Abstract
BACKGROUND
The first week of human embryonic development comprises a series of events that change highly specialized germ cells into undifferentiated human embryonic stem cells (hESCs) that display an extraordinarily broad developmental potential. The understanding of these events is crucial to the improvement of the success rate of in vitro fertilization (IVF). With the emergence of new technologies such as Omics, the gene expression profiling of human oocytes, embryos, and hESCs has been performed and generated a flood of data related to the molecular signature of early embryo development.
METHODS
In order to understand the complex genetic network that controls the first week of embryo development, we performed a systematic review and study of this issue. We performed a literature search using PubMed and EMBASE to identify all relevant studies published as original articles in English up to March 2010 (n=165). We also analyzed the transcriptome of human oocytes, embryos and hESCs.
RESULTS
Distinct sets of genes were revealed by comparing the expression profiles of oocytes, embryos on day-3 and hESCs, that are associated with totipotency, pluripotency and reprogramming properties, respectively. Known components of two signalling pathways (WNT and transforming growth factor-beta) were linked to oocyte maturation and early embryonic development.
CONCLUSION
Omics analysis provides tools for understanding the molecular mechanisms and signalling pathways controlling early embryonic development. Furthermore, we discuss the clinical relevance of using a non-invasive molecular approach to embryo selection for the single embryo transfer (SET) program.
Keywords: Embryonic Development; genetics; Embryonic Stem Cells; metabolism; Female; Gene Expression Profiling; Gene Expression Regulation, Developmental; Humans; Maternal Age; Oocytes; metabolism; Polycystic Ovary Syndrome; genetics; metabolism; Pregnancy; Pregnancy Outcome
Keywords: oocyte genes, embryo genes, embryo stem cells, microarray, non-invasive approach
Introduction
In humans, early embryo development is a complex process that consists of sequential maturation events of the oocyte, fertilization and embryo growth (4-cell, 8-cell, morula and blastocyst). Indeed, oocytes and spermatozoa are atypical, highly specialized cell types compared to somatic cells. Yet, after fertilization, a zygote is formed, the ultimate totipotent cell that can be considered to be the ultimate undifferentiated cell type, as it gives rise to all cell types and live offspring. Totipotency persists for the very first cell doublings, from the single cell and zygote to at least the 4-cell pre-embryo. Initiation of transcription in the newly-formed embryonic genome reportedly occurs at the 4- and 8-cell stage (Braude et al., 1988). A few studies have documented key events that follow fertilization in humans such as decreases in abundance of individual mRNAs (Taylor et al., 2001), overall patterns of gene expression in individual human oocyte and preimplantaion embryos stage (Dobson et al., 2004), and more generalizable transcription profiles of pooled morphologically normal human oocytes and embryos (Zhang et al., 2009). These data provide fundamental resources for understanding the genetic network controlling the early stages of human embryo development. At the morula stage, the human pre-embryo undergoes compaction, with the loss of the cellular distinction between the blastomeres. This is followed by the high expression of genes involved in tight intercellular junctions. Then, when the human embryo has reached the 35- to 65-cell stage, the trophoblast cells pump nutrients and water into the interior of the cell-sphere, forming a blastocyst, within which the inner cell mass (ICM) cells continue to proliferate. It is at the blastocyst stage that ICM cells are harvested for the derivation of embryonic stem (ES) cells (Reubinoff et al., 2000; Thomson et al., 1998). Pluripotent cells can be isolated, adapted and propagated indefinitely in vitro in an undifferentiated state as human embryonic stem cells (hESCs). hESCs are remarkable in their ability to generate virtually any cell type; hence they carry many hopes for cell therapy. Human mature MII oocytes, as well as hESCs, are able to achieve the feat of cell reprogramming towards pluripotency, either by somatic cell nuclear transfer or by cell fusion, respectively (Cowan et al., 2005; Hochedlinger et al., 2004; Saito et al., 2008; Sung et al., 2006). Knowledge gathered from the field of ESCs was at the heart of the groundbreaking discovery that both mouse and human somatic cells can be reprogrammed into a pluripotent state by defined factors. The field of induced pluripotent stem cells (iPSCs) has marked a new era in stem cell research, and has also provided data pertinent to improving the understanding of pluripotency (Takahashi et al., 2007; Yu et al., 2007). Since totipotency and pluripotency are at the center of early embryonic development, comprehending their molecular mechanisms is crucial to our understanding of reproductive biology and to regenerative medicine.
DNA microarray technology is one of the most widely used and potentially revolutionary research tools derived from the human genome project (Venter et al., 2001). This technology provides a unique tool for the determination of gene expression at the level of messenger RNA (mRNA) on a genomic scale. Its capacity has opened new paths for biological investigation and generated a large number of applications (Stoughton. 2005), including the analysis of the transcriptomic profiles from early embryo development and the identification of new prognostic biomarkers for use in the in vitro fertilization (IVF) program (Hamatani et al., 2004a; Assou et al., 2008). The application of microarray technology to the analysis of human oocyte and early embryo cleavage poses specific challenges associated with the picogram levels of mRNA in a single oocyte and embryo, the plasticity of the embryonic transcriptome, the scarcity of the material and ethical considerations.
In this review we analyzed data from published reports and include our own data to define the genomic profile during early embryonic development. Once the molecular signature is established, biomarkers can be identified on a large scale, validated and tested prior to clinical applications for embryo selection to improve single embryo transfer (SET) programs. Such a research workflow should provide an understanding of the molecular and cellular mechanisms of oocyte and embryo function, as well as important insights for the development of diagnostic tests for oocyte quality and embryo competence.
Methods
A search of English-language publications from 4 computerized databases (PubMed, EMBASE, Science Direct and Ingenta Connect) was undertaken. We built an expression compendium by combining U133 Plus 2.0 (Affymetrix, Santa Clara, USA) microarray data from the US National Center for Biotechnology Information, from the Gene Expression Omnibus (GEO) through the provisional accession numbers (the GSE7234, GSE7896, GSE11450, and GSE7896 series), and from 15 samples from our own laboratory (5 pooled oocytes, 2 embryos on day-3, and 8 hESC lines). This study received institutional review board (IRB) approval, as well as French authority: Agence de la Biomédecine (ABM). The information gathered was analyzed through the GCOS 1.2 software (Affymetrix), using the default analysis settings and global scaling as the first normalization method, with a trimmed mean target intensity value (TGT) for each array arbitrarily set to 100, in agreement with the MIAME recommendations (Brazma et al., 2001). Principle component analysis (PCA) was performed using GeneSpring® software to provide a global view of how the various sample groups were related. Hierarchical clustering was carried out with CLUSTER and TREEVIEW software (Eisen et al., 1998). To uncover functional biological networks, we imported gene expression signatures into the Ingenuity Pathways Analysis (IPA) Software (Ingenuity Systems, Redwood City, CA, USA).
Results
Global analysis of the human oocytes gene expression profile
The human oocyte is equipped with an extraordinary biological competence. It can be fertilized and, at the same time, is able to reprogram the sperm chromatin, giving rise to totipotency and driving towards early embryonic development. However, the molecular mechanisms underlying oocyte competence are still largely unknown. Throughout follicle growth, the nuclear maturation of oocytes is arrested at meiotic prophase I. However, the cytoplasm progresses through a series of maturational stages, accumulating mRNAs and proteins that enable the oocyte to be fertilized and to progress through the first cleavage divisions until embryonic genes begin to be expressed. To improve the understanding of oocyte maturation and competence, microarray technology permits the assessment of changes of the global gene expression profiles in human and mouse oocytes (see Table I).
Table I.
Microarray studies of oocytes and embryos
| human oocytes and embryos | ||||
|---|---|---|---|---|
| Techniques | Samples | Number of identified genes | Targets | Study |
| HG-U133 Plus 2.0 array (Affymetrix) | Oocytes | 1,361 transcripts expressed in oocytes | Study oocyte transcriptomes | (Bermudez et al., 2004) |
| HG-U133 Plus 2.0 array (Affymetrix) | Oocytes | 1,514 overexpressed in oocytes compared with cumulus cells | Understanding of the mechanisms regulating oocyte maturation | (Assou et al., 2006) |
| HG-U133 Plus 2.0 array (Affymetix) | Oocytes | 5,331 transcripts enriched in metaphase II oocytes relative to somatic cells | Comprehension of genes expressed in in vivo matured oocytes | (Kocabas et al., 2006) |
| HG-U133 Plus 2.0 array (Affymetrix) | Oocytes | 10,183 genes were expressed in germinal vesicle | Study of global gene expression in human oocytes at the later stages of folliculogenesis (germinal vesicle stage) | (Zhang et al., 2007) |
| HG-U133 Plus 2.0 array (Affymetrix) | Oocytes | Of the 8123 transcripts expressed in the oocytes, 374 genes showed significant differences in mRNA abundance in PCOS oocytes | Understanding of PCOS | (Wood et al., 2007) |
| HG-U133 Plus 2.0 array (Affymetix) | Oocytes | --- | Identify new potential regulators and marker genes which are involved in oocyte maturation. | (Gasca et al., 2007) |
| HG-U133 Plus 2.0 array (Affymetix) | Oocytes | 283 genes found in the case report sample | Identify molecular abnormalities in metaphase II (MII) oocytes | (Gasca et al., 2008) |
| Whole Genome Bioarrays printed with 54,840 discovery probes representing 18,055 human genes and an additional 29,378 human expressed sequence tags (EST) | Oocytes | 2,000 genes were identified as expressed at more than 2-fold higher levels in oocytes matured in vitro than those matured in vivo | Analysis of gene expression profile of oocytes following in vivo or in vitro maturation | (Jones et al., 2008b) |
| Applied Biosystems Human Genome Survey Microarray (32,878 60-mer oligonucleotide) | Oocytes | Germinal vesicle, in vivo-MII, and IVM-MII oocytes expressed 12,219; 9,735; and 8,510 genes, respectively | Characterized the patterns of gene expression in germinal vesicle stage and meiosis II oocytes matured in vitro or in vivo | (Wells and Patrizio. 2008) |
| HG-U133 Plus 2.0 array (Affymetix) | Oocytes | 342 genes showed a significantly different expression level between the two age groups (women aged, 36 years (younger) and women aged 37–39 years (older)) | Investigate the effect of age on gene expression profile in mature oocytes | Grondahl et al., 2010) |
| Two cDNA microarrays, each containing about 20,000 targets (representing in total ~29,778 independent genes according to Unigene Build 155). | Oocytes and embryos | 1,896 significant changes in expression following fertilization through day 3 of development | Global analysis of the pre-implantation embryo transcriptome | (Dobson et al., 2004) |
| cDNA microarrays containing 9,600 cDNA spots | Oocytes and embryos | 184; 29; and 65 genes were overexpressed in oocytes, 4-and 8-cell embryos, respectively | Identify differential expression profiles of genes in single oocytes, 4- and 8-cell preimplantation embryos | (Li et al., 2006) |
| Genome Survey Microarrays V2.0 (Applied Biosystems) | Oocytes and embryos | 107 DNA repair genes were detected in oocytes | Identify the DNA repair pathways that may be active pre- and post-embryonic genome activation by investigating mRNA in human in vitro matured oocytes and blastocysts | (Jaroudi et al., 2009) |
| HG-U133 Plus 2.0 array (Affymetrix) | Oocytes and embryos | 5,477 transcripts differentially expressed into transition from mature oocyte (MII) to 2-day embryo and 2,989 transcripts differentially expressed into transition from 2- to 3-day embryo | Study of global gene expression in human pre-implantation development | (Zhang et al., 2009) |
| Mouse oocytes and embryos | ||||
| NIA 60-mer oligo microarray (Agilent) | Oocytes | 530 genes showed statistically significant expression changes between young and old oocytes | Comparison of transcriptional profile of oocytes obtained from aged mice with oocytes from young mice | (Hamatani et al., 2004b) |
| Genome Array MOE430 A and B chips (Affymetrix) | Oocytes | --- | Identification of gene expression profiles of oocytes during the primordial follicle stage to the large antral follicle stage | (Pan et al., 2005) |
| 7.4 K GenePlore TwinChip | Oocytes | 214 genes that are upregulated between primordial and primary follicles. 396 upregulated genes between primary and secondary follicles |
Study of global gene expression profiles of early folliculogenesis in primordial, primary, and secondary follicles | (Yoon et al., 2006) |
| Affymetrix 430 v2.0 GeneChips | Oocytes | 3,002 genes changed significantly during GV-to-MII transition | Investigate the relative changes of transcripts present in GV-stage versus MII-stage oocytes | (Su et al., 2007) |
