Abstract
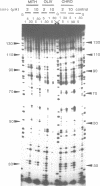
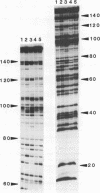
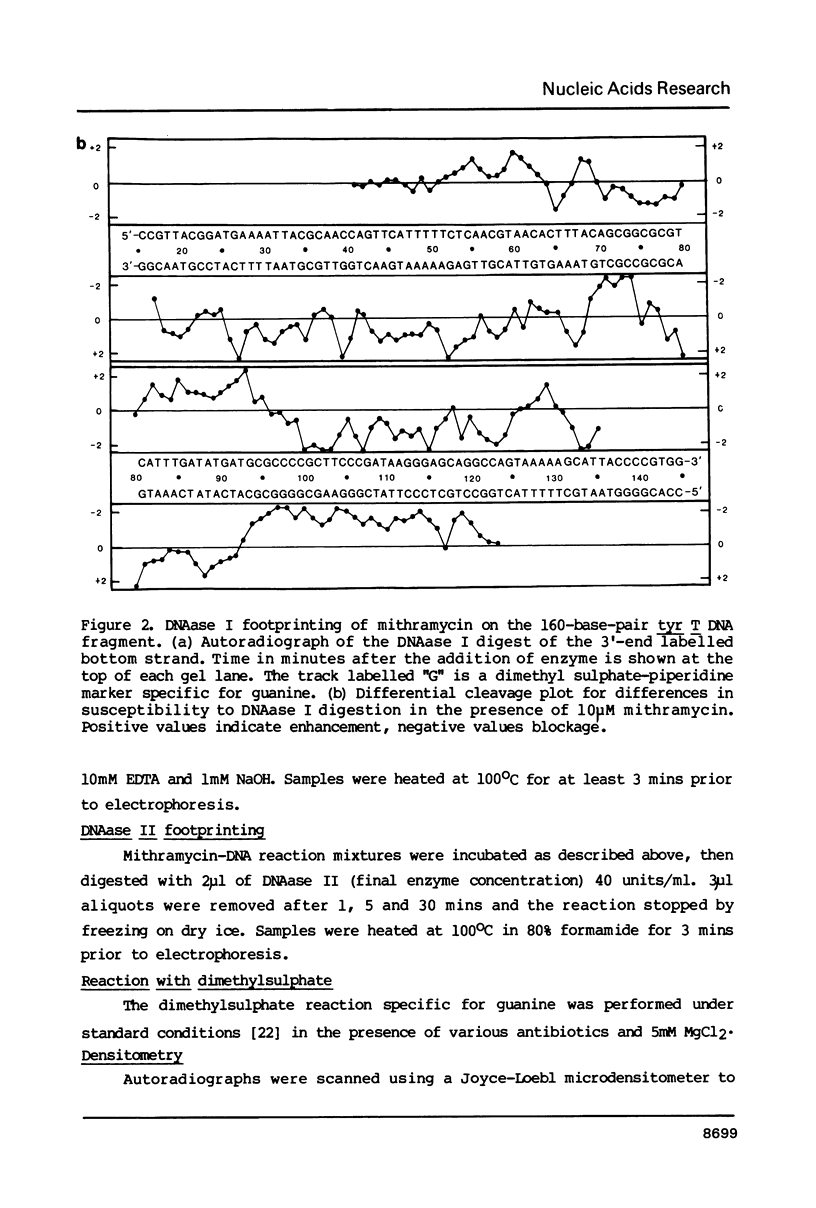
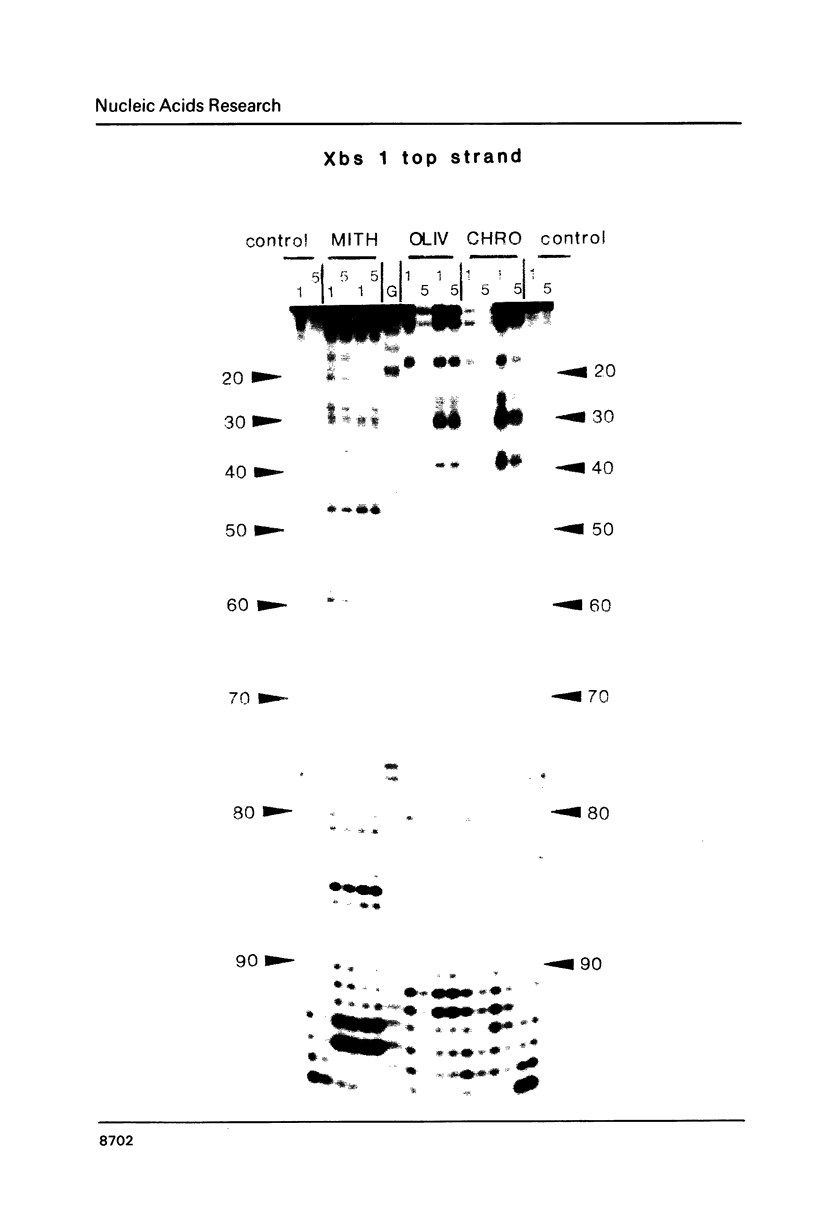
The preferred binding sites for mithramycin on four different DNA fragments have been investigated by DNAase I footprinting. Sites containing at least two contiguous GC base pairs are protected by the antibiotic, the preferred binding site consisting of the dinucleotide step GpG (or CpC). Related antibiotics chromomycin and olivomycin produce similar, but not identical footprinting patterns suggesting that they can recognize other sequences as well. All three antibiotics induce enhanced rates of enzyme cleavage at regions flanking some of their binding sites. These effects are generally observed in runs of A and T and are attributed to DNA structural variations induced in the vicinity of the ligand binding site. The reaction of dimethylsulphate with N7 of guanine was modified by the presence of mithramycin so that we cannot exclude the possibility that these antibiotics bind to DNA via the major groove.
