google Scholar]
- 14.Rozen J. G. 1970. Biology, immature stages, and phylogenetic relationships of fideliine bees, with the description of a new species of Neofidelia (Hymenoptera, Apoidea). Am. Mus. Novit. 2427, 1–25 [Google Scholar]
- 15.Michener C. 1964. Evolution of the nests of bees. Am. Zool. 4, 227–239 [Google Scholar]
- 16.Danforth B. N., Sauquet H., Packer L. 1999. Phylogeny of the bee genus Halictus (Hymenoptera: Halictidae) based on parsimony and likelihood analyses of nuclear EF-1 alpha sequence data. Mol. Phylogenet. Evol. 13, 605–618 10.1006/mpev.1999.0670 (doi:10.1006/mpev.1999.0670) [DOI] [PubMed] [Google Scholar]
- 17.Katoh K., Kazuharu M., Kuma K.-I., Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 10.1093/nar/gkf436 (doi:10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddison D. R., Maddison W. P. 2005. MacClade v. 4.08 for OSX. Sunderland, MA: Sinauer Associates, Inc [Google Scholar]
- 19.Kjer K. M. 1995. Use of rRNA secondary structure in phylogenetic studies to identify homologous positions: an example of alignment and data presentation from the frogs. Mol. Phylogenet. Evol. 4, 314–330 10.1006/mpev.1995.1028 (doi:10.1006/mpev.1995.1028) [DOI] [PubMed] [Google Scholar]
- 20.Gillespie J. J., Johnston J. S., Cannone J. J., Gutell R. R. 2006. Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): structure, organization, and retrotranposable elements. Insect Mol. Biol. 15, 657–686 10.1111/j.1365-2583.2006.00689.x (doi:10.1111/j.1365-2583.2006.00689.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huelsenbeck J. P., Ronquist F. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17, 754–755 10.1093/bioinformatics/17.8.754 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 22.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 23.Rambaut A., Drummond A. J. 2007. Tracer v. 1.4. Available from http://beast.bio.ed.ac.uk/Tracer
- 24.Drummond A., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A., Ludwig T., Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21, 456–463 10.1093/bioinformatics/bti191 (doi:10.1093/bioinformatics/bti191) [DOI] [PubMed] [Google Scholar]
- 26.Rambaut A., Charleston M. 2001. TreeEdit. Phylogenetic Tree Editor, v. 1.0a8 Oxford, UK: Oxford University Press [Google Scholar]
- 27.Melo G. A. R. 1999. Phylogenetic relationships and classification of the major lineages of Apoidea (Hymenoptera), with emphasis on the crabronid wasps. Sci. Pap. Nat. Hist. Mus. Univ. Kansas 14, 1–55 [Google Scholar]
- 28.Engel M. S. 2001. A monograph of the Baltic amber bees and evolution of the Apoidea (Hymenoptera). B. Am. Mus. Nat. Hist. 259, 1–192 (doi:10.1206/0003-0090(2001)259<0001:AMOTBA>2.0.CO;2) [DOI] [Google Scholar]
- 29.Michener C. D. 1979. Biogeography of the bees. Ann. Mo. Bot. Gard. 66, 277–347 10.2307/2398833 (doi:10.2307/2398833) [DOI] [Google Scholar]
- 30.Grimaldi D. A. 1999. The co-radiations of pollinating insects and angiosperms in the Cretaceous. Ann. Mo. Bot. Gard. 86, 373–406 10.2307/2666181 (doi:10.2307/2666181) [DOI] [Google Scholar]
- 31.Engel M. S. 2000. A new interpretation of the oldest fossil bee (Hymenoptera: Apidae). Am. Mus. Novit. 3296, 1–11 (doi:10.1206/0003-0082(2000)3296<0001:ANIOTO>2.0.CO;2) [DOI] [Google Scholar]
- 32.Smith S. A., Beaulieu J. M., Donoghue M. J. 2010. An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proc. Natl Acad. Sci. USA 107, 5897–5902 10.1073/pnas.1001225107 (doi:10.1073/pnas.1001225107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward P. S., Brady S. G., Fisher B. L., Schultz T. R. 2010. Phylogeny and biogeography of dolichoderine ants: effects of data partitioning and relict taxa on historical inference. Syst. Biol. 59, 342–362 10.1093/sysbio/syq012 (doi:10.1093/sysbio/syq012) [DOI] [PubMed] [Google Scholar]
- 34.Cardinal S., Straka J., Danforth B. N. 2010. Comprehensive phylogeny of apid bees reveals evolutionary origins and antiquity of cleptoparasitism. Proc. Natl Acad. Sci. USA 107, 16 207–16 211 10.1073/pnas.1006299107 (doi:10.1073/pnas.1006299107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y., Harris A. J., He X. 2010. S-DIVA (statistical dispersal-vicariance analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850 10.1016/j.ympev.2010.04.011 (doi:10.1016/j.ympev.2010.04.011) [DOI] [PubMed] [Google Scholar]