| Applied Biosystems Mouse Genome survey Array (32,996 60-mer oligonucleotide) | Oocytes | 1,682 genes with more highly expression in GV-stage oocytes than in MII-stage oocytes. 1,936 genes were more highly expressed in MII-stage oocytes | Comparison of gene expression profiles of germinal vesicle (GV) and metaphase II (MII)-stage oocytes | (Cui et al., 2007) |
| NIA 15k cDNA microarray | Embryos | 428 genes upregulated and 748 downregulated in blastocyst compared to morula | Identification of genes expressed differentially between morula and blastocyst | (Tanaka et al., 2004) |
| NIA 60-mer oligo microarray (Agilent) | Embryos | --- | Global gene expression profiling of all stages of pre-implantation embryos | (Hamatani et al., 2004a) |
| Affymetrix 25-mer DNA | Embryos | --- | Global gene expression analysis has also been applied to examine the effects of oxygen atmosphere on mouse pre-implantation embryo gene expression patterns | (Rinaudo, P. F. et al., 2006) |
| GenePlorer TwinChip Mouse 7.4 K elements (containing 7,410 different genes) | Embryos | 248 genes were differentially expressed between the two-cell embryos and two-cell block embryos | Analysis of differences in gene expression between two-cell and two-cell block embryos | (Jeong et al., 2006) |
| Affymetrix 430 2.0 GeneChips | Embryos | 1,912 was statistically different in the IVF cohort when compared with the in vivo control embryos | Global gene expression analysis has also been applied to contrast gene expression between cultured and in vivo derived mouse embryos | (Giritharan et al., 2007) |
| Affymetrix 25-mer DNA | Oocytes and embryos | 4,000 genes changed in expression over 5-fold | Analysis of gene activity during pre-implantation development | (Wang et al., 2004) |
| MG_U74Av2, MOE430A, and MOE430B GeneChips (Affymetrix) | Oocytes and embryos | 9,414 genes differentially expressed in oocytes | Analysis of global patterns of gene expression during pre-implantation development | (Zeng et al., 2004) |
| NIA 60-mer oligo microarray (Agilent) | Oocytes and embryos | --- | Identification of a large number of genes and multiple signaling pathways involved at each developmental stage of pre-implantaion embryos | (Hamatani et al., 2006) |
Gene expression of the oocyte during nuclear maturation
Nuclear maturation is a fundamental developmental event that has been extensively studied for several years. However, the molecular mechanisms underlying this critical process remain incompletely understood. Many exciting biological questions surrounding this event persist. Differences in the global mRNA transcript profile have been reported between mature oocytes (metaphase II (MII)) and immature oocytes (germinal vesicle (GV) and metaphase I (MI)) in both mice and human (Assou et al., 2006; Cui et al., 2007; Wang, Q. T. et al., 2004; Yoon et al., 2006; Zhang et al., 2007). The genome-wide gene expression of human immature and mature oocytes revealed minor modifications in the transcript profiles between GV and MI oocytes, while MII oocytes showed overexpression of more than 400 genes and a striking underexpression of more than 800 genes in comparison with less mature oocytes (Assou et al., 2006). The major modifications of the transcript profile appear to occur during the final stage of oocyte maturation. Components of the metaphase-promoting factor, the anaphase-promoting complex, and a number of oocyte specific genes were overexpressed in MII oocyte (Assou et al., 2006). Similarly, a global transcriptome comparison of GV-stage and MII-stage mouse oocytes revealed the expression of more than 12,000 genes by murine oocytes, more than 1,600 of which were upregulated in GV and 2,000 in MII oocytes (Cui et al., 2007). The authors reported a higher representation of genes associated with protein metabolism, the mitotic cell cycle, electron transport, fertilization, microtubule/cytoskeletal protein family, DNA replication, G protein-coupled receptors, and expression signalling in mature MII oocytes. The focus of our previously published study was to apply a microarray approach to identify new potential regulators and candidate genes involved in human oocyte maturation. In addition, (Assou et al., 2006) observed that genes involved in the proteasome pathway were over-represented during human oocyte maturation (Fig. 1), suggesting an important role in oogenesis for this gene family. This observation is of great interest in light of the recent findings on the role of the proteasome in transcription (Szutorisz et al., 2006). The proteasome is known to interact with chromatin during multiple stages of transcription, through both proteolytic and non-proteolytic activities (Collins and Tansey. 2006). Proteolytic activities of the proteasome are important for promoting the exchange of transcription factors on chromatin, and possibly for facilitating multiple rounds of transcription initiation. Non-proteolytic activities of the proteasome are important for transcriptional elongation checkpoint and histone modification (Collins and Tansey. 2006). Previous studies described the BARD1- BRCA1 heterodimer as an E3 ubiquitin ligase (Lorick et al., 1999; Ruffner et al., 2001). Gasca et al. (2007) observed that multiprotein E3 ubiquitin ligase complex containing BARD1, BRCA1 and BRCA2, termed BRCC are involved in human oocyte maturation. These proteins function predominantly in the nucleus to regulate cell cycle progression, DNA repair and gene transcription (Henderson. 2005). BARD1 is an important regulator of BRCA1 activity: the binding of BARD1 with BRCA1 maintains both proteins in the nucleus, consequently preventing apoptosis (Fabbro et al., 2004). Guillemin et al. (2009) reported that the apoptosis inhibitor protein BCL2L10 undergoes dynamic sub-cellular redistribution during human oocyte maturation. Knockdown of mouse Bcl2l10 using RNA interference (RNAi) in cultured oocytes resulted in maturation arrest at the MI stage accompanied by abnormal spindle and chromosome organizations (Yoon et al., 2009). These findings indicate that BCL2L10 may play a role in oocyte maturation.
Figure 1.
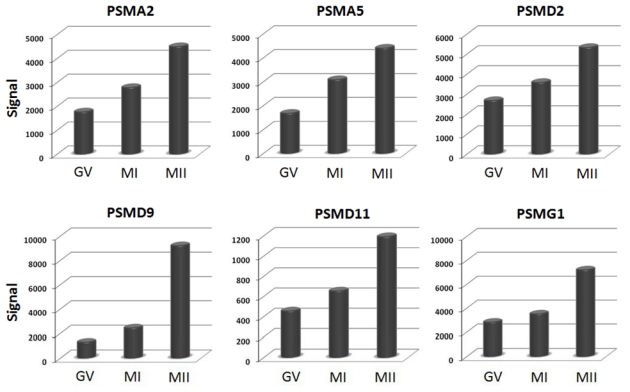
Upregulation of proteasome genes during oocyte maturation. Histograms show signal values of six proteasome genes (PSMA2, PSMA5, PSMD2, PSMD9, PSMD11 and PSMG1) in each stage of oocyte maturation. Gene expression is measured by pan-genomic HG-U133 Plus 2.0 Affymetrix oligonucleotides microarrays, and the signal intensity for each gene is shown on the Y-axis as arbitrary units determined by the GCOS 1.2 software (Affymetrix). GV; germinal vesicle; MI, metaphase I; MII, metaphase II.
Gene expression of oocytes according to female age
Oocyte quality is a main factor in human reproductive failure and has been strongly associated with advanced reproductive age. Recent studies have indicated differential gene expressions between younger and older oocytes in human (Steuerwald et al., 2007; Grondahl et al., 2010) and mouse (Hamatani et al., 2004b; Pan et al., 2008). Age affects the expression of human oocyte genes in a variety of major functional categories, including cell cycle regulation, cytoskeletal structure, energy pathways, transcription control, and stress responses (Steuerwald et al., 2007). Recently, Grondahl et al. (2010) performed full-genome microarray analysis on individual oocytes of women with advanced age in comparison with a younger patient. They observed a substantial difference between younger and older oocytes at the transcriptional level of genes involved in central biological functions, from establishment and organization of the meiotic spindle checkpoint, through protein metabolism and DNA repair to the regulation of meiotic division and cell cycle. In the mouse, gene profiles involved in oxidative stress, mitochondrial function, chromatin structure, DNA methylation and genome stability are altered with old age (Hamatani et al., 2004b). In addition, the expression profiling of young and old mouse oocytes revealed changes in the expression of several kinetochore components of the spindle assembly checkpoint (SAC); including protein kinases (e.g., Bub1, BubR1, Aurora kinase) and Cdc20, a critical activator of the Anaphase Promoting Complex (Pan et al., 2008).
Gene expression of oocytes under in vitro conditions
Intrinsic oocyte developmental competence, as assessed by development up to the blastocyst stage, has been positively associated with the site of oocyte maturation (Rizos et al., 2002). The site of oocyte maturation has a profound effect on oocyte quality; oocytes matured in vivo are of superior competence compared to those matured in vitro (Rizos et al., 2002). Microarray technologies were applied to humans and to mice to identify the differences between development in vitro versus in vivo matured oocytes (Jones et al., 2008; Pan et al., 2005; Wells and Patrizio. 2008). Jones et al. (2008) compared the transcriptome of oocytes matured in vivo (with relatively high developmental competence) with the transcriptome of oocytes under in vitro maturations (IVM) conditions (with low developmental competence) and identified an over-abundance of a large number of genes in human oocytes matured in vitro compared with in vivo. This study suggested, that the increase in gene expression detected in vitro could be due to either a deregulation in gene transcription or the posttranscriptional modification of genes, causing an inadequate temporal utilization of transcripts which could be translated as a developmental incompetence of any embryos resulting from these oocytes. Similarly, Zheng et al. (2005) indicated that the reduced developmental competence of non-human primate oocytes under IVM was a result of a failure of these oocytes to undergo the normal pattern of transcript silencing. Wells and Patrizio (2008) elegantly differentiated the mRNA expression in human oocytes at different maturational stages under IVM. They observed that high levels of mRNAs, proteins, substrates and nutrients are accumulated in the oocyte during maturation and are associated with oocyte developmental competence (Song et al., 2005; Watson. 2007). In addition, IVM seems to alter the abundance of certain MII oocyte mRNAs compared with in vivo MII oocytes (Lee et al., 2008; Zheng et al., 2005). The differences in gene expression detected may assist in the design of optimized protocols for ovarian stimulation and in further refining media formulations for IVM.
Gene expression of oocytes in PCOS patients
Polycystic ovary syndrome (PCOS) is a good model for studying the loss of oocyte quality. Studies comparing gene expression arrays in tissues from patients with PCOS compared with normal responders have reported similar pathways for differentially expressed genes in whole ovaries (Jansen et al., 2004; Oksjoki et al., 2005) and oocytes (Wood et al., 2007). Wood et al (2007) identified 374 genes with different mRNA transcript levels when analyzing morphologically indistinguishable oocytes from normal responders and PCOS patients (Wood et al., 2007). Many of the genes found to be differentially expressed in PCOS are involved in signal transduction, transcription, DNA and RNA processing, and the cell cycle. Of these, 68 are involved in chromosome alignment and segregation; other genes contain putative androgen receptors (Wood et al., 2007). These differences provide a partial explanation for the reduction of oocyte capacity observed in PCOS patients. Cumulus cells (CCs) play a major role in the control of oocyte metabolism, and therefore it is likely that dysfunction of these cells could play a role in PCOS.
Human oocyte-cumulus cells relationships
The oocyte is surrounded by several cell layers, CCs, which are tightly connected to each other and to the oocyte through gap junctions such as GJA1 (Cx43) and GJA4 (Cx37), that facilitate the bi-directional traffic between the oocyte and CCs (Tanghe et al., 2002). The oocytes create and control their microenvironment by promoting differentiation of the cumulus cell phenotype through secretion of paracrine signalling factors, such as growth and differentiation factor 9 (GDF9) and bone morphogenic protein-15 (BMP15), which are members of the transforming growth factor β (TGFβ) family (Eppig et al., 1997). Absence of these two oocyte-secreted factors has been shown to cause infertility (Dong et al., 1996; Galloway et al., 2000). GDF9 show increased cumulus production of hyaluronic acid synthetase (HAS2), an enzyme responsible for the hyaluronic acid synthesis which is the major structural backbone of the cumulus extracellular matrix (Elvin et al., 1999), as well as the proteoglycan versican, another important component of the cumulus matrix (Russell et al., 2006). Other genes were induced and regulated by GDF-9 such as Peroxiredoxin 2 (PRDX2) (Leyens et al., 2004) and SMAD family member 2 and 3 (SMAD2/3) (Dragovic et al., 2007). Studies on SMAD2/3 conditional knockout mice indicate that SMAD2/3 are indispensable for normal cumulus expansion (Li, Q. et al., 2008). Furthermore, CCs play an essential role, particularly for normal oocyte growth, ovulation, fertilization and embryo development (Chang et al., 2002). Active components of the cumulus matrix come from several sources: they are synthesized directly by CCs under the control of endocrine and oocyte-derived factors, secreted by mural granulosa cells (GCs), or enter the follicle from blood plasma (Eppig. 1982, 1991; Tanghe et al., 2002; Vanderhyden et al., 2003). In addition, CCs and oocytes have different metabolic needs. Oocytes themselves are unable to synthesize cholesterol and poorly metabolize glucose for energy production. Inversely, the CCs metabolize alternative substrates, such as cholesterol and glucose, which are essential for the development and function of oocyte (Eppig et al. 2005). The exact nature and diversity of oocyte-CC signalling molecules are complex and dynamic. Errors in the regulatory cumulus-oocyte complex (COC) may result in the production of oocytes unable to undergo embryo and pregnancy outcomes. The study of the CC gene expression profile offers the opportunity by a non-invasive method, to predict oocyte competence, because bidirectional communication between the CC and the oocyte are necessary for acquisition of this competence.
