SUMMARY
Glycoprotein-deleted (ΔG) rabies virus is a powerful tool for studies of neural circuit structure. Here we describe the development and demonstrate the utility of new resources that allow experiments directly investigating relationships between the structure and function of neural circuits. New methods and reagents allowed efficient production of twelve novel ΔG rabies variants from plasmid DNA. These new rabies viruses express useful neuroscience tools, including: the Ca++ indicator GCaMP3, for monitoring activity; Channelrhodopsin-2, for photoactivation; allatostatin receptor, for inactivation by ligand application; rtTA, ERT2CreERT2, or FLPo, for control of gene expression. These new tools allow neurons targeted based on their connectivity, to have their function assayed or their activity or gene expression manipulated. Combining these tools with in vivo imaging and optogenetic methods, and/or inducible gene expression in transgenic mice, will facilitate experiments investigating neural circuit development, plasticity, and function that have not been possible with existing reagents.
Keywords: channelrhodopsin, GCaMP3, allatostatin receptor, tTA, Cre, Flp
INTRODUCTION
Over many decades, neuroscientists have sought to understand how diverse neurons in the central nervous system generate perception and behavior. The functions of the brain are based on the activity patterns of large numbers of interconnected neurons which form neural circuits. Much progress has been made in electrophysiology, electron microscopy, optical imaging and molecular biology to understand how a neuron’s connectivity contributes to its function in the circuit. Genetic tools delivered by viral vectors or in transgenic animals have become a powerful resource for studies of the structure and function of neuronal networks (Arenkiel and Ehlers, 2009; Luo et al., 2008; Scanziani and Hausser, 2009). These tools can be used to change gene expression, to monitor or manipulate neural activity and to trace neuronal connectivity. Such studies are becoming increasingly sophisticated as a result of the combinatorial power allowed by the incorporation of multiple tools. For example, it is possible to identify the connections or function of specific cell types or a single cell, and by incorporating genetic tools into tracing viruses, it is possible to more directly link circuits and function (Boldogkoi et al., 2009; Choi et al., 2010; DeFalco et al., 2001; Haubensak et al., 2010; Luo et al., 2008; Marshel et al., 2010; Miyamichi et al., 2011; Rancz et al., 2011; Stepien et al., 2010; Wall et al., 2010; Wickersham et al., 2007b; Zhou et al., 2009).
Rabies virus is particularly useful for the study of neuronal circuits due to its ability to spread transsynaptically, exclusively in the retrograde direction (Callaway, 2008; Ugolini, 1995, 2010). Relative to other tracing viruses, such as one particular strain of herpes virus “pseudorabies virus” and other herpes viruses, it is unique in that infected cells remain viable for weeks (Wickersham et al., 2007a) and it can amplify from even a single viral particle (Coulon et al., 1982). It has been shown that a glycoprotein-deleted variant of the SAD-B19 strain of rabies virus encoding GFP (SADΔG-GFP) can be used to reveal the detailed morphology of infected projection neurons (Larsen et al., 2007; Nassi and Callaway, 2007; Wickersham et al., 2007a), that EnvA-pseudotyped SADΔG-GFP rabies viruses can be used to selectively label the direct inputs to a targeted neuronal population or even a single neuron (Haubensak et al., 2010; Marshel et al., 2010; Miyamichi et al., 2011; Rancz et al., 2011; Stepien et al., 2010; Wall et al., 2010; Wickersham et al., 2007b; Yonehara et al., 2011), and that a combination of EnvB-pseudotyped rabies viruses and a bridge protein with TVB can selectively target infection to specific neuron types which bind to the bridge protein (Choi et al., 2010).
While ΔG rabies viruses have already proven to be a powerful tool for revealing neural circuit structure, understanding how neural circuits develop and function requires direct links to be made between neural circuits, activity monitoring, and manipulation of activity or gene expression. We therefore aimed to extend the utility of a recombinant rabies virus by incorporating the potential to exploit other novel genetic technologies that have recently been pioneered. For example, much progress has been made at the interface of optical and genetic technologies (Luo et al., 2008; Scanziani and Hausser, 2009). In vivo 2-photon imaging of calcium transients in neurons labeled with indicator dyes allows monitoring of activity from many neurons simultaneously (Kerr et al., 2005; Ohki et al., 2005; Runyan et al., 2010; Svoboda and Yasuda, 2006), and the incorporation of genetically encoded calcium indicators allows the monitoring of genetically targeted neurons (Miyawaki, 2005; Tian et al., 2009). Genetic strategies for activating or inactivating selected neurons have also opened up new possibilities for understanding circuitry and behavior. In particular, optical stimulation, or “optogenetics”, has allowed for manipulation of the activity of genetically defined neurons with high temporal and spatial resolution (Boyden et al., 2005; Cardin et al., 2009; Sohal et al., 2009). Finally, the last decade has seen the development of a large number of “floxed”, “fretted”, or tTA-dependent mouse lines (Branda and Dymecki, 2004) and viral vectors (Kuhlman and Huang, 2008; Luo et al., 2008) to allow selective and inducible knockout of genes of interest, allowing investigations of the roles of particular genes in the development, plasticity, or function of defined components of the nervous system. By incorporating each of these classes of genetic tools into the ΔG rabies viruses, it is possible to combine their power with the ability to target connectionally defined neuronal networks.
Although rabies viruses are a powerful tool, the fact that rabies is a negative strand RNA virus presents both unique challenges and opportunities for the production and the use of new variants to interface with the growing arsenal of genetic tools and transgenic mouse lines. For example, DNA viruses, such as PRV, can be readily recovered following manipulation of the viral genome and expression of encoded genes can be made conditional upon interaction with recombinase expressed in transgenic mouse lines (DeFalco et al., 2001). In contrast, recovery of new rabies virus variants requires a more complex process (Inoue et al., 2003; Ito et al., 2003; Schnell et al., 1994; Wu and Rupprecht, 2008), and the ability to interface with transgenic mice requires development of novel strategies, as described below. Nevertheless, once a new ΔG rabies virus variant is successfully recovered, it can easily be propagated and amplified in a rabies glycoprotein-expressing cell line (Etessami et al., 2000; Mebatsion et al., 1996; Wickersham et al., 2007a; Wickersham et al., 2007b).
Here we establish reliable and efficient methods and reagents for recovery and amplification of rabies virus and describe the development and validation of the new SADΔG variants which we have produced. These variants include SADΔG rabies viruses encoding: red fluorescent proteins; blue fluorescent proteins; both red and green fluorescent proteins from the same genome; the calcium sensor GCaMP3 for monitoring neuronal activity (Tian et al., 2009); the light-gated cation channel Channelrhodposin-2 (ChR2) for the activation of neural activity (Boyden et al., 2005); the Drosophila Allatostatin receptor (AlstR) for silencing of neural activity (Lechner et al., 2002; Tan et al., 2006); the reverse tetracycline transactivator (rtTA), tamoxifen-inducible Cre-recombinase, and FLP-recombinase to allow control of gene expression in available transgenic mouse lines and viral vectors (Branda and Dymecki, 2004). We illustrate the utility of these variants and further discuss an even wider potential range of powerful applications.
RESULTS
System for recovery of glycoprotein-deleted rabies viruses from plasmid DNA
Although SADΔG-GFP can be recovered from DNA plasmids and amplified using previously established procedures (Buchholz et al., 1999), we aimed to improve the efficiency of ΔG rabies virus recovery from DNA. Here we generated new plasmids and cell lines, and tested various culture conditions to optimize recovery systems. Because rabies is a negative strand RNA virus but the tools that are available to manipulate genetic material work with DNA, it is necessary to generate and use several specialized reagents in order to recover new genetically modified rabies variants from a set of DNA plasmids. At least four different groups have developed and used such systems to recover various rabies strains (Inoue et al., 2003; Ito et al., 2003; Schnell et al., 1994; Wu and Rupprecht, 2008). The first published system recovered the SAD-B19 strain of rabies virus, utilized transcription from the T7 promoter, required T7 RNA polymerase provided by a vaccinia helper virus (Schnell et al., 1994), and in later developments was supported by a cell line expressing T7 polymerase (BSR T7/5)(Buchholz et al., 1999; Wickersham et al., 2007b) to replace vaccinia. This system was used in all previous studies recovering ΔG rabies virus. Intact HEP-Flury rabies virus has also been recovered using transcription from the CMV promoter, and did not require T7 polymerase (Inoue et al., 2003). Previous studies have not determined whether such a system can be used to efficiently recover ΔG rabies viruses or the SAD-B19 strain.
To generate the new rabies variants described here, we independently developed a hybrid system for recovery of SADΔG rabies virus, incorporating both CMV and T7 promoters, similar to a system for recovery of the ERA strain of rabies virus (Wu and Rupprecht, 2008). The recovery system is composed of a rabies virus genomic plasmid (pcDNA-SADΔG-GFP) containing the full-length genome of the SAD-B19 strain of rabies virus with the glycoprotein gene deleted and replaced with GFP (SADΔG-GFP), and a set of helper plasmids for expression of viral genes required to recover virus. In pcDNA-SADΔG-GFP, the genomic sequence was flanked by hammerhead ribozyme (HamRz) (Le Mercier et al., 2002) and hepatitis delta ribozyme (HdvRz) (Symons, 1992) and expression was under the control of both theCMV promoter and T7 RNA polymerase promoter. Helper plasmids encoded individual viral genes, also under the control of both CMV and T7 promoters. All of the above plasmids were constructed following reverse transcription from the parent virus SADΔG-GFP (Wickersham et al., 2007a).
