Abstract
The N-end rule relates the regulation of the in vivo half-life of a protein to the identity of its N-terminal residue. Degradation signals (degrons) that are targeted by the N-end rule pathway include a set called N-degrons. The main determinant of an N-degron is a destabilizing N-terminal residue of a protein. In eukaryotes, the N-end rule pathway is a part of the ubiquitin system and consists of two branches, the Ac/N-end rule and the Arg/N-end rule pathways. The Ac/N-end rule pathway targets proteins containing Nα-terminally acetylated (Nt-acetylated) residues. The Arg/N-end rule pathway recognizes unacetylated N-terminal residues and involves N-terminal arginylation. Together, these branches target for degradation a majority of cellular proteins. For example, more than 80% of human proteins are cotranslationally Nt-acetylated. Thus, most proteins harbor a specific degradation signal, termed AcN-degron, from the moment of their birth. Specific N-end rule pathways are also present in prokaryotes and in mitochondria. Enzymes that produce N-degrons include methionine-aminopeptidases, caspases, calpains, Nt-acetylases, Nt-amidases, arginyl-transferases, and leucyl-transferases. Regulated degradation of specific proteins by the N-end rule pathway mediates a legion of physiological functions, including the sensing of heme, oxygen, and nitric oxide; selective elimination of misfolded proteins; the regulation of DNA repair, segregation, and condensation; the signaling by G proteins; the regulation of peptide import, fat metabolism, viral and bacterial infections, apoptosis, meiosis, spermatogenesis, neurogenesis, and cardiovascular development; and the functioning of adult organs, including the pancreas and the brain. Discovered 25 years ago, this pathway continues to be a fount of biological insights.
Keywords: ubiquitin, N-end rule, N-degron, proteolysis, arginylation, leucylation
Glossary of Terms
“Sequelog” and “spalog” denote, respectively, a sequence that is similar, to a specified extent, to another sequence, and a three-dimensional (3D) structure that is similar, to a specified extent, to another 3D structure.1 Derivatives of these terms include “sequelogous” and “sequelogy” (sequence similarity); “spalogous” and “spalogy” (spatial similarity). In addition to their usefulness as separate terms for sequence and spatial similarities, the rigor-conferring advantage of “sequelog” and “spalog” is their evolutionary neutrality, in contrast to interpretation-laden terms such as “homolog,” “ortholog,” and “paralog.” The latter terms are compatible with the sequelog/spalog terminology and can be used to convey understanding about functions and common descent, if this (additional) information is available.1
Ubiquitin (Ub): a highly conserved 76-residue eukaryotic protein that can be enzymatically conjugated to other proteins, thereby marking them for processive degradation or other metabolic fates.
N-end rule: it relates the regulation of the in vivo half-life of a protein to the identity of its N-terminal residue.
N-end rule pathway: a proteolytic pathway whose physiological targets include proteins with destabilizing N-terminal residues.
Degron: a degradation signal.2
N-degron: one class of degradation signals that can be targeted by an N-end rule pathway. The main determinant of an N-degron is either a modified or unmodified destabilizing N-terminal residue of a protein.
Pro-N-degron: precursor of N-degron. A pro-N-degron is a specific sequence or conformational determinant of a polypeptide chain that can be cleaved or otherwise modified to produce a destabilizing N-terminal residue. This definition of a pro-N-degron implies that other determinants of an N-degron, for example, a “targetable” internal Lys residue of a substrate, are in place as well.
N-recognin: recognition component of an N-end rule pathway that recognizes (binds to) specific N-degrons.
Ndp residue: a primary (p) destabilizing N-terminal residue, that is, an unmodified N-terminal residue that is directly recognized by an N-recognin.
Nds residue: a secondary (s) destabilizing N-terminal residue, that is, a residue whose destabilizing activity requires a specific preliminary modification, such as, for example, N-terminal arginylation (Nt-arginylation).
Ndt residue: a tertiary (t) destabilizing N-terminal residue, that is, a residue whose destabilizing activity requires two preliminary modifications.
Arg/N-end rule pathway: a branch of the eukaryotic N-end rule pathway that involves the Nt- arginylation of protein substrates and also the targeting of unmodified bulky hydrophobic and basic N-terminal residues by an N-recognin E3 ubiquitin ligase.
Ac/N-end rule pathway: a branch of the eukaryotic N-end rule pathway that may also be present in archaeal prokaryotes. The Ac/N-end rule pathway involves the Nα-terminal acetylation (Nt-acetylation) of nascent proteins whose N-termini bear either Met or the small uncharged residues Ala, Val, Set, Thr, or Cys. These residues become N-terminal after the cotranslational removal of Met by Met-aminopeptidases. Nt-acetylated proteins are targeted for degradation by the Ac/N-end rule pathway.3
Leu/N-end rule pathway: a bacterial N-end rule pathway that involves Nt-leucylation of protein substrates by specific l-transferases and also the targeting of bulky hydrophobic N-terminal residues by the ClpS N-recognin, an adaptor protein that delivers bacterial N-end rule substrates to the ClpAP protease.
Introduction
The lifespans of protein molecules in a cell range from less than a minute to many days. Among the functions of intracellular proteolysis are the elimination of misfolded or otherwise abnormal proteins, the maintenance of amino acid pools in cells affected by stresses such as starvation, and the generation of protein fragments that act as hormones, antigens, or other effectors. One major role of proteolytic pathways is the selective destruction of regulatory proteins whose concentrations must vary with time and alterations in the state of a cell. A short in vivo half-life of a protein provides a way to generate its spatial gradient and to rapidly adjust its concentration or subunit composition through changes in the rate of its degradation. Proteolysis can also serve to activate protein molecules and specific circuits, by removing an autoinhibitory protein domain or by selectively destroying an inhibitory subunit of a protein complex. The regulated (and processive) degradation of intracellular proteins is carried out largely by the ubiquitin-proteasome system [Ub system; Fig. 1(A)], in conjunction with molecular chaperones, autophagy, and lysosomal proteolysis. Chaperones mediate in vivo protein folding and the assembly/disassembly of protein complexes. A meta-system that includes the Ub system and chaperones determines the time-dependent probability, for each protein, of being either in its “normal” (functional) state, or targeted for degradation, or perturbed in ways (including aggregation) that may or may not lead to degradation. Other mediators of intracellular proteolysis include cytosolic and nuclear proteases such as caspases and calpains. These and other nonprocessive proteases can function as “upstream” components of the Ub system, producing protein fragments that are often targeted and degraded to short peptides by Ub-mediated pathways. Proteins that are damaged, misfolded, or otherwise abnormal are often short-lived in vivo, with significant exceptions that include a subset of perturbed proteins (and/or their aggregates) that are harmful but cannot be efficaciously repaired or removed. The resulting proteotoxicity underlies both aging and specific diseases, including neurodegeneration.
Figure 1.
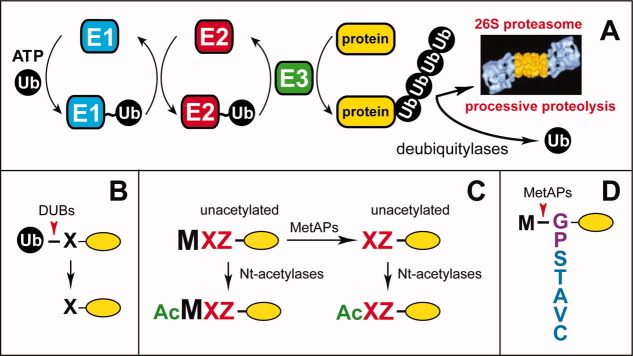
The ubiquitin-proteasome system, the ubiquitin fusion technique, and N-terminal processing of newly formed proteins. A: The ubiquitin-proteasome system (Ub system).6,45–65 The conjugation of Ub to other proteins involves a preliminary ATP-dependent step in which the last residue of Ub (Gly76) is joined, via a thioester bond, to a Cys residue of the E1 (Ub-activating) enzyme. The “activated” Ub moiety is transferred to a Cys residue in one of several Ub-conjugating (E2) enzymes, and from there, through an isopeptide bond, to a Lys residue of an ultimate acceptor, denoted as “protein”. E2 enzymes function as subunits of E2-E3 Ub ligase complexes that can produce substrate-linked poly-Ub chains. Such chains have specific Ub-Ub topologies, depending on the identity of a Lys residue of Ub (which contains several lysines) that forms an isopeptide bond with C-terminal Gly76 of the adjacent Ub moiety in a chain. Specific poly-Ub chains can confer the degradation of a substrate by the 26S proteasome or other metabolic fates. Monoubiquitylation of some protein substrates can also occur, and has specific functions. One role of E3 is the recognition of a substrate's degradation signal (degron). Individual mammalian genomes encode at least a 1,000 distinct E3 Ub ligases. B: The Ub fusion technique.4,213 In eukaryotes, linear fusions of Ub to other proteins are cotranslationally cleaved by deubiquitylases at the last residue of Ub, making it possible to produce, in vivo, different residues at the N-termini of otherwise identical proteins. C: N-terminal processing of nascent proteins by Nα-terminal acetylases (Nt-acetylases) and Met-aminopeptidases (MetAPs). “Ac” denotes the Nα-terminal acetyl moiety. M, Met. X and Z, single-letter abbreviations for any amino acid residue. Yellow ovals denote the rest of a protein. D: Met-aminopeptidases (MetAPs) cleave off the N-terminal Met residue if a residue at Position 2 belongs to the set of residues shown.101 Gly and Pro at Position 2 are depicted in a different color because these residues, in contrast to other small residues, are rarely Nt-acetylated after the removal of N-terminal Met [Fig. 2(B)]. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The N-end rule relates the regulation of the in vivo half-life of a protein to the identity of its N-terminal residue. The 1986 discovery of the N-end rule pathway identified the first specific pathway of the Ub system.4–6 It was also the discovery of the first primary degradation signals (degrons2) in short-lived proteins.4 Ub, a 76-residue protein, is a “secondary” degron in that Ub is conjugated to proteins that contain primary degradation signals. For accounts of the early history of the Ub field, see Refs. 6–8.
Overview of the N-End Rule Pathway
N-terminal degradation signals of the N-end rule pathway are called N-degrons. The main determinant of an N-degron is a destabilizing N-terminal residue of a protein3–6,9–44 (Figs. 2–5). In eukaryotes, the N-end rule pathway is a part of the Ub system, which mediates selective protein turnover through the conjugation of Ub to specific proteins, thereby marking them for degradation by the 26S proteasome, a multisubunit ATP-dependent protease6,45–65 [Fig. 1(A)]. Prokaryotes, that is, bacteria and archaea, contain Ub-like proteolytic pathways but lack the bona fide Ub system.66–70 Nevertheless, prokaryotes contain specific versions of the N-end rule pathway that do not involve ubiquitylation5,6,14–16,37,71–78 (Fig. 5).
Figure 2.
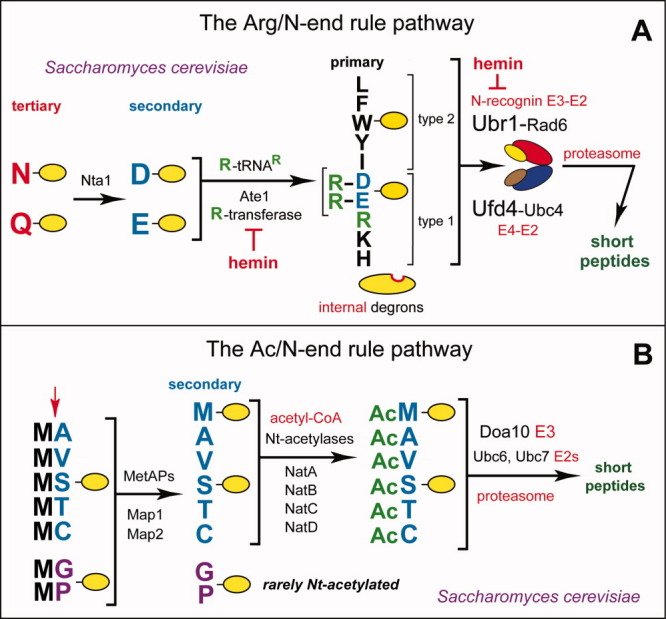
The N-end rule pathway in Saccharomyces cerevisiae. A: The Arg/N-end rule pathway.44 See the main text for details. Yellow ovals denote the rest of a protein substrate. “Primary”, “secondary” and “tertiary” denote mechanistically distinct subsets of destabilizing N-terminal residues. The physically associated Ubr1 (N-recognin) and Ufd4 E3s have substrate-binding sites that recognize internal (non-N-terminal) degrons in substrates of the Arg/N-end rule pathway that lack N-degrons. Ubr1 (but not Ufd4) recognizes N-degrons as well.44 B: The Ac/N-end rule pathway.3 Red arrow on the left indicates the removal of N-terminal Met by Met-aminopeptidases (MetAPs). This Met residue is retained if a residue at Position 2 is nonpermissive (too large) for Met-aminopeptidases [Fig. 1(D)]. If the (retained) N-terminal Met or N-terminal Ala, Val, Ser, Thr and Cys are followed by residues that allow Nt-acetylation (see the main text), these N-terminal residues are usually Nt-acetylated.91–93 The resulting N-degrons are called AcN-degrons. The term “secondary” refers to the necessity of modification (Nt-acetylation) of a destabilizing N-terminal residue before a protein can be recognized by a cognate Ub ligase. Proteins containing AcN-degrons are targeted for ubiquitylation and proteasome-mediated degradation by the Doa10 E3 N-recognin, in conjunction with the Ubc6 and Ubc7 E2 enzymes.44 Although Gly and Pro can be made N-terminal by MetAPs, and although Doa10 can recognize Nt-acetylated Gly and Pro, few proteins with N-terminal Gly or Pro are Nt-acetylated.91–93 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 5.