Global analysis of human embryo gene expression profile
Embryo survival requires that the oocyte be sufficiently equipped with stored maternal products and that embryonic gene expression start at the correct time. Human early embryos are highly sensitive to the environment under in vitro culture conditions. The culture conditions can alter the gene expression pattern (Lonergan et al., 2006). Several studies have explored global patterns of embryos gene expression in both mouse (Hamatani et al., 2004a; Rinaudo et al., 2004; Wang, Q. T. et al., 2004; Zeng et al., 2004) and human (Dobson et al., 2004; Li, S. S. et al., 2006; Wells et al., 2005; Zhang et al., 2009) embryos (Table I). Gene expression profiling of early mouse embryos showed characteristic patterns of maternal RNA depletion and revealed that embryonic genome activation (EGA) happens in two phases: the first phase is zygotic genome activation (ZGA), the second mid-preimplantation gene activation (MGA), which precedes the dynamic morphological and functional changes from the morula to the blastocyst stage (Hamatani et al., 2006). Two major groups of gene activity can be defined: firstly in oocytes and first early cleavage stages and the second consisting of 4-cell to blastocyst stages, which correlates with the transition from maternal to zygotic expression. Several transcripts of molecules involved in WNT or BMP signalling pathways were identified in the embryonic transcriptome, indicating that these critical regulators of cell fate and patterning are conserved and functional at these stages of pre-implantation development. Dobson et al. (2004) reported the global patterns of gene expression in human embryos on day 2 and 3 (n=8) by using DNA microarray analysis. This study revealed that the first few days of oocyte maturation and embryo development were characterized by a significant decrease in transcript levels, suggesting that decay of RNAs associated with gamete identity is integral to embryo development (Dobson et al., 2004). Furthermore, this study revealed that developmental arrest and activation of the embryonic genome are unrelated events. Wells et al. (2005) used reverse transcription and real-time fluorescent PCR to quantify the expression patterns of nine known genes implicated in DNA damage and cell division (BRCA1, BRCA2, ATM, TP53, RB1, MAD2, BUB1, APC, and beta-Actin) at the early stages of human embryonic development. The data suggest that embryos with patterns of gene expression appropriate for their developmental stage may have superior viability to those displaying atypical gene activities (Wells et al., 2005). Li et al. (2006) used a cDNA microarray containing 9,600 transcripts to investigate 631 differently expressed genes in oocytes, as well as the 4-cell and 8-cell human embryonic stages. Numerous genes were found to be expressed at a value double that of the median value of all genes expressed; 184 genes in oocytes, 29 in 4-cell embryos, and 65 genes in 8-cell embryos. These results indicate that the expression of some zygotic genes had already occurred at the 4-cell embryonic stage (Li et al., 2006). In addition, Chen et al. (2005) observed global gene expression changes during the hatching of mouse blastocysts, an essential process for implantation. This study observed that there were 85 genes which were upregulated in blastocysts at the hatching stage. These genes included cell adhesion molecules, epigenetic regulators, stress response regulators, and immunoresponse regulators (Chen et al., 2005). Furthermore, Adjaye et al. (2005) identified biomarker transcripts specific to inner cell mass (ICM) such as (OCT4/POU5F1, NANOG, HMGB1, and DPPA5) when comparing gene expression profiles of ICM and trophoectoderm cells (TE) from human blastocysts. This data indicates that the emergence of pluripotent ICM lineages from the morula is controlled by metabolic and signalling pathways including WNT, mitogen-activated protein kinase (MAPK), transforming growth factor-β (TGF-β), NOTCH, integrin-mediated cell adhesion and apoptosis-signalling pathways (Adjaye et al., 2005).
The molecular mechanism of early embryonic development is still unclear. To identify genes that are involved in early embryonic development, we have conducted an analysis related to the expression profiles of five pooled oocytes, two embryos on day-3 and eight human embryonic stem cell (hESC) lines. In order to ascertain whether there is a specific molecular signature corresponding to each stage of early embryonic development, we performed principal component analysis (PCA) and hierarchical clustering (Figure 2). All of the oocyte, embryo and hESC samples clustered as homogeneous groups (Figure 2A) in agreement with the robustness of the Affymetrix microarrays (Irizarry et al., 2005). Hierarchical clustering on 10,000 genes delineated five major clusters of genes (Figure 2B). The hierarchical clustering dendrogram showed that hESCs are distantly related to oocytes and embryos on day-3 (Figure 2C), revealing the distinctive patterns of maternal RNA degradation and embryonic gene activation, which contribute to the dramatic morphological changes during early embryonic development. We identified genes whose expression gradually increased or decreased during early embryonic development (Figure 3A–B). Taken together, these results seem to indicate that early embryonic development, particularly totipotent cells and highly pluripotent cells (hESCs), have quite distinct genetic programs. This supports the notion of a gradual decrease of developmental potential from the pre-implantation embryo stage to stem cells.
Figure 2.
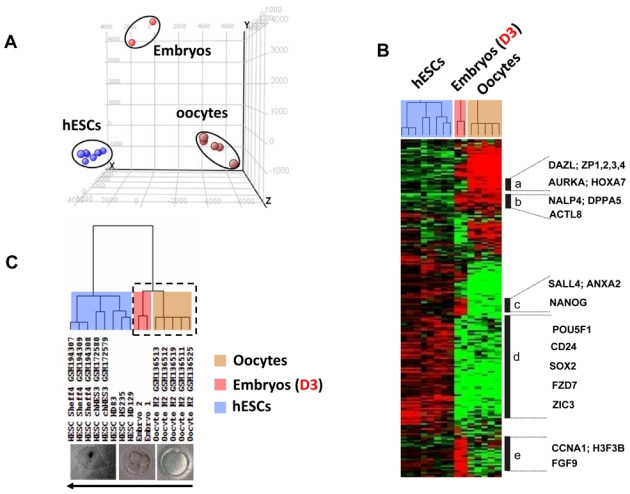
Principal component analysis (PCA) and hierarchical clustering of all samples from different developmental stages. (A) PCA distributes samples into a three-dimensional space based on the variances in gene expression; samples that have similar trends in their gene expression profiles will cluster together in the PCA plot. This analysis, using GeneSpring® software, resulted in a clear segregation of the fifteen replicated samples into three clusters, corresponding to human mature oocytes, human embryos, and hESCs. Each colored point represents a sample, characterized by the gene expression of all probe sets (54,675) on the Affymetrix HG U133 Plus 2 array. The first, second, and third principal components are displayed on the X-, Y- and Z-axes, respectively. (B) The expression signatures of samples were visualized by hierarchical clustering on the 10,000 probe sets with the highest variation coefficient. The colors indicate the relative expression levels of each gene, with red indicating an expression above median, green indicating expression under median and black representing median expression. Cluster (a) was a group of genes differentially overexpressed in oocytes, including DAZL, ZP1, ZP2, ZP3, ZP4, AURKA and HOXA7. Cluster (b) featured genes that were detected in both oocyte and embryo samples, such as the NALP4, DPPA5, and ACTL8. Cluster (c) grouped genes that were detected in both embryo and hESC samples, such as NANOG, SALL4, and ANXA2. Cluster (d) included genes differentially overexpressed in hESCs, such as POU5F1, CD24, SOX2, FZD7 and ZIC3. Cluster (e) assembled genes overexpressed in day-3 embryos, such as CCNA1, H3F3B and FGF9. (C) The dendogram shows that all replicates are clustered by their appropriate stage. All the replicate samples of the hESC group self-cluster into one branch. Both oocyte and embryo samples self-cluster into another branch (dotted box) that further divides into two major sub-branches and into which all the replicates from oocytes and embryos self-cluster.
Figure 3.
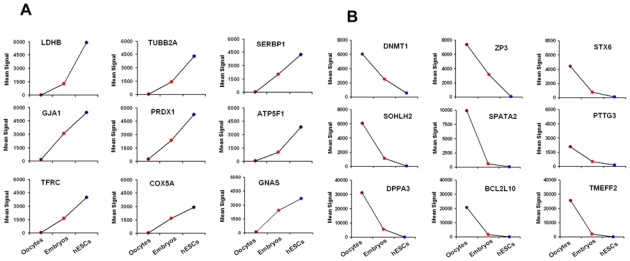
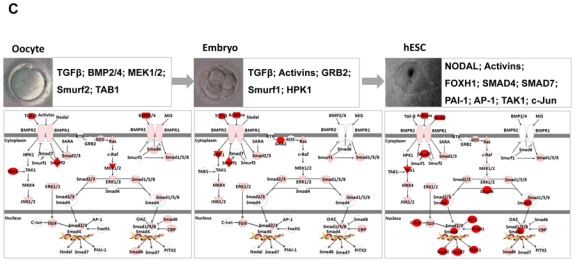
Expression of selected genes in human oocytes, and embryos on day-3 and hESCs. Figure show signal values of 18 genes that are gradually increased (A) or decreased (B) during early embryonic development. (C) Characterization of TGF-β signalling pathway during early embryonic development. Genes shown in red are upregulated in human oocytes or in embryos on day-3 or in hESCs. Examples of genes overexpressed (red) in each stage are indicated in boxes.
DNA damage can have severe cellular consequences, especially in cells which are the origin of all cells of the future human being. Hence, DNA repair systems play a crucial role in early embryonic development. The pathways and strategies for DNA repair in early embryos have been reviewed elsewhere (Jaroudi et al., 2007). The importance of DNA repair gene products during development is highlighted by the phenotypes observed for human genetic disorders associated with DNA damage response (Hales. 2005). Synthesis and repair of DNA, as well as methylation of DNA, is crucial in gametogenesis, fertilization and pregnancy. Recently, DNA repair gene expression was investigated in human oocytes and blastocysts to identify the pathways involved at these stages and to detect potential differences in DNA repair mechanisms of pre- and post-embryonic genome activation (Jaroudi et al., 2009). Large numbers of repair genes were detected indicating that all DNA repair pathways are potentially functional in human oocytes and blastocysts. Expression levels of DNA repair genes at the pre- and post-EGA transcriptional level suggest differences in DNA repair mechanisms at these developmental stages (Jaroudi et al., 2009).
Epigenetic regulation of human early embryonic development
Epigenetics refers to a collection of mechanisms that can cause a change in the phenotype of a cell without altering its DNA sequence (Goldberg et al., 2007). Early embryonic development is regulated epigenetically. During the development process, the epigenetic signature changes as the cell enters fertilization and/or differentiation. DNA methylation is an important epigenetic event, regulating gene expression and chromatin structure in any developmental processes including gene imprinting and embryogenesis (Kafri et al., 1992). To maintain methylated DNA, DNA methyltransferases (DNMTs) are necessary (Vassena et al., 2005). This family of enzymes is divided into two major classes: DNMT1 and DNMT3 (including DNMT3A and DNMT3B). DNMT1 enzyme is the main methyltransferase by far and its exceptional preference for hemimethylated DNA indicates its role in the maintenance of methylated status during DNA replication. Genetic studies have suggested that DNMT1 may play a role in the methylation of repeat elements in growing oocytes (Gaudet et al., 2004). Our genes expression data indicate that the DNMT1 mRNA was overexpressed in mature MII oocytes and decreased progressively in embryos on day-3 and in hESCs (Figure 3B), revealing that the oocyte-specific DNMT1 transcript is expressed in human oogenesis and persists in embryos at day-3. DNMT3B transcripts were detected in mature MII oocytes and hESCs (Assou et al., 2009), and the expression of DNMT3A was enriched in undifferentiated hESCs (Assou et al., 2007; Huntriss et al., 2004; Richards et al., 2004), indicating that the DNMT3 family methyltransferases are candidate epigenetic regulators during preimplantation development. Both DNMT3A and DNMT3B enzymes are involved in de novo methylation processes with different substrate preferences. DNMT3A acts preferentially on unmethylated DNA while DNMT3B can act on both hemimethylated and unmethylated DNA (Vassena et al., 2005). These enzymes are responsible for the establishment of DNA methylation during development (Okano et al., 1999; Watanabe et al., 2002).