We tested the importance of T7 RNA polymerase expression, the relative utility of three different cells lines, and two different transfection methods to quantitatively compare the success rates of different recovery strategies using these reagents. Conditions under which T7 polymerase was expressed included the use of the T7-expressing cell line BSR T7/5 (Buchholz et al., 1999), or HEK293t or BHK-21 cells transfected with a plasmid to induce expression of T7 RNA polymerase with a nuclear localization signal (Wu and Rupprecht, 2008). In these cases, we attempted to recover SADΔG-GFP by transfecting cells with the rabies genomic plasmid and helper plasmids using a Calcium phosphate method, and cells were cultured in 5% CO2 at 37°C. SADΔG-GFP rabies virus could be successfully recovered under all conditions in which T7 polymerase was expressed, but was never recovered in the absence of T7 polymerase. The highest rate of recovery was achieved using the BSR T7/5 cells (50.0%, 3 of 6 attempts), while success was less than half as likely using HEK293t (16.7%, 1 of 6) or BHK-21 cells (16.7%, 1 of 6) transfected with T7 polymerase. The requirement for T7 polymerase expression with our system contrasts with successful T7 polymerase-independent recovery of intact HEP-Flury rabies virus (Inoue et al., 2003). This difference could be due to differences in the strain of rabies virus, the presence of glycoprotein in the viral genome, or other differences in the reagents and methods used.
While the moderate success rates with this system were adequate for recovery of rabies viruses expressing fluorescent proteins, they were still relatively inefficient. Furthermore, we found that it was not possible to recover the ChR2-mCherry virus using these methods. We reasoned that because recovery efficiency is likely dependent on the expression of both T7 polymerase and B19G, it would be helpful to stably express both of these genes in producer cells. We therefore established new packaging cells expressing both T7 RNA polymerase and the rabies glycoprotein. BSR T7/5 cells expressing T7 RNA polymerase were infected with the HIV lentivirus encoding both Histone2B-tagged GFP and rabies glycoprotein B19G linked by an F2A self-cleaving element under the control of CMV promoter. Infected cells expressed GFP in their cell nuclei, and 6.2% of total cells were collected as a GFP-high+ fraction using FACS sorting (Figure S1A). The FACS-sorted cells expressed T7 RNA polymerase, B19G and GFP (referred to as “B7GG cells” hereafter. Figure S1B). Using the B7GG cells, we tested various parameters to increase the efficiency of recovery and amplification of ΔG rabies viruses: plasmid concentrations, transfection reagents, and culture conditions. Under 35°C and 3% CO2 conditions, B7GG cells decreased proliferation and remained healthier for about 1 week compared to standard culture conditions (37°C and 5% CO2). When B7GG cells were transfected with the rabies genomic plasmid carrying GFP (pSADΔG-GFP-F2) and helper plasmids carrying B19N, B19P, B19L and B19G with Lipofectamin2000 in a humidified atmosphere of 3% CO2 at 35°C, the success rate of recovery was 100% (6 wells were examined in one set of experiments. The reproducibility of the results was confirmed in four independent sets of experiments.), significantly higher than with the first established protocol (BSR T7/5 cells with a Calcium phosphate method under 37°C and 5% CO2 conditions; 37.4 ± 8.0 %; p<0.01, t-test). Furthermore, the titer of virus recovered by the new protocol (B7GG cells with Lipofectamine2000 under 35°C and 3% CO2 conditions) was more than ten times higher than with the earlier protocol (BSR T7/5 cells with a Calcium phosphate method under 37°C and 5% CO2 conditions). Therefore, 35°C and 3% CO2 conditions markedly increased both the recovery efficiency after transfection and further amplification, allowing for efficient recovery of viruses expressing membrane proteins, such ChR2 and AlstR (see below).
Generation of glycoprotein-deleted rabies viral vectors encoding multiple exogenous genes
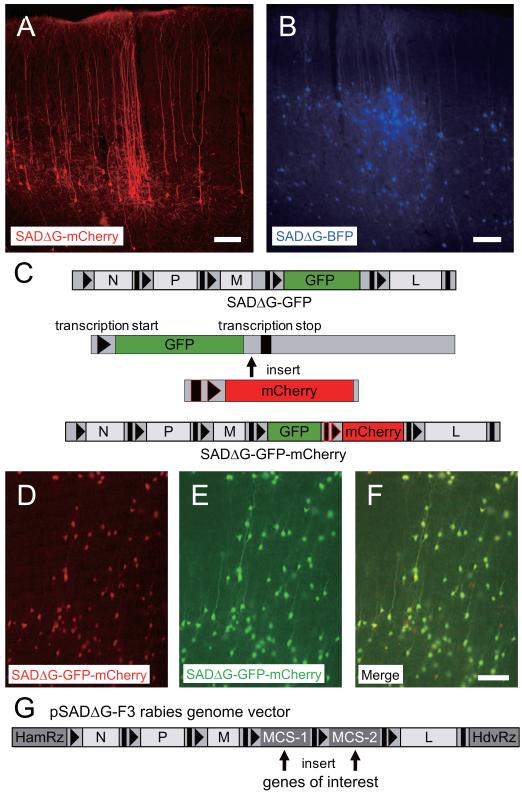
We next produced versions of ΔG rabies viruses expressing various fluorescent proteins because combinations of different colors are indispensable, for example, for interfacing with GFP-expressing mouse lines or for combined injections of different viruses. We replaced B19G in the rabies genome with a red fluorescent protein, DsRed2 (SADΔG-DsRed2), mCherry (Figure 1A, SADΔG-mCherry), c-myc epitope-tagged mCherry (SADΔG-mCherry-myc), or a blue fluorescent protein, BFP (Figure 1B, SADΔG-BFP). These viruses were recovered from DNA, amplified, and concentrated by ultracentrifugation. The titers to which these viruses could be grown and concentrated were indistinguishable from the original SADΔG-GFP (Table 1). Concentrated SADΔG-mCherry was tested in vivo by injection into the dorsal lateral geniculate nucleus (dLGN) of rats (Figure 1A) or into two strains of GAD-GFP mice which express GFP in cortical interneurons (GIN(Oliva et al., 2000) in Figure S2A and G30(Lopez-Bendito et al., 2004) in Figure S2B). As expected from previously published studies using SADΔG-GFP injections into the thalamus (Larsen et al., 2007), following 3 days survival, cortical neurons in locations known to project to the dLGN were infected and completely filled with mCherry, allowing clear visualization of detailed neuronal morphology, including dendritic spines and axons (Figure 1A). Injections of SADΔG-mCherry into dLGN of GAD-GFP mouse lines further demonstrated that infection was restricted from cortical inhibitory neurons which do not project to dLGN, and the GFP interneurons could be clearly distinguished from the rabies-infected, mCherry-expressing corticothalamic projection neurons in the primary visual cortex (V1) (Figure S2A,B). When we injected SADΔG-BFP into the rat V1, nearby neurons in layers 2/3 and 4 of the V1 were labeled with BFP (Figure 1B). Similar results were observed for SADΔG-mCherry-myc (Figure S3).
Figure 1. Production of rabies viral vectors encoding multiple genes.
(A, B) Generation of SADΔG-mCherry (A) and SADΔG-BFP (B) rabies viruses. (A) Neurons in layer 6 of the rat primary visual cortex were visualized following injection of SADΔG-mCherry into the dLGN. (B) Neurons in layers 2/3 and 4 of rat visual cortex were labeled with SADΔG-BFP by injection into nearby visual cortex. Scale bars, 100 μm. (C) Each open reading frame requires transcription start and stop sequences to be inserted before and after the open reading frame in the rabies genome. To insert an additional exogenous gene, mCherry with additional transcription start and stop sequences (black with red outlines) followed by mCherry was inserted between the last codon of GFP and its transcription end sequence. (D, E, F) Neurons in cortical slice cultures infected with SADΔG-GFP-mCherry. All of the neurons infected with SADΔG-GFP-mCherry express both mCherry (D, F) and GFP (E, F), indicating reliable transcriptional regulation and expression of both gene products. Scale bar, 100 μm. (G) Plasmid created for efficient introduction of one or two trasngenes into the rabies genome. The pSADΔG-F3 rabies genome vector has two multiple cloning sites (MCS-1 and MCS-2) for insertion of genes of interest. For expression of a single gene from rabies vectors, use of one site in MCS-1 and one site in MCS-2 enables insertion of a single ORF and deletion of stop and start transcription cassettes. For cloning of two transgenes, insertion of each gene in MCS-1 and MCS-2 allows reliable co-expression from the rabies vectors.
Table. 1.