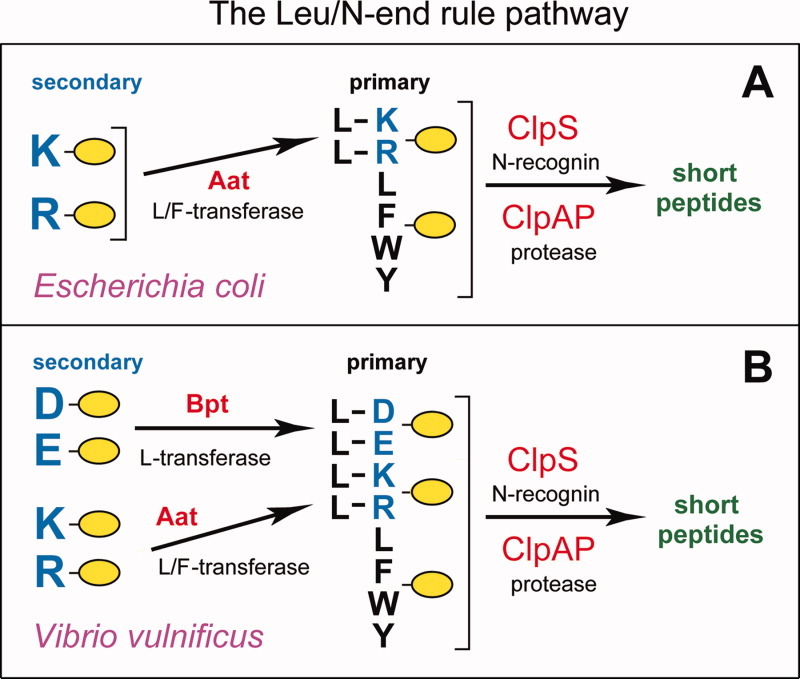
Bacterial N-end rule pathways. A: The E. coli Leu/N-end rule pathway.14,16,37 See Overview of the N-End Rule Pathway, Structure and Targeting of N-degrons, and Prokaryotic N-End Rule Pathways sections for details. The Aat L/F-transferase conjugates (largely) Leu to N-terminal Arg or Lys. N-end rule substrates bearing primary (bulky hydrophobic) destabilizing N-terminal residues are recognized by the ClpS N-recognin and are delivered for degradation to the ClpAP protease. B: The Leu/N-end rule pathway in another gram-negative bacterium, V. vulnificus, which contains both the Aat L/F-transferase and the Bpt L-transferase. As a result, N-terminal Asp and Glu, which are stabilizing (nondestabilizing) residues in E. coli, are secondary destabilizing residues in the V. vulnificus Leu/N-end rule pathway.37 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Recognition components of the N-end rule pathway are called N-recognins.5 In bacteria, the 12-kDa ClpS, identified as an N-recognin by the Bukau laboratory,72 binds to N-degrons of N-end rule substrates and delivers them to the ATP-dependent ClpAP protease16,72,74,76–82 [Figs. 5 and 6(B–D)]. In eukaryotes, N-recognins are E3 Ub ligases that bind to specific N-degrons3,5,6,22,30,38,39,43,83–85 (Figs. 7 and 8). A complex of an E3 N-recognin and its cognate E2 Ub-conjugating enzyme polyubiquitylates N-end rule substrates at their internal Lys residues, thereby targeting these proteins for degradation by the 26S proteasome9,11 (Figs. 2 and 3). The term “Ub ligase” denotes either an E2-E3 complex or its E3 component.6,54,57,62 In eukaryotes, the N-end rule pathway comprises two major branches, one of which is termed the Arg/N-end rule pathway. This branch involves the N-terminal arginylation (Nt-arginylation) of protein substrates and also the targeting of specific unmodified N-terminal residues by E3 N-recognins [Figs. 2(A) and 3]. The other branch is termed the Ac/N-end rule pathway.3 It involves the cotranslational Nα-terminal acetylation (Nt-acetylation) of nascent proteins86–95 whose N-termini bear either Met or the small uncharged residues Ala, Val, Ser, Thr, or Cys. These residues become N-terminal after the cotranslational removal of N-terminal Met by Met-aminopeptidases96–101 [Fig. 1(C,D)]. Nt-acetylated proteins are targeted for regulated degradation by the Ac/N-end rule pathway3 [Fig. 2(B)].
Figure 6.
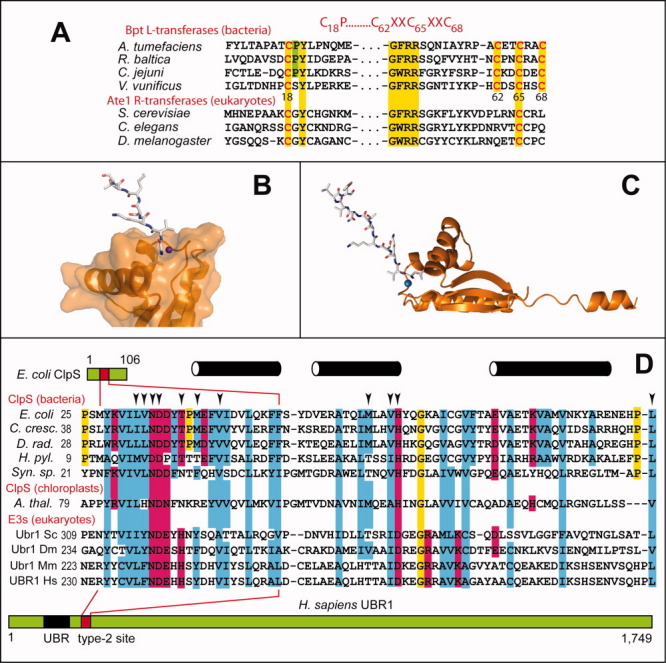
Bpt L-transferases and ClpS N-recognins. A: Sequence alignments of bacterial Bpt L-transferases and eukaryotic Ate1 R-transferases.37 The sequelogy (sequence similarity1) between Bpt and Ate1 encompasses more of their sequences than shown here.37 Ate1 R-transferases lack a sequence motif (its consensus, in red, is shown at the top of the diagram) that Bpt L-transferases uniformly contain. This motif is characteristic of proteins that bind to a Fe-S cluster319,320 (Prokaryotic N-End Rule Pathways section). B: Surface representation of the C-terminal domain of the E. coli ClpS N-recognin in a complex with an 11-mer peptide (shown as a stick model) that bears N-terminal Leu, a primary destabilizing (Ndp) residue in the Leu/N-end rule pathway.74 Blue sphere, water molecule. C: Ribbon representation of the full-length 12-kDa ClpS in the same complex. D: Sequence alignments of the ∼70-residue domain of bacterial ClpS N-recognins in Caulobacter crescentus, E. coli, Deinococcus radiodurans, Helicobacter pylori, Synechocystis sp. PCC6803 (the latter a photosynthesis-capable cyanobacterium), and in chloroplast (A. thaliana). This region of ClpS binds to N-terminal Ndp residues of the Leu/N-rule pathway. ClpS sequences are aligned with sequelogous regions of eukaryotic (S. cerevisiae, D. melanogaster, M. musculus, H. sapiens) Ubr1 N-recognins (they are ∼20-fold larger than ClpS) that encompass the Type-2 substrate-binding site of Ubr1 [Fig. 7(A)]. The specificity of this Ubr1 binding site for bulky hydrophobic N-terminal Ndp residues is nearly the same as the specificity of ClpS, except that the Ubr1 site binds to N-terminal Ile as well [Fig. 2(A)], in contrast to ClpS (see the main text). Arrowheads indicate the positions of crystallographically determined contacts between the ClpS of C. crescentus and an N-end rule peptide.73 Black cylinders indicate α-helices in this region of ClpS. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 7.
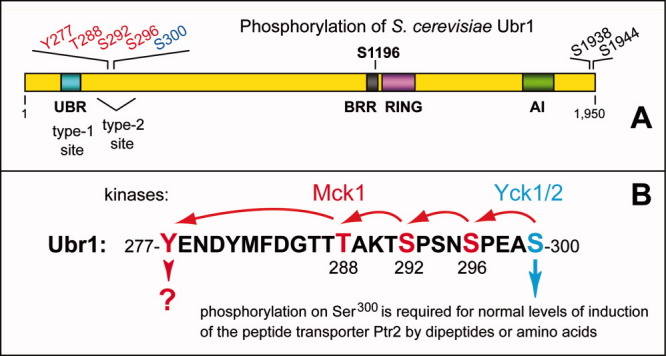
Structural organization and phosphorylation of the Ubr1 N-recognin. A: Phosphorylated residues of the S. cerevisiae Ubr1 E3 N-recognin of the Arg/N-end rule pathway are indicated above the diagram.39 The regions containing the Type-1 substrate-binding site (UBR domain), the Type-2 substrate-binding site, the BRR (basic residues-rich) domain, the Cys/His-rich RING domain and the AI (autoinhibitory) domain15,22,29,39,83–85 are also indicated. B: The “primed” cascade of Ubr1 phosphorylation.39 The initial phosphorylation of Ubr1 on Ser300 by the Yck1/Yck2 kinases of the casein kinase type-I family makes possible (primes) the subsequent, apparently sequential phosphorylation of Ubr1 by Mck1, a Gsk3-type kinase, on Ser,296 Ser,292 Thr,288 and Tyr.277 Also indicated is the identified function of the Ser300 phosphorylation of Ubr1 in the control of peptide import.39 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 8.
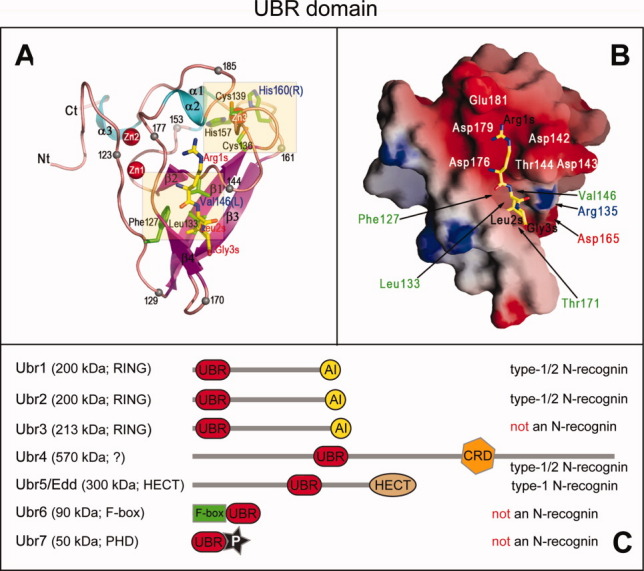
UBR domains in N-recognins and putative N-recognins of the mammalian Arg/N-end rule pathway. A: Ribbon diagram of the ∼80-residue S. cerevisiae UBR domain83 (Fig. 7A) in the complex with the RLGES peptide that bears N-terminal Arg, a Type-1 Ndp residue.28 The bound RLGES is shown as a stick model, with carbon atoms colored yellow. Several residues are marked with a black sphere and numbered to facilitate the tracing of the polypeptide chain. The names of residues of the RLGES peptide are in red, with the letter “s” (substrate) appended to their position numbers. Side chains of residues in the UBR domain that are present near missense mutations in UBR1 of patients with Johanson–Blizzard syndrome (JBS; C.-S. Hwang et al., unpublished data) are shown in a stick form, with carbon atoms colored green. Three coordinated zinc ions of the UBR domain83 are shown as red spheres. B: Molecular surface of the S. cerevisiae UBR domain. Negatively and positively charged surfaces are shaded red and blue, respectively. The bound RLGES peptide is shown in yellow. Some residues of Ubr1 that comprise the N-degron-binding cleft are labeled.83 C: Diagram of the mammalian UBR-domain family of E3 Ub ligases, showing both UBR and other domains of these E3s (RING, HECT, PHD, CRD and F-box) that contribute to recognition and ubiquitylation of protein substrates.31,43 Ubr1, Ubr2, Ubr4, and Ubr5/Edd of this set are operationally defined N-recognins of the mammalian Arg/N-end rule pathway in that they specifically bind to the Type-1 and/or Type-2 destabilizing N-terminal residues, whereas Ubr3, Ubr6, and Ubr7 are not N-recognins43,274 (The double-E3 design of the Arg/N-end rule pathway section). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 3.
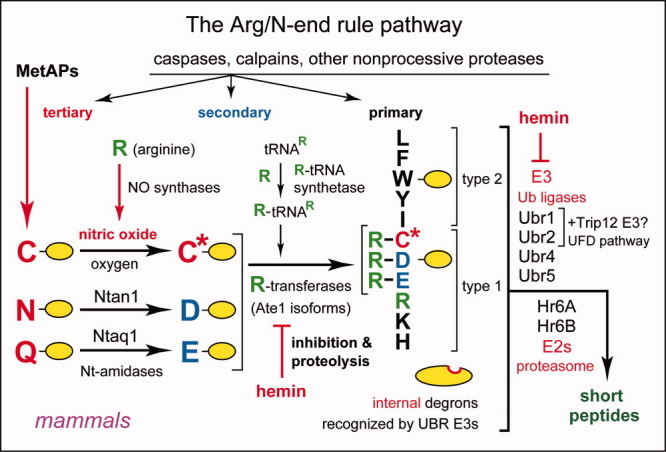
The mammalian Arg/N-end rule pathway. See the main text for details. N-terminal residues are indicated by single-letter abbreviations for amino acids. Yellow ovals denote the rest of a protein substrate. “Primary”, “secondary” and “tertiary” denote mechanistically distinct subsets of destabilizing N-terminal residues. C* denotes oxidized N-terminal Cys, either Cys-sulfinate or Cys-sulfonate, produced in vivo through reactions that require both nitric oxide (NO) and oxygen.32,33 The mammalian N-recognins Ubr1, Ubr2, Ubr4, and Ubr5 (Edd) have multiple substrate binding sites that also recognize internal (non-N-terminal) degrons in other substrates of the Arg/N-end rule pathway, the ones that lack N-degrons. A question mark after Trip12 (which mediates the mammalian UFD pathway258 and is a sequelog of the S. cerevisiae Ufd4 E3) denotes the untested possibility that mammalian Ubr1 and/or Ubr2 form complexes with Trip12, by analogy with the Ubr1–Ufd4 complex in S. cerevisiae [Fig. 2(A)]. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The Nt-acetylated Met, Ala, Val, Ser, Thr, and Cys residues of newly formed proteins comprise a specific class of N-degrons, termed AcN-degrons3 [Figs. 2(B) and 4(A)]. The cotranslational Nt-acetylation of nascent proteins87–95,102 [Fig. 1(C)] is both enzymatically and functionally distinct from the largely posttranslational acetylation of internal residues in many proteins.103,104 Nt-acetylation and internal acetylation are carried out by (mostly) nonoverlapping sets of specific acetylases. In addition, Nt-acetylation is apparently irreversible. No Nt-deacetylases have been identified, in contrast to a dynamic internal acetylation/deacetylation, with specific deacetylases removing internally conjugated acetyl groups (Ref. 104 and references therein). As described below, the proteolytic function of Nt-acetylation [Fig. 2(B)] is likely to be relevant to more than 80% of the entire proteome, that is, to thousands of Nt-acetylated proteins.3 In contrast, either an identified or inferred necessity of Nt-acetylation for other (nonproteolytic) functions involves, at present, only ∼10 Nt-acetylated proteins (Refs. 92,105–110 and references therein).