As imprinting is closely related to embryogenesis, we analyzed the expression of imprinted genes in oocytes, embryos on day-3 and hESCs. We looked at the expression of forty imprinted human genes (Morison et al., 2001) in our data (the list of imprinted genes is available at http://www.otago.ac.nz/IGC). The analysis of the expression levels of these imprinted genes compared with their expression levels in mature MII oocytes, embryos on day-3, and hESCs reveals their reprogramming time points in humans. Several imprinted genes (see Table II) were expressed to high levels in the oocytes, embryos, and hESCs. The study of the methylome and the epigenome is important because minor defects can lead to serious human diseases. Indeed, several syndromes are associated with imprinting defects during preimplantation development (van der Maarel. 2008; Yamazawa et al., 2008). Beyond transcription and epigenetics, the discovery of hundreds of microRNAs (miRNAs) has defined yet another level of mRNA regulation that could have a major impact on defining the mechanisms controlling early embryonic development (Blakaj et al., 2008; Kedde et al., 2008). These short mRNAs have the ability to target and regulate the translation and stability of target mRNAs, and thus are likely to have profound effects on embryonic gene expression patterns.
Table II.
Imprinted genes abundantly expressed in human oocytes, embryos on day-3 and hESCs
| Oocytes | Embryos (Day-3) | hESCs | |||
|---|---|---|---|---|---|
| paternally | Maternally | paternally | Maternally | paternally | Maternally |
| SNRPN, PLAGL1 | TP73L, GRB10, H19, KCNQ1, CDKN1C, OSBPL5, UBE3, PRIM2A, MEG3 | INPP5F, KCNQ1OT1, ZIM2, PEG3, PON1 | TP73L, GRB10, CPA4, KCNK9, PHLDA2, OSBPL5, UBE3A, ZNF597 | PEG10, SGCE, NDN, SNRPN, MAEGL2, DLK1, PEG3, SANG | H19, GRB10, PPP1R9A, CDKN1C, MEG3, SLC22A18 |
Expression of known components of two pathways in human early embryo development
Known components of two signalling pathways involved in human early embryonic development, (i) the WNT signalling pathway and (ii) the TGF-β superfamily pathway, have been observed.
i. WNT signalling
The WNT gene family consists of numerous conserved glycoproteins that regulate pattern formation during embryogenesis in a wide variety of tissues (Morkel et al., 2003). Limited data is available about the role of the WNT pathway in the reproduction process. It has been shown that this pathway is operational during human and mouse pre-implantation development stages (Adjaye et al., 1999). Furthermore, it has also been reported that activation of the canonical WNT pathway is sufficient to maintain self-renewal of both human and mouse ESCs (Sato et al., 2003). We observed that the main components of the WNT pathway such as DVL2, CTNNB1, CTNNA2, CTNND1, LEF1, CSNK1G3, and DAAM1 were overexpressed in MII oocytes; FZD4, GSK3B, CTNND1, APC, SENP5, SENP8, CSNK2A2, and CSNK1E to be overexpressed in embryos on day-3; FZD3, as well as FZD7, SNP6, CXXC1, AXIN, and SFRP1 in hESCs. These findings imply that the WNT pathway may participate in early embryonic development. On the other hand, we observed that expression of glycogen synthase 3 kinase (GSK-3β) is downregulated in the hESCs, corroborating the reported inactivation of GSK-3β, which leads to the activation of the WNT pathway and the maintaining of the undifferentiated state of ESCs (Sato et al., 2004). In the absence of a WNT signal, the GSK-3β kinase phosphorylates β-catenin and targets it for ubiquitin-mediated destruction. Activation of the WNT pathway by a WNT ligand inhibits GSK-3β activity, resulting in the accumulation of β-catenin. Stable β-catenin can interact with DNA-binding TCF factor, at the site where the complex activates transcription of target genes. Treating ESCs with WNT or inhibiting GSK-3 activity can stimulate ESC self-renewal and support pluripotency (Sato et al., 2004). Recently, two studies (Lluis et al., 2008; Marson et al., 2008) show that WNT-β-catenin signalling stimulates nuclear reprogramming. These studies, using distinct reprogramming methods, offer insights into the mechanisms underlying the acquisition and maintenance of pluripotency.
ii. TGF-β superfamily
TGF-β and its family members, including BMPs, NODAL, and ACTIVINS, have been implicated in the development of oocytes and embryos (Chang et al., 2002). Kocabas et al. (2006) confirmed the presence of TGF-β pathways in human oocytes, previously described only in mice (Mummery. 2001). TGF-β family signals play important roles in the maintenance of self-renewal and pluripotency in both human and mouse ESCs (Watabe et al., 2009). Inhibition of TGF-β/ACTIVIN/NODAL signalling by SB-431542, a chemical inhibitor of the kinases of type 1 receptors for TGF-β/ACTIVIN/NODAL (Inman et al., 2002), resulted in decreased expression of the markers of undifferentiated states (James et al., 2005). Using Ingenuity Software Knowledge Base (Redwood City, CA; http://www.ingenuity.com), we observed the expression of most members of the TGF-β superfamily in human oocytes, embryos on day-3, and hESCs (Figure 3C). However, some components of the TGF-β pathway, such as ACTIVIN/INHIBIN, are absent in oocytes but present in both embryos on day-3 and hESCs, whereas NODAL is present only in hESCs. It was recently shown that human iPSCs rely on ACTIVIN/NODAL signalling to control NANOG expression and thereby maintain pluripotency (Vallier et al., 2009). The transcription factors SMAD4 and SMAD7 are significantly overexpressed in hESCs but are underexpressed in both oocytes and embryos on day-3. In addition, it was shown that mouse embryos deficient in SMAD4 display defective epiblast proliferation and delayed outgrowth of the ICM (Sirard et al., 1998). Expression of the signalling receptors for TGF-β in human oocytes and embryo at blastocysts stage, suggests a role for TGF-β in early pre-implantation development.
The expression program of human embryonic stem cells
hESCs do not exist in nature but are derived from the inner cell mass (ICM) of the human blastocyst (Thomson et al., 1998) and are considered equivalent to ICM cells, having captured the phenotype of a cell type that exists only transiently in vivo. hESCs have the capacity to differentiate into the derivatives of all three germ layers: endoderm, mesoderm and ectoderm (Carpenter et al., 2003; Lu et al., 2007); thus hESCs have great potential in the field of regenerative medicine. Although artificial, hESCs represent an excellent model for investigating fundamental aspects of the early embryo development and the pluripotency. Numerous genomic studies have used microarray techniques to capture a detailed view of ESC gene expression (Abeyta et al., 2004; Assou et al., 2007; Beqqali et al., 2006; Bhattacharya et al., 2004; Cowan et al., 2005; Forsyth et al., 2008; Golan-Mashiach et al., 2005). These transcriptome data can generate molecular signatures of ESCs that define the individual components, and the pathways, that regulate pluripotency and maintain the undifferentiated state. We performed a meta-analysis of 38 papers on the topic (Assou et al., 2007). We determined that 1,076 genes were found to be upregulated in hESCs by at least three studies when compared to differentiated cell types. In fact, only a single gene, OCT3/4, was found to be in common to all 38 lists of hESC-specific genes. Our analysis shows that gene expression analyses are sensitive to experimental conditions, therefore requiring extreme caution during data interpretation. A cross-comparison of the gene lists generated by these efforts shows that OCT3/4, NANOG, SOX2, DNMT3B, LIN28, and CD24 are commonly expressed in the majority of hESCs (Assou et al., 2007). A “core” set of transcription factors, OCT3/4, SOX2 and NANOG, is common between the hESC lines and is considered to be a general criteria for hESC characterization and pluripotency (Hoffman et al., 2005; Zaehres et al., 2005). A list of OCT3/4, SOX2, and NANOG target genes has been determined by cross-species genomic analysis followed by chromatin immunoprecipitation and gene expression analysis (Boyer et al., 2005). Genes that are activated by OCT3/4, SOX2, and NANOG in ESCs are mostly upregulated and important for ESC self-renewal. Cross-species comparisons between human and mouse ESC transcriptomes revealed both distinct and similar features (Rao. 2004; Sato et al., 2003; Wei et al., 2005). Wei et al. (2005) compared gene expression profiles between hESC and mESC, and showed that LIFR was overexpressed in mice, but not in humans; FGF2 was overexpressed in humans, while FGF4 was abundant in mice. Epiblast derived mouse ESCs also maintain self-renewal by the dependence on FGF2, hence activated FGF signalling (Brons et al., 2007; Tesar et al., 2007). The accumulating ESC transcriptome data is now being aimed at identifying molecular markers that will be useful for quality control of ESCs for clinical applications.
Common molecular signatures between human metaphase II oocytes and ICM
ESCs and mature MII oocytes are able to reprogram the nuclei of fully differentiated human somatic cells and render these cells pluripotent (Cowan et al., 2005; Rideout et al., 2001). Through the comparative analysis of the transcriptome of such cells, it may be possible to achieve a better understanding of the mechanism of nuclear reprogramming, the molecular events which occur during the first week of human embryo development, and the identification of the genes involved in pluripotency initiation (Kocabas et al., 2006; Zhang et al., 2007; Assou et al., 2009). Kocabas et al. (2006) compared the transcriptome of human MII oocytes and hESCs with a reference sample consisting of a mixture of total RNA from different normal somatic cells. In this study, the authors identified 1,626 transcripts that were significantly upregulated in the hESs relative to somatic cells. Intersection of these 1,626 transcripts enriched in hESCs with the 5,331 transcripts enriched in human MII oocytes, identified 388 common genes that are hypothesized to play an important role in oocyte reprogramming and early events associated with human early embryo development (Kocabas et al., 2006). Zhang et al. (2007) obtained a large amount of information on gene expression in human oocytes at GV stage as compared with hESCs. In total, 1,629 genes were upregulated in GV oocytes when compared with hESCs and human foreskin fibroblasts (hFIBs). This list of genes, including MATER, ZAR1, NPM2, FIGLA, some imprinted genes, and some components of four signalling pathways such as MOS-MPF, NOTCH, WNT and TGF-beta (Zhang et al., 2007), may provide good candidate genes for understanding the oocyte maturation and the early embryonic development. On the other hand, Assou et al. (2009) compared the gene expression profiles of mature MII oocytes with hESCs, and then both to somatic tissues. There was a common oocyte/hESC gene expression profile, which included genes associated with pluripotency, such as LIN28 and TDGF1, and a large chromatin remodeling network (TOP2A, DNMT3B, JARID2, SMARCA5, CBX1, CBX5). Interestingly, a large set of genes was also found to code for proteins involved in the ubiquitination and proteasome pathway (Assou et al., 2009). This list provides reliable candidate genes for future studies on molecular mechanisms of pluripotency and nuclear reprograming. The zinc finger motif is a DNA binding domain dependent on a zinc ion frequently found in transcription factors. In seeking to identify genes encoding zinc finger domain proteins expressed in both mature MII oocytes and hESCs, we observed that the distribution of genes encoding these proteins on the genome is not random: they are significantly enriched in chromosomes 19 and 20 in mature MII oocytes, and in 16 and 17 in hESCs (Assou et al., 2009). As can be seen in Figure 4, seven zinc finger proteins were common to mature MII oocytes and hESCs, three were found in oocytes only (e.g. ZNF528, ZNF468, ZNF313), and three in hESCs only (e.g. ZNF268, ZNF232, ZNF423).
Figure 4.
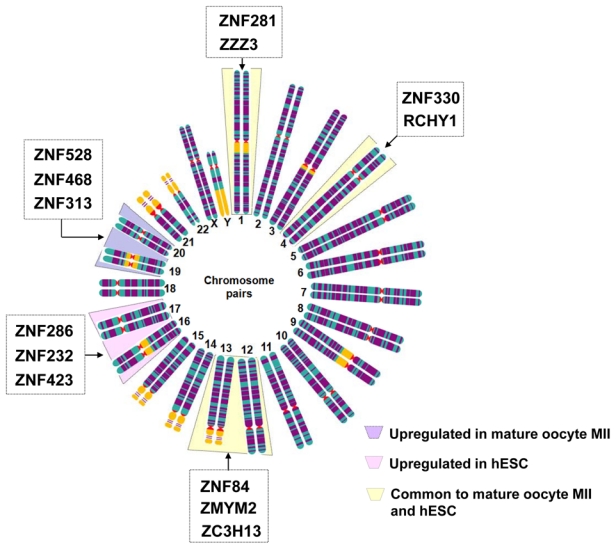
Chromosomal distribution of genes encoding zinc finger domain proteins were expressed significantly differently in human mature oocytes (light violet) as compared to hESCs (light pink) as well as to those commonly expressed in mature human oocytes and hESCs (light yellow). The selected genes were retrieved in lists previously published in (Assou et al., 2009).