Full text
PDF






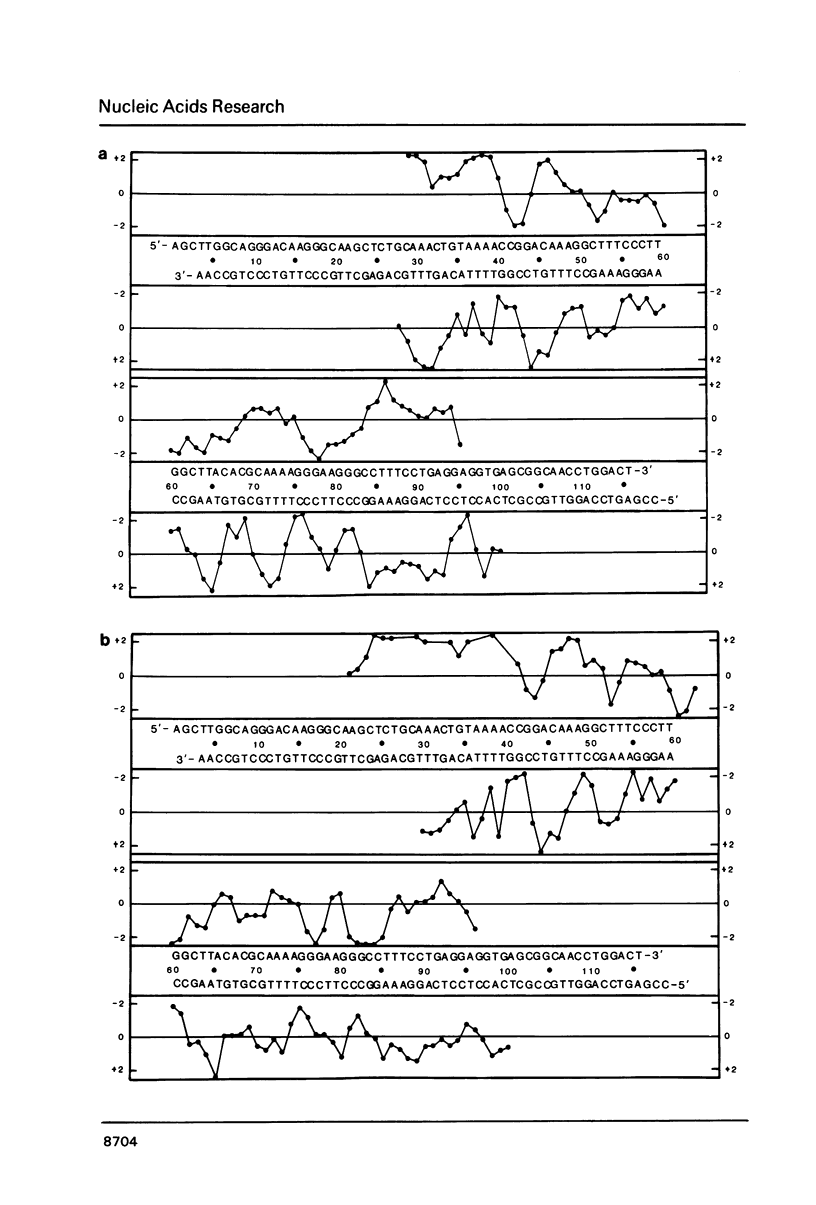
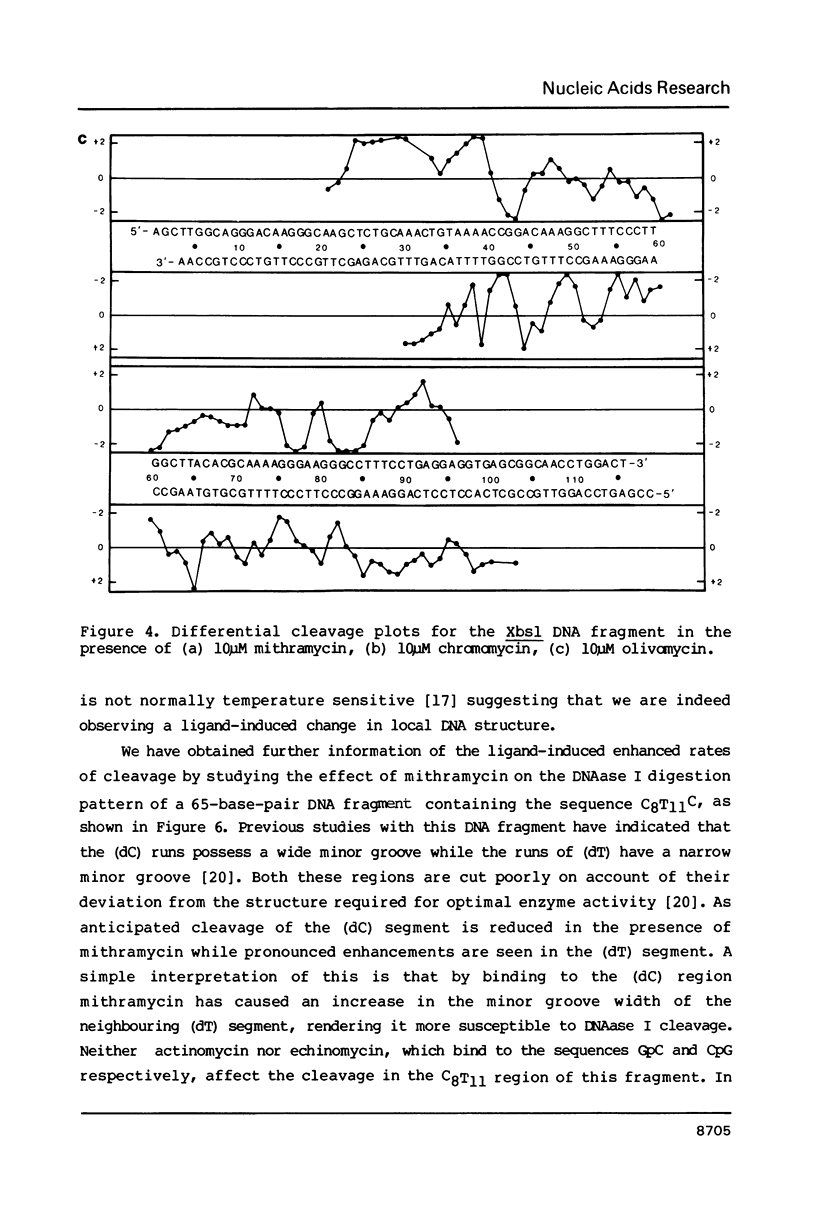
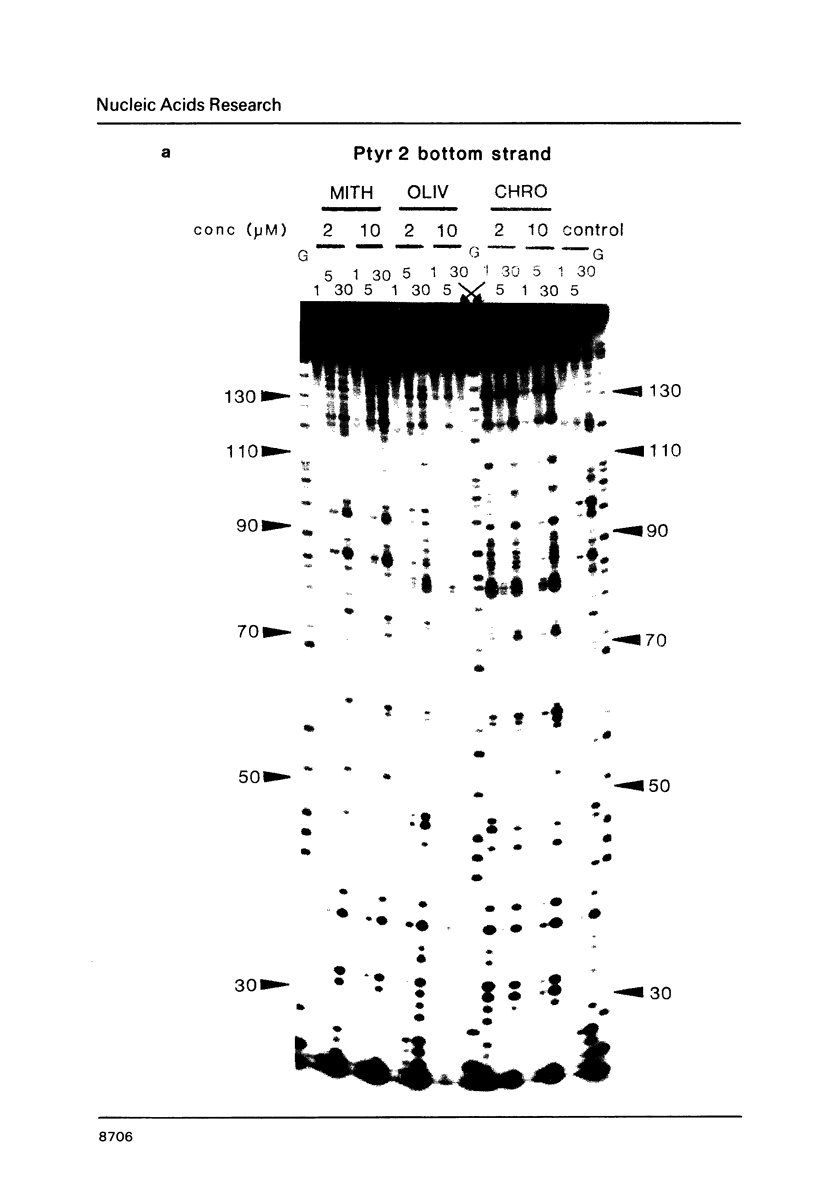
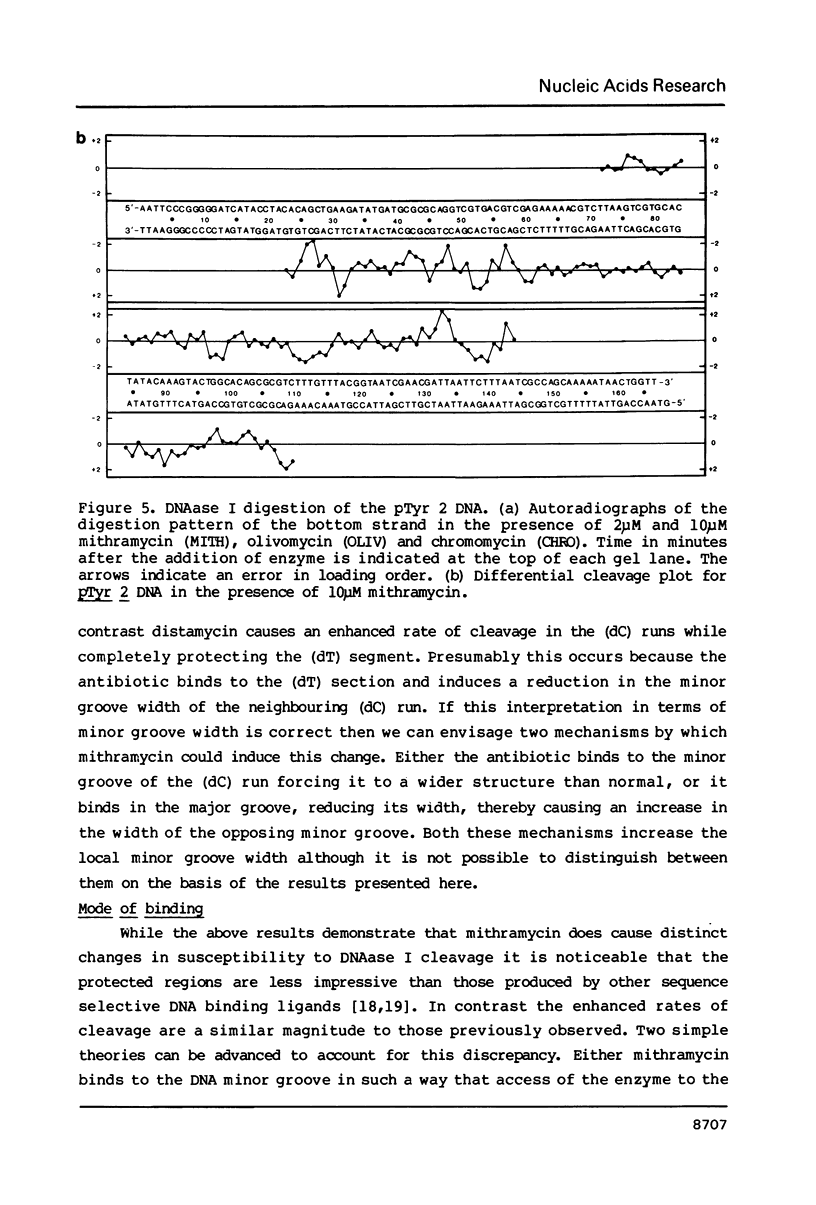





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cerami A., Reich E., Ward D. C., Goldberg I. H. The interaction of actinomycin with DNA: requirement for the 2-amino group of purines. Proc Natl Acad Sci U S A. 1967 Apr;57(4):1036–1042. doi: 10.1073/pnas.57.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta D., Shashiprabha B. K., Podder S. K. Mode of action of antitumour antibiotic: Part II--Evidence for intercalation of mithramycin between DNA bases in the presence of Mg2+. Indian J Biochem Biophys. 