- 36.Ree R., Smith S. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14 10.1080/10635150701883881 (doi:10.1080/10635150701883881) [DOI] [PubMed] [Google Scholar]
- 37.Michener C. D. 2007. The bees of the world. Baltimore, MD: The Johns Hopkins University Press [Google Scholar]
- 38.Pagel M. 1997. Inferring evolutionary processes from phylogenies. Zool. Scr. 26, 331–348 10.1111/j.1463-6409.1997.tb00423.x (doi:10.1111/j.1463-6409.1997.tb00423.x) [DOI] [Google Scholar]
- 39.Rozen J. G. 1977. Biology and immature stages of the bee genus Meganomia (Hymenoptera, Melittidae). Am. Mus. Novit. 2630, 1–14 [Google Scholar]
- 40.Rabosky D. L. 2006. LASER: a maximum likelihood toolkit for inferring temporal shifts in diversification rates from molecular phylogenies. Evol. Bioinf. Online 2, 257–260 [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team 2010. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]
- 42.Free Map Tools. 2010. Free map tools. See http://www.freemaptools.com/
- 43.Alfaro M., Santini F., Brock C., Alamillo H., Dornburg A., Rabosky D., Carnevale G., Harmon L. 2009. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl Acad. Sci. USA 106, 13 410–13 414 10.1073/pnas.0811087106 (doi:10.1073/pnas.0811087106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maddison W. P., Maddison D. R. 2010. Mesquite: a modular system for evoluionary analysis. Sunderland, MA: Sinauer Associates, Inc [Google Scholar]
- 45.Scotese C. R., Gahagan L. M., Larson R. L. 1988. Plate tectonic reconstructions of the Cretaceous and Cenozoic ocean basins. Tectonophysics 155, 27–48 10.1016/0040-1951(88)90259-4 (doi:10.1016/0040-1951(88)90259-4) [DOI] [Google Scholar]
- 46.Kass R. E., Raftery A. E. 1995. Bayes factors. J. Am. Stat. Assoc. 90, 773–795 10.2307/2291091 (doi:10.2307/2291091) [DOI] [Google Scholar]
- 47.Sanmartin I. 2002. A paleogeographic history of the Southern Hemisphere. Uppsala, Sweden: Uppsala University [Google Scholar]
- 48.Pitts-Singer T. L., Cane J. H. 2011. The alfalfa leafcutting bee, Megachile rotundata: the world's most intensively managed solitary bee. Annu. Rev. Entomol. 56, 221–237 10.1146/annurev-ento-120709-144836 (doi:10.1146/annurev-ento-120709-144836) [DOI] [PubMed] [Google Scholar]
- 49.Parent C. E., Crespi B. J. 2006. Sequential colonization and diversification of Galapagos endemic land snail genus Bulimulus (Gastropoda, Stylommatophora). Evolution 60, 2311–2328 [PubMed] [Google Scholar]
- 50.Rozen J. G. 1967. Review of the biology of panurgine bees, with observations on North American forms (Hymenoptera, Andrenidae). Am. Mus. Novit. 2297, 1–44 [Google Scholar]
- 51.Neff J. L. 1984. Observations on the biology of Eremapis parvula Ogloblin, an anthophorid bee with a metasomal scopa (Hymenoptera: Anthophoridae). Pan-Pac. Entomol. 60, 155–163 [Google Scholar]
- 52.Radchenko V. G., Pesenko Y. A. 1996. ‘Proto-bee’ and its nests: a new hypothesis concerning the early evolution of Apoidea (Hymenoptera). Entomol. Rev. 75, 140–162 [Google Scholar]
- 53.Bohart R. M., Menke A. S. 1976. Sphecid wasps of the world: a generic revision. Berkeley, CA: University of California Press [Google Scholar]
- 54.Evans H. E. 1966. The comparative ethology and evolution of the sand wasps. Cambridge, MA: Harvard University Press [Google Scholar]
- 55.Kaltenpoth M., Göttler W., Herzner G., Strohm E. 2005. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 15, 475–479 10.1016/j.cub.2004.12.084 (doi:10.1016/j.cub.2004.12.084) [DOI] [PubMed] [Google Scholar]
- 56.Farrell B. D. 1998. ‘Inordinate fondness’ explained: why are there so many beetles? Science 281, 555–559 10.1126/science.281.5376.555 (doi:10.1126/science.281.5376.555) [DOI] [PubMed] [Google Scholar]
- 57.Sedivy C., Praz C. J., Müller A., Widmer A., Dorn S. 2008. Patterns of host-plant choice in bees of the genus Chelostoma: the constraint hypothesis of host-range evolution in bees. Evolution 62, 2487–2507 10.1111/j.1558-5646.2008.00465.x (doi:10.1111/j.1558-5646.2008.00465.x) [DOI] [PubMed] [Google Scholar]
- 58.Müller A. 1996. Host-plant specialization in western Palearctic anthidiine bees (Hymenoptera: Apoidea: Megachilidae). Ecol. Monogr. 66, 235–257 10.2307/2963476 (doi:10.2307/2963476) [DOI] [Google Scholar]
- 59.Evans H. E. 1971. Observations on the nesting behaviour of wasps of the tribe Cercerini. J. Kansas Entomol. Soc. 44, 500–523 [Google Scholar]