Abstract
Aromatic polyketides are an important class of natural products that possess a wide range of biological activities. The cyclization of the polyketide chain is a critical control point in the biosynthesis of aromatic polyketides. The aromatase/cyclases (ARO/CYCs) are an important component of the Type II polyketide synthase (PKS) and help fold the polyketide for regiospecific cyclizations of the first ring and/or aromatization, promoting two commonly observed first-ring cyclization patterns for the bacterial Type II PKSs: C7–C12 and C9–C14. We had previously reported the crystal structure and enzymological analyses of the TcmN ARO/CYC, which promotes C9–C14 first-ring cyclization. However, how C7–C12 first-ring cyclization is controlled remains unresolved. In this work, we present the 2.4 Å crystal structure of ZhuI, a C7–C12-specific first-ring ARO/CYC from the Type II PKS pathway responsible for the production of the R1128 polyketides. Though ZhuI possesses a helix-grip fold shared by TcmN ARO/CYC, there are substantial differences in overall structure and pocket residue composition to implicate the preference for directing C7–C12 (rather than C9–C14) cyclization. Docking studies and site-directed mutagenesis coupled to an in vitro activity assay demonstrate that ZhuI pocket residues R66, H109, and D146 are important for enzyme function. The ZhuI crystal structure helps visualize the structure and putative dehydratase function of the di-domain ARO/CYCs from KR-containing Type II PKSs. The sequence-structure-function analysis described for ZhuI elucidates the molecular mechanisms that control C7–C12 first-ring polyketide cyclization and builds a foundation for future endeavors into directing cyclization patterns for engineered biosynthesis of aromatic polyketides.
The biosynthesis of aromatic polyketides by Streptomyces is accomplished by the Type II polyketide synthase (PKS), which consists of the ketosynthase-chain length factor (KS-CLF, also known as KSa-KSb) heterodimer for chain elongation, a dedicated acyl-carrier protein (ACP), and chain modifying enzymes such as ketoreductase (KR) and aromatase/cyclase (ARO/CYC). Previous studies have established the biosynthetic routes for aromatic polyketides in ‘reducing’ (KR present) or ‘non-reducing’ (KR absent) PKS systems (1–3). The cyclization of the poly-β-keto intermediate is a critical control point in the natural biosynthetic pathway and is a potential diversification point for engineered biosynthesis (Figure S1). Additionally, the multi-cyclic aromatic nature of the polyketides produced by Type II PKS is essential for their anticancer and antibiotic activities (4–7). Due to the inherent reactivity of poly-β-ketone compounds (8), the type II PKS needs to specifically restrain the polyketide intermediate to control cyclization and avoid aberrant cyclization events that would lead to unwanted end products. Interestingly, of all the possible pathways for first-ring cyclization, there are only two commonly observed patterns for type II PKS, C9–C14 and C7–C12 (9). Though the precise timing of cyclization has not been established, previous genetic and biochemical studies demonstrate that the ARO/CYCs can control the initial cyclization events in non-reducing PKS systems (10–16). First-ring cyclization between C9–C14 is often associated with the mono-domain ARO/CYCs from non-reducing systems such as TcmN (11, 12, 17), WhiE-ORFVI (18), and RemI (19, 20). C7–C12 first-ring cyclization is commonly associated with the di-domain ARO/CYCs such as ActVII (21) and Gris-ORF4 (22, 23) from reducing PKS systems, where the ARO/CYC may act as a dehydratase to aromatize a pre-cyclized first ring. However, C7–C12 cyclization may also be accomplished in non-reducing systems by mono-domain ARO/CYC such as ZhuI (24, 25) or di-domain ARO/CYCs such as MtmQ (26, 27). Our previous work on TcmN ARO/CYC (17) showed that the interior pocket of the ARO/CYC directs the C9–C14 cyclization specificity. However, little is known about how C7–C12 ARO/CYCs control their respective cyclization and aromatization events.
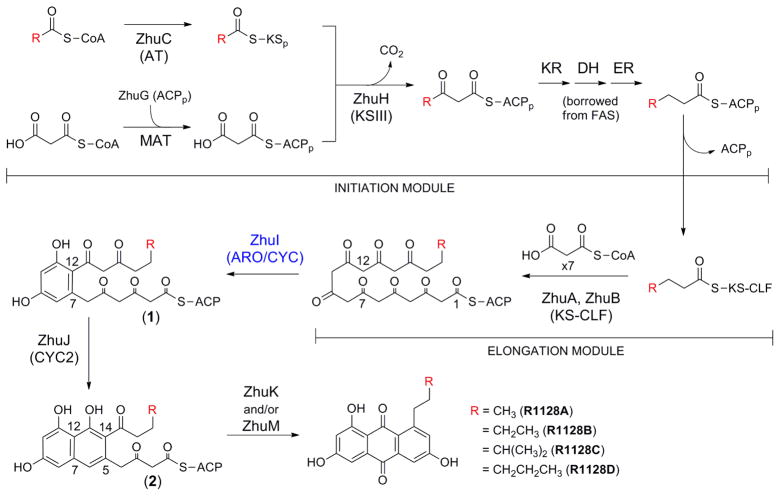
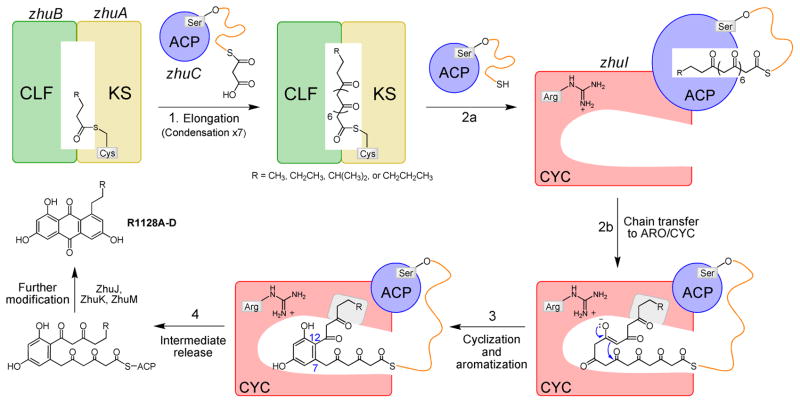
The zhu gene cluster of Streptomyces sp. R1128 is responsible for the biosynthesis of the R1128 family of polyketides (24). The R1128 polyketides are potent non-steroidal estrogen receptor antagonists that exhibit favorable selectivity and low toxicity compared to tamoxifen, the most widely used drug for anti-hormone treatment of estrogen receptor positive breast cancer (28). The R1128 gene cluster encodes proteins comprising a unique chain initiation module, an elongation module, and post-elongation tailoring enzymes (Figure 1). Three proteins, ZhuH (ketosynthase III homolog), ZhuG (ACP), and ZhuC (acyl transferase homolog), together with fatty acid KR, DH and ER (predicted to be recruited from the endogenous FAS), define the initiation module dedicated to the generation of four acyl-ACP units (butyryl-, valeryl-, 4-methylvaleryl-, and hexanoyl-ACP) used to ‘prime’ the downstream Type II octaketide synthase (Figure 1)(25, 29, 30). The Type II MinPKS elongation module is formed by ZhuA (CLF) and ZhuB (KS), which interact to form a heterodimer, and ZhuN (ACP). After the KS-CLF is primed by one of the four acyl groups provided by the initiation module, the MinPKS catalyzes seven rounds of decarboxylative condensation with malonyl units to produce a polyketide chain with a total number of carbons between 18 and 20. Following chain elongation, the polyketide intermediate then undergoes stepwise cyclization: ZhuI ARO/CYC is proposed to catalyze aldol condensation between C7 and C12 (followed by aromatization) to yield intermediate 1, which is then transferred to ZhuJ for second ring cyclization between C5 and C14 (followed by aromatization) to give intermediate 2. After third ring cyclization between C2 and C15, presumed to occur spontaneously, oxidation by ZhuK and/or ZhuM affords the final substituted-anthraquinone products R1128A-D (24).
Figure 1.
Proposed biosynthetic pathway of the zhu PKS from Streptomcyes sp. R1128.
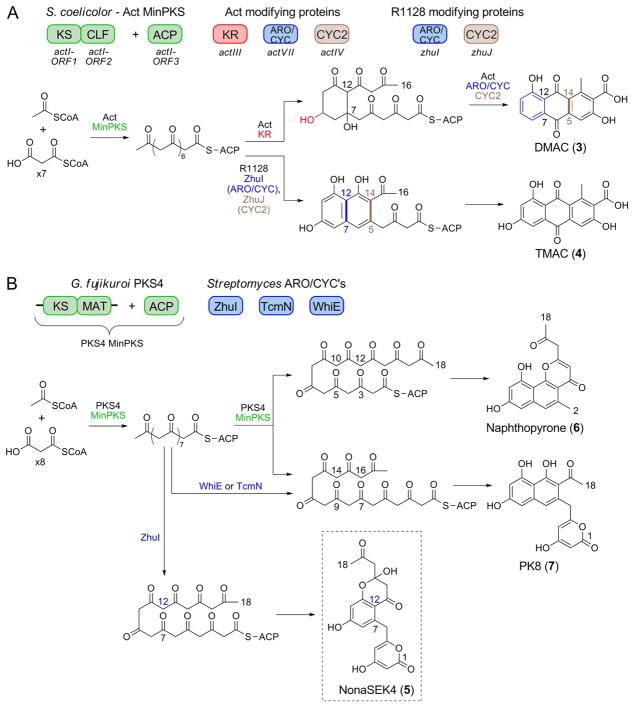
In vivo and in vitro PKS reconstitution studies validated that ZhuI and ZhuJ are responsible for first ring (between C7–C12, ZhuI) and second ring (between C5–C14, ZhuJ) cyclizations, respectively. For example, over-expression of the Act MinPKS, ZhuI, and ZhuJ in S. coelicolor resulted in controlled first- and second-ring cyclization of an unreduced octaketide chain for the production of TMAC (3) (Figure 2A), an analog of DMAC (4) (25). Further, incubation of purified ZhuI with the KS-MAT and ACP domains from the fungal PKS4 resulted in the biosynthesis of a novel 18-carbon, C7–C12 first-ring cyclized polyketide NonaSEK4 (5, Figure 2B). Compared to SEK4 (31), 5 has an acetyl group in place of the terminal methyl, reflecting the additional cycle of chain elongation to produce the C18 (vs. C16) polyketide chain (27). Importantly, the PKS4 MinPKS alone does not synthesize NonaSEK4 but instead produces two major nonaketide products with C10–C15 (naphthopyrone, 6) or C9–C14 (PK8, 7) first-ring cyclization patterns (27), while the inclusion of the C9–C14 cyclases Tcm or WhiE ARO/CYC promotes the exclusive production of PK8 (7) (Figure 2B). This in vitro study demonstrates the precise control of cyclization regiospecificity by different ARO/CYCs, and provides a convenient assay to monitor the C7–C12 cyclization activity of ZhuI ARO/CYC, which is otherwise masked by the intrinsic C7–C12 activities of other minimal PKSs.