Figure 4.
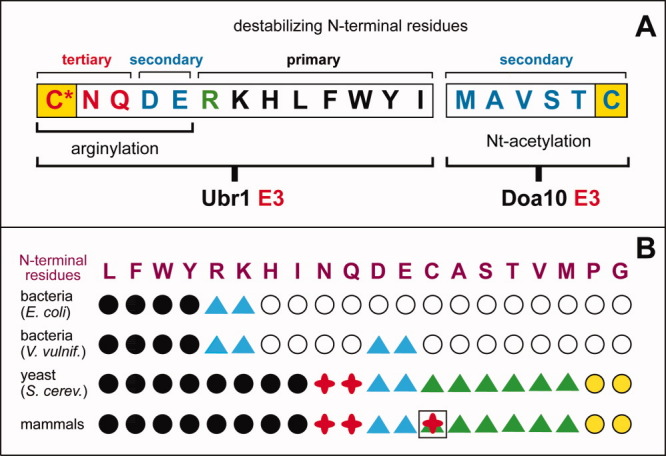
Rule books of N-end rules. A: The N-end rule in eukaryotes. It results from combined activities of the Arg/N-end rule and Ac/N-end rule pathways.3 In eukaryotes that produce NO, the N-terminal Cys residue (in yellow rectangles) can be targeted, alternatively, by either one of the two branches of the N-end rule pathway, with oxidized Cys marked by an asterisk (see Overview of the N-End Rule Pathway section). B: A comparison of rule books of N-end rule pathways in different organisms, indicated on the left. Black circles, blue or green triangles, and red crosses denote primary (Ndp), secondary (Nds) and tertiary (Ndt) destabilizing N-terminal residues, respectively. Blue triangles denote secondary destabilizing N-terminal residues that involve either Nt-leucylation (in bacteria) or Nt-arginylation (in eukaryotes). Green triangles denote secondary destabilizing N-terminal residues that involve Nt-acetylation.3 N-terminal Cys is denoted by both a green triangle and a red cross, given its alternative functioning as a part of NO/O2-mediated N-degrons or AcN-degrons. Open circles, in bacterial N-end rules, denote stabilizing (nondestabilizing) N-terminal residues. Yellow circles, in eukaryotic N-end rules, denote Pro and Gly. These N-terminal residues are rarely Nt-acetylated and therefore, operationally, are stabilizing (nondestabilizing) residues. But in some proteins with N-terminal Pro or Gly these residues can be Nt-acetylated. If other components of an AcN-degron are also in place (see The Ac/N-End Rule Pathway section), such proteins can become substrates of the Ac/N-end rule pathway.3 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Apart from expanding the N-end rule and its functions, the 2010 discovery of the Ac/N-end rule pathway3 has also revealed the main physiological roles of two classes of enzymes, Nt-acetylases, and Met-aminopeptidases. Specifically, Nt-acetylases produce AcN-degrons while the “upstream” Met-aminopeptidases, by cleaving off the N-terminal Met residue, make these degradation signals possible, all of them save for those AcN-degrons that contain the Nt-acetylated N-terminal Met [Figs. 1(C,D) and 2(B)]. Nt-acetylases and Met-aminopeptidases are essential and universally present enzymes86–101 whose physiological functions had been largely unknown. These enzymes are now specific components of the Ac/N-end rule pathway3 [Fig. 2(B)].
N-terminal Arg, Lys, His, Leu, Phe, Tyr, Trp, Ile, Asp, Glu, Asn, Gln, and Cys comprise the main determinants of N-degrons in the Arg/N-end rule pathway [Figs. 2(A) and 3]. Among these N-degrons, the unmodified basic (Arg, Lys, His) and bulky hydrophobic (Leu, Phe, Tyr, Trp, Ile) N-terminal residues are recognized directly by cognate E3 N-recognins [Figs. 2(A), 3, 7, and 8]. These E3s contain highly spalogous (spatially similar1) ∼80-residue regions called UBR domains or Type-1 binding sites.83–85 Folded around three zinc ions, a UBR domain binds to N-terminal Arg, Lys, or His, the Type-1 primary destabilizing residues of N-end rule substrates (Figs. 3 and 8). Another (usually adjacent) region of UBR-type N-recognins, called the Type-2 binding site, recognizes N-terminal Leu, Phe, Tyr, Trp, or Ile, which are called the Type-2 primary destabilizing residues [Fig. 7(A)]. Together, the directly recognized primary destabilizing N-terminal Arg, Lys, His, Leu, Phe, Tyr, Trp, and Ile are denoted as Ndp residues (N, N-terminal; d, destabilizing; p, primary).6,43,83–85 In contrast to these residues, the N-terminal Asp, Glu, Asn, Gln, and Cys function as destabilizing residues through their preliminary modifications. One of these modifications is Nt-arginylation. N-terminal Arg is an Ndp residue, that is, it can be recognized by E3 N-recognins of the Arg/N-end rule pathway [Figs. 2(A) and 3]. Arg-tRNA-protein transferase (R-transferase) conjugates Arg to N-terminal Asp, Glu, or oxidized Cys of proteins or short peptides, with Arg-tRNA as the cosubstrate and the donor of Arg. R-transferases are encoded by Ate1 and its sequelogs from yeast to mammals but are absent from examined prokaryotes32,35,36,111–114 (Fig. 9). In contrast to N-terminal Asp, Glu and oxidized Cys, the N-terminal Asn and Gln residues cannot be arginylated by R-transferase. However, the Arg/N-end rule pathway contains specific N-terminal amidases (Nt-amidases) that convert N-terminal Asn and Gln to Asp and Glu, respectively, followed by their Nt-arginylation23,25,42,115–117 [Figs. 2(A) and 3].
Figure 9.
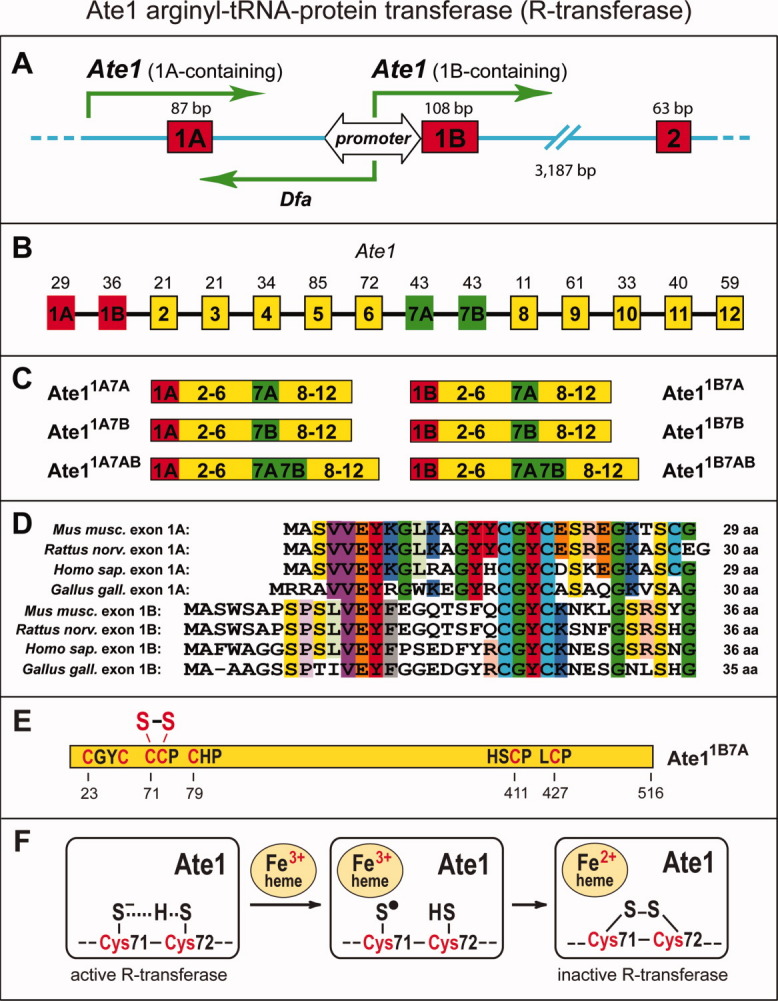
Splicing-derived isoforms of the Ate1-encoded Arg-tRNA-protein transferase (R-transferase) and its inhibition by hemin. A: The bidirectional DfaPAte1 promoter (containing a CpG island) upstream of exon 1B of the mouse Ate1 gene.35,282 Green arrows indicate transcriptional units oriented in both directions from DfaPAte1, and also from an unmapped “upstream” promoter that mediates the expression of Ate1 transcripts containing exon 1A. The locations and sizes of some Ate1 exons are shown as well. B: The exons, including alternative exons, of the mouse Ate1 gene, with deduced lengths of the corresponding polypeptide segments indicated on top. C: Mouse R-transferase isoforms (and their designations) that are produced through alternative splicing of Ate1 pre-mRNA. D: Sequence comparisons of translated vertebrate Ate1 exons 1A amongst themselves and with the set of longer but also sequelogous alternative exons 1B. Most of recurrent amino acid identities are highlighted by color. Mus musculus, mouse; Rattus norvegicus, rat; Homo sapiens, human; Gallus gallus, chicken. E: The mouse ATE11B7A isoform, with locations of significant Cys-containing motifs, including the vicinal Cys71 and Cys72 residues. A disulfide bond between them is the result of hemin-mediated oxidation and functional inactivation of R-transferase.35 F: Diagram of the previously proposed35 redox mechanism of the hemin-mediated disulfide formation between Cys71 and Cys72 of Ate1. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
N-terminal Cys residues can be Nt-acetylated (in proteins that contain the initially present N-terminal Met-Cys sequence) after the cotranslational removal of N-terminal Met by Met-aminopeptidases [Fig. 1(C,D)]. The same is true for other second-position (penultimate) small residues such as Ala, Val, Ser, or Thr. Nt-acetylation of Cys produces an AcN-degron of the Ac/N-end rule pathway and thereby precludes the (alternative) participation of N-terminal Cys in the Arg/N-end rule pathway3 [Fig. 2(B); cf. Fig. 3]. However, some sequence contexts, for example, a basic residue at Position 2, inhibit the Nt-acetylation of N-terminal Cys and other N-terminal residues. The mammalian proteins Rgs4, Rgs5, and Rgs16 are one example of this inhibition. They bind to Gα subunits of specific G proteins and increase the intrinsic GTPase activity of Gα, thereby downregulating the signaling by these G proteins. The N-terminal Cys residue of Rgs4, Rgs5, and Rgs16 is followed by a basic residue (Fig. 10), hence the absence of Nt-acetylation of these RGS proteins.32,33,118 (Whereas in the yeast Saccharomyces cerevisiae a basic residue at Position 2 suffices to block Nt-acetylation, some proteins with Position 2 basic residues can be Nt-acetylated in mammalian cells.91)
Figure 10.
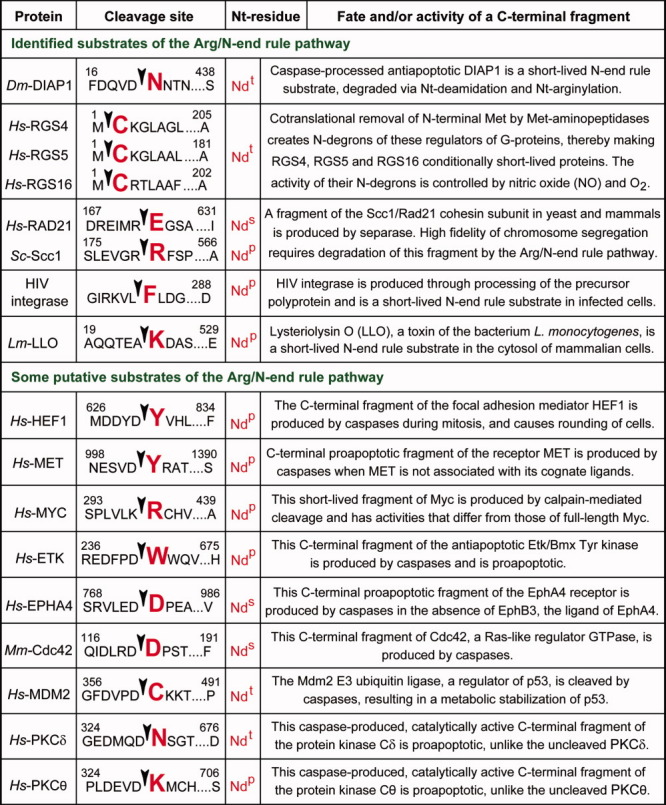
Confirmed and putative N-end rule substrates produced by caspases and other nonprocessive proteases. Amino acid residues are indicated by single-letter abbreviations. Arrowheads and enlarged residues, in red, indicate the cleavage sites and N-terminal residues of the corresponding C-terminal fragments. A number on the left represents the first residue of a protein (numbered as in the full-length protein) that is shown in the diagram. A number on the right represents the last residue of a full-length protein. The prefixes Dm, Hs, Mm, and Sc refer to proteins of D. melanogaster, H. sapiens, M. musculus and S. cerevisiae, respectively. See Substrates of the N-End Rule Pathway section for a description of specific protein fragments cited in this list. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In addition, conditional cleavages of cellular proteins by nonprocessive proteases such as caspases or calpains can produce C-terminal fragments that bear unmodified N-terminal Cys residues. If the protein's N-terminal Cys can be oxidized through (apparently nonenzymatic) reactions that require both nitric oxide (NO) and oxygen, and if these compounds are present in a cell at sufficient levels, the resulting N-terminal Cys-sulfinate or Cys-sulfonate (but not the original Cys) can be Nt-arginylated by the Ate1 R-transferase.32,33 The necessity of NO and oxygen for the destabilizing activity of N-terminal Cys makes the Arg/N-end rule pathway a sensor of both NO and oxygen (Functions of the N-end rule pathway vis-á-vis their mechanisms section). In sum, depending on specific protein substrates and in vivo conditions such as the presence of NO, the N-terminal Cys of a protein can function either as an NO/O2-mediated N-degron of the Arg/N-end rule pathway or, alternatively, as an AcN-degron of the Ac/N-end rule pathway3,32,33 [Figs. 2(B) and 3].