Human early embryo development transcriptome data visualization through an internet interface
Amazonia! (http://www.amazonia.transcriptome.eu) is a freely accessible web site dedicated to the visualization of large, publicly available human transcriptome data, including data on stem cells, human embryo and germinal cells, for facilitating the free exchange of information on genomics. Data is visualized as expression bar plots with a color code enabling the recognition of cell type (Assou et al., 2007; Le Carrour et al., 2010). Genes are accessed either by keywords or through lists. Most interestingly, when data is obtained using the same platform format, sample labeling, and data normalization, it is possible to combine different experiments in a single virtual experiment. Amazonia! generate bar plots that combine expression in oocytes, embryos and hESCs with more than 200 germinal and somatic tissues.
Omics: tools for the identification of reliable biomarkers for oocyte and embryo selection
Since the birth of Louise Brown, the first baby conceived by IVF in the UK in 1978, more than 2 million children in the world have been born as a result of assisted reproductive technology (ART). Since then, these techniques have advanced considerably. ART protocols continue to evolve with the aim of achieving higher pregnancy rates, reducing multiple pregnancies and obtaining healthier babies. However, despite the incontestable advances, the pregnancy rate is still relatively low and has not increased significantly in the last decade (Andersen et al., 2007). The selection of embryos with higher implantation potential is one of the major challenges in ART. Initially, multiple embryo transfer was used to maximize pregnancy rates. However, improved embryo quality and rising multiple pregnancy rates has resulted in the decrease in number of embryos for replacement. Therefore, selection of the “best” embryo has become crucial, particularly with elective SET being strongly recommended. There is therefore a need to develop new objective approaches for embryo selection. The classical methods to select healthy embryos under IVF and ICSI conditions are based on morphological criteria such as early embryonic cleavage, the number and size of blastomeres, fragmentation degree, and the presence of multi-nucleation at the 4- or 8-cell stages (Fenwick et al., 2002). However, most studies suggest, that embryos with proper morphological appearance alone are not sufficient to predict a successful implantation. Beyond the criteria of embryo selection, defining oocyte quality remains one of the most difficult challenges (Wang, Q. et al., 2007). Many morphologically normal embryos do not achieve implantation or spontaneously abort during pregnancy. Considering the limitation of morphologic evaluation and cytogenetic screening methods, there is now a movement towards more sophisticated, high-performance technologies and the emerging “omics” science, such as genomics, transcriptomics, proteomics, and metabolomics (Hillier. 2008). These approaches may contribute to the design of a non-invasive viability assay to assist in the embryos selection in IVF or ICSI programs. It is necessary to distinguish the invasive approaches, which are based on direct analysis of the oocyte and embryo, from non-invasive approaches (Figure 5). Can omics approaches help shape the future of ART?
Figure 5.
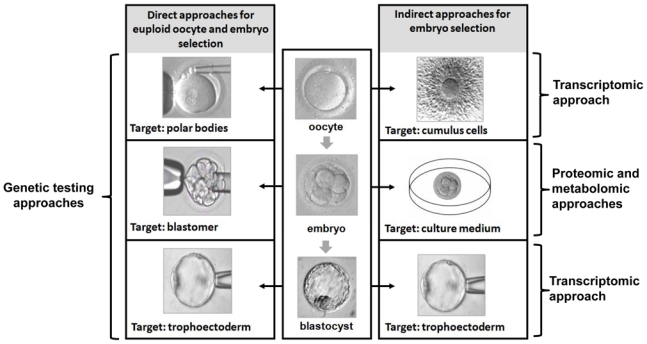
Different direct or indirect approaches suggested for oocyte or embryo selection (transcriptomic, proteomic and metabolomic approaches).
Transcriptomic approach
Several investigations using reverse transcription followed by real-time PCR have found that specific genes display alterations in activity apparently related to oocyte or embryo quality (Dode et al., 2006; Fair et al., 2004). Despite the accuracy and sensitivity of real-time PCR, it is still limited by the restricted number of genes that can be assessed per single oocyte or embryo. Microarray technology permits the simultaneous analysis of tens of thousands of genes, and provides greater hope for the identification of new biomarkers. Transcriptomics represent a valuable approach to biomarkers development. Both technical and statistical advances are currently facilitating the integration of this approach to IVF programs. The transcriptomic approach has been used experimentally in humans on throphoblastic biopsies of blastocysts (Jones et al., 2008a). Analysis of the transcriptome of aneuploid and normal oocytes, as determined by comparative genomic hybridization (CGH) analysis of the corresponding polar bodies, has identified several differentially expressed genes with roles in chromosome segregation during meiosis, including genes affecting cell cycle checkpoints, spindle dynamics, and chromosomal movement (Fragouli et al., 2006b; Fragouli et al., 2006a; Fragouli et al., 2009; Gutierrez-Mateo et al., 2004). An indirect and attractive approach for predicting embryo and pregnancy outcomes has been recently reported by using transcriptomic data of CCs gene expression (Assou et al., 2008). The CCs are abundant and easily accessible, which makes them an ideal material to use for the potential assessment of oocyte and embryo quality. Data concerning biomarker expression in these cells should yield more precise information, improving the embryologists’ capacity to select competent embryos either for fresh replacement or for embryo freezing. By studying genes expression profile of CCs or GCs using microarrays or reverse transcription-polymerase chain reaction (RT-PCR) and quantitative RT-PCR some studies have identified candidate biomarkers for (i) oocyte quality and competence (Zhang et al., 2005; Cillo et al., 2007; Hamel et al., 2008; Adriaenssens et al., 2010), (ii) early embryo development (Anderson et al., 2009; McKenzie et al., 2004; van Montfoort et al., 2008) and (iii) embryo quality and pregnancy outcome (Assou et al., 2008; Hamel et al., 2010) (Table III). Zhang et al. (2005) reported that the expression of pentraxin-3 (PTX3) in CCs was positively correlated with oocyte competence. PTX3 was initially studied for its possible role in inflammatory reaction processes (Gewurz et al., 1995). Fertilization in vivo is very low in PTX3 knoukout mice (Varani et al., 2002). Feuerstein et al (2007) reported that a number of genes including prostaglandin-endoperoxide syntase-2 (PTGS2) and steroidogenic acute regulatory protein (STAR) were also negatively associated with oocyte competence. In addition, the expression of gremlin 1 (GREM1) is also indicative of embryo quality (McKenzie et al., 2004; Cillo et al., 2007; Anderson et al., 2009, Adriaenssens et al., 2010). GREM1 is an antagonist of BMPs and TGF-β signalling (Hsu et al., 1998). It has been proposed that GREM1 regulates the GDF9 and BMP15 balance (Pangas et al., 2004). By using microarrays, our group reported a specific transcriptomic signature in CCs including 630 genes correlated with embryo and pregnancy outcomes. Among this gene list, we identified BCL2L11, PCK1, and NFIB (Assou et al., 2008). BCL2L11 is involved in triggering cell death in response to abnormalities, PCK1 is associated with energy production, and NFIB regulates some of the earliest processes in embryonic development. Other studies associated the gene expression profile in follicular cells (CCs or GCs) with pregnancy (Hamel et al., 2010). They found that variation of PGK1, a transferase enzyme involved in the process of glycolysis and regulation of G-protein signalling 2 (RGS2) in follicular cells are likely candidate genes that could have a utility in the prediction of pregnancy. All these studies provide supporting evidence to the concept that CCs are a promising source of reliable biomarkers for predicting oocyte quality and/or embryo and/or pregnancy outcome (Table III). These efforts will ultimately lead to improved efficiency outcomes of ART.
Table III.
Potential follicular cells biomarkers correlated to oocyte quality, embryo competence or pregnancy outcome. CCs: cumulus cells, GCs: granulosa cells
| Biomarkers | Name | Function | Samples (individual or pooled) | Approches | Outcome | Reference |
|---|---|---|---|---|---|---|
| PTX3 | Pentraxin-3 | involved in inflammatory reaction processes and acts to stabilize the expanded CCs | Pooled CCs from eggs | Microarrays (4 chips) | Positively correlated with oocyte competence | (Zhang, X. et al., 2005) |
| PCK1, BCL2L11, NFIB | Phosphoenolpyru vate carboxykinase 1, BCL2-like 11 (apoptosis facilitator), Nuclear factor I/B | PCK1: associated with energy production BCL2L11: involved in triggering cell death in response to abnormalities NFIB: regulates some of the earliest processes in embryonic development |
CCs from individual eggs | Microarrays (50 chips) | Predict embryo and pregnancy outcomes | (Assou et al., 2008) |
| CYP19A1, SERPINE2, CDC42, FDX1 | Cytochrome P450, family 19, subfamily A, polypeptide 1, Serpin peptidase inhibitor clade E member 2, Cell division cycle 42, Ferredoxin 1 | CYP19A1: metabolizes androgen into estradiol- 17β SERPINE2: involved in apoptosis and chromatin condensation CDC42: involved in apoptosis, transcriptional activation, cell proliferation and cell polarity FDX1 and HSD3B1: responsible for progesterone synthesis |
Mural GCs and CCs (pooled) | Microarrays (4 chips) | Positively associated with pregnancy | (Hamel et al., 2008) |
| CCND2, CXCR4, GPX3, HSPB1, DVL3, DHCR7, CTNND1, TRIM28 | Cyclin D2, Chemokine (C-X- C motif) receptor 4, Glutathione peroxidase 3, Heat shock protein 1, Dishevelled, dsh homolog 3 7- dehydrocholestero l reductase, Catenin, delta 1, Tripartite motif- containing 28 | CCND2: plays an role in proliferation of cells (cell cycle regulator) CXCR4 and GPX3: have a role in relieve hypoxic stress HSPB1: act as co- repressor of estrogen signaling DVL3: plays a role in angiogenesis DHCR7: involved in progesterone and estrogen synthesis CTNND1: has a role in adhesion TRIM28: has a role in DNA repair |
CCs from individual eggs | Microarrays (16 chips) | Negatively associated with oocyte competence | (van Montfoort et al., 2008) |
| HAS2, GREM1 | Hyaluronan synthase 2, Gremlin 1 | HAS2: involved in extracellular matrix (ECM) formation GREM1: antagonist of BMP and TGF-β |
CCs from individual eggs | RT-PCR | Positively associated with oocyte developmental competence | (Cillo et al., 2007) |
| STAR, AREG, Cx43, PTGS2, SCD1 and SCD5 | Steroidogenic acute regulatory protein, Amphiregulin, Connexin 43, Prostaglandin- endoperoxide synthase 2 Stearoyl-CoA desaturase 1 and 5 | STAR: regulated the cholesterol transport into the inner mitochondrial AREG: act as mediators of LH Cx43: permit the transfer of metabolits for growth and development and maintenance of meiotic arrest of the oocyte PTGS2: involved in inflammation and mitogenesis SCD: involved in biosynthesis of monounsaturated fatty acids from saturated acids |
CCs from individual eggs | RT-PCR | Negatively associated with oocyte competence | (Feuerstein et al., 2007) |
| HAS2, PTGS2, GREM1 | Hyaluronan synthase 2, Prostaglandin- endoperoxide synthase 2, Gremlin 1 | CCs from individual eggs | Quantitative RT-PCR | Positively associated with Oocyte competence and embryo development | (McKenzie et al., 2004) | |
| BDNF, GREM1 | Brain-derived neurotrophic factor, Gremlin 1 | BDNF: neurotrophic factor playing a role in regulation of stress response | CCs from individual eggs | Quantitative RT-PCR | negative and positive predictors of embryo quality respectively | Anderson et al., 2009) |
| PGK1, RGS2 and RGS3, CDC42 | Phosphoglycerate kinase 1, Regulator of G- protein signaling 2 and 3, Cell division cycle 42 | PGK1: involved in glycolysis RGS: hydrolyzed GTP to GDP |
Mural GCs and CCs (individual) | Quantitative RT-PCR | Associated with pregnancy | Hamel et al., 2010) |
| VCAN, RPS6KA2, ALCAM, GREM1 | Versican, Ribosomal protein S6 kinase polypeptide 2, Activated leukocyte cell adhesion molecule, Gremlin 1 | VCAN: plays a central role in tissue morphogenesis and maintenance RPSK6KA2: involved in the EGF signaling cascade ALCAM: involved in immune response |
Pooled CCs from eggs | Quantitative RT-PCR | Correlated with oocyte maturity, low fragmentation and embryo development | Adriaenssen s et al., 2010) |
Proteomic approach
Proteomics describes changes in the expression of proteins translated from a single genome. The human proteome, estimated at over a million proteins, is highly diverse and dynamic. The analysis of the protein composition of the embryos poses a particular challenge because of the low concentration and wide dynamic range of the population of molecules which need to be analyzed; unlike mRNA, proteins cannot be amplified for analysis. Recent advances in mass spectrometry have led to the development of methods sensitive enough to allow the examination of the secretome of single oocytes or embryos. The secretome has been defined as the subset of secreted or consumed proteins that can be found in the environment where the oocyte or embryo grows. It is now possible to obtain comprehensive protein profiles from limited amounts of complex biological fluids including IVF spent culture mediums. The secretome of an individual human embryo has been previously analyzed by surface-enhanced laser desorption and ionization time-of-flight (SELDI-TOF) mass spectrometry (MS) technology, and distinctive secretome signatures were observed at each embryonic developmental stage from fertilization to the blastocyst stage (Katz-Jaffe et al., 2006; Katz- Jaffe et al., 2009). However, no clear biomarkers of embryo viability and implantation have yet been reported. Dominguez et al. (2010) compared blastocyst conditioned media with control medium using protein microarrays, thus revealing increased expression of interleukin 10 (IL-10) and soluble tumor necrosis factor (TNF) receptor 1 (TNFR1) and decreased expression of stem cell factor (SCF) and chemokine ligand 13 (CXCL13) (Dominguez et al., 2010). Other studies have reported a link between the level of cytokines in human follicular fluid (hFF) and the implantation potential of the embryo derived from the oocyte of this follicle. Using a bead-based multiplex sandwich immunoassay (Luminex), Ledee et al. (2008) demonstrated that only granulocyte colony-stimulating factors (G-CSF) measured in individual hFF samples together with 26 other cytokines was particularly elevated in the hFF corresponding to the embryos with highest implantation potential (Ledee et al., 2008). In addition, significantly higher levels of interleukin 10 (IL10) and interferon (IFN-γ) were found in hFF from oocytes that generated early cleavage embryos. However, Luminex technology is complex and expensive. Further studies will help to confirm the advantageous applications of proteomics testing in the field of IVF.