Titer and transgene size of rabies virus variants
| Name | Gene 1 (Size) | Gene2 (Size) | Titer |
|---|---|---|---|
| SADΔG-GFP | GFP (0.7 kb) | ------- | 5.8 × 10^8 – 7.5 × 10^9 (RG) |
| 8.8 × 10^7 – 6.2 × 10^8 (EnvA) | |||
| SADΔG-mCherry | mCherry (0.7 kb) | ------- | 5.5 × 10^8 – 1.6 × 10^10 (RG) |
| 1.3 × 10^8 – 6.9 × 10^8 (EnvA) | |||
| SADΔG-mCherry-myc | mCherry-myc (1.0 kb) | ------ | 1.5 × 10^8 – 5.6×10^8 (RG) |
| SADΔG-DsRed2 | DsRed2 (0.7 kb) | ------- | 2.0 × 10^8 – 3.6 × 10^8 (RG) |
| SADΔG-BFP | BFP (0.7 kb) | ------- | 1.8 × 10^8 – 3.7 × 10^9 (RG) |
| SADΔG-GFP-mCherry | GFP (0.7 kb) | mCherry (0.7 kb) | 5.7 × 10^5 – 8.7 × 10^6 (RG, Supernatant) |
| SADΔG-GCaMP3 | GCaMP3 (1.4 kb) | ------- | 3.5 × 10^8 – 9.1 × 10^8 (RG) |
| SADΔG-GCaMP3-DsRedX | GCaMP3(1.4 kb) | DsRedX (0.7 kb) | 3.7 × 10^8 – 7.5 × 10^8 (RG) |
| SADΔG-ChR2-mCherry | Channelrhodopsin2-mCherry (1.6 kb) | ------- | 1.3 × 10^8 – 4.1 × 10^9 (RG) |
| SADΔG-GFP-AlstR | GFP (0.7 kb) | Allatostatin receptor (1.2 kb) | 4.3 × 10^8 – 6.7 × 10^9 (RG) |
| SADΔG-GFP-rtTA | GFP (0.7 kb) | rtTA (0.8 kb) | 7.8 × 10^8 – 8.1 × 10^9 (RG) |
| 2.9 × 10^5 – 4.9 × 10^5 (EnvA Supernatant) | |||
| SADΔG-GFP-ERT2CreERT2 | GFP (0.7 kb) | ERT2CreERT2 (2.9 kb) | 3.1 × 10^8 – 8.9 × 10^8 (RG) |
| SADΔG-FLPo-DsRedX | FLPo (1.3 kb) | DsRedX (0.7 kb) | 7.5 × 10^8 – 2.1 × 10^9 (RG) |
Twelve new rabies virus variants and SADΔGFP were produced in the present study. The transgenes, their size and the titer of each ΔG-rabies virus used are listed. Unpseudotyped rabies viruses and EnvA-pseudotyped rabies viruses were titrated with HEK293t cells and HEK293-TVA cells, respectively. Titers are expressed as infectious units/ml.
For many applications, it is important to be able to mark infected neurons with one gene and to also express a second gene that, for example, controls cell function. This strategy is often preferable to the use of fusion proteins, where the incorporation of a marker protein can prevent normal function, or in cases where the gene product might be targeted to membrane compartments so that cell morphology is not easily visualized. Because G-deleted rabies viruses expressing multiple exogenous genes have not been reported previously and the genome location of our inserts differed from that in previous “two-gene” viruses (McGettigan et al., 2003; Ohara et al., 2009; Schnell et al., 2000), we were concerned that possible complications related to transcriptional regulation or viral capacity might prevent successful recovery and/or co-expression of both genes. We therefore sought to recover ΔG rabies virus encoding two different fluorescent proteins GFP and mCherry. This allowed direct visualization of whether both genes were reliably co-expressed. Two-gene expression was achieved by introduction of a second gene into an independent open reading frame in the SADΔG-GFP genome. The novel genes were introduced along with endogenous transcriptional control elements to take advantage of the rabies virus RNA-dependent RNA polymerase, which assures segmentation of separate genomic transcripts. Transcription stop/start signals derived from between the N and P coding sequences in the SAD-B19 genome (McGettigan et al., 2003; Ohara et al., 2009; Schnell et al., 2000) were placed upstream of an mCherry ORF and this was then inserted into pcDNA-SADΔG-GFP between the stop codon of GFP and transcription stop signal of GFP to create pcDNA-SADΔG-GFP-mCherry (Figure 1C). We recovered the GFP- and mCherry-expressing rabies virus (SADΔG-GFP-mCherry) from the plasmid pcDNA-SADΔG-GFP-mCherry, and then tested the virus in cortical slice cultures. GFP and mCherry expression patterns confirmed that the rabies virus expressed both genes reliably and at high levels in the same neurons (Figure 1D,E,F). Thus, to express two different transgenes from the rabies genome, we used the same transcription stop/start sequences in subsequent experiments requiring expression of multiple genes. Finally, we designed a new rabies genome vector to efficiently introduce one gene or two genes (pSADΔG-F3). The pSADΔG-F3 has two multiple-cloning sites (MCS-1 and MCS-2), which flank the transcription stop/start sequence cassette in the rabies genome (Figure 1G).
Monitoring of neural activity with genetically-encoded calcium indicators
Genetically-encoded calcium indicators enable neuroscientists to examine the function of genetically-defined neuronal populations (Luo et al., 2008; Miyawaki, 2005; Tian et al., 2009). To facilitate studies linking circuit structure and function, we produced G-deleted rabies viruses expressing the genetically-encoded calcium sensor GCaMP3 (Tian et al., 2009). As described above for viruses expressing fluorescent proteins, we produced two new rabies virus variants encoding GCaMP3. The first of these viruses expresses only GCaMP3 in place of B19G (SADΔG-GCaMP3), while the second expresses both GCaMP3 and DsRedX (SADΔG-GCaMP3-DsRedX). We recovered and amplified both SADΔG-GCaMP3 and SADΔG-GCaMP3-DsRedX in B7GG cells under 35°C, 3% CO2 conditions, and concentrated the viruses for in vivo injection. Again, titers of these viruses were indistinguishable from viruses encoding GFP only (Table 1). To test whether SADΔG-GCaMP3 is functional, we stereotaxically injected concentrated SADΔG-GCaMP3 (Table 1) into the V1 of mice and later analyzed visual responses of GCaMP3-expressing V1 neurons in vivo using two-photon imaging. Supplementary Figure 4A shows an anatomical reconstruction of a selected V1 neuron imaged in vivo 11 days after infection. Drifting gratings were presented in 8 directions in 45 degree orientation steps in random order. Fluorescence changes in the same cell were monitored during presentation of visual stimuli. As illustrated in Supplementary Figures 4B and 4C, visual stimulation resulted in robust increases in fluorescence, the strength of which depended on the grating orientation. (A movie of the response to the preferred orientation is available as Supplementary Movie 1.). These results are similar to those described in mouse V1 using the calcium indicator dye Oregon Green BAPTA (OGB), but the percent changes in fluorescence are much greater than are typically observed with indicator dyes (Kerlin et al., 2010; Runyan et al., 2010).
Although SADΔG-GCaMP3 proved to be effective for monitoring activity of infected neurons, the numbers of infected neurons that could be identified during in vivo 2-photon imaging was lower than expected from subsequent post-mortem examination (data not shown). We suspected that the difficulty in identifying infected neurons in vivo resulted from relatively dim baseline fluorescence, as expected from the dependence of fluorescent intensity on baseline calcium levels (Kerlin et al., 2010; Kerr et al., 2005; Ohki et al., 2005; Runyan et al., 2010). Therefore, to facilitate in vivo identification of infected neurons, we used SADΔG-GCaMP3-DsRedX in subsequent studies, and first searched for expression of DsRedX before subsequent characterization of functional responses based on the co-expression of GCaMP3. Furthermore, to allow functional characterization of an identified subset of V1 neurons making connections to another visual cortical area, we injected the SADΔG-GCaMP3-DsRedX into the lateral extrastriate cortical area AL of mice and assessed visual responses of retrogradely infected neurons in V1.
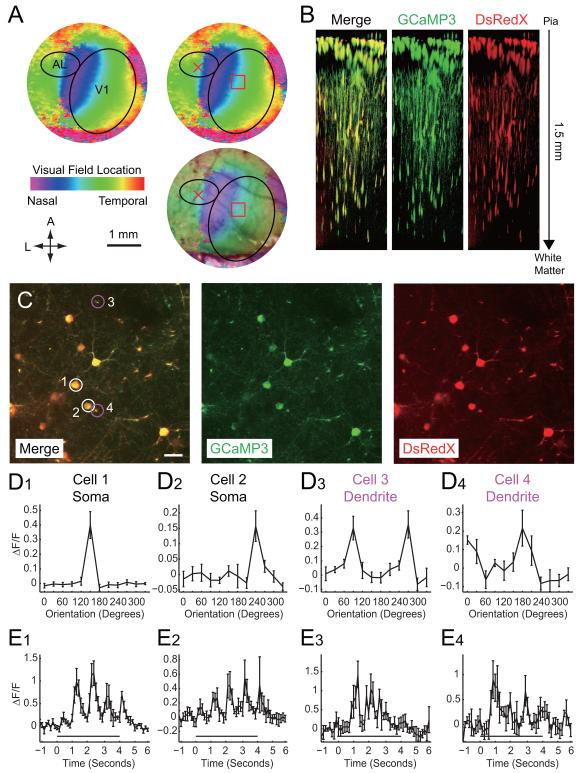
Injection of rabies virus into AL, subsequent identification of V1 and alignment of 2-photon imaging with the expected location of retrogradely-infected neurons was facilitated by intrinsic signal imaging to map retinotopy in V1 (Figure 2A) ((Kalatsky and Stryker, 2003), see Methods for further details.). Nine days after virus infection, blood vessel patterns were used to select a location in V1 expected to provide input to the virus injection location in AL (Figure 2A). Two-photon imaging at wavelengths sensitive to detection of DsRedX revealed a large field of infected neurons in V1. Remarkably, infected neurons could be clearly visualized to depths of more than a millimeter below the pial surface of the cortex (Figure 2B; The stack movie of SADΔG-GCaMP3-DsRedX-infected neurons in the V1 is available as Supplementary Movie 3.), far deeper than is typically possible with imaging using OGB (Kerlin et al., 2010; Kerr et al., 2005; Ohki et al., 2005; Runyan et al., 2010). GCaMP3 was also visualized in all DsRedX-positive cells and processes (Figure 2B).
Figure 2. Monitoring of neural activity with GCaMP3-expressing ΔG rabies virus.