N-terminal Asp, Glu, Asn, Gln, and Cys that are targeted by the Arg/N-end rule pathway are termed “secondary” (Nds) or “tertiary” (Ndt) destabilizing residues, depending on the number of specific modifications (arginylation of Asp and Glu; deamidation/arginylation of Asn and Gln; oxidation/arginylation of Cys) that precede their targeting by N-recognins.15,32,33,36,41,42 Analogously, the N-terminal Met, Ala, Val, Ser, Thr, and Cys residues that become AcN-degrons after the Nt-acetylation of these residues are classed as Nds (secondary destabilizing N-terminal) residues, because they must be Nt-acetylated before their targeting by N-recognins of the Ac/N-end rule pathway3 [Fig. 2(B)].
Together, the Arg/N-end rule and Ac/N-end rule pathways target a majority of cellular proteins for regulated degradation. For example, more than 80% of human proteins are cotranslationally Nt-acetylated by a family of ribosome-associated Nt-acetylases that act after Met-aminopeptidases, which are also bound to the ribosomes89,91,92,95 [Fig. 2(B)]. Thus, remarkably, most proteins harbor a specific degradation signal (AcN-degron) from the moment of their birth.3 Posttranslational Nt-acetylation of proteins can occur as well (J.-H. Oh and A. Varshavsky, unpublished data), presumably because there is a significant pool of Nt-acetylases that are not bound to ribosomes.
In sum, N-degrons of the Ac/N-end rule and Arg/N-end rule pathways can be produced either cotranslationally or posttranslationally (and conditionally), by enzymes that include caspases, calpains, separases, other nonprocessive proteases, Nt-acetylases, Nt-amidases, and R-transferases. These enzymes function as upstream components of the N-end rule pathway, preparing its substrates for targeting and polyubiquitylation by N-recognins6,15,16 (Figs. 2 and 3). In contrast to Nt-arginylation in eukaryotes, N-end rule substrates that bear Nds residues in bacteria are Nt-leucylated by the Aat L/F-transferase or the Bpt L-transferase, which conjugate (largely) Leu, an Ndp residue, to N-terminal Nds residues of bacterial proteins, before their recognition by the ClpS N-recognin16,37 (Fig. 5).
Physiological functions of the N-end rule pathway are strikingly broad and continue to be discovered. Regulated degradation of proteins by the eukaryotic Arg/N-end rule pathway [Figs. 2(A) and 3] mediates the sensing of heme, NO, oxygen, and short peptides; the selective elimination of misfolded proteins; the regulation of DNA repair (through degradation of Mgt1, a DNA repair protein); the cohesion/segregation of chromosomes (through degradation of a subunit of cohesin); the signaling by transmembrane receptors (through degradation of the G-protein regulators Rgs4, Rgs5, and Rgs16); the control of peptide import (through degradation of Cup9, the import's transcriptional repressor); the regulation of apoptosis, meiosis, viral and bacterial infections, fat metabolism, cell migration, actin filaments, cardiovascular development, spermatogenesis, neurogenesis, and memory; the functioning of adult organs, including the brain, muscle, testis, and pancreas; and the regulation of leaf and shoot development, leaf senescence, and seed germination in plants (Refs. 3,6,15,16,18,26,32,34,36,39–42,113,119–136 and references therein). Mutations in UBR1, an E3 N-recognin of the human Arg/N-end rule pathway, cause Johanson–Blizzard syndrome (JBS). It comprises physical malformations, insufficiency and inflammation of the exocrine pancreas, frequent mental retardation, and deafness.34 Remarkably, an N-recognin such as mammalian UBR2 can also function to protect specific proteins from degradation.137 The recently discovered Ac/N-end rule pathway [Fig. 2(B)] is likely to mediate, among other things, protein quality control, the regulation of in vivo stoichiometry of proteins that form multisubunit complexes, and the degradation of long-lived proteins.3 Physiological roles of the bacterial (E. coli) N-end rule pathway include the regulated degradation of the Dps nucleoid-condensing protein and the YgjG putrescine aminotransferase.76,77
Terminology and Definitions
The terms used in this review are defined above and in Glossary (Glossary of Terms Section). The notations “Arg/N-end rule pathway” and “Ac/N-end rule pathway” (Figs. 2 and 3) should be applicable to any eukaryote, as they bring up a key modification, either Nt-arginylation (Arg) or Nt-acetylation (Ac) but do not invoke specific genes or proteins. In this terminology, the bacterial N-end rule pathway is called the Leu/N-end rule pathway, given its similarity (despite the absence of ubiquitylation) to the eukaryotic Arg/N-end rule pathway, with Nt-leucylation in bacteria versus Nt-arginylation in eukaryotes (Fig. 5).
As an experimentally observed but formal (nonmechanistic) relation between the regulation of the in vivo half-life of a protein and the identity of its N-terminal residue, the N-end rule does not place constraints on the nature of processing steps (e.g., proteolytic cleavages) or specific enzymes that produce N-degrons and implement the N-end rule pathway.5,6,15,16 An N-degron is classified as such if an unmodified or covalently modified N-terminal residue of a protein is an essential determinant of that protein's degradation signal. (A protein may contain, and often does, other degrons as well.) This function-based definition does not specify molecular devices that produce, recognize, or regulate N-degrons. It is also compatible with any route through which a destabilizing residue becomes N-terminal in a polypeptide. In sum, a feature that suffices to demarcate a processive proteolytic pathway as a branch of the N-end rule pathway is its ability to target specific N-terminal residues, unmodified or covalently modified.
Substrate-binding sites of an E3 N-recognin that targets N-degrons of protein substrates are apparently always accessible, whereas other sites of the same E3, the ones that target internal degrons of other protein substrates, can be autoinhibited. This autoinhibition can be allosterically reversed by ligands such as short peptides that bind to the sites of N-recognin that target N-degrons.27,29,38,138 Given these functionally important connections among different degron-recognizing sites of N-recognins, an intracellular protein is classified as a substrate of the N-end rule pathway if it is targeted by an N-recognin, irrespective of whether the targeting involves an internal degron of the protein or an N-degron.6 This hardware-centric (N-recognin-based) definition of substrates of the N-end rule pathway bypasses the semantically intractable issue of multiple-binding sites in N-recognins.
Substrates of the N-End Rule Pathway
An N-degron can be produced from a pro-N-degron (precursor of N-degron) through a cotranslational or posttranslational proteolytic cleavage. Ribosome-associated Met-aminopeptidases cleave off the Met residue from the N-terminus of a nascent protein if the residue at Position 2, to become N-terminal after cleavage, has a small enough side chain100,101 [Fig. 1(D)]. Consequently, of the 13 residues that are destabilizing in the mammalian Arg/N-end rule pathway, only Cys can be made N-terminal by Met-aminopeptidases [Figs. 1(D) and 3]. (Any destabilizing residue, including Cys, can be made N-terminal through posttranslational cleavages of proteins by other nonprocessive proteases.6,28) In contrast to larger residues at Position 2, the second-position Ala, Val, Ser, Thr, or Cys can be made N-terminal by Met-aminopeptidases3 [Fig. 1(D)]. These residues, which are usually Nt-acetylated, are the secondary destabilizing (Nds) residues of the Ac/N-end rule pathway3 [Fig. 2(B)]. The initial N-terminal Met of a nascent protein is also an Nds residue of the Ac/N-end rule pathway, the only such residue that does not require a preliminary proteolytic cleavage to form an AcN-degron. If N-terminal Met is followed by a bulky residue, this Met is not cleaved off, and is usually Nt-acetylated3 [Figs. 1(C,D) and 2(B)].
More than 80% of mammalian proteins are cotranslationally Nt-acetylated.91–93 About 20 Nt-acetylated S. cerevisiae and mammalian proteins, chosen nearly at random, have been examined, thus far, for the presence of AcN-degrons, using methods that included pulse-chase and cycloheximide-chase assays, as well as genetic techniques. Nearly every one of the tested Nt-acetylated proteins was found to contain an AcN-degron (Ref. 3; A. Shemorry et al., unpublished data). Given these results and the pervasiveness of Nt-acetylation, our current premise is that most cellular proteins can become substrates of the Ac/N-end rule pathway, either during their synthesis, or immediately afterwards, or significantly later [Fig. 2(B)]. In contrast, the Arg/N-end rule pathway appears to target fewer substrates (but still hundreds of them; see below), in part because most N-degrons of this pathway are produced posttranslationally, through cleavages by proteases other than Met-aminopeptidases. That is so because second-position residues that are destabilizing in the Arg/N-end rule pathway are too large to be made N-terminal by Met-aminopeptidases [Figs. 1(D), 2(A), and 3]. The sole exception is Cys [Fig. 4(A)]. Specifically, the N-degrons of the mammalian Rgs4, Rgs5, and Rgs16 proteins (Overview of the N-End Rule Pathway and Functions of the N-end rule pathway vis-á-vis their mechanisms sections) would be expected to form cotranslationally or nearly so in the presence of NO and other conditions that are conducive to oxidation of N-terminal Cys (Figs. 3 and 10). N-degrons also form cotranslationally in engineered N-end rule substrates that are expressed in vivo as Ub-X-protein fusions (in which X is a destabilizing residue), because deubiquitylases cotranslationally remove the fusion's Ub moiety.9,24,139 In addition, UBR-type E3 N-recognins of the Arg/N-end rule pathway recognize not only N-degrons but also internal (non-N-terminal) degradation signals [Figs. 2(A) and 3]. Proteins that lack N-degrons but contain these internal degrons comprise yet another set of substrates of the Arg/N-end rule pathway. Only a few (out of probably many) such substrates have been identified so far (Functions of the N-end rule pathway vis-á-vis their mechanisms section).
Physiological substrates of the eukaryotic Arg/N-end rule pathway include the Drosophila melanogaster DIAP1 regulator of apoptosis140,141; the mammalian Rgs4, Rgs5, and Rgs16 regulators of G proteins32,33,118; the C-terminal fragments of the Scc1/Rad21 cohesin subunit that are produced by separase in eukaryotes from yeast to mammals (ref. 28; J. Zhou et al., unpublished data); the human immunodeficiency virus-1 (HIV-1) integrase132,133; and the Listeria monocytogenes listeriolysin (Lys-LLO), which is secreted by this bacterium into the cytosol of infected mammalian cells124 (Fig. 10 and Functions of the N-end rule pathway vis-á-vis their mechanisms section). In addition, the S. cerevisiae Mgt1 DNA repair protein and the Cup9 transcriptional repressor are targeted by the Arg/N-end rule pathway through their internal degrons (Functions of the N-end rule pathway vis-á-vis their mechanism section). Figure 10 and a brief description below cite the currently known substrates of the Arg/N-end rule pathway (other than Mgt1 and Cup9) and a few putative (unverified) N-end rule substrates that are a part of a much larger set of such substrates (A. Varshavsky, unpublished data). For some of these protein fragments, published evidence suggests their metabolic instability; other fragments have not been examined in this regard.
C-terminal fragment of HEF1 (a focal adhesion-associated docking protein) that bears N-terminal Tyr (an Ndp residue) and is produced by caspases not only during apoptosis but also during normal mitosis142,143 (Fig. 10).
C-terminal fragment of the MET tyrosine kinase, a transmembrane receptor of HGF/SF, a hepatocyte growth factor-scatter factor. This fragment of MET is produced by caspases (if MET is not bound to HGF/SF) and bears N-terminal Tyr, an Ndp residue.144–147 MET is a member of the family of more than 10 mammalian “dependence” receptors (DRs). These transmembrane receptors are usually not related by sequence or structure but are functionally analogous because of their ability to mediate two opposite physiological outcomes. In the presence of its cognate ligand, a DR receptor activates signaling pathways that mediate cell survival, migration, and differentiation. However, in the absence of a cognate ligand, a DR receptor acquires an “opposite” activity, that is, it produces an apoptotic signal, often through the formation, by caspases or other nonprocessive proteases, of a proapoptotic C-terminal cytosolic fragment(s) that functions in the cytosol and/or the nucleus.146,147
C-terminal fragment of the MYC oncoprotein, termed MYC-nick,148 that is produced by calpain(s), bears N-terminal Arg (an Ndp residue), and exhibits physiological activities that are different from those of full-length MYC.