Metabolomic approach
Metabolomics aims at the comprehensive and quantitative analysis of metabolites in biological samples. This approach is presently being applied to human embryos (Botros et al., 2008; Seli et al., 2008, 2010) and oocytes (Singh et al., 2007). Because of the chemical diversity of cellular metabolites, no single analytical platform can measure the metabolome in a cell. This is due to the absence of methods to amplify metabolites, the labile nature of many metabolites, and their chemical heterogeneity and complexity. Thus, measuring the metabolome is a considerable analytical challenge. Rapid technological advances in MS and nuclear magnetic resonance (NMR) have enabled the simultaneous detection of a wide range of small molecules. Recently, the development of a screening technology using Raman and near infrared (NIR) spectroscopy were used to detect biomarkers in culture medium with algorithms generated for positive, as compared to negative, IVF outcomes (Scott et al., 2008; Seli et al., 2007). NIR analysis is not typically used for target metabolite identification, but is used for overall spectral profile comparisons. Recent studies using a SET program on days-2 and -3 showed higher mean viability scores for embryos that resulted in a pregnancy with fetal heart activity, compared with those without (Seli et al., 2008; Vergouw et al., 2008). They showed that metabolomic profiling of culture media from embryos was independent of morphology. Large-scale, prospective studies are necessary to relate the metabolic profile of medium to the developmental potential of human oocytes and embryos. In addition, the ideal metabolomic profile should be confirmed by several independent platforms.
Conclusion
This review highlights the recent advances in elucidating the molecular complexity in early embryonic development. The knowledge of the global pattern of gene expression is important for understanding critical regulatory pathways involved in the crucial steps of early embryonic development. Certain gene expression profiles help to unravel the mystery of these developmental stages. Further understanding of the biological role of these genes may expand our knowledge of the oocyte maturation, fertilization, chromatin remodeling, totipotency, pluripotency, and early morphogenesis steps. The practical implications of compiling gene expression information on human oocytes and embryos would be enormous since it could potentially help us to understand and solve problems related to infertility.
In recent years, the emergence of new technologies ‘Omics’ such as microarrays have already started to be used in the IVF/ICSI program. The use of microarray technology in the analysis of early embryonic development poses specific challenges associated with the picogram levels of mRNA in a single cell/embryo. Another challenge for applying microarrays to early embryonic development is the high degree of expression plasticity seen in early stage embryos.
The ideal DNA microarrays would potentially give rise to diagnostics for assessing human embryo quality in IVF program. The application of transcriptomic, proteomic and metabolomic approaches have greatly broadened our understanding of early human development. The challenge now is to correlate gene/protein/metabolite function and regulation to specific events in early embryonic development. These tools may ultimately lead to non-invasive tests for oocyte or embryo quality revealing previously hidden information concerning both oocyte and embryonic developmental competence. Once fully validated, these new approaches are expected to improve oocyte and embryo selection, leading to increased implantation rates and higher success in elective SET. A multitude of advantages may ensue from the use of a rapid, non-invasive and reliable technology as an adjunct to embryo assessment for clinical applications. An improved understanding of embryo viability should help to identify the healthy embryos that will most likely result in pregnancy and allow more accurate decisions in selecting the best embryo for the SET program.
Acknowledgments
Funding
We thank the direction of the University-Hospital of Montpellier, the Association Française contre les Myopathies (AFM), Vitrolife, Genevrier, and Ferring Pharmaceutical Companies for their support.
References
- Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RA, Firpo MT. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–8. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- Adjaye J, Bolton V, Monk M. Developmental expression of specific genes detected in high-quality cDNA libraries from single human preimplantation embryos. Gene. 1999;237:373–83. doi: 10.1016/s0378-1119(99)00329-7. [DOI] [PubMed] [Google Scholar]
- Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, et al. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. 2005;23:1514–25. doi: 10.1634/stemcells.2005-0113. [DOI] [PubMed] [Google Scholar]
- Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, Coucke W, Devroey P, Smitz J. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod. 2010;25:1259–70. doi: 10.1093/humrep/deq049. [DOI] [PubMed] [Google Scholar]
- Andersen AN, Goossens V, Gianaroli L, Felberbaum R, de Mouzon J, Nygren KG. Assisted reproductive technology in Europe, 2003. Results generated from European registers by ESHRE. Hum Reprod. 2007;22:1513–25. doi: 10.1093/humrep/dem053. [DOI] [PubMed] [Google Scholar]
- Anderson RA, Sciorio R, Kinnell H, Bayne RA, Thong KJ, de Sousa PA, Pickering S. Cumulus gene expression as a predictor of human oocyte fertilisation, embryo development and competence to establish a pregnancy. Reproduction. 2009;138:629–37. doi: 10.1530/REP-09-0144. [DOI] [PubMed] [Google Scholar]
- Assou S, Cerecedo D, Tondeur S, Pantesco V, Hovatta O, Klein B, Hamamah S, De Vos J. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Anahory T, Pantesco V, Le Carrour T, Pellestor F, Klein B, Reyftmann L, Dechaud H, De Vos J, Hamamah S. The human cumulus--oocyte complex gene-expression profile. Hum Reprod. 2006;21:1705–19. doi: 10.1093/humrep/del065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Haouzi D, Mahmoud K, Aouacheria A, Guillemin Y, Pantesco V, Reme T, Dechaud H, De Vos J, Hamamah S. A non-invasive test for assessing embryo potential by gene expression profiles of human cumulus cells: a proof of concept study. Mol Hum Reprod. 2008;14:711–9. doi: 10.1093/molehr/gan067. [DOI] [PubMed] [Google Scholar]
- Assou S, Le Carrour T, Tondeur S, Strom S, Gabelle A, Marty S, Nadal L, Pantesco V, Reme T, Hugnot JP, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–73. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beqqali A, Kloots J, Ward-van Oostwaard D, Mummery C, Passier R. Genome-wide transcriptional profiling of human embryonic stem cells differentiating to cardiomyocytes. Stem Cells. 2006;24:1956–67. doi: 10.1634/stemcells.2006-0054. [DOI] [PubMed] [Google Scholar]
- Bermudez MG, Wells D, Malter H, Munne S, Cohen J, Steuerwald NM. Expression profiles of individual human oocytes using microarray technology. Reprod Biomed Online. 2004;8:325–37. doi: 10.1016/s1472-6483(10)60913-3. [DOI] [PubMed] [Google Scholar]
- Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, Yang AX, Joshi BH, Ginis I, Thies RS, Amit M, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–64. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- Blakaj A, Lin H. Piecing together the mosaic of early mammalian development through microRNAs. J Biol Chem. 2008;283:9505–8. doi: 10.1074/jbc.R800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botros L, Sakkas D, Seli E. Metabolomics and its application for non-invasive embryo assessment in IVF. Mol Hum Reprod. 2008;14:679–90. doi: 10.1093/molehr/gan066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332:459–61. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Carpenter MK, Rosler E, Rao MS. Characterization and differentiation of human embryonic stem cells. Cloning Stem Cells. 2003;5:79–88. doi: 10.1089/153623003321512193. [DOI] [PubMed] [Google Scholar]
- Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor790 beta superfamily. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen JJ, Yu SL, Li HN, Yang PC, Su CM, Au HK, Chang CW, Chien LW, Chen CS, et al. Transcriptome analysis in blastocyst hatching by cDNA microarray. Hum Reprod. 2005;20:2492–501. doi: 10.1093/humrep/dei084. [DOI] [PubMed] [Google Scholar]
- Cillo F, Brevini TA, Antonini S, Paffoni A, Ragni G, Gandolfi F. Association between human oocyte developmental competence and expression levels of some cumulus genes. Reproduction. 2007;134:645–50. doi: 10.1530/REP-07-0182. [DOI] [PubMed] [Google Scholar]
- Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–73. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- Cui XS, Li XY, Yin XJ, Kong IK, Kang JJ, Kim NH. Maternal gene transcription in mouse oocytes: genes implicated in oocyte maturation and fertilization. J Reprod Dev. 2007;53:405–18. doi: 10.1262/jrd.18113. [DOI] [PubMed] [Google Scholar]
- Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RA. The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet. 2004;13:1461–70. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- Dode MA, Dufort I, Massicotte L, Sirard MA. Quantitative expression of candidate genes for developmental competence in bovine two-cell embryos. Mol Reprod Dev. 2006;73:288–97. doi: 10.1002/mrd.20427. [DOI] [PubMed] [Google Scholar]
- Dominguez F, Gadea B, Mercader A, Esteban FJ, Pellicer A, Simon C. Embryologic outcome and secretome profile of implanted blastocysts obtained after coculture in human endometrial epithelial cells versus the sequential system. Fertil Steril. 2010;93:774–782. doi: 10.1016/j.fertnstert.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–5. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- Dragovic RA, Ritter LJ, Schulz SJ, Amato F, Thompson JG, Armstrong DT, Gilchrist RB. Oocyte-secreted factor activation of SMAD 2/3 signaling enables initiation of mouse cumulus cell expansion. Biol Reprod. 2007;76:848–57. doi: 10.1095/biolreprod.106.057471. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin JA, Clark AT, Wang P, Wolfman NM, Matzuk MM. Paracrine actions of growth differentiation factor-9 in the mammalian ovary. Mol Endocrinol. 1999;13:1035–48. doi: 10.1210/mend.13.6.0310. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. The relationship between cumulus cell-oocyte coupling, oocyte meiotic maturation, and cumulus expansion. Dev Biol. 1982;89:268–72. doi: 10.1016/0012-1606(82)90314-1. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. Intercommunication between mammalian oocytes and companion somatic cells. Bioessays. 1991;13:569–74. doi: 10.1002/bies.950131105. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Chesnel F, Hirao Y, O’Brien MJ, Pendola FL, Watanabe S, Wigglesworth K. Oocyte control of granulosa cell development: how and why. Hum Reprod. 1997;12:127–32. [PubMed] [Google Scholar]
- Eppig J. Mouse oocytes control metabolic co-operativity between oocytes and cumulus cells. Reprod Fertil Dev. 2005;17:1–2. doi: 10.1071/rdv17n2paperabs. [DOI] [PubMed] [Google Scholar]
- Fabbro M, Schuechner S, Au WW, Henderson BR. BARD1 regulates BRCA1 apoptotic function by a mechanism involving nuclear retention. Exp Cell Res. 2004;298:661–73. doi: 10.1016/j.yexcr.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Fair T, Murphy M, Rizos D, Moss C, Martin F, Boland MP, Lonergan P. Analysis of differential maternal mRNA expression in developmentally competent and incompetent bovine two-cell embryos. Mol Reprod Dev. 2004;67:136–44. doi: 10.1002/mrd.10385. [DOI] [PubMed] [Google Scholar]
- Fenwick J, Platteau P, Murdoch AP, Herbert M. Time from insemination to first cleavage predicts developmental competence of human preimplantation embryos in vitro. Hum Reprod. 2002;17:407–12. doi: 10.1093/humrep/17.2.407. [DOI] [PubMed] [Google Scholar]