Figure 2.
The influence of ZhuI on first-ring cyclization specificity in engineered biosynthesis. (A) Combination of the Act MinPKS, KR, ARO/CYC, and CYC proteins from S. coelicolor results in production of the octaketide DMAC. In comparison, when ZhuI and ZhuJ are combined with the Act MinPKS they direct C7–C12 and C5–C14 cyclization of an unreduced octaketide chain to yield TMAC (4). (B) In vitro reconstitution of polyketide biosynthesis using the PKS4 MinPKS from Gibberella fujikuroi results in the production of napthopyrone 6 and PK8 (7). The addition of the C9–C14 cyclases TcmN or WhiE results in the exclusive production of 7. In contrast, the addition of ZhuI directs the regiospecific C7–C12 cyclization of the nonaketide chain to yield a novel polyketide, NonaSEK4 (5). Note that TcmN and WhiE direct two consecutive cyclizations (C9–C14 and C7–C16), whereas ZhuI promotes only a single cyclization event between C7–C12.
ZhuI is a highly unique ARO/CYC in the following aspects: 1) A C7–C12 cyclase. ZhuI is a mono-domain ARO/CYC that promotes the regiospecific C7–C12 first-ring cyclization for unreduced polyketide chains of variable length (C16, C18, C19, and C20). C7–C12 specificity for first-ring cyclization is unique among non-reducing Type II PKS systems, given that other mono-domain ARO/CYCs associated with non-reducing systems catalyze C9–C14 first-ring closure. C7–C12 first-ring cyclization is normally associated with the reducing Type II systems, where the di-domain ARO/CYCs are predicted to act on a precyclized intermediate to catalyze aromatization (32). 2) A single-ring cyclase. ZhuI catalyzes only one aldol condensation event to yield a monocyclic intermediate that is then transferred to the downstream enzyme ZhuJ for second-ring cyclization. This is similar to the reducing Type II systems where, following first ring cyclization and aromatization, a distinct and dedicated enzyme is required for second-ring cyclization (such as ActIV that works downstream of the di-domain aromatase ActVII, resulting in DMAC production (33)). In contrast, TcmN ARO/CYC catalyzes at least two consecutive aldol condensations for intermediates of varying chain lengths en route to Tcm F2 (10), RM80 (34), PK8 (35), and pBR3-#4 (17). Similar ‘multi-ring cyclase’ functionality is also observed for WhiE ARO/CYC (18) and RemI (19). Therefore, though ZhuI possesses mono-domain ARO/CYC architecture, its functionality lies somewhere between that observed for the majority of mono-domain and di-domain ARO/CYCs. How ZhuI specifically promotes the C7–C12 first ring cyclization is not well understood.
In this work, we present the 2.4 Å crystal structure of ZhuI, functional analysis of ZhuI pocket residue mutants, and polyketide docking simulations. These results provide the first structure of a C7–C12 ARO/CYC from a non-reducing Type II PKS and demonstrate the ability of the interior pocket of ZhuI to accommodate a polyketide intermediate to direct C7–C12 cyclization. In addition, in vitro activity assay results further substantiate the importance of interior pocket residues in ARO/CYC functionality, and provide a means to understand the structure and activity of the functionally distinct di-domain ARO/CYC proteins from reducing Type II PKS systems.
MATERIALS AND METHODS
Protein expression and purification
Plasmid pYT224 containing the zhuI gene from Streptomyces sp. R1128 was cloned into the NdeI and HindIII sites of pET28, allowing recombinant protein expression with an N-terminal 6x-His tag. WT and mutant ZhuI proteins were produced in Escherichia coli strain BL21(DE3) and purified in a similar manner: 2 × 1 L LB cultures containing kanamycin (50 μg/mL) were grown at 37°C until A600 reached ~0.6, and protein expression was induced by addition of 1 mM IPTG followed by overnight growth (~18 hrs) at 18°C. Following incubation, cultures were harvested and the cell pellets frozen and stored at −80°C. Cell pellets were thawed at room temperature and suspended in a total volume of 40 mL lysis buffer (50 mM Tris·HCl, pH 7.5, 300 mM NaCl, 10 mM imidazole, 10% glycerol). Cells were then lysed by repeated sonication and the lysate clarified by centriguation (21,000 × g for 60 min). The resulting supernatant was then added to Ni2+ metal-chelate affinity resin (Bio-Rad) and incubated at 4°C for one hour for batch binding of His-tagged protein. The protein-Ni2+ slurry was then applied to a gravity-flow column, washed with lysis buffer (25 mL), then lysis buffer containing 20 and 30 mM imidazole (25 mL’s each). Six 5 mL volumes containing 50 to 500 mM imidazole were then applied to the column to elute His-tagged protein. The elutions containing ZhuI protein were concentrated using a 9,000 kDa MWCO centrifugal device (iCON, Pierce) until the final volume reached 2.5 mL. A PD-10 column (GE Healthcare) was used to buffer exchange the concentrated protein into Q-Buffer A (20 mM Tris·HCl, pH 7.0, 1mM EDTA). The buffer exchanged ZhuI protein was used directly for in vitro enzyme activity assays.
For ZhuI protein crystallization, the His-tag was removed by adding bovine thrombin (Sigma) at 10 units thrombin per mg ZhuI protein and incubated overnight (~18 hrs) at 15°C. Following thrombin cleavage, the protein was applied to a 5 mL anion-exchange column (Hitrap SP FF, GE Healthcare), and eluted using a linear gradient from 0–1M NaCl in Q-Buffer A over 20 column volumes. The eluent was concentrated to 1 mL and further purified and buffer exchanged into crystallization buffer (20 mM HEPES, pH 7.0) by gel filtration chromatography (Superdex 200 10/300 GL column, GE Healthcare). Protein purity was assessed using SDS-PAGE with Coomassie staining, and protein concentration was determined using the Bradford method, with BSA as a standard.
Selenomethionine-substituted (SeMet) ZhuI protein was produced in E. coli strain BL21 (DE3) in M9 minimal media using metabolic inhibition of the methionine pathway (36). A 50 μl glycerol stock of protein was used to inoculate a 5 mL culture of LB for overnight growth at 37°C in the presence of 50 μg/mL kanamycin. The resulting cells were pelleted and washed twice by suspending in 10 mL M9 media, then used to inoculate 2 × 1L M9 media with 50 μg/mL kanamycin. Cultures were incubated at 37°C until A600 reached ~0.6, at which point the temperature was reduced to 25°C and the following amino acids were added as solids to each 1L culture: lysine, phenylalanine, threonine (100 mg each); isoleucine, leucine, valine (50 mg each); L-selenomethionine (30 mg) (Sigma). After a 30-minute incubation period, 0.1 mM IPTG was added to induce protein expression. Cultures were allowed to grow at 25°C overnight (~16 hrs) and harvested by centrifugation. The purification protocol for the SeMet protein followed the WT procedure but with the addition of 5 mM DTT to all buffers. The incorporation of selenomethionine (total of three residues) was confirmed by MALDI-TOF mass spectrometry; native-6x-His (observed: 20,528 m/z), SeMet-6x-His (observed: 20,662 m/z); expected Δmass 141 Da, observed Δmass 134 Da.
Expression and purification of the PKS4 KS-MAT di-domain construct and the standalone PKS4 ACP (together constituting the PKS4 ‘MinPKS’ for use in the in vitro reconstitution assay) was performed as previously described (27).