(A) Retinotopic organization of the striate and extrastriate cortical regions. The retinotopic map from intrinsic imaging was overlaid on the image of surface blood vessels. Location of the border between V1 and AL was identified based on the representation of the vertical meridien (nasalmost visual fields) to allow targetting of viral injections to AL (red X). SADΔG-GCaMP3-DsRedX was injected into the lateral extrastriate cortical area AL of the mice and the corresponding retinotopic location in V1 was noted as the expected location of retrogradely infected neurons (red square). (B) Z-stack of SADΔG-GCaMP3-DsRedX-infected neurons visualized in vivo with two-photon imaging of V1, 9 days after rabies injection. AL-projecting V1 neuronal cell bodies and processes could seen in imaging planes extending from the cortical surface to a depth of 1.5 mm (C) Top view of two-photon laser-scanning images of SADΔG-GCaMP3-DsRedX-infected neurons at depth of 370 μm from the cortical surface. V1 neurons were retrogradely labeled with SADΔG-GCaMP3-DsRedX and co-expressed GCaMP3 (green) and DsRedX (red). Note that GCaMP3 could be seen in dendrites and axons, as well as cell bodies. Scale bar, 25 μm. (D) Orientation selectivity of SADΔG-GCaMP3-DsRedX-infected V1 neurons. Panels in D1-D4 correspond to neuronal cell bodies (D1-D2) or dendrites (D3-D4) labeled 1-4 in panel C. Orientation tuning curves are plotted as the mean change in fluorescence of the cell body (D1 and D2) or dendritic segments (D3 and D4) during the entire stimulus period, in response to square-wave gratings presented at various orientations in a random order. (E) Changes in fluorescence over time, in response to drifting gratings at the preferred orientation. Time 0 indicates the onset of the visual stimulus, which lasted for 4 seconds, as indicated by the black bar. Note that fluorescence was modulated at temporal frequencies that correspond to the temporal frequencies of the drifting gratings. These temporal modulations in phase with the visual stimuli can also be seen in supplemental movies. Values in D and E represent means ± S.E.M. of ΔF/F values across 5 repetitions of the visual stimulus.
We next selected planes of interest in the Z-axis for imaging of visually evoked functional changes in GCaMP3 fluorescence. Figure 2C shows anatomical images at a depth of 370 μm from the pial surface, while Figures 2D and 2E illustrate fluorescence changes of GCaMP3 in selected cell bodies or dendrites in response to visual stimuli. For visual stimulation, square-wave gratings were drifted at 12 directions in 30 degree orientation steps in random order. Infected V1 neurons exhibited significant increases in the GCaMP3 fluorescence at particular grating orientations. The two neuronal somata illustrated had direction selective visual responses (Figures 2D1 and 2D2; A movie of the response to the preferred orientation in Figure 2D1 is available as Supplementary Movie 3.). Interestingly, orientation selective responses were also detected from GCaMP3-labeled dendrites (Figures 2D3 and 2D4). It is likely that the combination of high expression levels and sparse labeling with GCaMP3 (compared to dense OGB labeling), contribute importantly to the ability to determine visual responses of distinct neuronal processes.
The surprisingly clear label that could be observed in neurons deep in the cortex suggested that these reagents might make it possible to also measure their visual responses. We therefore selected additional imaging planes at 520-535 μm below the pial surface and assayed orientation selectivity of fluorescence changes from labeled neurons in deeper cortical layers (Supplementary Figure 5A-5F). We obtained clear changes in the GCaMP3 fluorescence in response to the preferred directions, comparable to those typically observed in more superficial layers. However, unlike in superficial layers, fluorescence changes could be observed in identified neuronal cell bodies but not in dendritic processes. This likely reflects weaker fluorescence in the dendrites versus cell bodies and poorer imaging signal at greater depths.
To demonstrate the prolonged viability and visual responsiveness of rabies-infected neurons, the same animal was imaged again 2 days later, 11 days after the initial rabies injection into AL. Although we did not attempt to identify the same neurons that were imaged at 9 days, we again identified a field of DsRedX-expressing neurons and monitored their visual responses based on changes in GCaMP3 fluorescence. We performed the same set of experiments as described above to obtain orientation selectivity tuning curves on day 9 (Supplementary Figure 5G-5L). The infected cells in the V1 showed robust orientation selective fluorescence changes on day 11 from both GCaMP3-labeled soma and GCaMP3-labeled dendrites at a depth of 370 μm.
From these results, we conclude that rabies virus-mediated expression of GCaMP3 can be used to monitor activity of neurons targeted based on their connections to more distant neurons. Fluorescence changes can be monitored in vivo at either the soma or the dendrites, at depths greater than 500 μm, and virus infection does not prevent functional characterization even 11 days post-infection.
Control of neural activity with G-deleted rabies viruses encoding ChR2 or AlstR
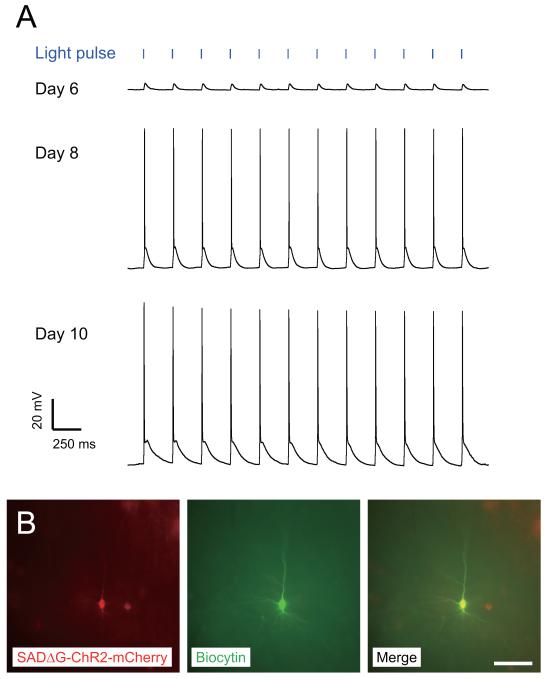
We next developed rabies virus variants which can control neural activity in targeted neuronal populations. To allow fast, light-controlled neuronal activation, we used the light-activated ion channel ChR2 fused to mCherry (Boyden et al., 2005; Nagel et al., 2003). We recovered and amplified SADΔG-ChR2-mCherry in B7GG cells under 3% CO2, 35°C conditions. It should be noted that SADΔG-ChR2-mCherry was difficult to recover with our original recovery system and did not grow well under more standard culture conditions. Nevertheless, using optimized procedures it could be grown at titers indistinguishable from GFP-expressing virus (Table 1). This difference is more likely to relate to the gene product expressed rather than the insert size, as similar difficulties were observed with another membrane protien (AlstR, see below) but not with several other viruses with even larger genomes (Table 1). To test the functionality of SADΔG-ChR2-mCherry, it was injected into the S1 barrel cortex of P9 mice. As expected from infection of neurons with axon terminals at the injection site, 6-10 days after injection, numerous infected neurons were observed in the neighboring barrel cortex, as well as other structures projecting to S1, such as the contralateral S1 cortex, M1 cortex and the thalamic nucleus VPM. We performed fluorescence-targeted whole-cell recordings from ChR2-mCherry-positive neurons in acute slices of injected mice and assayed their responses to photoactivation with a blue-light emitting diode (LED) connected to a light fiber. We selected cortical brain slices with relatively sparse labeling of ChR2-mCherry-positive neurons in order to minimize possible network effects that could result from simultaneous activation of large populations of neurons. We recorded from slices prepared from different animals at 3 different timepoints 6, 8 and 10 days post-infection. Although numerous mCherry neurons where clearly visible in the brain slices at all timepoints, functional activation was weaker at 6 days post-infection than at 8 or 10 days (Figure 3A). Figure 3A shows results of current-clamp recordings from representative mCherry-expressing pyramidal neurons located near the viral injection sites in S1, at 6, 8 and 10 days after virus injection. After recording, neurons were filled with biocytin included in the patch pipette to confirm with later streptavidin-Cy2 staining that the recorded cell expressed mCherry derived from the rabies virus genome (Figure 3B). We found that blue light pulses at 5 Hz with 2 ms duration induced depolarization in every ChR2-mCherry-positive cell (n=14) regardless of time after infection (Figure 3A), as expected from previous characterization of ChR2 (Boyden et al., 2005). However, on day 6 after rabies infection, recordings demonstrated that light-induced depolarizing currents using these light levels and pulse duration were below threshold for action potential generation in all neurons sampled (n=5/5). 8-10 days after virus injection, however, the same blue light stimuli invariably induced action potentials in ChR2-expressing neurons (n=5/5 on day 8; n=4/4 on day 10). These results indicate that rabies virus-mediated expression of ChR2 allows reliable optical control of infected neurons, but expression levels and/or membrane localization at early timepoints are likely to be lower, resulting in reduced sensitivity.
Figure 3. Photoactivation of neurons infected with ChR2-mCherry-expressing ΔG rabies virus.
(A) Generation of action potentials in ChR2-mCherry-expressing neurons by blue light pulses. Intracellular recordings were made from a layer 5 neuron in S1 barrel cortex 6, 8 and 10 days after injection of SADΔG-ChR2-mCherry in postnatal nine-day-old mice. During current-clamp, action potentials were reliably generated by blue light pulses of 5 Hz and 2 ms duration on day 8 and 10, but not day 6. (B) The recorded neuron, filled with biocytin and stained with streptavidin-Cy2 (green), was positive for mCherry (red). Scale bar, 30 μm.