1979 Feb;16(1):18–21. [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. Structural junctions in DNA: the influence of flanking sequence on nuclease digestion specificities. Nucleic Acids Res. 1985 Jun 25;13(12):4445–4467. doi: 10.1093/nar/13.12.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias E. G., Evans J. T. Mithramycin in the treatment of Paget's disease of bone. J Bone Joint Surg Am. 1972 Dec;54(8):1730–1736. [PubMed] [Google Scholar]
- Fok J., Waring M. Breakdown of pulse-labeled ribonucleic acid in Bacillus megaterium, revealed by exposure to the antibiotics mithramycin, chromomycin, and nogalamycin. Mol Pharmacol. 1972 Jan;8(1):65–74. [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. Kinetic evidence for redistribution of actinomycin molecules between potential DNA-binding sites. Eur J Biochem. 1984 Dec 17;145(3):579–586. doi: 10.1111/j.1432-1033.1984.tb08596.x. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. Kinetic evidence for redistribution of actinomycin molecules between potential DNA-binding sites. Eur J Biochem. 1984 Dec 17;145(3):579–586. doi: 10.1111/j.1432-1033.1984.tb08596.x. [DOI] [PubMed] [Google Scholar]
- Hartmann G., Behr W., Beissner K. A., Honikel K., Sippel A. Antibiotics as inhibitors of nucleic acid and protein synthesis. Angew Chem Int Ed Engl. 1968 Sep;7(9):693–701. doi: 10.1002/anie.196806931. [DOI] [PubMed] [Google Scholar]
- Kennedy B. J., Yarbro J. W., Kickertz V., Sandberg-Wollheim M. Effect of mithramycin on a mouse glioma. Cancer Res. 1968 Jan;28(1):91–97. [PubMed] [Google Scholar]
- Kersten W., Kersten H., Szybalski W. Physicochemical properties of complexes between deoxyribonucleic acid and antibiotics which affect ribonucleic acid synthesis (actinomycin, daunomycin, cinerubin, nogalamycin, chormomycin, mithramycin, and olivomycin). Biochemistry. 1966 Jan;5(1):236–244. doi: 10.1021/bi00865a031. [DOI] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Sequence-specific binding of echinomycin to DNA: evidence for conformational changes affecting flanking sequences. Nucleic Acids Res. 1984 Jun 25;12(12):4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Jain S. C. Stereochemistry of actinomycin binding to DNA. II. Detailed molecular model of actinomycin-DNA complex and its implications. J Mol Biol. 1972 Jul 14;68(1):21–34. doi: 10.1016/0022-2836(72)90259-8. [DOI] [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]