Site-Directed Mutagenesis
The QuikChange II Site-Directed Mutagenesis Kit (Stratagene) was used to introduce ZhuI pocket-residue mutations, which were confirmed through automated DNA sequencing. The primers used for mutagenesis are as follow (mutated nucleotides are underlined):
T34A, 5′ GACATCTTCCCGCCGGCGGAGAAGGTCGAGATC 3′
S64A, 5′ CAACACCTGGACCGCGCGCCGCGACC 3′
R66A, 5′ CTGGACCTCGCGCGCGGACCTGGACCCG 3′
R66Q, 5′ CTGGACCTCGCGCCAGGACCTGGACCCGG 3′
R66K, 5′ CACCTGGACCTCGCGCAAAGACCTGGACCCGGCCC 3′
R66E, 5′ CACCTGGACCTCGCGCGAAGACCTGGACCCGGCCC 3′
H109A, 5′ CAACTGGTGCTCACCGCGGACTTCGTGACGCG 3′
D146A, 5′ CAACAGCGTCGCCGCGCTGAACGCGGTGCTC 3′
D146 N, 5′ GAGCGCAACAGCGTCGCCAATCTGAACGCGGTGCTCGGC 3′
Crystallization and data collection
Native and SeMet ZhuI protein (4 μL at 20 mg/mL) was combined with with 1 μL each of 1M guanidine HCl and 1M sodium citrate (as additives), then mixed with 4 μL of well solution (1.8M NaH2PO4/K2PO4, 0.1M CAPS, and 0.2M Li2SO4) and the mixture allowed to equilibrate over a well volume of 500 μL using sitting drop vapor diffusion. Crystal formation was optimal after ~1 week at room temperature. Crystals were cryoprotected in well solution containing 1.3 M Li2SO4 prior to flash freezing in liquid nitrogen. Monochromatic x-ray diffraction data (λ = 0.97607 Å, 300 frames at 0.6° oscillation, 5 sec exposure) for native ZhuI was collected to 2.4 Å resolution on beamline 7–1 of the Stanford Synchrotron Radiation Laboratory (SSRL). Multiwavelength anomalous dispersion (MAD) data were collected to 3.3 Å for SeMet ZhuI at λ = 0.97903 Å (selenium peak), λ = 0.97923 Å (inflection), and λ = 0.96104 Å (remote) on beamline 8.2.2 of the Advanced Light Source (ALS). For MAD data collection, the exposure time was set to 1 sec, and 360 frames were collected using inverse beam geometry at 1° oscillation width for both peak and inflection wavelengths, while 180 frames at 1° oscillation were collected for the remote wavelength. All data were processed using HKL2000 (37), MAD data were scaled as both ‘merged’ or ‘unmerged’ (regarding treatment of the Friedel pairs) as required for utilization of downstream phasing and refinement programs. Data collection and processing statistics for merged native and SeMet data are provided in Table 1.
Table 1.
Data collection and refinement statistics
| ZhuI ARO/CYC
|
||||
|---|---|---|---|---|
| Native* | SeMet derivative* |
|||
| Peak | Inflection | Remote | ||
| Crystallographic Data: | ||||
| Wavelength (Å) | 0.97607 | 0.97903 | 0.97923 | 0.96104 |
| Resolution (Å) | 50–2.4 | 50–3.3 | 50–3.3 | 50–3.4 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit-cell parameters | ||||
| a (Å) | 50.016 | 49.523 | 49.523 | 49.482 |
| b (Å) | 135.731 | 136.598 | 136.587 | 136.497 |
| c (Å) | 175.496 | 175.921 | 176.001 | 175.950 |
| α = β = γ (°) | 90 | 90 | 90 | 90 |
| Total observations | 334640 | 267978 | 268231 | 123683 |
| Unique reflections | 48168 | 19016 | 19044 | 17599 |
| High res. shell (Å) | 2.49–2.40 | 3.42–3.30 | 3.42–3.30 | 3.52–3.40 |
| Average redundancy† | 7.0 (6.3) | 14.1 (14.4) | 14.1 (14.3) | 7.0 (7.2) |
| Completeness (%) | 99.8 (99.0) | 100 (100) | 100 (100) | 100 (100) |
| Mean I/σ(I) | 16.4 (3.7) | 18.1 (7.0) | 17.6 (6.6) | 11.6 (4.2) |
| Rsym (%) | 12.8 (46.6) | 18.2 (40.6) | 18.1 (43.1) | 19.2 (48.3) |
| Refinement: | ||||
| Resolution (Å) | 50–2.4 | |||
| No. of reflections | 45669 | |||
| No. of protein atoms | 7679 | |||
| No. of water atoms | 167 | |||
| No. of heteroatoms | 101 | |||
| No. of TLS groups | 24 | |||
| Rcrys (%) | 19.3 | |||
| Rfree (%) | 26.1 | |||
| Geometry: | ||||
| rmsd bonds (Å) | 0.028 | |||
| rmsd angles (°) | 2.5 | |||
| Ave. B factors (Å2) | 21.0 | |||
| Ramachandran (%) | ||||
| Most favored | 89.1 | |||
| Additional allowed | 10.9 | |||
| Generously allowed | 0 | |||
Single crystals were used to collect the corresponding monochromatic and multiwavelength data.
Values in parentheses represent the highest resolution shell
Phasing, model building, and refinement
The results from Matthews coefficient analysis (38) suggested that six ZhuI molecules constituted the crystallographic asymmetric unit (Vm of 2.6 Å3/Da, solvent content of 53.4%). Therefore, the program SHELX_C/D (39) was used to search for a total of 18 selenium sites to be used for subsequent MAD phasing. The top 13 sites located by SHELX_C/D were converted to fractional coordinates and input into SOLVE (40) to search for additional sites (one additional site found) and for calculation of experimental phases to 3.8 Å (mean figure of merit = 0.58, overall Z-score = 47.8). Density modification and automatic model building were performed with RESOLVE (40), providing an initial electron density map and fragmented poly-alanine models for all six molecules in the asymmetric unit. Though significant gaps existed in the model output from RESOLVE, the backbone was complete enough to recognize the helix-grip fold of the six ZhuI monomers. The electron density map from RESOLVE was used for manual model building performed in COOT (41); wherein the complete backbone for a single ZhuI monomer unit was built (as a poly-alanine model) into the clearest section of electron density, and this basic unit was copied and manually placed into the electron density representing the other five monomers. Rigid body refinement was applied in REFMAC (42), and the resulting model was used for density modification and phase extension to 3.3 Å (using SeMet peak data) by CNS (43) and DM (CCP4) (44, 45). The appropriate side chain groups were then added to a single monomer unit and this model was placed by Phaser (CCP4) (46) to complete the asymmetric unit. The full six-molecule ensemble was then transferred to the native data and refined to 2.4 Å with REFMAC (42). Iterative rounds of manual model building in COOT and restrained Maximum-Likelihood refinement in REFMAC were performed until the Rwork reached 24.4% (Rfree = 29.7%). At this stage waters were added automatically in COOT and manually edited (inserted/deleted) where appropriate, followed by addition of seven CAPS molecules (one in each ZhuI pocket and an additional located at a protein-protein interface region) and four potassium ions. The final model of ZhuI was refined to an Rwork of 19.3% (Rfree = 26.1%) and includes residues 1–167 (chain A), 1–164 (chain B), 1–163 (chain D), 1–163 (chain D), 1–162 (chain E), 1–160 (chain F), and up to three amino acids (Gly-Ser-His) present at the N-terminus leftover as cloning artifacts; 167 water molecules; and heteroatoms consisting of 7 CAPS molecules and 4 K+ ions. Refinement and model statistics are listed in Table 1.
Small-molecule docking
The program GOLD (47, 48) was used for docking between ZhuI ARO/CYC and the polyketide portion of the putative monocyclic intermediate 1 (Figure 1). The protein input file was prepared by removing waters and adding hydrogens then saved as a Mol2 file. Ligands were generated and energy-minimized using ChemOffice (CambridgeSoft Corp.), and saved as Mol2 files. The ZhuI ligand-binding pocket was defined as all residues within 15 Å of atom 491 (indole nitrogen of Trp62), and docking was performed using the default settings for the GA parameters with 20 docking trials performed for each ligand.
In vitro activity assay
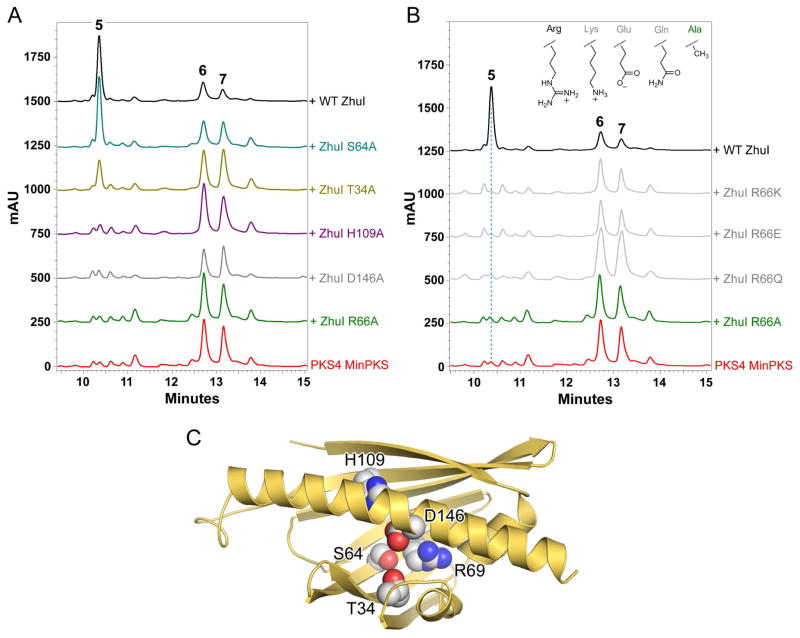
To measure the enzyme activity of WT and mutant ZhuI ARO/CYC we performed a PKS4-based in vitro reconstitution assay similar to that described previously (27). Briefly, 250 μL reactions were prepared containing 10 μM PKS4 KS-MAT, 50 μM PKS4 ACP, 5 mM malonyl-CoA (Sigma), and (when included) 50 μM WT or mutant ZhuI in 0.05 M Tris-HCl, pH 7.0. Reactions were incubated in the dark at room temperature overnight (16 hours) and then extracted twice by the addition of 95% EtOAc/4.9% MeOH/0.1% formic acid. The organic phase was separated, evaporated under vacuum, and the resulting residue was dissolved in 100 μL of 100% DMSO (Sigma). Product analysis was performed using reverse-phase HPLC, where 20 μL injections were separated at a flow rate of 1.0 mL/min on a Beckman Ultrasphere C18 column (5 μm, 80 Å, 4.6 × 150mm) by running a gradient from 5–50% MeCN in H20/0.1% formic acid over 15 minutes followed by 50–95% MeCN in H20/0.1% formic acid over 5 minutes. Under these conditions, the HPLC retention times were observed to be: NonaSEK4 (5), 10.4 min; PK8 (7), 12.7 min; and naphthopyrone 6, 13.2 min. Product elution was monitored by diode-array detection (from 190–600 nm) and trace data presented in Figure 7 was generated by selecting for absorbance at 270 nm. Product identity was confirmed by UV/Vis, ESI-MS, and by matching retentions times with authentic standards.