- Feuerstein P, Cadoret V, Dalbies-Tran R, Guerif F, Bidault R, Royere D. Gene expression in human cumulus cells: one approach to oocyte competence. Hum Reprod. 2007;22:3069–77. doi: 10.1093/humrep/dem336. [DOI] [PubMed] [Google Scholar]
- Forsyth NR, Kay A, Hampson K, Downing A, Talbot R, McWhir J. Transcriptome alterations due to physiological normoxic (2% O2) culture of human embryonic stem cells. Regen Med. 2008;3:817–33. doi: 10.2217/17460751.3.6.817. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Wells D, Whalley KM, Mills JA, Faed MJ, Delhanty JD. Increased susceptibility to maternal aneuploidy demonstrated by comparative genomic hybridization analysis of human MII oocytes and first polar bodies. Cytogenet Genome Res. 2006a;114:30–8. doi: 10.1159/000091925. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Wells D, Thornhill A, Serhal P, Faed MJ, Harper JC, Delhanty JD. Comparative genomic hybridization analysis of human oocytes and polar bodies. Hum Reprod. 2006b;21:2319–28. doi: 10.1093/humrep/del157. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Escalona A, Gutierrez-Mateo C, Tormasi S, Alfarawati S, Sepulveda S, Noriega L, Garcia J, Wells D, Munne S. Comparative genomic hybridization of oocytes and first polar bodies from young donors. Reprod Biomed Online. 2009;19:228–37. doi: 10.1016/s1472-6483(10)60078-8. [DOI] [PubMed] [Google Scholar]
- Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta TS, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet. 2000;25:279–83. doi: 10.1038/77033. [DOI] [PubMed] [Google Scholar]
- Gasca S, Pellestor F, Assou S, Loup V, Anahory T, Dechaud H, De Vos J, Hamamah S. Identifying new human oocyte marker genes: a microarray approach. Reprod Biomed Online. 2007;14:175–83. doi: 10.1016/s1472-6483(10)60785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasca S, Reyftmann L, Pellestor F, Reme T, Assou S, Anahory T, Dechaud H, Klein B, De Vos J, Hamamah S. Total fertilization failure and molecular abnormalities in metaphase II oocytes. Reprod Biomed Online. 2008;17:772–81. doi: 10.1016/s1472-6483(10)60404-x. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Rideout WM, 3rd, Meissner A, Dausman J, Leonhardt H, Jaenisch R. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol Cell Biol. 2004;24:1640–8. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewurz H, Zhang XH, Lint TF. Structure and function of the pentraxins. Curr Opin Immunol. 1995;7:54–64. doi: 10.1016/0952-7915(95)80029-8. [DOI] [PubMed] [Google Scholar]
- Giritharan G, Talbi S, Donjacour A, Di Sebastiano F, Dobson AT, Rinaudo PF. Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction. 2007;134:63–72. doi: 10.1530/REP-06-0247. [DOI] [PubMed] [Google Scholar]
- Golan-Mashiach M, Dazard JE, Gerecht-Nir S, Amariglio N, Fisher T, Jacob-Hirsch J, Bielorai B, Osenberg S, Barad O, Getz G, et al. Design principle of gene expression used by human stem cells: implication for pluripotency. Faseb J. 2005;19:147–9. doi: 10.1096/fj.04-2417fje. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Grondahl ML, Yding Andersen C, Bogstad J, Nielsen FC, Meinertz H, Borup R. Gene expression profiles of single human mature oocytes in relation to age. Hum Reprod. 2010;25:957–68. doi: 10.1093/humrep/deq014. [DOI] [PubMed] [Google Scholar]
- Guillemin Y, Lalle P, Gillet G, Guerin JF, Hamamah S, Aouacheria A. Oocytes and early embryos selectively express the survival factor BCL2L10. J Mol Med. 2009;87:923–40. doi: 10.1007/s00109-009-0495-7. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Mateo C, Wells D, Benet J, Sanchez-Garcia JF, Bermudez MG, Belil I, Egozcue J, Munne S, Navarro J. Reliability of comparative genomic hybridization to detect chromosome abnormalities in first polar bodies and metaphase II oocytes. Hum Reprod. 2004;19:2118–25. doi: 10.1093/humrep/deh367. [DOI] [PubMed] [Google Scholar]
- Hales BF. DNA repair disorders causing malformations. Curr Opin Genet Dev. 2005;15:234–40. doi: 10.1016/j.gde.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004a;6:117–31. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MS. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet. 2004b;13:2263–78. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Ko M, Yamada M, Kuji N, Mizusawa Y, Shoji M, Hada T, Asada H, Maruyama T, Yoshimura Y. Global gene expression profiling of preimplantation embryos. Hum Cell. 2006;19:98–117. doi: 10.1111/j.1749-0774.2006.00018.x. [DOI] [PubMed] [Google Scholar]
- Hamel M, Dufort I, Robert C, Gravel C, Leveille MC, Leader A, Sirard MA. Identification of differentially expressed markers in human follicular cells associated with competent oocytes. Hum Reprod. 2008;23:1118–27. doi: 10.1093/humrep/den048. [DOI] [PubMed] [Google Scholar]
- Hamel M, Dufort I, Robert C, Leveille MC, Leader A, Sirard MA. Genomic assessment of follicular marker genes as pregnancy predictors for human IVF. Mol Hum Reprod. 2010;16:87–96. doi: 10.1093/molehr/gap079. [DOI] [PubMed] [Google Scholar]
- Henderson BR. Regulation of BRCA1, BRCA2 and BARD1 intracellular trafficking. Bioessays. 2005;27:884–93. doi: 10.1002/bies.20277. [DOI] [PubMed] [Google Scholar]
- Hillier SG. Research challenge: what is the best non-invasive test of oocyte/embryo competence? Mol Hum Reprod. 2008;14:665. doi: 10.1093/molehr/gan068. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Blelloch R, Brennan C, Yamada Y, Kim M, Chin L, Jaenisch R. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18:1875–85. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23:699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- Hsu DR, Economides AN, Wang X, Eimon PM, Harland RM. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol Cell. 1998;1:673–83. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- Huntriss J, Hinkins M, Oliver B, Harris SE, Beazley JC, Rutherford AJ, Gosden RG, Lanzendorf SE, Picton HM. Expression of mRNAs for DNA methyltransferases and methyl-CpG-binding proteins in the human female germ line, preimplantation embryos, and embryonic stem cells. Mol Reprod Dev. 2004;67:323–36. doi: 10.1002/mrd.20030. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, Gabrielson E, Garcia JG, Geoghegan J, Germino G, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–50. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–82. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Jansen E, Laven JS, Dommerholt HB, Polman J, van Rijt C, van den Hurk C, Westland J, Mosselman S, Fauser BC. Abnormal gene expression profiles in human ovaries from polycystic ovary syndrome patients. Mol Endocrinol. 2004;18:3050–63. doi: 10.1210/me.2004-0074. [DOI] [PubMed] [Google Scholar]
- Jaroudi S, SenGupta S. DNA repair in mammalian embryos. Mutat Res. 2007;635:53–77. doi: 10.1016/j.mrrev.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Jaroudi S, Kakourou G, Cawood S, Doshi A, Ranieri DM, Serhal P, Harper JC, SenGupta SB. Expression profiling of DNA repair genes in human oocytes and blastocysts using microarrays. Hum Reprod. 2009;24:2649–55. doi: 10.1093/humrep/dep224. [DOI] [PubMed] [Google Scholar]
- Jeong HJ, Kim HJ, Lee SH, Kwack K, Ahn SY, Choi YJ, Kim HG, Lee KW, Lee CN, Cha KY. Gene expression profiling of the pre-implantation mouse embryo by microarray analysis: comparison of the two-cell stage and two-cell block. Theriogenology. 2006;66:785–96. doi: 10.1016/j.theriogenology.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Jones GM, Cram DS, Song B, Kokkali G, Pantos K, Trounson AO. Novel strategy with potential to identify developmentally competent IVF blastocysts. Hum Reprod. 2008a;23:1748–59. doi: 10.1093/humrep/den123. [DOI] [PubMed] [Google Scholar]
- Jones GM, Cram DS, Song B, Magli MC, Gianaroli L, Lacham-Kaplan O, Findlay JK, Jenkin G, Trounson AO. Gene expression profiling of human oocytes following in vivo or in vitro maturation. Hum Reprod. 2008b;23:1138–44. doi: 10.1093/humrep/den085. [DOI] [PubMed] [Google Scholar]
- Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–14. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe MG, Schoolcraft WB, Gardner DK. Analysis of protein expression (secretome) by human and mouse preimplantation embryos. Fertil Steril. 2006;86:678–85. doi: 10.1016/j.fertnstert.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe MG, McReynolds S, Gardner DK, Schoolcraft WB. The role of proteomics in defining the human embryonic secretome. Mol Hum Reprod. 2009;15:271–7. doi: 10.1093/molehr/gap012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Agami R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle. 2008;7:899–903. doi: 10.4161/cc.7.7.5644. [DOI] [PubMed] [Google Scholar]
- Kocabas AM, Crosby J, Ross PJ, Otu HH, Beyhan Z, Can H, Tam WL, Rosa GJ, Halgren RG, Lim B, et al. The transcriptome of human oocytes. Proc Natl Acad Sci U S A. 2006;103:14027–32. doi: 10.1073/pnas.0603227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Carrour T, Assou S, Tondeur S, Lhermitte L, Lamb N, Reme T, Pantesco V, Hamamah S, Klein B, De Vos J. Amazonia!: An Online Resource to Google and Visualize Public Human whole Genome Expression Data. The Open Bioinformatics Journal. 2010;4:5–10. [Google Scholar]
- Ledee N, Lombroso R, Lombardelli L, Selva J, Dubanchet S, Chaouat G, Frankenne F, Foidart JM, Maggi E, Romagnani S, et al. Cytokines and chemokines in follicular fluids and potential of the corresponding embryo: the role of granulocyte colony-stimulating factor. Hum Reprod. 2008;23:2001–9. doi: 10.1093/humrep/den192. [DOI] [PubMed] [Google Scholar]
- Lee YS, Latham KE, Vandevoort CA. Effects of in vitro maturation on gene expression in rhesus monkey oocytes. Physiol Genomics. 2008;35:145–58. doi: 10.1152/physiolgenomics.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyens G, Verhaeghe B, Landtmeters M, Marchandise J, Knoops B, Donnay I. Peroxiredoxin 6 is upregulated in bovine oocytes and cumulus cells during in vitro maturation: role of intercellular communication. Biol Reprod. 2004;71:1646–51. doi: 10.1095/biolreprod.104.030155. [DOI] [PubMed] [Google Scholar]
- Li Q, Pangas SA, Jorgez CJ, Graff JM, Weinstein M, Matzuk MM. Redundant roles of SMAD2 and SMAD3 in ovarian granulosa cells in vivo. Mol Cell Biol. 2008;28:7001–11. doi: 10.1128/MCB.00732-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS, Liu YH, Tseng CN, Singh S. Analysis of gene expression in single human oocytes and preimplantation embryos. Biochem Biophys Res Commun. 2006;340:48–53. doi: 10.1016/j.bbrc.2005.11.149. [DOI] [PubMed] [Google Scholar]
- Lluis F, Pedone E, Pepe S, Cosma MP. Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell. 2008;3:493–507. doi: 10.1016/j.stem.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Lonergan P, Fair T, Corcoran D, Evans AC. Effect of culture environment on gene expression and developmental characteristics in IVF-derived embryos. Theriogenology. 2006;65:137–52. doi: 10.1016/j.theriogenology.2005.09.028. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin960 conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SJ, Feng Q, Caballero S, Chen Y, Moore MA, Grant MB, Lanza R. Generation of functional hemangioblasts from human embryonic stem cells. Nat Methods. 2007;4:501–9. doi: 10.1038/nmeth1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–5. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, Amato P, Matzuk MM. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004;19:2869–74. doi: 10.1093/humrep/deh535. [DOI] [PubMed] [Google Scholar]
- Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 2001;29:275–6. doi: 10.1093/nar/29.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morkel M, Huelsken J, Wakamiya M, Ding J, van de Wetering M, Clevers H, Taketo MM, Behringer RR, Shen MM, Birchmeier W. Beta-catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development. 2003;130:6283–94. doi: 10.1242/dev.00859. [DOI] [PubMed] [Google Scholar]