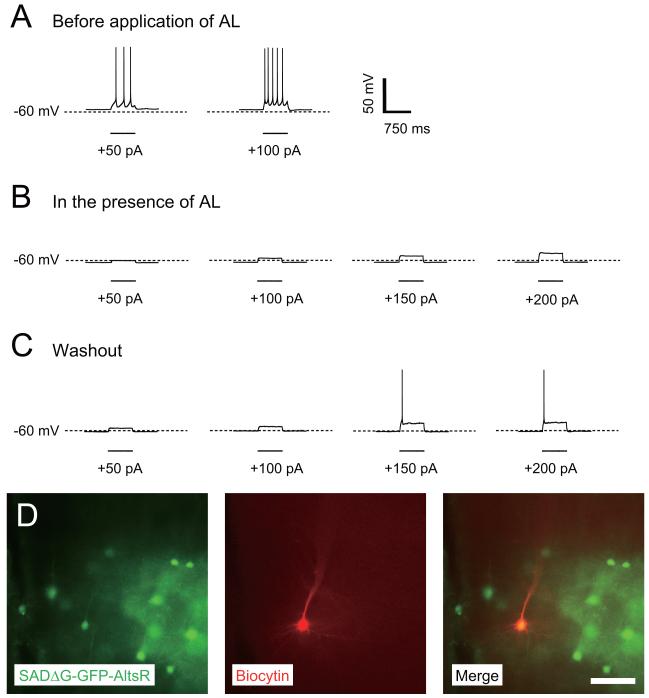
We also generated AlstR-encoding ΔG rabies to allow selective and reversible pharmacological silencing of neural activity (Lechner et al., 2002; Tan et al., 2006; Tan et al., 2008; Zhou et al., 2009). Expression of AlstR in mammalian neurons and application of the ligand allatostatin (AL) results in opening G protein-coupled inward rectifier K+ channels (Birgul et al., 1999), which hyperpolarizes neurons and decreases their input resistance, thus suppressing their ability to generate action potentials (Lechner et al., 2002). To allow visualization of infected neurons as well as control of their activity, we generated ΔG rabies encoding both GFP and AlstR (SADΔG-GFP-AlstR). We recovered and amplified SADΔG-GFP-AlstR in B7GG cells, again at 35°C and with 3% CO2. Although titers of virus grown under standard culture conditions were somewhat lower than for SADΔG-GFP, under these modified culture conditions this virus could be grown to high titers, indistinguishable from other G-deleted rabies viruses (Table 1). The concentrated SADΔG-GFP-AlstR was injected into the barrel field of S1 cortex of P18 mice. We prepared acute cortical slices of the injected mice 7 days after injection, and targeted whole-cell recordings to GFP-positive neurons. Recordings characterized the membrane potential, input resistance, and excitability of the SADΔG-GFP-AlstR-infected neurons. As exemplified for the neuron in Figure 4, before application of AL, resting membrane potential was −56.1 mV and input resistance was 87.0 MΩ Spike threshold was determined by injection of a series of depolarizing current pulses, gradually increasing in amplitude and the initial spike threshold was less than +50 pA (Figure 4A). Application of AL (1 μM) decreased membrane potential to −62.2 mV and input resistance to 37.3 MΩ. In the presence of AL, +50 pA current pulses were no longer sufficient to induce an action potential, and even pulses of +220 pA were still insufficient, indicating greatly reduced excitability (Figure 4B). At 30 min after AL washout, resting membrane potential and input resistance in the AlstR-expressing neuron partially recovered to −59.8 mV and 53.1 MΩ, respectively. The amplitude of depolarizing current pulses necessary to elicit an action potential was +150 pA, also indicating partial recovery (Figure 4C). After recording, we confirmed that the recorded cell was infected with SADΔG-GFP-AlstR by co-labeling with GFP and biocytin (stained with streptavidin-Cy3) (Figure 4D). Incomplete recovery is likely due to failure to completely remove AL from the recording chamber during washout. Silencing effects by AL were also observed 5 days after injection of SADΔG-GFP-AlstR (n=2/2). At 13-15 days post-injection, however, infected cells exhibited relatively depolarized resting membrane potentials (from −30 mV to −40mV), although cell morphology appeared intact (n=10/10). Poor cell health at 13-15 days is expected, as some rabies infected cells are likely killed by this timepoint (Wickersham et al., 2007a). It is possible that such effects are also exacerbated by expression of high levels of a membrane protein such as AlstR. These results indicate that AlstR expressed from the rabies virus is functional, and all results are consistent with far more rigorous characterizations that have been conducted previously (Lechner et al., 2002; Tan et al., 2006).
Figure 4. Allatostatin-induced silencing of neural activity with AlstR-expressing ΔG rabies virus.
(A-C) Whole-cell current clamp recordings from a pyramidal neuron in a brain slice from the barrel cortex of an 18-day-old mouse, 7 days after injection of SADΔG-GFP-AlstR. (A) Before application of allatostatin (AL), representative traces of responses to depolarizing current pulses (+50, +100, +150, and +200 pA, 750 ms duration) show that multiple action potentials are generated with as little as +50pA. (B) Application of the ligand AL at 1 μM inactivated the SADΔG-GFP-AlstR-infected neuron. +200 pA of current could no longer generate action potentials in the presence of AL. (C) The effects of AL were partially reversed by washout of AL with normal ACSF. +150 pA induced action potentials after washout of AL. (D) Photographs of the recorded neuron, as well as neighboring infected neurons. The recorded neuron was filled with biocytin and stained with streptavidin-Cy3 (red), and was positive for GFP derived from SADΔG-GFP-AlstR (green). Scale bar, 30 μm.
Temporal control of gene expression and transsynaptic viral tracing from a genetically targeted single postsynaptic neuron
Genetic methods for the control of gene expression, such as the Cre-lox, the FLP-frt, and the TetO/rtTA (Tet-On) systems, have widespread and powerful applications. We envision numerous applications for rabies viruses that encode Cre recombinase, FLP recombinase or rtTA in their genomes. These include: temporal control of the spread of virus across multiple synaptic steps and the ability to interface with Cre and tTA responder mouse lines or viral vectors. For example, using a Cre-expressing rabies in Brainbow mice (Livet et al., 2007) could allow labeling of the inputs to single neurons (Marshel et al., 2010; Wickersham et al., 2007b), with the individual presynaptic cells each uniquely marked by expression of different combinations of fluorescent protein. Interfacing with responder mice that conditionally express rabies glycoprotein (Weible et al., 2010) could potentially allow temporal control of the spread of ΔG rabies iteratively across multiple synaptic steps. While we do not explore all of these possibilities here, we describe the development of G-deleted rabies viruses expressing genes that allow control of gene expression, demonstrate that these viruses can be grown to high titer, and that the gene products are functional in rabies-infected cells.
Using methods described above, we made three new variants of rabies virus, each encoding both a fluorescent marker gene and another protein for control of gene expression. These include reverse tTA (rtTA) (Urlinger et al., 2000), which requires doxycycline (dox) for activation, tamoxifen-inducible Cre-recombinase (ERT2CreERT2) (Matsuda and Cepko, 2007; Young et al., 2008), and FLP-recombinase (FLPo) (Raymond and Soriano, 2007; Zhu and Sadowski, 1995), respectively. The resulting viruses are called SADΔG-GFP-rtTA, SADΔG-GFP-ERT2CreERT2, and SADΔG-FLPo-DsRedX, respectively, and all could be grown to high titers (Table 1).
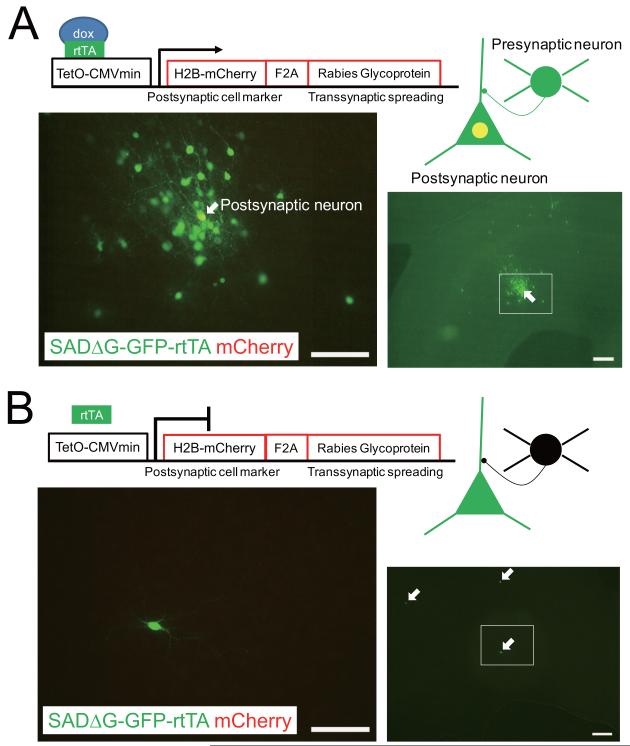
To illustrate the utility of SADΔG-GFP-rtTA for analyzing neural circuits, we demonstrate dox-dependent transsynaptic spread from genetically targeted neurons. Previously we established a tracing method which uses EnvA-pseudotyped SADΔG-GFP to selectively label the neurons which are directly presynaptic to a single, genetically-targeted postsynaptic neuron (Wickersham et al., 2007b). With this system, the transsynaptic spread of ΔG rabies virus was dependent on complementation by rabies glycoprotein expressed constitutively in the targeted postsynaptic neuron. Here we demonstrate conditional and temporal control of viral spread by placing rabies glycoprotein expression under the control of the TetO promoter and using the SADΔG-GFP-rtTA rabies virus. We constructed a plasmid conditionally expressing both nuclear-localized mCherry and rabies glycoprotein B19G linked by an F2A self-cleaving element and a GSG linker (Ryan and Drew, 1994; Szymczak et al., 2004), under the control of the TetO promoter (TetO-H2B-mCherry-2A-rabies glycoprotein) (Figure 5). Biolistics was used to cotransfect neurons in organotypic cortical slice cultures with both pTetO-H2B-mCherry-2A-rabies glycoprotein and another plasmid expressing TVA, a cognate receptor for the envelope protein EnvA, driven by the CMV promoter (Wickersham et al., 2007b). We then applied EnvA-pseudotyped SADΔG-GFP-rtTA to the transfected slice cultures either in the presence or absence of dox, an analogue of tetracycline (1.0 μg/ml). As expected from previous studies (Wickersham et al., 2007b) as well as dox and rtTA-dependent expression of mCherry and rabies glycoprotein, in the presence of dox, the EnvA-pseudotyped virus selectively infected TVA-transfected neurons, which subsequently expressed mCherry as well as rabies glycoprotein, which allowed transcomplementation so that the rabies virus spread to numerous nearby presynaptic neurons, which expressed GFP from the rabies genome (Figure 5A). In contrast, in the absence of dox, GFP expression from the rabies genome was restricted to isolated neurons that were presumably transfected and expressed TVA and they did not express mCherry (Figure 5B). This result demonstrated the requirement for the presence of dox to allow the originally-infected, isolated neurons to express mCherry and rabies glycoprotein, which is required for transsynaptic labeling of neighboring neurons.