Figure 7.
Results of ZhuI pocket residue mutagenesis as tested in vitro. (A) HPLC analysis (traces displayed at 270 nm) of the product profiles obtained by in vitro reconstitution of polyketide biosynthesis utilizing the PKS4 MinPKS plus WT or mutant ZhuI ARO/CYC. (B) The product profiles of additional pocket arginine mutants (R66Q, R66E, and R66K). Inset, amino acid side chain groups at residue position 66 for WT and mutant ZhuI as assayed. (C) Structure of ZhuI showing the pocket residues mutated as spheres.
RESULTS AND DISCUSSION
Sequence analysis of ZhuI ARO/CYC showed high homology to di-domain ARO/CYCs
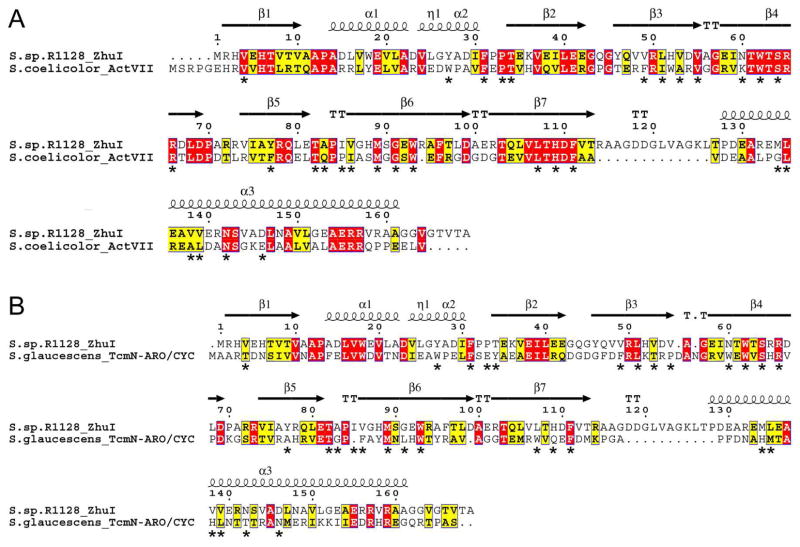
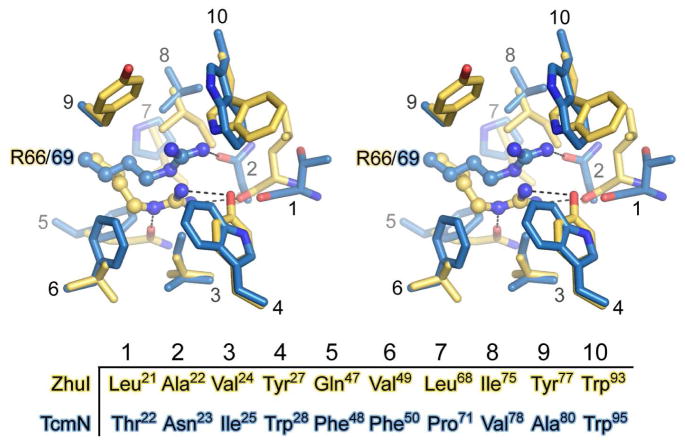
Amino acid sequence comparison shows that ZhuI bears the highest similarity to the N-terminal domain of the di-domain ARO/CYCs, its closest functionally characterized homolog is ActVII from S. coelicolor (Figure 3A) with an overall identity/similarity of 31%/45% (compared to a 17%/36% identity/similarity between ZhuI and the ARO/CYC domain of TcmN, Figure 3B). Alignment of pocket-defining residues shows 57% identity between ZhuI and ActVII, and 27% identity between ZhuI and TcmN, with seven pocket residues found in common between the aligned sequences of all three proteins (numbered according to ZhuI: F31, W62, S64, R66, P84, M89, and W94). Sequence alignment illustrates that there is a significant switch in pocket residue composition between the C9–C14 mono-domain versus the C7–C12 di-domain ARO/CYCs, with ZhuI bearing the highest similarity to the di-domain ARO/CYCs (Figure S2) (17). This pocket residue ‘switch’ may be relevant in understanding ZhuI cyclization specificity. Based on the high sequence similarity of ZhuI and di-domain ARO/CYCs, the interior pocket of ZhuI may promote the C7–C12 folding pattern for both first-ring cyclization and aromatization of the ACP-bound polyketide chain.
Figure 3.
Sequence alignment of ZhuI ARO/CYC with (A) the N-half ARO/CYC domain (residues 1–156) of the di-domain protein ActVII from S. coelicolor and (B) the ARO/CYC domain (residues 1–157) of TcmN from S. glaucescens. ActVII is associated with dehydration and aromatization of a C7–C12 cyclized first ring, whereas TcmN ARO/CYC is associated with C9–C14 first-ring cyclization. Secondary structure elements shown on the top of each alignment are according to the ZhuI structure and asterisks below the alignments denote pocket-defining residues.
Overall structure of ZhuI: dimeric, and much smaller pocket than that of TcmN ARO/CYC
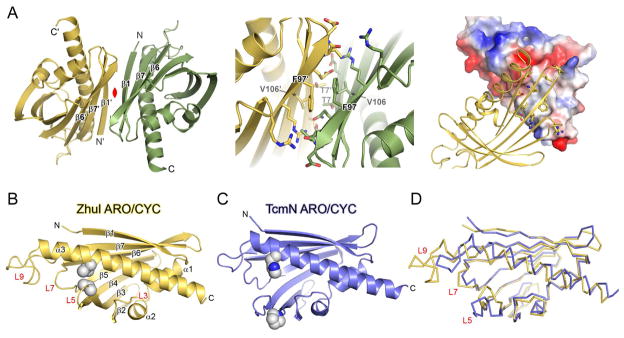
The 2.4 Å-resolution ZhuI crystal structure was solved by SeMet MAD, revealing that ZhuI is dimeric with each monomer consisting of one helix-grip fold (49) (Figure 4). Molecular replacement using TcmN ARO/CYC (or other helix-grip fold structures) as search model templates was unsuccessful, which is indicative of structural differences between ZhuI and the search models. TcmN ARO/CYC was solved as a monomer, with one molecule per asymmetric unit. In comparison, the ZhuI crystal structure has six molecules per asymmetric unit. Each of the six ZhuI molecules overlay very well with one another, with an average RMSD of 0.70 Å for backbone atoms and RMSD of 0.33 Å for core residues. Structure solution reveals that three essentially equivalent dimer pairs constitute the six molecules in the asymmetric unit, an unanticipated finding with the knowledge that TcmN and WhiE ARO/CYC both crystallized as monomers. Size exclusion chromatography confirms that ZhuI exists as a dimer in solution (Figure S3). The ZhuI dimer does not resemble the iodide-induced dimer of TcmN ARO/CYC (17), the double-hotdog dimer of FabZ (50), or the dimerization mode adopted by some helix-grip fold proteins (Figure S4). The ZhuI dimer interface possesses a modest protein contact surface area of ~750 Å2 consisting of a hydrophobic central patch (T7, V74, F96, L98, and the methylene groups of the Q104 side chain) surrounded by polar and charged residues that form hydrogen bonding and ionic interactions (E5, T9, R72, R94, Q104, and the backbone carbonyl of L98) (Figure 4A). A non-crystallographic two-fold rotation axis relates each molecule of the ZhuI dimer. As a result, the residues involved in forming the dimer interface are symmetrical for each dimer pair (Figure 4A). The dimer interface residues are not conserved among ARO/CYCs, suggesting that the dimerization mode may be a unique property of ZhuI only. Therefore, for putative interaction with KS-CLF and/or ACP during aromatic polyketide biosynthesis it is possible that ZhuI utilizes interaction surfaces distinct from other mono-domain ARO/CYCs such as TcmN or WhiE ARO/CYC.
Figure 4.
Overall structure of ZhuI and a comparison with TcmN ARO/CYC. (A) Left, the ZhuI dimer structure. β-strands at the dimer interface are labeled, and the non-crystallographic 2-fold rotation axis relating the two monomers is in red. Middle, close-up view of the dimer interface. Right, illustration of complementarity and chemical nature of the dimer contact surface; one monomer is represented as electrostatic surface (red = electronegative, blue = electropositive, and grey = neutral) and the other is drawn as cartoon with interacting residues in sticks. (B) Monomer structure of ZhuI with secondary structure elements labeled and two interacting residues (V55 and V138) shown as spheres. (C) The crystal structure of TcmN ARO/CYC with residues P56 and H128 (structurally equivalent to those listed for (B)) drawn as spheres, see the sequence alignment in Figure 4–3B. (C) Backbone overlay of ZhuI and TcmN ARO/CYC
Superimposition of the ZhuI and TcmN ARO/CYC structures (overall RMSD of 2.7 Å and an RMSD of 1.3 Å for core residues) reveals several significant differences (Figure 4B–D): 1) An extended loop (L9, Figure 4B) in ZhuI arises from an insertion of 11 residues (117 – 128) into the region connecting β7 and αC. Sequence alignment between ZhuI and other ARO/CYCs shows that this loop insertion is unique to ZhuI (Figure S2). 2) Helix αC of ZhuI is extended by one complete turn at the N-terminal end, and is moved down toward loops 3 and 5 relative to the equivalent helix in TcmN ARO/CYC. 3) The space defining the entrance to the interior pocket of ZhuI is dramatically different (and constricted) compared to TcmN ARO/CYC. The major structural difference that gives rise to this change is the movement of loop 5 towards the central helix due to hydrophobic interaction between V55 & A56 (loop 5, Figure 4B) and M134 & V138 (from αC, Figure 4C). Interestingly, neither residue is conserved among other ARO/CYCs, indicating that the interaction between loop 5 and αC may be specific to ZhuI. An additional structural feature leading to the narrowing of the pocket-entrance includes the conformational change of loop 7. Consequently, the ZhuI pocket entrance is defined by the open space between loops 5, 7, 9, and the first two turns of αC. The approximate dimensions of the pocket entrance dimensions are 7 Å wide by 6 Å tall, while these dimensions could accommodate a polyketide intermediate (e.g. a first-ring cyclized/aromatized product which is ≈ 6 Å by 2 Å), it is likely that movement in loops 5, 7, and 9 is an integral part of substrate binding and product release. The structural differences between ZhuI and TcmN ARO/CYC may contribute to the observed C7–C12 onering-only cyclization specificity of ZhuI, compared to the C9–C14 specificity of TcmN ARO/CYC. As elaborated below, the composition and geometry of the interior pocket residues, in addition to the pocket entrance size discussed above, exert physical restraints that potentially dictate cyclization specificity.