- Mummery CL. Transforming growth factor beta and mouse development. Microsc Res Tech. 2001;52:374–86. doi: 10.1002/1097-0029(20010215)52:4<374::AID-JEMT1022>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oksjoki S, Soderstrom M, Inki P, Vuorio E, Anttila L. Molecular profiling of polycystic ovaries for markers of cell invasion and matrix turnover. Fertil Steril. 2005;83:937–44. doi: 10.1016/j.fertnstert.2004.10.037. [DOI] [PubMed] [Google Scholar]
- Pan H, Ma P, Zhu W, Schultz RM. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev Biol. 2008;316:397–407. doi: 10.1016/j.ydbio.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, O’Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Pangas SA, Jorgez CJ, Matzuk MM. Growth differentiation factor 9 regulates expression of the bone morphogenetic protein antagonist gremlin. J Biol Chem. 2004;279:32281–6. doi: 10.1074/jbc.M403212200. [DOI] [PubMed] [Google Scholar]
- Rao RR, Calhoun JD, Qin X, Rekaya R, Clark JK, Stice SL. Comparative transcriptional profiling of two human embryonic stem cell lines. Biotechnol Bioeng. 2004;88:273–86. doi: 10.1002/bit.20245. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Richards M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–8. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- Rinaudo P, Schultz RM. Effects of embryo culture on global pattern of gene expression in preimplantation mouse embryos. Reproduction. 2004;128:301–11. doi: 10.1530/rep.1.00297. [DOI] [PubMed] [Google Scholar]
- Rinaudo PF, Giritharan G, Talbi S, Dobson AT, Schultz RM. Effects of oxygen tension on gene expression in preimplantation mouse embryos. Fertil Steril. 2006;86:1252–65. 65 e1–36. doi: 10.1016/j.fertnstert.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Rizos D, Ward F, Duffy P, Boland MP, Lonergan P. Consequences of bovine oocyte maturation, fertilization or early embryo development in vitro versus in vivo: implications for blastocyst yield and blastocyst quality. Mol Reprod Dev. 2002;61:234–48. doi: 10.1002/mrd.1153. [DOI] [PubMed] [Google Scholar]
- Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci U S A. 2001;98:5134–9. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DL, Salustri A. Extracellular matrix of the cumulus-oocyte complex. Semin Reprod Med. 2006;24:217–27. doi: 10.1055/s-2006-948551. [DOI] [PubMed] [Google Scholar]
- Saito S, Sawai K, Murayama Y, Fukuda K, Yokoyama K. Nuclear transfer to study the nuclear reprogramming of human stem cells. Methods Mol Biol. 2008;438:151–69. doi: 10.1007/978-1-59745-133-8_13. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–13. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Scott R, Seli E, Miller K, Sakkas D, Scott K, Burns DH. Noninvasive metabolomic profiling of human embryo culture media using Raman spectroscopy predicts embryonic reproductive potential: a prospective blinded pilot study. Fertil Steril. 2008;90:77–83. doi: 10.1016/j.fertnstert.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Seli E, Botros L, Sakkas D, Burns DH. Noninvasive metabolomic profiling of embryo culture media using proton nuclear magnetic resonance correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2008;90:2183–9. doi: 10.1016/j.fertnstert.2008.07.1739. [DOI] [PubMed] [Google Scholar]
- Seli E, Sakkas D, Scott R, Kwok SC, Rosendahl SM, Burns DH. Noninvasive metabolomic profiling of embryo culture media using Raman and near-infrared spectroscopy correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2007;88:1350–7. doi: 10.1016/j.fertnstert.2007.07.1390. [DOI] [PubMed] [Google Scholar]
- Seli E, Vergouw CG, Morita H, Botros L, Roos P, Lambalk CB, Yamashita N, Kato O, Sakkas D. Noninvasive metabolomic profiling as an adjunct to morphology for noninvasive embryo assessment in women undergoing single embryo transfer. Fertil Steril. 2010;94:535–542. doi: 10.1016/j.fertnstert.2009.03.078. [DOI] [PubMed] [Google Scholar]
- Singh R, Sinclair KD. Metabolomics: approaches to assessing oocyte and embryo quality. Theriogenology. 2007;68 (Suppl 1):S56–62. doi: 10.1016/j.theriogenology.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Sirard C, de la Pompa JL, Elia A, Itie A, Mirtsos C, Cheung A, Hahn S, Wakeham A, Schwartz L, Kern SE, et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12:107–19. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JL, Wessel GM. How to make an egg: transcriptional regulation in oocytes. Differentiation. 2005;73:1–17. doi: 10.1111/j.1432-0436.2005.07301005.x. [DOI] [PubMed] [Google Scholar]
- Steuerwald NM, Bermudez MG, Wells D, Munne S, Cohen J. Maternal age-related differential global expression profiles observed in human oocytes. Reprod Biomed Online. 2007;14:700–8. doi: 10.1016/s1472-6483(10)60671-2. [DOI] [PubMed] [Google Scholar]
- Stoughton RB. Applications of DNA microarrays in biology. Annu Rev Biochem. 2005;74:53–82. doi: 10.1146/annurev.biochem.74.082803.133212. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302:104–17. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung LY, Gao S, Shen H, Yu H, Song Y, Smith SL, Chang CC, Inoue K, Kuo L, Lian J, et al. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat Genet. 2006;38:1323–8. doi: 10.1038/ng1895. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue1044 specific gene loci in embryonic stem cells. Cell. 2006;127:1375–88. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Ko MS. A global view of gene expression in the preimplantation mouse embryo: morula versus blastocyst. Eur J Obstet Gynecol Reprod Biol. 2004;115 (Suppl 1):S85–91. doi: 10.1016/j.ejogrb.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Tanghe S, Van Soom A, Nauwynck H, Coryn M, de Kruif A. Minireview: Functions of the cumulus oophorus during oocyte maturation, ovulation, and fertilization. Mol Reprod Dev. 2002;61:414–24. doi: 10.1002/mrd.10102. [DOI] [PubMed] [Google Scholar]
- Taylor DM, Handyside AH, Ray PF, Dibb NJ, Winston RM, Ao A. Quantitative measurement of transcript levels throughout human preimplantation development: analysis of hypoxanthine phosphoribosyl transferase. Mol Hum Reprod. 2001;7:147–54. doi: 10.1093/molehr/7.2.147. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vallier L, Touboul T, Brown S, Cho C, Bilican B, Alexander M, Cedervall J, Chandran S, Ahrlund- Richter L, Weber A, et al. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells. 2009;27:2655–66. doi: 10.1002/stem.199. [DOI] [PubMed] [Google Scholar]
- van der Maarel SM. Epigenetic mechanisms in health and disease. Ann Rheum Dis. 2008;67(Suppl 3):iii97–100. doi: 10.1136/ard.2008.098392. [DOI] [PubMed] [Google Scholar]
- van Montfoort AP, Geraedts JP, Dumoulin JC, Stassen AP, Evers JL, Ayoubi TA. Differential gene expression in cumulus cells as a prognostic indicator of embryo viability: a microarray analysis. Mol Hum Reprod. 2008;14:157–68. doi: 10.1093/molehr/gam088. [DOI] [PubMed] [Google Scholar]
- Vanderhyden BC, Macdonald EA, Nagyova E, Dhawan A. Evaluation of members of the TGFbeta superfamily as candidates for the oocyte factors that control mouse cumulus expansion and steroidogenesis. Reprod Suppl. 2003;61:55–70. [PubMed] [Google Scholar]
- Varani S, Elvin JA, Yan C, DeMayo J, DeMayo FJ, Horton HF, Byrne MC, Matzuk MM. Knockout of pentraxin 3, a downstream target of growth differentiation factor-9, causes female subfertility. Mol Endocrinol. 2002;16:1154–67. doi: 10.1210/mend.16.6.0859. [DOI] [PubMed] [Google Scholar]
- Vassena R, Dee Schramm R, Latham KE. Species-dependent expression patterns of DNA methyltransferase genes in mammalian oocytes and preimplantation embryos. Mol Reprod Dev. 2005;72:430–6. doi: 10.1002/mrd.20375. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vergouw CG, Botros LL, Roos P, Lens JW, Schats R, Hompes PG, Burns DH, Lambalk CB. Metabolomic profiling by near-infrared spectroscopy as a tool to assess embryo viability: a novel, non1082 invasive method for embryo selection. Hum Reprod. 2008;23:1499–504. doi: 10.1093/humrep/den111. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sun QY. Evaluation of oocyte quality: morphological, cellular and molecular predictors. Reprod Fertil Dev. 2007;19:1–12. doi: 10.1071/rd06103. [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133–44. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Watabe T, Miyazono K. Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–15. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Suetake I, Tada T, Tajima S. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech Dev. 2002;118:187–90. doi: 10.1016/s0925-4773(02)00242-3. [DOI] [PubMed] [Google Scholar]
- Watson AJ. Oocyte cytoplasmic maturation: a key mediator of oocyte and embryo developmental competence. J Anim Sci. 2007;85:E1–3. doi: 10.2527/jas.2006-432. [DOI] [PubMed] [Google Scholar]
- Wei CL, Miura T, Robson P, Lim SK, Xu XQ, Lee MY, Gupta S, Stanton L, Luo Y, Schmitt J, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–85. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- Wells D, Patrizio P. Gene expression profiling of human oocytes at different maturational stages and after in vitro maturation. Am J Obstet Gynecol. 2008;198:455 e1–9. doi: 10.1016/j.ajog.2007.12.030. discussion 55 e9–11. [DOI] [PubMed] [Google Scholar]
- Wells D, Bermudez MG, Steuerwald N, Thornhill AR, Walker DL, Malter H, Delhanty JD, Cohen J. Expression of genes regulating chromosome segregation, the cell cycle and apoptosis during human preimplantation development. Hum Reprod. 2005;20:1339–48. doi: 10.1093/humrep/deh778. [DOI] [PubMed] [Google Scholar]
- Wood JR, Dumesic DA, Abbott DH, Strauss JF., 3rd Molecular abnormalities in oocytes from women with polycystic ovary syndrome revealed by microarray analysis. J Clin Endocrinol Metab. 2007;92:705–13. doi: 10.1210/jc.2006-2123. [DOI] [PubMed] [Google Scholar]
- Yamazawa K, Kagami M, Fukami M, Matsubara K, Ogata T. Monozygotic female twins discordant for Silver-Russell syndrome and hypomethylation of the H19-DMR. J Hum Genet. 2008;53:950–5. doi: 10.1007/s10038-008-0329-4. [DOI] [PubMed] [Google Scholar]
- Yoon SJ, Kim KH, Chung HM, Choi DH, Lee WS, Cha KY, Lee KA. Gene expression profiling of early follicular development in primordial, primary, and secondary follicles. Fertil Steril. 2006;85:193–203. doi: 10.1016/j.fertnstert.2005.07.1296. [DOI] [PubMed] [Google Scholar]
- Yoon SJ, Kim EY, Kim YS, Lee HS, Kim KH, Bae J, Lee KA. Role of Bcl2-like 10 (Bcl2l10) in Regulating Mouse Oocyte Maturation. Biol Reprod. 2009;81:497–506. doi: 10.1095/biolreprod.108.073759. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005;23:299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–96. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zhang P, Kerkela E, Skottman H, Levkov L, Kivinen K, Lahesmaa R, Hovatta O, Kere J. Distinct sets of developmentally regulated genes that are expressed by human oocytes and human embryonic stem cells. Fertil Steril. 2007;87:677–90. doi: 10.1016/j.fertnstert.2006.07.1509. [DOI] [PubMed] [Google Scholar]
- Zhang P, Zucchelli M, Bruce S, Hambiliki F, Stavreus-Evers A, Levkov L, Skottman H, Kerkela E, Kere J, Hovatta O. Transcriptome profiling of human pre-implantation development. PLoS One. 2009;4:e7844. doi: 10.1371/journal.pone.0007844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jafari N, Barnes RB, Confino E, Milad M, Kazer RR. Studies of gene expression in human cumulus cells indicate pentraxin 3 as a possible marker for oocyte quality. Fertil Steril. 2005;83 (Suppl 1):1169–79. doi: 10.1016/j.fertnstert.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Zheng P, Patel B, McMenamin M, Moran E, Paprocki AM, Kihara M, Schramm RD, Latham KE. Effects of follicle size and oocyte maturation conditions on maternal messenger RNA regulation and gene expression in rhesus monkey oocytes and embryos. Biol Reprod. 2005;72:890–7. doi: 10.1095/biolreprod.104.035881. [DOI] [PubMed] [Google Scholar]