Figure 5. Doxycycline-dependent gene expression and conditional transsynaptic spread with SADΔG-GFP-rtTA rabies virus.
(A, B) Following Biolistic transfection of isolated neurons in rat cortical slice culture with both pTetO-CMVmin-Histone2B-mCherry-F2A-B19G and pCMMP-TVA, application of EnvA-pseudotyped SADΔG-GFP-rtTA resulted in selective infection of transfected neurons. (A) The presence of both dox and rtTA expressed from the rabies virus allowed mCherry expression in EnvA-SADΔG-GFP-rtTA-infected neurons as shown by the yellow (GFP and mCherry) appearance of the soma of the “postsynaptic neuron”. The expression of rabies glycoprotein in this same postsynaptic neuron allowed complementation of the ΔG rabies so that the virus could spread to and express GFP in numerous presynaptic neurons. (B) Under the same conditions in the absence of dox (1.0 μg/ml), the EnvA-SADΔG-GFP-rtTA rabies virus also selectively infected scattered, isolated TVA-expressing neurons. But without dox there was no mCherry or rabies glycoprotein expression and therefore no complementation or transsynaptic spread of the rabies virus. Only three transfected and primarily infected neurons (green cells indicated by arrows) could be found in the brain slice in the absence of dox. Scale bars, 100 μm (right figures), 50 μm (left figures).
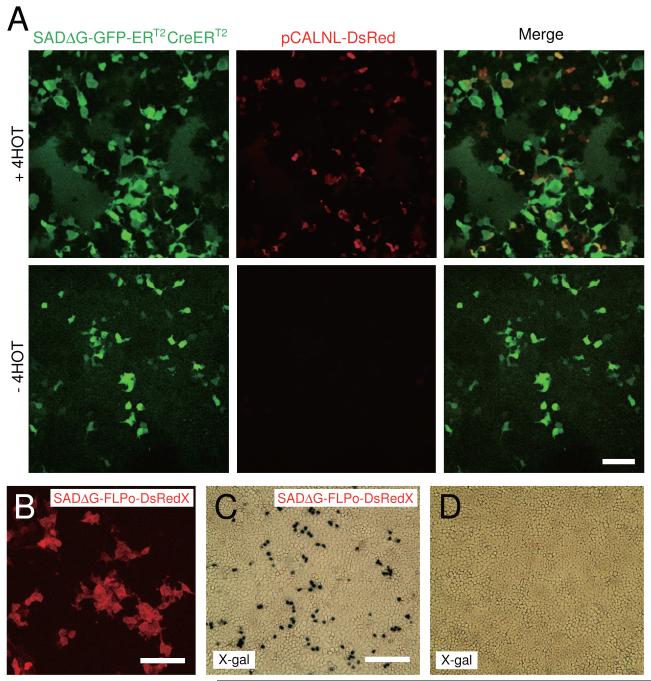
To test rabies virus expressing tamoxifen-inducible Cre-recombinase (SADΔG-GFP-ERT2CreERT2), HEK293t cells were transfected with a reporter plasmid expressing a loxP-STOP-loxP DsRed casette (pCALNL-DsRed) and then infected with SADΔG-GFP-ERT2CreERT2. In the presence of 4-hydroxytamoxifen (4-HOT), Cre recombined pCALNL-DsRed and co-expression of both GFP and DsRed was observed. However, without tamoxifen, only GFP expression was observed. These results indicate that tamoxifen controls Cre-dependent recombination in virus-infected cells (Figure 6A).
Figure 6. Recombination-dependent gene expression by ERT2CreERT2 or FLPo ΔG rabies viruses.
(A) Temporal regulation of gene expression using inducible Cre recombinase. SADΔG-GFP-ERT2CreERT2 activates tamoxifen-dependent expression of DsRed in HEK293t cells transfected with pCALNL-DsRed (recombination indicator). Top and bottom panels show results in the presence and absence of tamoxifen (+4HOT, 1 nM), respectively. Left panels show GFP expression from the rabies genome, middle panels show Cre-induced DsRed expression in the presence of 4HOT (top) but not in the absence of 4HOT (bottom); and overlays in right panels. Scale bar, 100 μm. (B) SADΔG-FLPo-DsRedX-infected cells expressed a red fluorescent DsRedX in HEK293t cells 3 days after virus infection. Scale bar, 100 μm. (C, D) Recombination induced in SADΔG-FLPo-DsRedX-infected cells. X-gal staining showed that HeLa cells stably expressing frt-STOP-frt LacZ cassette (recombination indicator) expressed LacZ in the presence of SADΔG-FLPo-DsRedX (C), but not in the absence of SADΔG-FLPo-DsRedX (D). Scale bar, 300 μm.
Similarly, we tested SADΔG-FLPo-DsRedX rabies virus using both HEK293t cells and HeLa cells stably expressing a frt-STOP-frt nuclear-localized LacZ casette. SADΔG-FLPo-DsRedX-infected HEK293t cells had red fluorescence (Figure 6B). Only cells that express FLPo from the rabies genome excise a frt-flanked STOP signal and activate constitutive LacZ expression. X-gal staining demonstrated that the SADΔG-FLPo-DsRedX virus caused recombination and LacZ expression in the HeLa cells (Figure 6C), whereas no LacZ expression was detected in the absence of the virus expressing FLPo (Figure 6D). These observations indicate functional recombination by FLPo derived from rabies virus.
DISCUSSION
We have described the development of a set of DNA plasmids suitable for generation of new genetically modified variants of SADΔG rabies viruses. We have used these reagents to generate twelve new SADΔG rabies virus variants that express transgenes of broad utility. These include ΔG rabies virus variants expressing two exogenous genes from the same rabies genome, as well as viruses to combine rabies labeling with monitoring or manipulating neuronal activity, and finally new variants expressing genes to allow control of gene expression within infected neurons. Furthermore, we have demonstrated the utility of these viruses for directly correlating connectivity with function in the intact brain and for manipulating activity or controlling gene expression in live cells. These new variants can now be propagated indefinitely and are available for use. These tools are expected to be an important component of a growing arsenal of genetic tools for the study of neural circuits (Luo et al., 2008) and open the door to new approaches for circuit research, whereby neuronal connectivity can be directly related to circuit function.
Strengths and limitations of G-deleted rabies virus
Many recently published studies have demonstrated the tremendous advantages of G-deleted rabies viruses for tracing neural circuits at high resolution. For example: it is possible to retrogradely infect neurons and express fluorescent proteins at high levels for detailed anatomical investigations (Larsen et al., 2007; Nassi and Callaway, 2007; Wickersham et al., 2007a); to identify neurons that are directly presynaptic to specific types of projection neurons (Stepien et al., 2010; Yonehara et al., 2011); to identify neurons that are directly presynaptic to specific genetically defined cell types (Haubensak et al., 2010; Miyamichi et al., 2011; Wall et al., 2010); and to identify neurons that are directly presynaptic to single, functionally-characterized neurons (Marshel et al., 2010; Rancz et al., 2011).
All of these studies traced connections in vivo, suggesting that they could be combined with methods for imaging or manipulating live neurons in the intact nervous system, as we have demonstrated here. The utility of these viruses for such studies depends on the ability of G-deleted rabies virus to infect cells and drive high levels of viral gene expression without killing them. Previously it has been shown that basic properties of infected neurons are not altered by infection after 7 days, but that many neurons are killed by about 14 days after infection (Wickersham et al., 2007a). We have shown here that there exists a working time window between about 5 and 11 days post-infection when functional studies of infected neurons are feasible. Nevertheless it is important for users to consider possible unwanted affects of rabies infection that may be unique to any of the various experimental conditions that might be used. For example, rabies virus infection can reduce expression of genes from the infected cells (Weible et al., 2010) and our experiments with AlstR expressing rabies virus show that at 13-15 days post-infection, live neurons display altered physiological properties. It is therefore crucial for potential users of these reagents to test and to control for any adverse affects of rabies virus infection. Despite these limitations, rabies virus is the only neurotropic virus we are aware of in which such studies directly linking connectivity to function over a time window as long as 6 days would be possible, as other viruses such as HSV and PRV kill neurons much more rapidly (Brittle et al., 2004; Card and Enquist, 2001).