ZhuI interior pocket residues and identification of three polar residues for catalysis
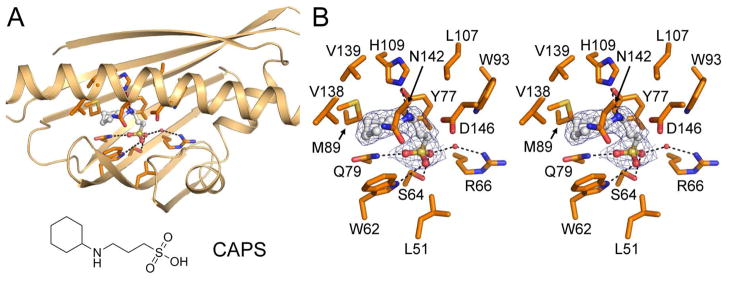
The crystal structure of ZhuI reveals that a component of the crystallization solution, N-cyclohexyl-3-aminopropanesulfonic acid (CAPS), is bound to the interior pocket of all six ZhuI molecules in the asymmetric unit (Figure 5). Although the ligand-binding mode of CAPS is somewhat variable, the sulfonic group is consistently positioned within hydrogen bond distance to W62, S64, Q79, and a water molecule that forms a hydrogen bond with R66. The 3-aminopropane moiety of CAPS interacts with Y77 and L107 through van der Waals contact, and may form a hydrogen bond with H109. Additionally, the cyclohexyl moiety of CAPS is surrounded by the hydrophobic residues W62, M89, V138, C139, and the methylene group of N142. Although CAPS is chemically different from a polyketide, the binding of CAPS to the interior pocket of ZhuI demonstrates the ability of the ZhuI interior pocket to bind small molecule ligands and provides an experimental glimpse into the distribution of hydrophobic and polar contact between ZhuI and a bound ligand.
Figure 5.
Structure of ZhuI with CAPS bound. (A) Overall view with protein backbone rendered as cartoon and residues within 4 Å of the CAPS molecule drawn as sticks. Potential hydrogen bonds are indicated by dashed lines. The structure of CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) is drawn below ZhuI. (B) Stereoview of interacting residues. Blue mesh around the bound ligand is derived from the 2.4 Å |2Fo – Fc| electron density map contoured to 0.8 σ.
Similar to TcmN ARO/CYC, the interior pocket of ZhuI is amphipathic, defined by residues exhibiting hydrophobic (F31, P33, V49, L51, V53, V55, W62, I75, A83, I85, V86, M89, W93, L105, L107, F111, M134, L135, V138, V139), polar (Y27, T34, N60, S64, Y77, Q79, T82, G91, N142), and potentially charged (R66, H109, D146) character. Nearly two thirds of the pocket residues are hydrophobic and very few pocket residues contain hydrophilic side chains that may be charged at physiological pH. The majority of ZhuI pocket residues may be candidates for substrate/product binding and only a few could serve as the active site acid/base for aldol condensation. The polar or charged residues could be involved in enolate stabilization through partial charge or ionic interactions. Of the pocket residues listed, F31, W62, S64, R66, M89, W93, N142, and the hydrophobic side chain groups of L135 and V139 are highly conserved among both mono- and di-domain ARO/CYC proteins; while additional residues P33, T34, Q79, V86, L107, H109, and the side chain character of L51, Y77, and D146 are conserved between ZhuI and the di-domain ARO/CYCs (Figure 3, Figure S2).
If pocket residues are important for ARO/CYC function, the position and conservation of pocket residues may provide insight into the mechanism of first-ring cyclization by ARO/CYCs. A major consideration for first-ring cyclization is whether the interior pocket of ARO/CYC acts as a mold by bringing the appropriate substituents into proximity for spontaneous chemistry, or if the pocket residues are directly involved in activating the substrate for aldol condensation. If the second scenario is operant, the position and conservation of the ZhuI pocket residues R66, H109, and D146 make these residues candidates for enolate activation (R66, H109) or acid-base chemistry (H109, D146) for first-ring cyclization and aromatization. The crystal structure of ZhuI shows that the side chains of H109 and D146 are separated by ~5.5 Å and are ~11 Å and 9 Å, respectively, from the guanidinium of R66 (Figure 5B). Although the cyclization and aromatization of an unreduced polyketide has been found to occur favorably in aqueous solution near neutral pH (51), the equivalent pocket Arg of TcmN ARO/CYC has been shown to be critical for enzyme function (17). Both ZhuI H109 and D146 are found mainly as Gln among other mono-domain ARO/CYCs from non-reducing Type II PKSs, but are conserved as His and Glu respectively among di-domain ARO/CYCs (Figure S2). Of relevance is that structural and functional analysis of the product template (PT) domain of the norsolorinic acid PksA has demonstrated that pocket His and Asp residues are essential for the cyclizations and aromatization of an unreduced polyketide chain (52). Further, His-Asp/Glu constitutes the conserved catalytic dyad of the fatty acid and polyketide DHs for the acid/base chemistry of dehydration (53–55). Despite low sequence homology, the PksA PT domain, and either the Type I PKS DH (DEBS module 4) (54) or Type I mFAS DH (55), share a similar fold and active site residue geometry. Although the sequence, fold, and active site geometry of ARO/CYC is distinct from PT or DH, the precedent for acid/base catalysis by a His-Asp/Glu pair may be extended for analysis of ZhuI and di-domain ARO/CYC function. ZhuI may require the His-Asp pair for cyclization, dehydration, and aromatization of the unreduced polyketide chain. In comparison, the di-domain ARO/CYCs may utilize the conserved pocket His-Glu pair to catalyze dehydration(s) of a C7–C12 pre-cyclized (and C-9 carbonyl-reduced) polyketide intermediate en route to first ring aromatization. In summary, the ZhuI crystal structure leads to the identification of active site residues (H109, D146, R66) in the interior pocket, and the role of these residues (or functionally conservative switches, such as D146 → E) in catalysis may be extended to the di-domain ARO/CYC as well.
Docking simulation of ZhuI and polyketide substrates elucidates its cyclization specificity
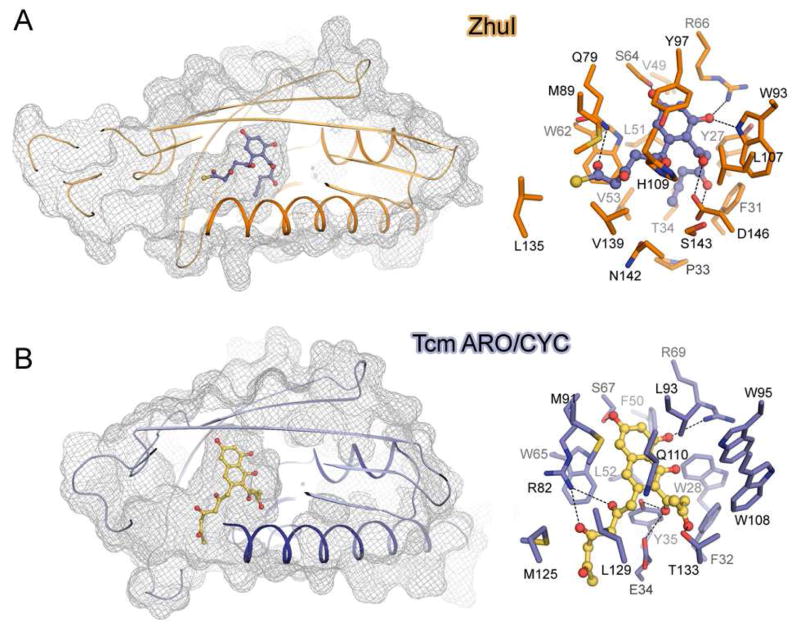
To understand how ZhuI promotes the C7–C12 first-ring-only cyclization we conducted computer simulated docking studies. The putative 18-carbon (butyryl-ACP primed), C7–C12 first-ring cyclized intermediate 1 (with R=CH3, Figure 1) is consistently docked in ZhuI, and shows that when the C9 and C11 hydroxyl functional groups are anchored near the highly conserved S64 and R66, the butyryl terminus favorably docks in a hydrophobic region of the pocket defined by Y27, F31, P33, T34, V53, and L51 (Figure 6A, Figure S5A). Additional docking results indicate that there is sufficient space in this region to accommodate up to 20 carbons, the longest chain intermediate in the R1128 pathway (hexanoyl-ACP primed), without causing steric clash. Therefore, this region may help sequester the alkyl end of the polyketide intermediate to discourage close contact between reactive centers and prevent additional cyclization events, leading to the functionality of ZhuI as a first-ring only ARO/CYC. In comparison, this space does not exist in TcmN ARO/CYC due to the presence of I25, W28, E34, and Y35. Consequently, in TcmN ARO/CYC, the methyl end of the polyketide intermediate docks toward the entrance of the pocket in proximity to the opposing end of the polyketide chain, effectively setting up the C7 and C16 centers for subsequent second-ring cyclization (Figure 6B, Figures S5B).