C-terminal fragment of the ETK/BMX tyrosine protein kinase that is produced by caspases, bears N-terminal Trp (an Ndp residue), and is proapoptotic.149
C-terminal fragment of the transmembrane EPHA4 “dependence” receptor (item 2 above146,147) that is produced by caspases (if EPHA4 is not bound to its ligand EPHB3) and bears N-terminal Asp, an Nds residue.150
C-terminal fragment of the mouse Cdc42 GTP-binding protein that is produced by caspases and bears N-terminal Asp, an Nds residue.151
C-terminal fragment of the MDM2 E3 Ub ligase (whose targets include p53) that is produced by caspases and bears N-terminal Cys, an Ndt residue.152
C-terminal fragment of the protein kinase Cδ (PKCδ) that is produced by caspases, bears N-terminal Asn (an Ndt residue), and is proapoptotic, in contrast to the full-length PKCδ kinase.153,154
C-terminal fragment of the protein kinase Cθ (PKCθ) that is produced by caspases, bears N-terminal Lys (an Ndp residue), and is proapoptotic, in contrast to the full-length PKCθ kinase153,155 (Fig. 10).
Many more substrates of the Arg/N-end rule pathway that contain N-degrons are likely to exist in mammals, but they remain either putative or unknown, given the logistics and uncertainties of current proteome-scale assays, and also because of insufficient knowledge about nonprocessive (and conditional) cleavages of intracellular proteins that yield in vivo N-end rule substrates. The expected multitude of such substrates stems from the existence of nonprocessive proteases that function, in particular, in the nucleus and/or cytosol and are known or expected to produce C-terminal fragments of specific proteins that bear destabilizing N-terminal residues of the Arg/N-end rule pathway. These proteases include Met-aminopeptidases,96–101 caspases,156–160 calpains, separase,161,162 taspase,163 MALT1 protease,164 γ-secretase,165 proteinase-3 (PR3),166 and viral proteases.131–133 It should be emphasized that caspases cleave specific intracellular proteins not only in settings that lead to apoptotic cell death but also in pathways of cell differentiation that do not result in cell death.158,159 In addition, if the cleavage of a protein by a caspase produces a proapoptotic C-terminal fragment, it being a short-lived N-end rule substrate would counteract apoptosis, and therefore would “buffer” a cell against toxicity of caspases that become active owing to a significant level of noise in caspase-activation circuits (Functions of the N-end rule pathway vis-á-vis their mechanisms section).
In addition to the substrates cited in Figure 10, several other likely in vivo substrates of the Ate1 R-transferase (Figs. 2A and 3) include protein disulfide isomerase (PDI), glucose-regulated protein 78 (Grp78), β-actin, γ-actin, and calreticulin.35,127,128,130,135,136 Although the Ate1 R-transferase and the rest of the Arg/N-end rule pathway are apparently confined to the cytosol and the nucleus, and although calreticulin, Grp78 and PDI are present largely in the lumen of the endoplasmic reticulum (ER), a variety of evidence suggests that these proteins (lacking their cleaved-off signal sequences and bearing N-terminal Nds residues) are also present in the cytosol and other non-ER compartments, where they may become R-transferase substrates.35,136 Partial Nt-arginylation of apparently long-lived proteins such as β-actin and calreticulin128,135,136 suggests that Nt-arginylation may have nonproteolytic functions as well.
Hamilton et al.167 characterized Go/Gi heterotrimeric G proteins purified from bovine brains. The bulk of the Gγ2 subunit of the Go protein had the expected N-terminal sequence Ac-ASNNTASIA, produced through the removal of N-terminal Met by Met-aminopeptidases and the (presumably) cotranslational Nt-acetylation of N-terminal Ala [Overview of the N-End Rule Pathway and The Ac/N-End Rule Pathway sections, and Fig. 2(B)]. However, a minor but significant fraction of the Gγ2 subunit of Go protein had the N-terminal sequence RDTASIA. Such a sequence would be produced from ASNNTASIA via the removal (through a single cleavage or sequential proteolysis) of the first three residues (after initial Met) by an unknown protease, deamidation of the resulting N-terminal Asn by the Ntan1 NtN-amidase (Fig. 3 and N-terminal deamidation in the Arg/N-end rule pathway section) and Nt-arginylation of the resulting N-terminal Asp by the Ate1 R-transferase.167 The Arg-Gγ2 protein produced from engineered Ub-Arg-Gγ2 using the Ub fusion technique [Fig. 1(B)] was a short-lived N-end rule substrate in reticulocyte extract.167 It remains to be determined whether Arg-Gγ2 is a physiological N-end rule substrate, because the still unexcluded possibility is that Arg-Gγ2 might be produced through in vitro proteolysis, Nt-deamidation, and Nt-arginylation in crude extracts during purification of G proteins.167
The currently known physiological substrates of the bacterial Leu/N-end rule pathway (Fig. 5) are discussed in Prokaryotic N-End Rule Pathways section.
Structure and Targeting of N-degrons
Mechanistic aspects of the N-end rule pathway that are critical for its functions include the regulation of E3 N-recognins, for example, through a specific phosphorylation cascade39 (Fig. 7) and also through changes in the activity/accessibility of degrons in N-end rule substrates.27,29,138 A key mechanistic capability of the N-end rule pathway is its subunit selectivity,13,168 that is, the ability to selectively target and destroy a subunit of a protein complex while sparing the rest of it [Fig. 11(A-D)]. Examples of subunit-selective protein remodeling by the Arg/N-end rule pathway are described in Functions of the N-end rule pathway vis-á-vis their mechanisms section. Although degrons that are targeted by the Ub system are many and varied, their design is fundamentally similar to the multideterminant organization of N-degrons, the first primary degradation signals in short-lived proteins to be discovered and analyzed4,5,9,17,19,24 [Fig. 11(A)].
Figure 11.
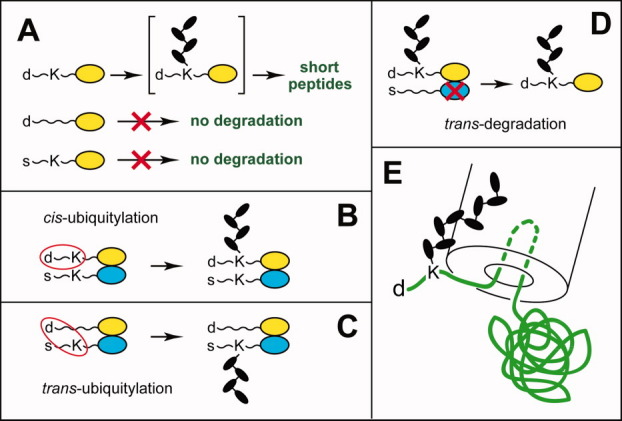
Organization and cis-trans targeting of eukaryotic N-degrons. A: Three determinants of N-degron. d, a destabilizing N-terminal residue. K, a “ubiquitylatable” internal Lys residue. The absence of one of these determinants abrogates polyubiquitylation of a protein, despite the presence of another determinant. The third determinant of N-degron is an unstructured region that is required for polyubiquitylation and/or the initiation of degradation of a polyubiquitylated N-end rule substrate by the 26S proteasome. See Substrates of the N-End Rule Pathway section for references and details. B and C: cis versus trans polyubiquitylation of an oligomeric N-end rule substrate that results in the degradation of a subunit that becomes linked to a poly-Ub chain13 (see Substrates of the N-End Rule Pathway section). D: trans-degradation, in which a specific subunit of oligomeric protein is polyubiquitylated but is not degraded by the 26 proteasome, for example, because it lacks an unstructured region that is required for the initiation of degradation. Instead, a subunit-selective degradation of another, nonubiquitylated subunit takes place. This mode of degradation was demonstrated by the Matouschek laboratory168 for oligomeric substrates of the UFD pathway. It remains to be determined whether the analogous (hypothetical) trans-degradation of an oligomeric N-end rule substrate can also occur. E: The 1989–1996 hairpin insertion model of protein targeting by the 26S proteasome.5 No details of the 26S proteasome structure (such as the 19S regulatory particle (RP)) are shown in this 1996 diagram,5 and the sizes of specific components such as Ub moieties, the poly-Ub chain and the proteasome, are not to scale (see Substrates of the N-End Rule Pathway section). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The activity of N-degrons and other Ub-mediated degradation signals is a function of several variables.5,9,17,19,168–171 One of them is the efficacy of a degron's first determinant, that is, a region of a protein substrate that is recognized by a cognate E3 Ub ligase. In the case of an N-degron that is targeted by the Arg/N-end rule pathway, this would be either an original or “acquired” primary destabilizing (Ndp) residue [Figs. 2(A) and 3]. Residues downstream from a substrate's N-terminal Ndp residue, and particularly a residue at Position 2, can modulate the binding of an E3 N-recognin to an N-end rule substrate.38,83,84 If an initially formed N-degron (produced from a pre-N-degron) contains a secondary (Nds) or a tertiary (Ndt) destabilizing residue, the efficacy of N-degron's first determinant would be determined, in addition, by the rate(s) of covalent modification(s) of the initial N-terminal residue that eventually yields an Ndp residue that can be bound by a cognate N-recognin. Thus, the levels and activity of Nt-amidases and/or R-transferase [Figs. 2(A) and 3] would be expected to influence the corresponding Nds/Ndt-based N-degrons. Yet another parameter that influences these preliminary stages of N-degron's targeting is the extent of steric exposure of an Ndp residue and the extent of flexibility of a protein's N-terminal region that would be expected to facilitate the binding of an Ndp residue by an N-recognin.5,9
Once the N-terminal Ndp residue of a substrate is bound by a complex of N-recognin E3 and its associated Ub-conjugating (E2) enzyme, a race against time begins, given the transiency of the bound state and the necessity to produce a substrate-linked poly-Ub chain that is required for downstream targeting steps. The synthesis of a poly-Ub chain (usually but not always a Lys48-type chain) is initiated, in most cases, at an internal Lys residue of the substrate.9,11,168 In eukaryotes, this internal lysine is the second determinant of an N-degron [Fig. 11(A)]. In some N-end rule substrates, only one internal lysine may be appropriately positioned for a kinetically efficacious attack on the thioester bond (denoted as ∼) between E2 and Ub in the substrate-associated E3/E2∼Ub complex. In other cases, including engineered N-end rule substrates,9,20,24,168 more than one Lys residue of a substrate is capable of such an attack. Its successful completion usually preempts participation by alternative lysines and is followed by a processive synthesis of a substrate-linked poly-Ub by a substrate-bound N-recognin E2/E3 Ub ligase. For example, there are two efficaciously “ubiquitylatable” Lys residues, at Positions 15 and 17, in an unstructured ∼40-residue N-terminal extension of the 21-kDa mouse dihydrofolate reductase (DHFR), an engineered N-end rule substrate.9 Either one of those lysines could be polyubiquitylated during the targeting, that is, just one of two lysines (either one) had to be present for the activity of N-degron.9 The otherwise identical but lysine-lacking N-terminal extension (with both lysines converted to Arg) did not result in the reporter's degradation by the Arg/N-end rule pathway, despite the presence of a destabilizing N-terminal residue9 [Fig. 11(A)].
In sum, the efficacy of a second-determinant Lys residue of a substrate's N-degron is determined by the lysine's spatial proximity to E2∼Ub thioester of the targeting complex, and also by the extent of flexibility of a region containing the requisite lysine or lysines. This understanding of the necessity of an unstructured region (a requirement that is relevant to other Ub-dependent degrons as well) was produced by Bachmair and myself9 in 1989, in conjunction with the discovery, by Chau et al.11 in our laboratory, of the Lys48-type poly-Ub chains and their necessity for protein degradation. We suggested that the main function of a substrate-linked poly-Ub chain is its physical binding to a specific site of the proteasome. By decreasing the rate of substrate dissociation, the resulting retention, through a poly-Ub chain, of a targeted substrate at the 26S proteasome would increase the probability of substrate unfolding and degradation by other components of the proteasome.9,11 In 1989, specific “downstream” degradation steps and their mediator, the 26S proteasome, were just beginning to be defined experimentally, by several laboratories. With the above understanding of targeting in place by that time, it remained to be determined whether the demonstrated requirement for an unstructured region in N-end rule substrates9 was important only in the context of that region's polyubiquitylation. In an alternative mechanism, the same region, or a similar (unstructured) but separate region of a substrate would also be required for a proteasome-mediated step that would initiate the processive degradation of a substrate that had been captured through its poly-Ub chain.
As predicted in 1989 (Refs. 9 and11), the 19S regulatory particle (RP) of the 26S proteasome was eventually shown to contain specific subunits that bind to a poly-Ub chain (ref. 54 and references therein). In one of possible models, the interaction of the 26S proteasome with a substrate-linked poly-Ub chain and the resulting delay in dissociation of the substrate from the proteasome would allow the ATP-dependent unfoldases of RP to unfold a previously structured region of the substrate and to insert it into the proteolytic chamber, thereby initiating processive proteolysis. An unfolded region could be, for example, the C-terminus or the N-terminus of substrate or, nonalternatively, an internal region, in which case it would be a hairpin loop whose insertion into the chamber would initiate processive proteolysis. The hairpin-insertion model of the proteasome-substrate interaction was suggested by us in 1989, and more explicitly in 1996. Figure 11(E) illustrates the original model,5,9,11 which was proposed before the structural understanding of RP, its poly-Ub-binding subunits and other aspects of the 26S proteasome.