It is also important to consider possible effects of high levels of transgene expression. For many experiments, the high levels of gene expression that are obtained with rabies viruses, relative to replication incompetent viruses (e.g. (Wickersham et al., 2007a)) are advantageous: GFP expressed at high levels allows detailed anatomical reconstructions (Larsen et al., 2007; Nassi and Callaway, 2007); ChR2 must be expressed at high levels for optogenetic control of activity (Figure 3); and high levels of fluorescent protein likely facilitate 2-photon imaging of neurons deep within live brain tissue (Figure 2B). While some transgenes have been reported to have toxicity at high expression levels, successful generation of transgenic and knock-in animals expressing GFP, mCherry, GCaMP, ChR2, AlstR, rtTA, tTA, Cre, or FLP (Arenkiel et al., 2007; Diez-Garcia et al., 2007; Gosgnach et al., 2006; Hippenmeyer et al., 2005; Tsien et al., 1996) suggest that moderate expression of these genes is well-tolerated for long time periods. It is therefore important for users to consider possible effects of high-level transgene expression from ΔG rabies viruses, however, effects over long time periods are likely to be moot, as the virus will likely kill neurons of interest before such issues are relevant. In cases where high levels of expression of a particularly toxic gene product are a concern during the limited period when rabies virus-infected neurons are viable, it may be possible to drive transgene expression from a less efficient means, such as in a transgenic animal, under the control of rtTA, Cre-ER or FLPo expressed from the rabies genome (e.g. Figure 5).
The utility of the novel rabies variants we have described here also depends on the degree to which they behave similarly to the better characterized ΔG rabies viruses expressing GFP or mCherry. For example, efficient infection is an important feature that is likely determined primarily by the titers at which these viruses can be grown and purified. We observed that ChR2 and AlstR-expressing ΔG rabies viruses were more difficult to grow than GFP-expressing virus, but using modified culture conditions they could be made at high titers that were indistinguishable from GFP-expressing viruses (Table 1). Within the limited range of insert sizes that we tested, there was no consistent relationship or apparent affect on viral titers (Table 1). For example, the largest genome we have recovered is for SADΔG-GFP-ERT2CreERT2, which includes GFP (0.7 kb) and ERT2CreERT2 (3.4 kb) as well as four native viral genes (N, P, M and L) for total of 13.6 kb, which is 1.7 kb larger than the native SAD-B19 genome of 11.9Kb (Conzelmann et al., 1990). Following recovery, this rabies virus was amplified as efficiently as the SADΔG-GFP rabies virus (10.2 kb genome). While in our hands neither the transgene expressed or the size of the viral geneome prevented production of high-titer ΔG rabies viruses, it is likely that the utility of these viruses will depend on the skill and care taken by those who grow them as well as careful adherence to the established protocols we have developed.
Linking connectivity to function
One of the main goals of systems neuroscience is to understand the architecture and function of neural circuits. Understanding how neural circuits function will require resolving the connectivity of the components, correlating the function of components with their connectivity, manipulating the activity of selected components and monitoring the activity of other components within the networks, and finally assessing the behavioral outcome. Techniques for achieving these goals, however, are limited. The rabies tools we have described here provide many new opportunities to allow the combination of rabies virus-based circuit tracing with functional studies.
For example, expression of the calcium sensor GCaMP3 in neurons that have been infected as a result of their connectivity with specific cell types or a single neuron could allow observations of direct correlations between connectivity and function in a single living preparation. Here we have explicitly demonstrated this type of approach by combining retrograde infection with GCaMP3-expressing ΔG rabies virus with in vivo two-photon imaging of visual responses. This allowed measurements of the visual receptive fields of a specific subset mouse V1 neurons selected based on their connectivity to area AL.
Similarly, expression of ChR2 and AlstR should allow control of neural activities in vitro and in vivo to facilitate tests of the causal relationships between connectivity and function within defined neural circuits. It should also be possible to test possible postsynaptic targets of connectionally-targeted rabies virus-infected neurons for functional connectivity with potential postsynaptic neurons through intracellular recording combined with photoactivation of axons from neurons expressing ChR2 from the rabies genome (Petreanu et al., 2007).
Targeting infection and transsynaptic labeling with GCaMP3-ΔG rabies, ChR2-ΔG rabies and AlstR-ΔG rabies in defined cell types or single cells using retrograde infection (Stepien et al., 2010; Wickersham et al., 2007a; Wickersham et al., 2007b; Yonehara et al., 2011), Cre-dependent TVA transduction (Haubensak et al., 2010; Wall et al., 2010), bridge proteins with TVB (Choi et al., 2010), or single cell electroporation of TVA (Marshel et al., 2010; Rancz et al., 2011) will be extremely useful for functional studies of identified neural circuits. In particular, by taking advantage of an increasing number of Cre-expressing mouse lines for targeting particular cell types (Driver lines are available in GENSAT and the Allen Institute for Brain Science), there will be an unprecedented opportunity to reveal detailed connectivity and function of specific cell types in mice (Madisen et al., 2010; Wall et al., 2010).
Finally, the expression of tamoxifen-inducible Cre, FLPo, or rtTA from the rabies genome will allow conditional expression of transgenes, such as transcription factors. In particular, interfacing ΔG rabies viruses with the increasing number of mouse lines and viral vectors that express rabies glycoprotein in a Cre, FLPo or tTA dependent manner (Weible et al., 2010) might allow for temporally-controlled tracing across multiple synaptic steps by administration of tamoxifen or doxcycline. In conclusion, the new reagents that we have developed are expected to facilitate future studies of nervous system function by allowing neuronal connectivity to be directly related to function.
Method Summary
Production of G-deleted rabies virus
Genomic RNA of SADΔG-GFP rabies virus was purified and reverse transcribed to obtain partial cDNA fragments of the rabies virus genome. Rabies nucleocapsid (pcDNA-SADB19N), rabies viral RNA polymerases (pcDNA-SADB19P and pcDNA-SADB19L) or rabies glycoprotein (pcDNA-SADB19G) were cloned using PCR. To construct the rabies virus genomic cDNA, several pieces of rabies genomic cDNA were ligated and flanked by HamRz and HdvRz in pcDNA3.1, becoming pcDNA-SADΔG-GFP. To establish a two gene expression system in the rabies genome, transcription stop and start sequences and 6 unique restriction enzyme sites were synthesized and cloned to produce pSADΔG-F3.
For recovery of ΔG rabies virus, B7GG cells were transfected with the rabies genome pSADΔG vector, pcDNA-SADB19N, pcDNA-SADB19P, pcDNA-SADB19L and pcDNA-SADB19G and maintained in a humidified atmosphere of 3% CO2 at 35°C. For pseudotyping with EnvA, BHK-EnvA cells were infected with unpseudotyped SADΔG rabies viruses, washed with PBS, reacted with 0.25% trypsin-EDTA and replated on new dishes. For in vivo injection, ΔG rabies viruses were amplified in 10, 15 cm dishes in a humidified atmosphere of 3% CO2 at 35°C, filtrated with 0.45 μm filter and concentrated by two rounds of ultracentrifugation. Unpseudotyped rabies viruses and EnvA-pseudotyped rabies viruses were titrated with HEK293t cells and HEK293-TVA cells, respectively. The titers and transgene size of viruses are shown in Table 1.
Characterization of rabies virus variants
ΔG rabies viruses were injected into the LGN, V1, AL or S1 of mice or rats. SADΔG-GCaMP3 and SADΔG-GCaMP3-DsRedX were injected in V1 and AL of adult mice, respectively. GCaMP3 signals in the V1 were imaged with a two-photon microscope. Visual receptive fields were assayed using drifting square-wave gratings moving in various directions. SADΔG-ChR2-mCherry and SADΔG-GFP-AlstR were injected into the barrel cortex of mice aged from postnatal day 8 and day 18, respectively. Whole-cell recordings of infected neurons were performed on brain slices. For photostimulation of ChR2-expressing neurons, light stimuli were delivered at 0.2-5 Hz from a blue LED. For inactivation of AlstR-expressing neurons, the peptide ligand AL (Ser-Arg-Pro-Tyr-Ser-Phe-Gly-Leu-NH2) was applied by perfusion.
Organotypic brain slice cultures were used for testing SADΔG-GFP-rtTA. After Biolistic transfection of both pCMMP-TVA800 and pTetO-CMVmin-Histone2B-mCherry-F2A-B19G, slices were infected with EnvA-SADΔG-GFP-rtTA and maintained in the absence or presence of dox (1.0 μg/ml). For testing SADΔG-GFP-ERT2CreERT2, HEK293t cells were transfected with the Cre-dependent plasmid pCALNL-DsRed, infected with the rabies virus and maintained in the absence or presence of 4-HOT (1.0 μM). For testing SADΔG-FLPo-DsRedX, HeLa cells stably expressing a frt-STOP-frt nuclear-localized LacZ cassette were infected with the virus and then processed for X-gal staining.
Supplementary Material
Highlights.