Figure 6.
Docking of putative polyketide intermediates into the ARO/CYC pocket. (A) Docking of the 18-carbon intermediate 1 (Figure 1), the putative product of ZhuI. (B) Docking of a putative 20-carbon bicyclic aromatic intermediate with TcmN ARO/CYC. For both panels: left, a cutaway mesh surface view showing the positions of the docked molecules; right, a view of pocket residue interactions with the docked compounds.
Although the putative alkyl end-binding region of ZhuI can help rationalize the observed first-ring-only cyclization activity of ZhuI, docking simulations do not provide any conclusive evidence as to why ZhuI promotes C7–C12 (rather than C9–C14) first-ring cyclization. However, compared to TcmN ARO/CYC, there are several changes in ZhuI pocket residues that provide more space in the interior portion of the pocket, including (Tcm → ZhuI): W28 → Y27, E34 → P33, Y35 → T34, W108 → L107. This additional space provided by the ZhuI pocket may alleviate the steric clash observed when docking a C7–C12 cyclized polyketide intermediate into the TcmN ARO/CYC pocket (17), thus providing ZhuI with the ability to bind and fold a polyketide chain for C7–C12 first-ring cyclization. Further, residues defining the ZhuI interior pocket are much more conserved with residues of the di-domain ARO/CYCs (N-half, residues 1–160) than those of the C9–C14 mono-domain ARO/CYCs. With the knowledge that the functionality of the di-domain enzymes is to aromatize a C7–C12 cyclized first ring, ZhuI may have evolved from the di-domain ARO/CYCs so that the interior pocket is essentially ‘made’ to accommodate and to promote C7–C12 first-ring cyclization of an unreduced polyketide chain.
Pocket residue mutations and in vitro activity assay confirmed hypothesis about key catalytic residues
To test the postulated importantance of ZhuI pocket residues for activity and cyclization specificity, the following mutants were generated and assayed using a PKS4-based in vitro reconstitution assay (27): T34A, S64A, R66A/Q/E/K, H109A, and D146A/N (Figure 7C). At the foundation of this activity assay is the ability of WT (and functionally active mutant) ZhuI ARO/CYC to direct the C7–C12 first-ring cyclization of a nonaketide chain to biosynthesize NonaSEK4 (5, Figure 2). In contrast, the PKS4 MinPKS alone produces two nonaketide products with different first-ring cyclization regiospecificity: PK8 (7, C9–C14) and naphthopyrone 6 (C5–C10) (Figure 2). The PKS4 MinPKS and WT or mutant ZhuI ARO/CYC were incubated in vitro, followed by product extraction and HPLC analysis. An overlay of the HPLC trace data for different combinations of enzymes is shown in Figure 7, and the relative product ratio was calculated from HPLC peak integration (Table 2).
Table 2.
Production of polyketides by in vitro reconstitution of PKS proteins
| ZhuI ARO/CYC | Products isolated (peak area %) | Ratio (5/[6+7]) | |
|---|---|---|---|
| NonaSEK4 (5) | PK8 (7) + naphthopyrone (6) | ||
| —* | 2.7 % | 67 % | 0.040 |
| WT | 50 % | 33 % | 1.5 |
| T34A | 16 % | 60 % | 0.27 |
| S64A | 36 % | 40 % | 0.90 |
| R66A | 2.8 % | 64 % | 0.044 |
| R66Q | 2.8 % | 74 % | 0.038 |
| R66E | 2.1 % | 63 % | 0.033 |
| R66K | 2.1 % | 60 % | 0.035 |
| H109A | 5.3 % | 70 % | 0.075 |
| D146A | 5.0 % | 66 % | 0.075 |
| D146N | 4.8 % | 72 % | 0.067 |
This row represents the reaction containing the PKS4 MinPKS (KS-MAT and ACP) alone. All others listed below this row contain the PKS4 MinPKS plus WT or mutant ZhuI ARO/CYC protein as indicated.
Alanine mutation of R66 abolishes ZhuI activity, while mutation of H109 or D146 to alanine decreases the activity of ZhuI by approximately 20-fold (Figure 7A, Table 2). Although crystallization of these mutants in order to assess local conformational change was not attainable, CD spectroscopy reveals that these mutations do not significantly affect global structure (Figure S6). The abolished enzyme activity for ZhuI R66A is consistent with similar active-site arginine mutations of TcmN ARO/CYC (17). The severely compromised activity observed for D146A or H109A indicates important contributions from D146 and H109 in catalysis, possibly through participation in acid-base chemistry (D146) and/or charge-transfer activation (H109) for first-ring cyclization via aldol condensation. Importantly, the D146N mutant is inactive, suggesting that ionizability and/or negative charge of the D146 side-chain is necessary for function (Table 2, trace data not shown).
ZhuI T34 and S64 are proposed to be important catalytic residues based on their high sequence conservation among ARO/CYCs, as well as docking simulations of ZhuI or TcmN ARO/CYC (Figure 6, Figure S5) (17). We found that T34A decreases enzyme activity while S64A retains near WT level activity, indicating that they are not essential for enzyme activity, that other pocket residues may compensate for the loss of their binding potential.
To further explore the side-chain requirement at residue position 66 for ZhuI functionality we generated a series of mutants at this position, including R66Q, R66E, and R66K (Figure 7B). All of the additional R66 mutants generated were inactive illustrating that arginine at position 66 is required for ZhuI function. The inactivity of R66Q suggests that R66 may play a role in catalysis that requires more than simply polar interaction or the formation of hydrogen bonds for “polyketide-steering” functionality (where the active-site would simply bind and orient the polyketide chain to promote cyclization). Significantly, the R66K mutant did not produce NonaSEK4. Lysine possesses a positively charged ε-amino group that is similar in chain length to that of Arg. If R66 serves only an anchor to form hydrogen bonds with the polyketide substrate it is anticipated that R66K would retain some activity. However, if R66 is acting as a Lewis acid to stabilize the enolate negative charge at the carbonyl oxygen of C-11, the charge distribution of the guanidinium (Arg) compared to that of the primary amino group of Lys may prevent a Lys substitution from functioning appropriately for catalysis. Alternatively, we recognize that the inactivity of R66K could simply reflect incorrect positioning of the Lys side chain in the active site of the mutant protein. Taken together, the mutagenesis studies involving R66 suggest that the side-chain functionality and delocalized positive charge of Arg at this position are required for ZhuI catalyzed intramolecular aldol condensation of the polyketide chain. The above results strongly support the structure-derived hypothesis that R66, H109 and D146 are key catalytic residues of ZhuI.
Proposed mechanism of ZhuI
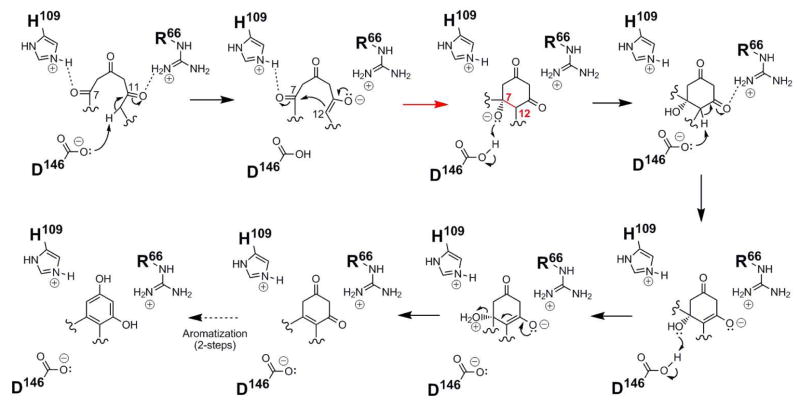
Based on the ZhuI structure, polyketide docking, and in vitro assay results, we propose the following mechanism for the C7–C12 cyclization of the first ring (Figure 8): residue D146 could serve as the general base for proton abstraction at C12, while R66 activates the C-11 carbonyl to promote base catalysis and stabilize the resulting C-11 oxyanion. Residue H109 may then help promote first-ring cyclization by activating the C-7 carbonyl for nucleophilic attack by the C-11 enolate. The now protonated carboxylic acid of D146 could act as a general acid to generate an alcohol at C-7. Dehydration across the newly formed C7-C12 bond could begin via deprotonation by D146 at C12 to form an enolate, again activated and stabilized by R66. Transfer of a proton from D146 to the C-7 hydroxyl affords a good leaving group to promote the collapse of the enolate for the loss of water and a double bond formation between C7–C12. Rearrangement of the cyclized (and dehydrated) first ring by proton shuffling to generate a conjugated aromatic ring system is favored and could occur in the enzyme active site or spontaneously following the product release.
Figure 8.
Proposed mechanism for C7–C12 cyclization catalyzed by ZhuI ARO/CYC.
Different position and chemical environment of the essential and conserved pocket arginine
The results of site-directed mutagenesis and reconstitution of polyketide biosynthesis in vitro support the critical role of residue R66 for ZhuI function. Because this residue is highly conserved among ARO/CYCs and is critical for the function of TcmN, WhiE, and ZhuI ARO/CYCs, a closer look at this residue was merited. Structure superimposition reveals that the position of this pocket arginine is notably different between Tcm (R69) and ZhuI (R66) ARO/CYCs. Both arginines are found deep in the pocket interior, approximately 15 Å from the pocket entrance. Although the α-carbon for R69/66 is located in nearly identical positions, the position of the guanidinium group is shifted by 2.8 Å (Figure 9). Differences between the two ARO/CYCs, such as neighboring residues, can account for the displacement of the Arg guanidinium side chain, including changes in sterics and polar interactions for the respective Arg of each structure. Structure analysis and docking results indicate that the positioning of this Arg may be precisely controlled to account for the different pocket dimensions between Tcm and ZhuI ARO/CYCs, thus allowing for appropriate binding of their respective natural polyketide substrates for productive access to this pocket arginine.