In this mechanism, an unstructured region of a substrate that encompasses its polyubiquitylated lysine may perform a “double” duty of being important not only for polyubiquitylation but also for downstream, proteasome-mediated steps. A priori, it was also possible that the 26S proteasome might not strictly require an unstructured region in a substrate that had been captured through its poly-Ub chain, because unfoldases of RP might be able to initiate, efficiently enough, the ATP-mediated unfolding of a substrate (held by its poly-Ub chain) through thermal fluctuations alone, even in the absence of unstructured regions. Recent studies of N-degrons and other Ub-dependent degrons by the Matouschek, Jentsch, Dantuma, Coffino, and other laboratories showed that the Ub-mediated protein degradation by the 26S proteasome does require the presence of an unstructured “initiation” region in a substrate that has been captured by the proteasome through its poly-Ub chain. It was also shown that although the location of such a region in a polyubiquitylated substrate is not determined rigidly by the location of a branch point containing poly-Ub, the unstructured region should reside at an optimal distance from the branch point.19,54,169,171–176
Thus, the degradation, by the Arg/N-end rule pathway, of a protein containing an N-degron requires (i) the first determinant of N-degron, that is, an N-terminal Ndp residue that can be recognized by a cognate E3 N-recognin (an Ndp residue is exposed by a proteolytic cut either directly or after modifications of initially exposed Nds or Ndt residues); (ii) the second determinant of N-degron, that is, a Lys residue(s), which functions as the site of formation of a poly-Ub chain and usually resides in an unstructured region of a substrate; and (iii) a sterically “suitable” unstructured region (either the same region that encompasses the Ub attachment site or another region) that serves as the initiation site for the unfolding of a captured substrate by the 26S proteasome.9,169 The latter requirement defines the third determinant of an N-degron. Yet another mechanistic aspect of N-degrons involves a complex of the chaperone-like ATPase Cdc48 (p97) with specific accessory proteins. This complex binds to several components of the Ub system, interacts with polyubiquitylated proteins “upstream” of the 26S proteasome, and facilitates protein degradation in ways that are incompletely understood.177–179 Despite these and other complexities, an N-degron can be an efficacious and portable degradation signal, capable of conferring extremely short (1–2 min) in vivo half-lives on either newly formed or conformationally mature proteins.4,9,21,24
It is likely (but remains to be verified) that the degradation of naturally unstructured (disordered) eukaryotic N-end rule substrates, while still requiring the targeting by a cognate N-recognin E3, may not require ubiquitylation. This possibility is made particularly likely by the previously demonstrated physical affinity of both the Ubr1 E3 (N-recognin) and its associated Ufd4 E3 [Fig. 2(A)] for specific subunits of the 26S proteasome.44,180,181 Another setting in which eukaryotic N-degrons may also act analogously to Ub-independent prokaryotic N-degrons is the previously analyzed Ubr1-mediated cotranslational degradation of N-end rule substrates as they are being made, that is, directly at the ribosome,139 apparently under conditions where the polypeptide chain that is being destroyed is targeted as a peptidyl-tRNA. The extent and mechanisms of the in vivo degradation of nascent proteins (i.e., of specific peptidyl-tRNAs) and newly formed (just completed) proteins, in comparison with degradation of the same proteins significantly after their synthesis21,24,139,182–185 is an extensively investigated but insufficiently understood subject. Given this problem's spatiotemporal and technical complexity, definitive advances in such studies would be likely to require new methods.
The understanding of degradation of N-end rule substrates by the Ub-independent bacterial Leu/N-end rule pathway (Fig. 5) is summarized in Figure 12. It describes a detailed model proposed by Román-Hernandez et al.82 and based on studies by the Baker, Sauer, Bukau, Maurizi, and other laboratories.16,75,76,78,81,82 Most propositions of this model are supported by specific evidence, including crystal structures of ClpS, its mutant derivatives, and complexes of ClpS with peptide mimics of N-end rule substrates73–75,79,186–189; crystal structures of ClpA and the ClpAP protease190–192; and single-molecule measurements of protein translocation and degradation by the ClpAP-like ClpXP protease.193,194 The targeting begins when the ClpS N-recognin binds to an Ndp residue of an N-end rule substrate (with Kd of ∼1 μM) and delivers the substrate to one of six ClpA subunits of the ClpAP protease. ClpAP consists of the ClpA6 unfoldase and the ClpP14 protease, in a complex that includes the axial pore in the ClpA6 hexamer that leads to the proteolytic chamber of the ClpA6-associated ClpP14 (Fig. 12).
Figure 12.
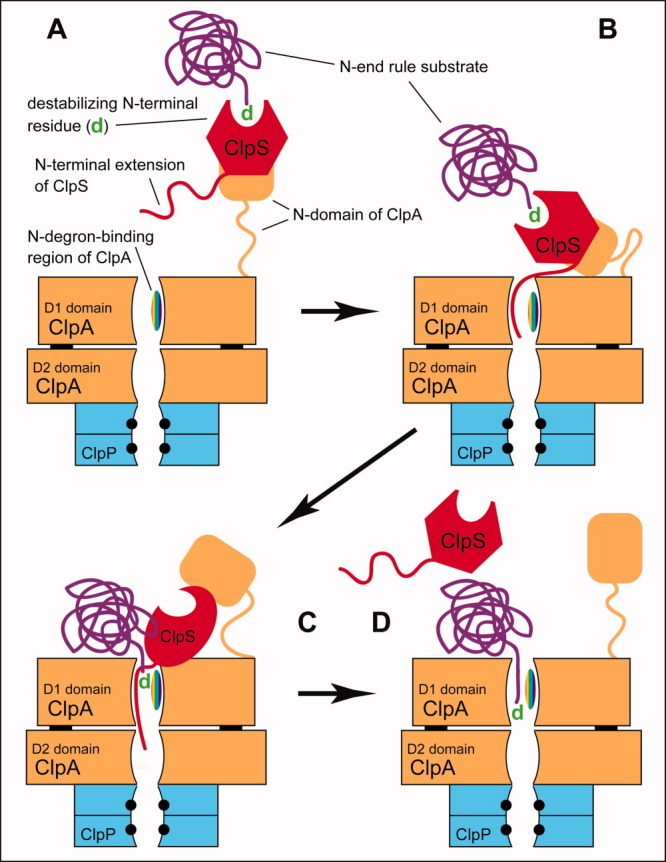
Targeting and degradation of N-end rule substrates by the bacterial Leu/N-end rule pathway. This model, proposed by Román-Hernandez et al.82 and based on studies by the Baker, Sauer, Bukau, Maurizi, and other laboratories,16,75,76,78,81,82 is described in the main text. A folded polypeptide chain of an N-end rule substrate (in purple color) is depicted “explicitly”, in contrast to solid-body renderings of ClpS and ClpAP. Black circles in the ClpP moiety indicate its proteolytic active sites. See Substrates of the N-End Rule Pathway section for details. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
A substrate-bound ClpS monomer interacts with ClpA of the ClpA6 hexamer ∼10-fold more tightly than free ClpS does.82 This binding preference “solves” the problem of competition between free and substrate-bound ClpS for ClpS-binding sites on ClpA6. In addition, the interaction between ClpS and ClpA6 exhibits negative cooperativity, that is, only one molecule of ClpS binds to ClpA6 with high affinity.78 The 12-kDa ClpS consists of the flexible N-terminal (Nt) extension and the folded core domain, which binds to an N-degron (the N-terminal Ndp residue) of an N-end rule substrate [Fig. 6(B–D)]. The substrate-bound ClpS interacts with ClpA at the N-domain of ClpA. This domain is connected to the rest of ClpA by a flexible linker region. A key feature of the model is a conversion of the initial “low-affinity” ternary complex [Fig. 12(A)] to a “high-affinity” delivery complex [Fig. 12(B,C)], in which a part of the previously free Nt-extension of ClpS becomes bound to the ClpA6 axial pore (near its entrance), thereby initiating the transfer of the ClpS-bound N-end rule substrate (its N-degron) from ClpS to a region inside the ClpA6 pore. Previous evidence (ref. 195 and references therein) suggests that this region of the ClpA6 pore exhibits a ClpS-like (but significantly weaker) affinity for a bulky hydrophobic (Ndp) N-terminal residue. In the resulting arrangement, a conformational change of ClpA, fueled by ATP hydrolysis, pulls on the pore-bound Nt-extension of ClpS, perturbs the conformation of the core domain of ClpS and thereby both weakens its binding to N-degron of a substrate and facilitates the transfer of substrate (its N-degron region) from ClpS to a site in the ClpA6 pore [Fig. 12(C,D)]. As is also the case with eukaryotic N-end rule substrates, “optimal” substrates of the bacterial Leu/N-end rule pathway contain features that are additional to the presence of a bulky hydrophobic N-terminal residue. These features include an unstructured region near the N-terminal destabilizing residue, an under-representation of acidic residues in that region, and a stretch of two to three hydrophobic residues 6–12 residues from the substrate's N-terminus.72,76,77
One experimentally supported assumption of the model is the possibility of more than one polypeptide chain occupying the pore of a ClpA-type unfoldase at the same time196,197 [Fig. 12(C)]. Another aspect of the model82 is resistance of the folded core of the ClpA-bound ClpS to unfolding by the ATP-fueled pull on the Nt-extension of ClpS [Fig. 12(C)]. This resistance, accompanied by a conformational perturbation of ClpS, prevents degradation of the Nt-extension and the rest of ClpS and, in addition, facilitates the dissociation of ClpS from ClpA, thereby leaving the Ndp residue (N-degron) of the substrate inside the pore and completing the targeting cycle [Fig. 12(D)]. Motor proteins operate through power strokes, through a biased Brownian motion (Brownian ratchet), or through a combination of these mechanisms.193,198–200 Experimental evidence favors a mechanism of the ClpA6 motor that relies predominantly on a power stroke.193,194 Once the N-terminal region of an N-end rule substrate is brought inside the pore of the ClpA6 motor [Fig. 12(C,D)], the ATP-dependent, presumably cyclic conformational changes of the pore region lead to a stepwise propulsion of the polypeptide chain of a captured substrate (accompanied by its unfolding) into the proteolytic chamber of the ClpA6-associated ClpP14. The result is processive degradation of the substrate to short peptides.16,75,76,78,81,82
Ub-lacking proteolytic circuits such as, for example, the bacterial Leu-N-end rule pathway (Figs. 5, 6, and 12) are significantly less complicated, composition-wise, and apparently also mechanism-wise, than Ub-dependent degrons and the targeting/degradation machinery in eukaryotes. The remarkable complexity (including compositional complexity) of the Ub system raises the question of how prokaryotes, which are obviously sophisticated about processive proteolysis,200,201 get by largely without Ub and ubiquitylation. Both bacteria and archaea have proteins that are spalogous (spatially similar1) to Ub in that they contain the β-grasp Ub fold.202 Most archaea and some bacteria contain proteolytic pathways that involve conjugation of Ub-like proteins to other proteins.66–70,203–207 However, a large fraction of processive proteolysis in extant bacteria does not appear to be mediated by Ub-like pathways. For example, in the bacterial Leu/N-end rule pathway, the ClpS N-recognin binds to N-end rule substrates and delivers them to the ClpAP protease for processive degradation without involving a ubiquitylation-like mechanism (Figs. 5 and 12).
One possibility is that the mechanistic complexity of the eukaryotic Ub system stems, at least in part, from its additional capabilities that are required in eukaryotes but not in prokaryotes. For example, some features of the subunit-selective proteolysis by the Ub system that are physiologically important in eukaryotes (Functions of the N-end rule pathway vis-á-vis their mechanisms section) might not be necessary in bacteria.13,168,208 This conjecture remains to be verified. In addition, the ER-associated degradation of proteins that travel through or reside in the secretory pathway is functionally essential and Ub-mediated in eukaryotes52,59,63,209 but a counterpart of this process in bacteria is Ub-independent. Specifically, bacteria use sophisticated protein quality-control pathways in the periplasmic space (analogous to the ER lumen in eukaryotes) but do not appear to use ubiquitylation-like mechanisms.
It is also possible that an aspect of folding of some eukaryotic proteins (vs. the presumed absence of such proteins in bacteria) presents a sufficient challenge to the eukaryotic 26S proteasome to require devices such as poly-Ub chains and associated machinery for destruction of such proteins. In other words, the Ub/proteasome-mediated processive proteolysis may have a greater protein-unfolding “power” than analogous mechanisms that lack polyubiquitylation. This conjecture is not precluded but is unlikely to be a sufficient explanation, in part because the degradation of one and the same N-end rule substrate can be shown to be Ub-dependent in eukaryotes but Ub-independent in bacteria.14 Specifically, Arg-eK-β-galactosidase (Arg-eK-βgal) is an engineered ∼110-kDa (as a monomer) N-end rule substrate derived from E. coli βgal (LacZ) and produced, using the Ub fusion technique [Fig. 1(B)], through the cotranslational in vivo deubiquitylation of Ub-Arg-eK-βgal.4,9,21,24 Arg-eK-βgal bears N-terminal Arg, an Ndp residue in eukaryotes, and an Nds residue in bacteria [Fig. 2(A) and 4]. Arg-eK-βgal also contains, between N-terminal Arg and the βgal moiety, a ∼40-residue region termed eK [extension (e) containing lysine (K)]. The apparently unstructured eK extension9,21,24 has the technically valuable property of lacking internal degrons while containing two “ubiquitylatable” Lys residues, Lys15 and Lys17. Arg-eK-βgal is polyubiquitylated and short-lived in S. cerevisiae, provided that its eK extension contains at least one of these two lysines. Specifically, Arg-eΔK-βgal, in which both Lys15 and Lys17 were converted to (nonubiquitylatable) Arg residues, is long-lived in S. cerevisiae, despite the presence of the N-terminal Arg residue.9,24 In contrast, both Arg-eK-βgal and Arg-eΔK-βgal are degraded at similar rates by the E. coli Leu/N-end rule pathway.14 Thus, at least in this case, the ubiquitylation/proteasome machinery of eukaryotes “imposes” the requirement of ubiquitylation that is unnecessary in bacteria, vis-á-vis the same reporter protein.