New methods allowed efficient production of ΔG rabies viruses from plasmid DNA
Twelve new rabies variants express useful genetically-encoded neuroscience tools
Encoded genes monitor or control activity or gene expression in defined circuits
These reagents facilitate studies directly linking circuit structure to function
ACKNOWLEDGEMENTS
We thank I. Wickersham and J. Choi for helpful discussions, K. Roby, M. De La Parra, and K. von Bochmann for technical assistance, members of the Callaway laboratory for stimulating discussions, K. Conzelmann for the BSR T7/5 cell line, O. Britz and M. Goulding for the HeLa cells expressing frt-STOP-frt-nLacZ, I. Verma for HIV lentivirus packaging plasmids, X. Wu for the pNLST7, R. Tsien for the mCherry plasmid, K. Deisseroth for the ChR2-mCherry plasmid, L. Looger for the GCaMP3 plasmid, and C. Cepko for the pCAG-ERT2CreERT2 and pCALNL-DsRed. F.O. is thankful to Noriko Osakada for constant encouragement and support. We are grateful for support from the National Institutes of Health (MH063912, NS069464, and EY010742: E.M.C.), the Kavli Institute for Brain and Mind at UC San Diego (E.M.C.), the Japan Society for the Promotion of Science (F.O.), the Kanae Foundation for the Promotion of Medical Science (F.O.), the Uehara Memorial Foundation (F.O.), and the Naito Foundation (F.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Full methods are available in the supplementary text on the web
REFERENCES
- Arenkiel BR, Ehlers MD. Molecular genetics and imaging technologies for circuit-based neuroanatomy. Nature. 2009;461:900–907. doi: 10.1038/nature08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgul N, Weise C, Kreienkamp HJ, Richter D. Reverse physiology in drosophila: identification of a novel allatostatin-like neuropeptide and its cognate receptor structurally related to the mammalian somatostatin/galanin/opioid receptor family. EMBO J. 1999;18:5892–5900. doi: 10.1093/emboj/18.21.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogkoi Z, Balint K, Awatramani GB, Balya D, Busskamp V, Viney TJ, Lagali PS, Duebel J, Pasti E, Tombacz D, et al. Genetically timed, activity-sensor and rainbow transsynaptic viral tools. Nat Methods. 2009;6:127–130. doi: 10.1038/nmeth.1292. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Brittle EE, Reynolds AE, Enquist LW. Two modes of pseudorabies virus neuroinvasion and lethality in mice. J Virol. 2004;78:12951–12963. doi: 10.1128/JVI.78.23.12951-12963.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18:617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card JP, Enquist LW. Transneuronal circuit analysis with pseudorabies viruses. Curr Protoc Neurosci. 2001 doi: 10.1002/0471142301.ns0105s09. Chapter 1, Unit1 5. [DOI] [PubMed] [Google Scholar]
- Cardin JA, Carlen M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Young JA, Callaway EM. Selective viral vector transduction of ErbB4 expressing cortical interneurons in vivo with a viral receptor-ligand bridge protein. Proc Natl Acad Sci U S A. 2010;107:16703–16708. doi: 10.1073/pnas.1006233107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann KK, Cox JH, Schneider LG, Thiel HJ. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- Coulon P, Rollin P, Aubert M, Flamand A. Molecular basis of rabies virus virulence. I. Selection of avirulent mutants of the CVS strain with anti-G monoclonal antibodies. J Gen Virol. 1982;61(Pt l):97–100. doi: 10.1099/0022-1317-61-1-97. [DOI] [PubMed] [Google Scholar]
- DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, Friedman JM. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- Diez-Garcia J, Akemann W, Knopfel T. In vivo calcium imaging from genetically specified target cells in mouse cerebellum. Neuroimage. 2007;34:859–869. doi: 10.1016/j.neuroimage.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Etessami R, Conzelmann KK, Fadai-Ghotbi B, Natelson B, Tsiang H, Ceccaldi PE. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol. 2000;81:2147–2153. doi: 10.1099/0022-1317-81-9-2147. [DOI] [PubMed] [Google Scholar]
- Gosgnach S, Lanuza GM, Butt SJ, Saueressig H, Zhang Y, Velasquez T, Riethmacher D, Callaway EM, Kiehn O, Goulding M. V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature. 2006;440:215–219. doi: 10.1038/nature04545. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, Ponnusamy R, Biag J, Dong HW, Deisseroth K, Callaway EM, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Shoji Y, Kurane I, Iijima T, Sakai T, Morimoto K. An improved method for recovering rabies virus from cloned cDNA. J Virol Methods. 2003;107:229–236. doi: 10.1016/s0166-0934(02)00249-5. [DOI] [PubMed] [Google Scholar]
- Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiology and immunology. 2003;47:613–617. doi: 10.1111/j.1348-0421.2003.tb03424.x. [DOI] [PubMed] [Google Scholar]
- Kalatsky VA, Stryker MP. New paradigm for optical imaging: temporally encoded maps of intrinsic signal. Neuron. 2003;38:529–545. doi: 10.1016/s0896-6273(03)00286-1. [DOI] [PubMed] [Google Scholar]
- Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JN, Greenberg D, Helmchen F. Imaging input and output of neocortical networks in vivo. Proc Natl Acad Sci U S A. 2005;102:14063–14068. doi: 10.1073/pnas.0506029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One. 2008;3:e2005. doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DD, Wickersham IR, Callaway EM. Retrograde tracing with recombinant rabies virus reveals correlations between projection targets and dendritic architecture in layer 5 of mouse barrel cortex. Front Neural Circuits. 2007;1:5. doi: 10.3389/neuro.04.005.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mercier P, Jacob Y, Tanner K, Tordo N. A novel expression cassette of lyssavirus shows that the distantly related Mokola virus can rescue a defective rabies virus genome. J Virol. 2002;76:2024–2027. doi: 10.1128/JVI.76.4.2024-2027.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner HA, Lein ES, Callaway EM. A genetic method for selective and quickly reversible silencing of Mammalian neurons. J Neurosci. 2002;22:5287–5290. doi: 10.1523/JNEUROSCI.22-13-05287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lopez-Bendito G, Sturgess K, Erdelyi F, Szabo G, Molnar Z, Paulsen O. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb Cortex. 2004;14:1122–1133. doi: 10.1093/cercor/bhh072. [DOI] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettigan JP, Naper K, Orenstein J, Koser M, McKenna PM, Schnell MJ. Functional human immunodeficiency virus type 1 (HIV-1) Gag-Pol or HIV-1 Gag-Pol and env expressed from a single rhabdovirus-based vaccine vector genome. J Virol. 2003;77:10889–10899. doi: 10.1128/JVI.77.20.10889-10899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T, Konig M, Conzelmann KK. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A. Innovations in the imaging of brain functions using fluorescent proteins. Neuron. 2005;48:189–199. doi: 10.1016/j.neuron.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Callaway EM. Specialized circuits from primary visual cortex to V2 and area MT. Neuron. 2007;55:799–808. doi: 10.1016/j.neuron.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara S, Inoue K, Yamada M, Yamawaki T, Koganezawa N, Tsutsui K, Witter MP, Iijima T. Dual transneuronal tracing in the rat entorhinal-hippocampal circuit by intracerebral injection of recombinant rabies virus vectors. Front Neuroanat. 2009;3:1. doi: 10.3389/neuro.05.001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki K, Chung S, Ch’ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- Oliva AA, Jr., Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20:3354–3368. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Rancz EA, Franks KM, Schwarz MK, Pichler B, Schaefer AT, Margrie TW. Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nat Neurosci. 2011;14:527–532. doi: 10.1038/nn.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyan CA, Schummers J, Van Wart A, Kuhlman SJ, Wilson NR, Huang ZJ, Sur M. Response features of parvalbumin-expressing interneurons suggest precise roles for subtypes of inhibition in visual cortex. Neuron. 2010;67:847–857. doi: 10.1016/j.neuron.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MD, Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. Embo J. 1994;13:928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanziani M, Hausser M. Electrophysiology in the age of light. Nature. 2009;461:930–939. doi: 10.1038/nature08540. [DOI] [PubMed] [Google Scholar]
- Schnell MJ, Foley HD, Siler CA, McGettigan JP, Dietzschold B, Pomerantz RJ. Recombinant rabies virus as potential live-viral vaccines for HIV-1. Proc Natl Acad Sci U S A. 2000;97:3544–3549. doi: 10.1073/pnas.050589197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. Embo J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien AE, Tripodi M, Arber S. Monosynaptic rabies virus reveals premotor network organization and synaptic specificity of cholinergic partition cells. Neuron. 2010;68:456–472. doi: 10.1016/j.neuron.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Symons RH. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Tan EM, Yamaguchi Y, Horwitz GD, Gosgnach S, Lein ES, Goulding M, Albright TD, Callaway EM. Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron. 2006;51:157–170. doi: 10.1016/j.neuron.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Tan W, Janczewski WA, Yang P, Shao XM, Callaway EM, Feldman JL. Silencing preBotzinger complex somatostatin-expressing neurons induces persistent apnea in awake rat. Nat Neurosci. 2008;11:538–540. doi: 10.1038/nn.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J Comp Neurol. 1995;356:457–480. doi: 10.1002/cne.903560312. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Advances in viral transneuronal tracing. J Neurosci Methods. 2010;194:2–20. doi: 10.1016/j.jneumeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci U S A. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci U S A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weible AP, Schwarcz L, Wickersham IR, Deblander L, Wu H, Callaway EM, Seung HS, Kentros CG. Transgenic targeting of recombinant rabies virus reveals monosynaptic connectivity of specific neurons. J Neurosci. 2010;30:16509–16513. doi: 10.1523/JNEUROSCI.2442-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007a;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007b;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Rupprecht CE. Glycoprotein gene relocation in rabies virus. Virus Res. 2008;131:95–99. doi: 10.1016/j.virusres.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yonehara K, Balint K, Noda M, Nagel G, Bamberg E, Roska B. Spatially asymmetric reorganization of inhibition establishes a motion-sensitive circuit. Nature. 2011;469:407–410. doi: 10.1038/nature09711. [DOI] [PubMed] [Google Scholar]
- Young P, Qiu L, Wang D, Zhao S, Gross J, Feng G. Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nat Neurosci. 2008;11:721–728. doi: 10.1038/nn.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, Balaji J, Neve R, Poirazi P, Silva AJ. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci. 2009;12:1438–1443. doi: 10.1038/nn.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XD, Sadowski PD. Cleavage-dependent ligation by the FLP recombinase. Characterization of a mutant FLP protein with an alteration in a catalytic amino acid. J Biol Chem. 1995;270:23044–23054. doi: 10.1074/jbc.270.39.23044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.