Figure 9.
Stereoview comparison of residues around the enzymatically essential pocket arginine of ZhuI (gold) and TcmN (blue) ARO/CYC. Dashed lines represent potential hydrogen bonding interactions.
Changes in the chemical environment around the critical pocket arginine are also of interest, with R66 of ZhuI in a significantly more polar environment than the structurally equivalent R69 of Tcm ARO (Figure 9). The guanidinium group of ZhuI R66 is within hydrogen-bond distance to the side chain –C=O of Q47 (functionally conserved as either Q or E among di-domain ARO/CYCs), the backbone carbonyl of L21, and the phenolic –OH of Y27 (unique, normally a Trp at this position), while four aliphatic hydrophobic residues are within Van der Waals contact on three sides of the R66 side chain. In contrast, the side chain of R69 in TcmN ARO/CYC is surrounded by three aliphatic hydrophobic and four aromatic hydrophobic residues, while its polar interaction is more limited, including one hydrogen-bond between the R69 guanidinium and the side chain –C=O group of N23 (conserved among mono-domain ARO/CYCs), as well as perpendicular cation-π interactions with the indole face of W95. Whereas the hydrophobic environment around R69 of TcmN ARO/CYC may serve to lower its pKa (56), the environment around R66 of ZhuI ARO/CYC may have the opposite effect to stabilize the delocalized positive charge of the gaunidinium and raise its pKa.
A model for ZhuI catalyzed cyclization of the polyketide chain
Combining the above results with previous work discussing the structure and function of the Type II chain elongation enzymes KS-CLF (57), ACP (58–61), and the modifying proteins TcmN (17) and WhiE ARO/CYC (62), a general model for the assembly of aromatic polyketides is proposed to account for ZhuI’s ability to promote C7–C12 first-ring cyclization in the biosynthesis of the R1128A-D polyketides (Figure 10): 1) Following priming of KS-CLF with an acyl group, chain elongation generates a polyketide intermediate by iterative rounds of decarboxylative condensation with malonyl units provided by malonyl-ACP. 2) The fully elongated and linear polyketide chain is then transferred from the KS-CLF polyketide tunnel to the interior pocket of ARO/CYC. The reactive poly-β-keto intermediate may be sequestered in ACP (the ‘switch-blade’ mechanism) (61, 63), or at the interface between KS-CLF and ACP, where the protein can bind and protect the polyketide chain from spontaneous chemistry during substrate shuttling. Recent protein NMR experiments by Crump et al. showed that a linear fatty acyl chain is not bound to the interior of ACP (64), which would disfavor the switch-blade mechanism. 3) Substrate delivery by ACP, in combination with the precisely defined ‘U-shaped’ interior pocket of ARO/CYC, directs a specific chain folding pattern where enolate formation of the appropriate ketone is promoted by the positively charged pocket arginine. The collapse of the enolate and nucleophilic attack on the opposing carbonyl forms the first six-membered ring. For ZhuI, the interior pocket is designed to promote the regiospecific C7–C12 cyclization of the first ring only, where a unique ‘alkyl binding region’ (shown in grey in Figure 10) may sequester the end of the polyketide chain to prevent subsequent cyclization events from occurring. In ZhuI, the active-site base to generate the enol(ate) may be D146 or water, and in TcmN ARO/CYC the base has been proposed to be a tyrosinate (Y35) (17). Subsequently, the aromatization of the first ring may occur spontaneously in the enzyme active site or following intermediate release. 4) Release of the intermediate from the ZhuI pocket to ACP, which is supported by the recent protein NMR study that the interior of a type II polyketide ACP is perturbed by the presence of a tricyclic aromatic polyketide (65). Finally, 5) downstream polyketide modification converts the partially cyclized intermediate into the final product.
Figure 10.
Model for the ability of the ZhuI pocket to direct first-ring cyclization specificity in the biosynthesis of the R1128 polyketides.
Biological significance
In terms of sequence, structure, and function, ZhuI is at the interface between mono-domain and di-domain ARO/CYCs. The work described herein demonstrates the following distinct properties of ZhuI: 1) ZhuI possesses mono-domain architecture but has a primary sequence more similar to di-domain ARO/CYCs. 2) The di-domain-like pocket of ZhuI may influence the binding and folding of an unreduced polyketide chain resulting in C7–C12 (rather than C9–C14) first-ring cyclization specificity. 3) ZhuI catalyzes cyclization (and subsequent aromatization) of only the first ring to yield a monocyclic intermediate rather than consecutive first- and second-ring cyclization events. Differences in residue composition and protein conformation between ZhuI and TcmN ARO/CYC effectively constrains the entrance to the interior pocket, while also creating a specific groove in the pocket interior where the alkyl-end of the R1128 intermediate docks. Both structural affects may combine to prevent interaction between reactive centers for additional cyclization events. 4) The sequence similarity between ZhuI and the di-domain ARO/CYCs helps visualize how the di-domain ARO/CYCs may bind C7–C12 cyclized polyketide intermediates. The structure of ZhuI identifies two conserved pocket residues, H109 and D146 (conserved as His and Glu in di-domain ARO/CYCs), that may serve as the catalytic dyad for first-ring dehydration/aromatization.
Mutagenesis studies found that three ZhuI pocket residues, R66, H109, and D146, are critical for the regiospecific C7–C12 cyclization (and aromatization) of a nonaketide chain en route to the formation of NonaSEK4. ZhuI D146 is proposed to be involved in acid/base chemistry for aldol condensation, while H109 may be involved in activation of the C-7 carbonyl to promote first-ring cyclization. The essential nature of the conserved pocket arginine (R66) corroborates previous studies performed for TcmN ARO/CYC (17). Based on the above results it is concluded that the ARO/CYC may require this arginine in the pocket, where its position, size, and positive charge is likely required for enol(ate) activation and stabilization at C-11 (ZhuI ARO/CYC) or perhaps for acid/base chemistry (as proposed for TcmN ARO/CYC (17)). The pKa of this pocket arginine may be tuned by the different chemical nature of interacting residues.
The structure of ZhuI provides the first molecular-level insight into an ARO/CYC enzyme from a non-reducing Type II PKS system that is responsible for the regiospecific C7–C12 first-ring cyclization of the nascent polyketide chain. Combining the information provided by the structure and function studies of ZhuI with that of the C9–C14 ARO/CYCs (17) may extend the potential of ARO/CYCs for engineered biosynthesis. A comparison of pocket residue composition and structure should allow for structure-based mutagenesis of pocket residues to control first-ring cyclization specificity.
Supplementary Material
Acknowledgments
We are grateful for the assistance of Dr. John Greaves, Director of the UCI Mass Spectrometry Facility, for product characterization. Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) and the Advanced Light Source (ALS).
Abbreviations
- ACP
acyl carrier protein
- ARO/CYC
aromatase/cyclase
- BSA
bovine serum albumin
- CAPS
N-cyclohexyl-3-aminopropanesulfonic acid
- CLF
chain length factor
- DH
dehydratase
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EtOAc
ethyl acetate
- ESI-MS
eletrospray ionization mass spectrometry
- FAS
fatty acid synthase
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- KS
ketosynthase
- LB
Luria-Bertani media
- MAD
multi-wavelength anomalous dispersion
- MeCN
acetonitrile
- MWCO
molecular weight cutoff
- MeOH
methanol
- PKS
polyketide synthase
- PT
product template domain
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Tris
tris(hydroxymethyl)aminomethane
Footnotes
SCT, BDA, and MYL are supported by the Pew Foundation and National Institute of General Medicinal Sciences (NIGMS R01GM076330). YT is supported by a CAREER proposal from the NSF CBET division.
The atomic coordinates have been deposited in the Protein Data Bank (accession code 3TFZ).