Yet another, nonalternative possibility is that the Ub system might have indeed been “overdesigned” in the course of eukaryotic evolution, that is, that bacterial-type mechanisms, with fewer components and without extensive use of ubiquitylation, can accomplish what the Ub system does. An overdesign might have happened for reasons, suggested by Lynch,210 that are generally relevant to evolution of eukaryotes. A population genetics-based argument can be made that many aspects of molecular circuits in eukaryotic cells have evolved, at least initially, through a quasi-random, recurrent genetic drift. The relative importance of evolution by this route (as distinguished from adaptive Darwinian evolution through positive selection) depends on the breeding system of a species, on the organization of its genome, and on its history of long-term population bottlenecks. A small population size is likely to have been a recurrent characteristic of early eukaryotic evolution, at the time of emergence of a “stem” eukaryote, roughly 1.5 billion years ago. Most new alleles of a genetic locus are either deleterious or nearly neutral. The probability of fixation of a new allele (vis-à-vis the probability of its disappearance from a population) is higher in a smaller population.210 Under such conditions, complicated circuits might have evolved, to a significant extent, through occasional fixations of mildly deleterious mutations that were eventually “compensated” by suppressor mutations that often had their own fitness costs and were compensated by yet additional suppressors.
In sum, complexity in designs of biological circuits is not always a sign of adaptive evolution alone, because complexity can also result from a recurrent, long-term, sometimes deleterious genetic drift. (The relatively large sizes and numbers of introns in mammals, vis-à-vis much lower sizes and numbers of introns in fungi and their absence in prokaryotes are one example of evolution, in multicellular eukaryotes, that is partially drift-based and population size-dependent.210) The history of a species contains, in differing proportions, both a drift mode and an adaptive (Darwinian) mode. The population sizes of prokaryotes are (and were) vastly larger than those of eukaryotes, indicating that populations of prokaryotes are much more resistant than eukaryotes to evolutionary changes that stem from genetic drift, as distinguished from positive selection.210 If the primordial Ub system emerged initially to a large extent through a genetic drift, this would account, at least in part, for the absence of the bona fide Ub system in extant prokaryotes. The S. cerevisiae and human Ub systems are remarkably similar, differing mostly by a larger number of human E3 Ub ligases and associated machinery. Thus, the design of the Ub system must have reached its essentially modern state in early eukaryotes, before the separation of fungal and animal lineages. In sum, although an adaptive Darwinian evolution undoubtedly played a major role in making the extant Ub pathways what they are, it is likely that the Ub system emerged and became “entrenched” in primordial eukaryotes to a large extent through a genetic drift unaccompanied (at the beginning) by a significant positive selection,80 a general pattern of evolution discussed by Lynch.210
Because an N-degron consists of several determinants, subunit-selective polyubiquitylation and degradation of an oligomeric protein can occur not only in cis (with all determinants of an N-degron in the same polypeptide chain) but also in trans13 [Fig. 11(B,C)]. In the resulting subunit-selective protein degradation, discovered in 1990 in the context of the Arg/N-end rule pathway and found to be characteristic of the entire Ub system as well, a destabilizing N-terminal residue of one protein subunit can be engaged by an N-recognin to direct selective degradation of another subunit in the same oligomeric protein.13 This capability makes possible protein remodeling, a major functional attribute of the Ub system. Regulatory circuits wield subunit-selective proteolysis for either positive or negative control, including transitions in the cell cycle, the control of transcription and DNA replication, and many other processes. Among specific examples are the activation of a major transcription factor NF-κB, through the degradation of its inhibitory subunit IκB, and the inactivation/alteration of cyclin-dependent kinases (which drive the cell cycle) through the subunit-selective degradation of their cyclin subunits.211,212 Matouschek and coworkers19,168 have shown that the location and properties of an unstructured region of a protein substrate that serves as the proteasome-initiation site can determine which subunit is degraded. For example, a polyubiquitylated subunit of an oligomeric protein that is delivered to the 26S proteasome via its poly-Ub chain can be resistant to degradation and direct the subunit-selective destruction of a nonubiquitylated subunit in the same oligomeric protein.168 In the cited study, this trans-degradation effect168 was demonstrated with substrates of the Ub-fusion degradation (UFD) pathway (see The double-E3 design of the Arg/N-end rule pathway section). Given the likely generality of this mechanism, Figure 11(D) illustrates the (hypothetical) possibility of a trans-degradation of this kind for an oligomeric substrate of the N-end rule pathway.
Another important property of N-end rule circuits is exemplified by the S. cerevisiae Ubr1 E3 N-recognin. The 225 kDa Ubr1 [Fig. 7(A)] contains at least four substrate-binding sites. Two of them recognize specific N-degrons, whereas the other sites recognize internal (non-N-terminal) degrons. The occupancy of Ubr1 sites that recognize N-degrons has been shown to regulate, in physiologically relevant ways, the activity of another binding site of the Ubr1 N-recognin27,29,38–40 (Functions of the N-end rule pathway vis-á-vis their mechanisms section). The multiplicity of substrate-binding sites in Ubr1 and their allosteric dependencies are likely to recur in other E3 N-recognins as well.
Applications of the Ubiquitin Fusion Technique and N-degrons
The Arg/N-end rule pathway [Fig. 2(A)] was discovered through the invention of the Ub fusion technique4,9 [Fig. 1(B)]. This strategy was later used to identify the bacterial Leu/N-end rule pathway as well, via expression of the yeast Ubp1 deubiquitylase in E. coli.14 The Ub fusion technique is still the method of choice for expressing, in vivo, a protein of interest that bears a desired N-terminal residue20,24,213 [Fig. 1(B)]. Over the last two decades, this approach gave rise to several “descendant” technologies, in the Ub field and beyond.6,139,213–219 One example is a heat-activated, portable N-degron that allows construction of temperature-sensitive (ts) variants of specific proteins without changing their amino acid sequences.216,220 Another example is the Ub sandwich assay, in which a linear fusion of three reporter domains that bears N-terminal Ub as well as Ub moieties between the domains is designed to detect and measure cotranslational protein degradation in living cells.139 Yet another technique, the Ub translocation assay, uses Ub fusions that contain N-terminal signal sequences to probe kinetic aspects of protein translocation across membranes in vivo.217 In 1994, Johnsson and I developed a method, later termed the protein fragment complementation assay (PCA), that detects protein interactions through the conditional in vivo reconstitution of a single-domain protein from its fragments.218 The first in vivo PCA involved a split-Ub design in which two halves of a mutant Ub moiety were configured as sensors of interactions between two proteins of interest that were linked to Ub halves.218,221 The split-Ub PCA led to other useful PCAs by other laboratories, including split GFP (green fluorescent protein), split DHFR, and split β-lactamase (Refs. 222–225 and references therein).
The Ub fusion technique also led to the concept of signal regulated, cleavage-mediated toxins (sitoxins), in which an initially short-lived (or otherwise downregulated) toxin is activated through a cleavage (e.g., by a viral protease in a virus-infected cell) that removes a degron or activates a toxin otherwise, thereby increasing the level of active toxin.226 Variants of this strategy were used by others to develop protein-based conditional reagents, including antiviral drugs.227–233 Muir and coworkers234 combined a version of the sitoxin approach226 with the split-Ub PCA method218 to develop the SURF (split Ub for the rescue of function) technique, in which a protein of interest that is made short-lived in vivo through a link to a portable degron can be metabolically stabilized by a small compound that causes a cleavage-mediated removal of degron. Using mouse DHFR as a reporter, we showed that the Ub-dependent proteolysis of a degron-bearing protein can be slowed down or halted through the addition of a compound (e.g., methotrexate, a tight-binding DHFR inhibitor) that increases thermodynamic stability of a protein.235,236 Wandless and coworkers237 employed this approach, using E. coli DHFR and its inhibitor trimethoprim, to develop a method for regulating protein degradation in cultured mammalian cells and intact animals, including their neurons. Another recent technique uses a conditionally expressed viral TEV protease to cleave, in vivo, a protein of interest that had been engineered to contain a TEV cleavage site at a desired position.238 This method makes it possible, among other things, to create an N-degron posttranslationally, as distinguished from the cotranslational production of N-degrons through the Ub fusion technique.4,9,213
The Ac/N-End Rule Pathway
The Ac/N-end rule pathway3 [Figs. 2(B) and 4(A)] was discovered 24 years after the Arg/N-end rule pathway4 [Fig. 2(A)]. In S. cerevisiae, AcN-degrons of the Ac/N-end rule pathway are recognized by the Doa10 E3 Ub ligase3 and apparently by other E3s as well (C.-S. Hwang et al., unpublished data). The 151-kDa RING-type Doa10 E3 is a multispanning integral membrane protein located in the ER membrane and in the inner nuclear membrane.239–241 The Doa10 E3 functions together with the Ubc6 and Ubc7 E2 enzymes and targets both “soluble” (nuclear and cytosolic) and transmembrane proteins.3,242 Isolated S. cerevisiae Doa10 selectively binds to the Nt-acetylated N-terminal Met, Ala, Val, Ser, Thr, Cys, Gly, and Pro residues of model peptides.3 Remarkably, the binding of Doa10 to Nt-acetylated residues of these peptides is precluded by the presence of a basic residue such as Lys at Position 2 (ref. 3). Thus, the S. cerevisiae Ac/N-end rule pathway avoids the targeting of proteins with a basic residue at Position 2 through two independent constraints: first, such proteins are not Nt-acetylated in yeast91,92; and second, the cognate Doa10 E3 apparently does not recognize Nt-acetylated proteins that bear a second-position basic residue.3 Evolutionary and mechanistic reasons for a “double” filter against potential substrates of this class are unclear at present, because the absence of Nt-acetylation of such proteins in S. cerevisiae should suffice. Mammalian E3s that are sequelogous (similar in sequence1) to S. cerevisiae Doa10 include the human TEB4 E3 Ub ligase.243 This and other evidence (C.-S. Hwang et al., unpublished data) indicate the presence of the Ac/N-end rule pathway in multicellular eukaryotes as well.
Nt-acetylation is largely cotranslational, apparently irreversible, and involves a majority of cellular proteins. What functions are subserved by such a massive production of degradation signals (AcN-degrons) in nascent proteins, if many of these proteins are destined for long half-lives? We suggested that a major role of these degradation signals involves quality-control mechanisms and the regulation of protein stoichiometries in a cell.3 A key feature of such mechanisms would be conditionality of AcN-degrons. If a nascent Nt-acetylated protein can fold its N-terminal domain rapidly enough, or if this protein interacts with a “protective” chaperone such as Hsp90, or becomes assembled into a cognate multisubunit complex, the cotranslationally created AcN-degron of this protein may become inaccessible to a cognate Ub ligase such as Doa10. Consequently, this Nt-acetylated protein would not be targeted for degradation via its AcN-degron. In contrast, delayed or defective folding of a protein's N-terminal domain (because of oxidative, heat or other stresses; or a conformation-perturbing mutation; or nonstoichiometric levels of cognate protein ligands) would keep an AcN-degron exposed (active) and thereby would increase the probability of the protein's destruction.
Might the conditionality of AcN-degrons underlie the regulation of protein assembly into multiprotein complexes? For example, histones and ribosomal proteins tend to be short-lived until they become integrated into larger assemblies, the nucleosomes (in chromosomes) and the ribosomes, respectively.244,245 Selective degradation of these proteins in their unassembled states makes possible the regulation of their levels vis-á-vis the rates of their production through transcription and translation. Little is known about degrons of ribosomal proteins or histones that mediate their conditional degradation. At the same time, most histones and ribosomal proteins are Nt-acetylated.91,92 We suggested that the regulation of in vivo protein stoichiometries in oligomeric proteins and nucleoproteins (including chromosomes and ribosomes) through the assembly-controlled protein degradation may be mediated, to a large extent, by AcN-degrons that form cotranslationally and are accessible to a cognate Ub ligase in “free” proteins but not in their “assembled” counterparts.3
Aneuploidy, in which the chromosome number in a cell is not an exact multiple of the haploid number, is a frequent property of cancer cells and a cause of birth defects such as Down syndrome (a trisomy of chromosome 21). Physiological defects in aneuploid cells are likely to be caused, at least in part, by maladaptive molar ratios of newly formed proteins in such cells, given their deviations from wild-type gene dosages on over-represented or under-represented chromosomes. Homeostatic responses in such cells would have to destroy a higher than normal load of unassembled proteins. This may account for the known hypersensitivity of aneuploid cells to proteasome inhibitors.246 If so, and if AcN-degrons contribute to proteolysis that regulates in vivo protein stoichiometries, aneuploid cancer cells may prove to be hypersensitive to inhibition of the Ac/N-end rule pathway, a mode of therapy that would be more selective than proteasome inhibitors, which downregulate the Ub system at large.
Nonlinear effects of in vivo protein degradation are a rule rather than exception. For example, if a homodimeric protein is significantly longer-lived in vivo in comparison to its monomer subunit (because a degron accessible in a monomer becomes buried or otherwise less active in a dimer), the steady-state concentration of this protein in a cell can be a strongly nonlinear (sigmoid) function of the rate of its synthesis. Such effects would occur with both homooligomers and heterooligomers. In what follows, I mention a potentially important aspect of this disposition that has not been pointed out previously, to my knowledge. Specifically, one should expect, in aneuploid and other “disbalanced” settings, a major difference in physiological outcomes for homooligomers versus heterooligomers. Consider, for example, an aneuploid cell that overproduces at least some proteins from its over-represented chomosome. If a specific overproduced protein forms a heterodimer with a protein encoded by another chromosome that is present at the normal dosage, an excess of overproduced protein would have its degron, for example, an AcN-degron, exposed (active) and would be preferentially destroyed by the Ac/N-end rule pathway. As a result, the in vivo concentration of this overexpressed protein would be at most marginally higher than it would have been in the absence of aneuploidy. By contrast, if an overproduced protein forms a homodimer (or a homooligomer) in which an AcN-degron is inactive (sterically shielded), the Ac/N-end rule pathway would be unable to “detect” an overexpression of this (self-associated) protein, with the consequent failure of proteolysis-based compensatory mechanisms. The same argument applies to degradation signals distinct from AcN-degrons, as long as a degron is rendered inactive (or less active) through the formation of a homooligomeric protein. This argument can also apply if a heterodimer is formed from subunits encoded by the same over-represented chromosome. In sum, it is possible (this conjecture remains to be verified) that fitness-reducing effects of aneuploidy stem, to a significant degree, not only from a disbalanced (nonstoichiometric) production of proteins, but also specifically from the impossibility, for an otherwise protective proteolytic circuit, to “detect” an overproduction of proteins that form homooligomers.