Supporting Information Available on the sequence alignment, stereo view of protein models, and detailed structural comparisons. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Natural Product Reports. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 2.Rawlings BJ. Biosynthesis of polyketides (other than actinomycete macrolides) Natural Product Reports. 1999;16:425–484. doi: 10.1039/a900566h. [DOI] [PubMed] [Google Scholar]
- 3.Hopwood DA. Genetic contributions to understanding polyketide synthases. Chemical Reviews. 1997;97:2465–2498. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 4.Rabbani A, Finn RM, Ausio J. The anthracycline antibiotics: antitumor drugs that alter chromatin structure. Bioessays. 2005;27:50–56. doi: 10.1002/bies.20160. [DOI] [PubMed] [Google Scholar]
- 5.Frederick CA, Williams LD, Ughetto G, van der Marel GA, van Boom JH. Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin. Biochemistry. 1990;29:2538–2549. [PubMed] [Google Scholar]
- 6.Chopra I, Roberts M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol Mol Biol Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, Li Y, Tang Y. Cyclization of aromatic polyketides from bacteria and fungi. Nat Prod Rep. 27:839–868. doi: 10.1039/b911518h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris TM, Harris CM. Biomimetic syntheses of aromatic polyketide metabolites. Pure & Appl Chem. 1986;58:283–294. [Google Scholar]
- 9.Rawlings BJ. Biosynthesis of polyketides (other than actinomycete macrolides) Nat Prod Rep. 1999;16:425–484. doi: 10.1039/a900566h. [DOI] [PubMed] [Google Scholar]
- 10.Shen B, Hutchinson CR. Deciphering the mechanism for the assembly of aromatic polyketides by a bacterial polyketide synthase. Proc Natl Acad Sci. 1996;93:6600–6604. doi: 10.1073/pnas.93.13.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDaniel R, Hutchinson CR, Khosla C. Engineered biosynthesis of novel polyketides: analysis of tcmN function in tetracenomycin biosynthesis. J Am Chem Soc. 1995;177:6805–6810. [Google Scholar]
- 12.Shen B, Summers RG, Wendt-Pienkowski E, Hutchinson CR. The Streptomyces glaucescens tcmKL polyketide synthase and tcmN polyketide cyclase genes govern the size and shape of aromatic polyketides. J Am Chem Soc. 1995;117:6811–6821. [Google Scholar]
- 13.Kramer PJ, Zawada RJX, McDaniel R, Hutchinson CR, Hopwood DA, Khosla C. Rational design and engineered biosynthesis of a novel 18-carbon aromatic polyketide. J Am Chem Soc. 1997;119:635–639. [Google Scholar]
- 14.Alvarez MA, Fu H, Khosla C, Hopwood DA, Bailey JE. Engineered biosynthesis of novel polyketides: properties of the whiE aromatase/cyclase. Nat Biotnol. 1996;14:335–338. doi: 10.1038/nbt0396-335. [DOI] [PubMed] [Google Scholar]
- 15.Yu T-W, Shen Y, McDaniel R, Floss HG, Khosla C, Hopwood DA, Moore BS. Engineered biosynthesis of novel polyketides from Streptomyces spore pigment polyketide synthase. J Am Chem Soc. 1998;120:7749–7759. [Google Scholar]
- 16.Zawada RJX, Khosla C. Heterologous expression, purification, reconstitution and kinetic analysis of an extended type II polyketide synthase. Chem Biol. 1999;6:607–615. doi: 10.1016/s1074-5521(99)80112-4. [DOI] [PubMed] [Google Scholar]
- 17.Ames BD, Korman TP, Zhang W, Smith P, Vu T, Tang Y, Tsai SC. Crystal structure and functional analysis of tetracenomycin ARO/CYC: Implications for cyclization specificity of aromatic polyketides. Proceedings of the National Academy of Sciences, USA. 2008;105:5349–5354. doi: 10.1073/pnas.0709223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu TW, Shen Y, McDaniel R, Floss HG, Khosla C, Hopwood DA, Moore BS. Engineered Biosynthesis of Novel Polyketides from Streptomyces Spore Pigment Polyketide Synthases. Journal of the American Chemical Society. 1998;120:7749–7759. [Google Scholar]
- 19.Fritzsche K, Ishida K, Hertweck C. Orchestration of Discoid Polyketide Cyclization in the Resistomycin Pathway. Journal of the American Chemical Society. 2008;130:8307–8316. doi: 10.1021/ja800251m. [DOI] [PubMed] [Google Scholar]
- 20.Jakobi K, Hertweck C. A gene cluster encoding resistomycin biosynthesis in Streptomyces resistomycificus; exploring polyketide cyclization beyond linear and angucyclic patterns. J Am Chem Soc. 2004;126:2298–2299. doi: 10.1021/ja0390698. [DOI] [PubMed] [Google Scholar]
- 21.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: actVII and actIV genes encode aromatase and cyclase enzymes, respectively. J Am Chem Soc. 1994;116:10855–10859. [Google Scholar]
- 22.Zawada RJX, Khosla C. Domain analysis of the molecular recognition features of aromatic polyketide synthase subunits. J Biol Chem. 1997;272:16184–16188. doi: 10.1074/jbc.272.26.16184. [DOI] [PubMed] [Google Scholar]
- 23.Yu TW, Bibb MJ, Revill WP, Hopwood DA. Cloning, sequencing, and analysis of the griseusin polyketide synthase gene cluster from Streptomyces griseus. Journal of Bacteriology. 1994;176:2627–2634. doi: 10.1128/jb.176.9.2627-2634.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marti T, Hu Z, Pohl NL, Shah AN, Khosla C. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist. Insights into an unusual priming mechanism. The Journal of biological chemistry. 2000;275:33443–33448. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- 25.Tang Y, Lee TS, Khosla C. Engineered biosynthesis of regioselectively modified aromatic polyketides using bimodular polyketide synthases. PLoS Biology. 2004;2:e31. doi: 10.1371/journal.pbio.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombó F, Blanco G, Fernández E, Méndez C, Salas J. Characterization of Streptomyces argillaceus genes encoding a polyketide synthase involved in the biosynthesis of the antitumor mithramycin. Gene. 1996;172:87–91. doi: 10.1016/0378-1119(96)00029-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Li Y, Tang Y. Engineered biosynthesis of bacterial aromatic polyketides in Escherichia coli. Proceedings of the National Academy of Sciences, USA. 2008;105:20683–20688. doi: 10.1073/pnas.0809084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher B, Costantino JP, Wickerham DL, Cecchini RS, Cronin WM, Robidoux A, Bevers TB, Kavanah MT, Atkins JN, Margolese RG, Runowicz CD, James JM, Ford LG, Wolmark N. Tamoxifen for the Prevention of Breast Cancer: Current Status of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 2005;97:1652–1662. doi: 10.1093/jnci/dji372. [DOI] [PubMed] [Google Scholar]
- 29.Meadows ES, Khosla C. In vitro reconstitution and analysis of the chain initiating enzymes of the R1128 polyketide synthase. Biochemistry. 2001;40:14855–14861. doi: 10.1021/bi0113723. [DOI] [PubMed] [Google Scholar]
- 30.Tang Y, Koppisch AT, Khosla C. The acyltransferase homologue from the initiation module of the R1128 polyketide synthase is an acyl-ACP thioesterase that edits acetyl primer units. Biochemistry. 2004;43:9546–9555. doi: 10.1021/bi049157k. [DOI] [PubMed] [Google Scholar]
- 31.Fu H, Ebertkhosla S, Hopwood DA, Khosla C. Engineered Biosynthesis of Novel Polyketides - Dissection of the Catalytic Specificity of the Act Ketoreductase. Journal of the American Chemical Society. 1994;116:4166–4170. [Google Scholar]
- 32.Korman TP, Tan YH, Wong J, Luo R, Tsai SC. Inhibition kinetics and emodin cocrystal structure of a Type II polyketide ketoreductase. Biochemistry. 2008;47:1837–1847. doi: 10.1021/bi7016427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered Biosynthesis of Novel Polyketides: actVII and actIV Genes Encode Aromatase and Cyclase Enzymes, Respectively. Journal of the American Chemical Society. 1994;116:10855–10859. [Google Scholar]
- 34.McDaniel R, Hutchinson CR, Khosla C. Engineered Biosynthesis of Novel Polyketides: Analysis of tcmN Function in Tetracenomycin Biosynthesis. Journal of the American Chemical Society. 1995;117:6805–6810. [Google Scholar]
- 35.Kramer PJ, Zawada RJX, McDaniel R, Hutchinson CR, Hopwood DA, Khosla C. Rational Design and Engineered Biosynthesis of a Novel 18-Carbon Aromatic Polyketide. Journal of the American Chemical Society. 1997;119:635–639. [Google Scholar]
- 36.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. Journal of Molecular Biology. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Katherine A, Kantardjieff BR. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Science. 2003;12:1865–1871. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallog D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. Journal of synchrotron radiation. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D - Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallog D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallog D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Cowtan KD. ‘DM’:An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 45.Collaborative. The CCP4 suite: programs for protein crystallography. Acta Crystallographica Section D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallographica Section D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 47.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 48.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 49.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 50.Kimber MS, Martin F, Lu Y, Houston S, Vedadi M, Dharamsi A, Fiebig K, Schmid M, Rock CO. The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J Biol Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]
- 51.Harris TM, Harris CM. Biomimetic syntheses of aromatic polyketide metabolites. Pure Appl Chem. 1986;58:283–294. [Google Scholar]
- 52.Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White SW, Zheng J, Zhang YM, Rock CO. The structural biology of type II fatty acid biosynthesis. Annual Review of Biochemistry. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 54.Keatinge-Clay A. Crystal Structure of the Erythromycin Polyketide Synthase Dehydratase. Journal of Molecular Biology. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 56.Harris TK, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 57.Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C, Stroud RM. An antibiotic factory caught in action. Nature Structural and Molecular Biology. 2004;11:888–893. doi: 10.1038/nsmb808. [DOI] [PubMed] [Google Scholar]
- 58.Crump MP, Crosby J, Dempsey CE, Parkinson JA, Murray M, Hopwood DA, Simpson TJ. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2) Biochemistry. 1997;36:6000–6008. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- 59.Li Q, Khosla C, Puglisi JD, Liu CW. Solution Structure and Backbone Dynamics of the Holo Form of the Frenolicin Acyl Carrier Protein. Biochemistry. 2003;42:4648–4657. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- 60.Roujeinikova A, Simon WJ, Gilroy J, Rice DW, Rafferty JB, Slabas AR. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. Journal of Molecular Biology. 2007;365:135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 61.Leibundgut M, Jenni S, Frick C, Ban N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 62.Tsai S, Ames BD. Chapter 2: Structural enzymology of polyketide synthases. Methods in Enzymology. 2009;459:17–47. doi: 10.1016/S0076-6879(09)04602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Natural Product Reports. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans SE, Williams C, Arthur CJ, Ploskon E, Wattana-amorn P, Cox RJ, Crosby J, Willis CL, Simpson TJ, Crump MP. Probing the Interactions of early polyketide intermediates with the Actinorhodin ACP from S. coelicolor A3(2) J Mol Biol. 2009;389:511–528. doi: 10.1016/j.jmb.2009.03.072. [DOI] [PubMed] [Google Scholar]
- 65.Haushalter RW, Filipp FV, Ko KS, Yu R, Opella SJ, Burkart MD. Binding and pK(a) Modulation of a Polycyclic Substrate Analogue in a Type II Polyketide Acyl Carrier Protein. ACS Chem Biol. 2010 doi: 10.1021/cb200004k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.