The irreversibility of Nt-acetylation may underlie a putative role of AcN-degrons in the degradation of relatively long-lived intracellular proteins, with half-lives of many hours or days. The initial burst of degradation, through an AcN-degron, of some among the molecules of a newly formed, not yet “assembled” Nt-acetylated protein is followed by the formation of a cognate oligomeric complex in which the protein's AcN-degron becomes inactive (sterically shielded). Whether or not this AcN-degron is activated (becomes accessible) later would be determined by the probability of dissociation of the complex over time, either a spontaneous dissociation or an “induced” one. The latter can be caused, for example, by a physiologically relevant phosphorylation of the complex, or by events such as heat or oxidative stress. The resulting transient accessibility of the (now exposed) AcN-degron of a protein would immediately and strongly increase the probability of the protein's destruction, until the cognate complex can reassemble and thereby again protect the protein. In contrast to the notion of a low and time-invariant probability of destruction of a long-lived protein, the above model posits recurrent transitions between at least two states. In one state, the probability of protein's destruction is high (AcN-degron is exposed). In the other state, in which the protein is a part of a complex, the probability of protein's destruction is low or negligible. The kinetics of reversible transitions between these states and the probability of protein's destruction in the (transient) absence of protective complex would determine, together, an experimentally measured in vivo decay curve of such a protein. Thus, an apparently first-order degradation kinetics may result, mechanistically, from transitions between different structural states of a protein and different probabilities of degradation. This model, which remains to be verified for AcN-degrons, is relevant to other degrons as well.
In the recently determined crystal structure, by the Barford laboratory, of a complex between the Hcn1 and Cut9 subunits of the yeast Schizosaccharomyces pombe APC/C Ub ligase, the Nt-acetylated Met residue of Hcn1 was found to be enclosed within a chamber created by the Cut9 subunit247 (Fig. 13). The authors interpreted this result as a likely example of the assembly-mediated shielding of AcN-degrons.3 Although Protein Data Bank (PDB) and other databases of protein structures contain thousands of proteins, the Hcn1-Cut9 complex247 is one of very few (<10) structures that show details of a protein-bound Nt-acetylated residue. Another structure of a complex with a known spatial location of Nt-acetylated N-terminus contains the tandem PHD1/2 finger (a part of the human DPF3b chromosomal protein) that is bound to the Nt-acetylated peptide of histone H4 (residues 1–22) and specifically interacts with several residues of this H4 peptide, including its Nt-acetylated Ser1 residue.248 The paucity of such structures in current databases stems from often unstructured N-terminal regions of studied proteins, from frequent N-terminal truncations of proteins (to facilitate their crystallization), and from the still prevalent use, for crystallization, of eukaryotic proteins expressed in bacteria, in which Nt-acetylation is not efficacious. Hcn1-Cut9 and analogous complexes in which the shielding of specific Nt-acetylated N-termini can be demonstrated at a near-atomic resolution,247 can now be used to explore physiological functions of AcN-degrons.
Figure 13.
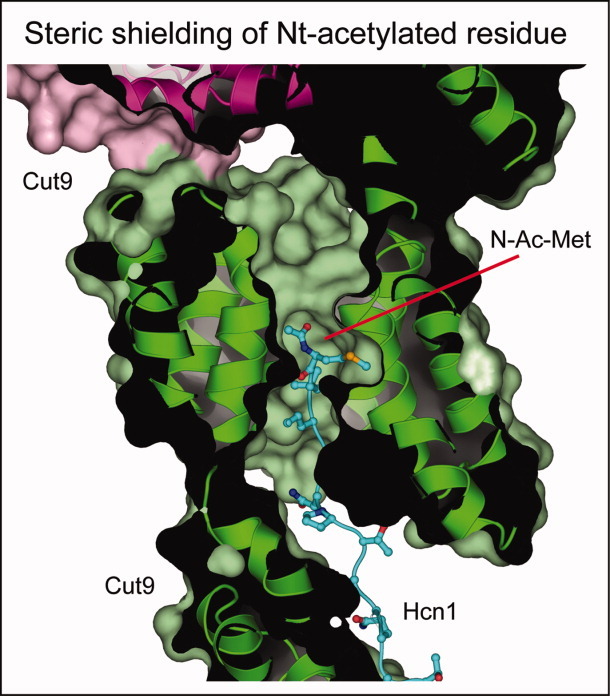
Steric shielding of the Nt-acetylated N-terminal residue of a subunit in a protein complex. Shown here is a part of the crystal structure, by the Barford laboratory, of a complex between the Hcn1 and Cut9 subunits of the S. pombe APC/C Ub ligase.247 In this structure, the Nt-acetylated N-terminal Met residue of Hcn1 is enclosed within a chamber formed by the Cut9 subunit, including its interface with the other Cut9 subunit in the heterotetramer of Hcn1 and Cut9. N-terminal region of Hcn1 is shown in cyan as a stick model, and Cut9 is depicted as a cut-out surface representation, to show the chamber's interior.247 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
For reasons described above, AcN-degrons comprise the most prevalent class of specific degradation signals in cellular proteins3 [Figs. 2(B) and 4]. Although the Nt-acetylation of cellular proteins by ribosome-associated Nt-acetylases is largely cotranslational,89,95 the (currently unknown) molar concentrations of specific Nt-acetylases in vivo are likely to be significantly lower than the molar concentration of ribosomes. This, and the inherent stochasticity of cotranslational enzymatic reactions make it virtually certain that Nt-acetylation of proteins is less than quantitative in most cases. Indeed, many proteins are known to be incompletely Nt-acetylated,91,110 despite the existence of posttranslational Nt-acetylation (J.-H. Oh and A. Varshavsky, unpublished data). The latter may not be able to compensate for the incompleteness of cotranslational Nt-acetylation because of rapid formation of cognate complexes in which the N-termini of proteins, either Nt-acetylated (Fig. 13) or unacetylated, would become sterically inaccessible. Given this and other aspects of Nt-acetylation, the emerging field of the Ac/N-end rule pathway may prove to be a source of new answers to several outstanding questions. For example, the phenomenon of proteotoxicity might stem, in part, from the incomplete Nt-acetylation of normally expressed proteins91–93,102 and the resulting presence, in a cell, of otherwise identical protein molecules that either lack or contain an AcN-degron. Given this possibility3 and putative functions of AcN-degrons, the Ac/N-end rule pathway may also play a role in processes that alter the rate of cellular and organismal aging.
The Arg/N-End Rule Pathway
The Arg/N-end rule pathway involves the Nt-arginylation of N-end rule substrates and also the targeting of specific unmodified N-terminal residues by UBR-type E3 N-recognins, which can recognize not only N-degrons but also internal (non-N-terminal) degrons [Figs. 2(A) and 3]. Despite their broad range, the known functions of the Arg/N-end rule pathway (Overview of the N-End Rule Pathway section) are still the tip of the iceberg. Some of these functions are understood, in part, both physiologically and mechanistically (Functions of the N-end rule pathway vis-á-vis their mechanisms section), but many roles of the Arg/N-end rule pathway were discovered through genetic approaches and therefore remain to be explicated in terms of specific substrates and mechanisms. The known physiological substrates of the Arg/N-end rule pathway are the beginning of a much longer list. It already contains a number of putative (unverified) substrates and may require new methods for its systematic elucidation (Substrates of the N-End Rule Pathway section and Fig. 10).
The double-E3 design of the Arg/N-end rule pathway
Until recently, polyubiquitylation of substrates by the S. cerevisiae Arg/N-end rule pathway was thought to be mediated by a dimer comprising the 225-kDa Ubr1 E3 and the Ubr1-bound 20-kDa Rad6 E2 enzyme.5,6,38,39,249 In 2010, it was discovered that the targeting ensemble is more elaborate. It comprises a physical complex of the RING-type Ubr1 E3 (N-recognin) and the HECT-type Ufd4 E3, in association with their cognate E2s Rad6 and Ubc4/Ubc5, respectively40,44 [Fig. 2(A)]. Ufd4 is the 168 kDa HECT-type E3 of the Ub-fusion degradation (UFD) pathway that recognizes a “nonremovable” N-terminal Ub moiety of a Ub fusion as a primary degron and polyubiquitylates the Ub moiety, a prerequisite for the fusion's degradation by the 26S proteasome.180,181,250–255 The UFD pathway was discovered through analyses of N-terminal Ub fusions in which an alteration of either the Ub moiety or a junctional amino acid residue inhibits the cleavage of a fusion by deubiquitylases and thereby results in the fusion's degradation by the UFD pathway.4,250,256,257 The UFD pathway is present in both yeast and mammals,258 suggesting that the Ubr1-Ufd4 double-E3 organization of the S. cerevisiae Arg/N-end rule pathway is universal among eukaryotes.44
Ufd4 is not an N-recognin, that is, it does not, by itself, recognize N-degrons, in contrast to Ubr1. However, through its physical interaction with the Ubr1 E3 [Fig. 2(A)], the Ufd4 E3 functions as a novel component of the Arg/N-end rule pathway that increases the efficacy of Ubr1, at least in part by augmenting the processivity of polyubiquitylation of N-end rule substrates.44 The function of Ufd4 in the Arg/N-end rule pathway is not confined to processivity enhancement, as Ufd4 can also recognize the internal degron of the Mgt1 DNA repair protein (a substrate of the Arg/N-end rule pathway) even in ubr1Δ cells,40 that is, in the absence of Ubr1. Thus, the sets of internal degrons recognized by Ubr1 and Ufd4 are partially overlapping. Although Ufd4 is not strictly essential for the ability of Ubr1 to mediate the Arg/N-end rule pathway, this pathway is detectably impaired in ufd4Δ cells.44
Earlier studies introduced the operationally defined concept of an E4 as an E3-like enzyme that cooperates with substrate-specific ubiquitylation machinery to increase the efficacy (including processivity) of polyubiquitylation, and in some cases alters topology of a growing poly-Ub chain, that is, the locations of Gly-Lys isopeptide bonds between the adjacent Ub moieties in a chain.257,259–261 Remarkably, the increased processivity of the Ubr1-dependent polyubiquitylation of N-end rule substrates by the Ubr1-bound Ufd4 Ub ligase44 is “reciprocated” in the context of the UFD pathway. Specifically, the Ufd4-bound Ubr1 Ub ligase increases the processivity of polyubiquitylation of UFD substrates.44 Thus, operationally, the complex of Ubr1 and Ufd4 [Fig. 2(A)] functions as an E3–E4 pair in which the “assignment” of an E3 or E4 function depends on a substrate and the nature of its degron. It was also found that Ubr1, similarly to Ufd4, contains a domain that specifically binds to the N-terminal Ub moiety of protein fusions but not to free Ub, indicating that Ubr1 can target UFD substrates independently of the Ufd4 Ub ligase.44 Because Ubr1 is apparently ∼10-fold less abundant than Ufd4 in wild-type yeast,44 the “double” E2-E3 Ubr1/Rad6-Ufd4/Ubc4 complex is expected to mediate the bulk of the Arg/N-end rule pathway [Fig. 2(A)], whereas the same complex mediates only a part of the UFD pathway. In vitro, Ubr1 and Ufd4 bind, separately and independently, to specific (partially overlapping) sets of subunits of the 26S proteasome.180 Now that the proteasome-binding in vivo entity turned out to be, most likely, the Ubr1–Ufd4 complex (with associated E2 enzymes), it is possible that the targeting of N-end rule and UFD substrates is mediated, at least in part, by this complex that is “prebound” to the 26S proteasome. These are just some of the ramifications of the discovery of the Ubr1–Ufd4 complex, a functional and mechanistic advance that unified two proteolytic systems that have been studied separately for two decades.44
Purified S. cerevisiae Ubr1 binds to either Type-1 (basic) or Type-2 (bulky hydrophobic) destabilizing N-terminal residues of 11-residue peptides with a Kd of ∼1 μM, whereas no binding (Kd > 0.1 mM) was observed with otherwise identical peptides bearing the N-terminal Gly residue.38,43 Crystal structures of the ∼80-residue UBR domains (Type-1 binding sites) of S. cerevisiae Ubr1 and its mammalian counterpart have been determined [Fig. 8(A,B)],83–85 but no structural information is available about the Type-2 binding site, let alone the entire ∼200-kDa mammalian or yeast Ubr1, despite crystallization attempts by more than one laboratory. The discovery of interaction between the Ubr1 and Ufd4 E3s suggests that their complex44 might be easier to crystallize than its individual components. Although the Type-1 and Type-2 binding sites are adjacent in S. cerevisiae Ubr1 [Fig. 7(A)], a screen for mutations in UBR1 that inactivate one but not the other binding site has readily yielded Ubr1 mutants with such properties,38 suggesting a structurally autonomous folding not only of the Type-1 (UBR) domain [Fig. 8(A,B)] but of a Type-2 domain as well.
A spatial proximity of the Type-1 and Type-2 binding sites of Ubr1 was made use of in designing a bivalent inhibitor of the N-end rule pathway that was expected to bind simultaneously to both of these sites and therefore to exhibit a higher efficacy than a “monovalent” inhibitor. In a proof-of-principle study, a tetramer of X-βgal, produced from Ub-X-βgal [Fig. 1(B); X=Arg or Leu], was produced as a set of homotetramers and heterotetramers by coexpressing, in S. cerevisiae, Arg-βgal (a Type-1 N-end rule substrate) together with Leu-βgal (a type-2 N-end rule substrate).262 These engineered proteins contained a ∼40-residue extension between N-terminal residue and the βgal moiety. This sequence, termed the eΔK extension (see Structure and Targeting of N-degrons section), lacked Lys residues, and the first 200 residues of the βgal moiety also lacked lysines. As a result, these βgal-based reporters were long-lived in vivo, despite their destabilizing N-terminal residues.9,24,213 In the 3D structure of E. coli βgal, two of its N-termini (with the other pair of N-termini on the opposite side of the tetramer) are spatially close and oriented in the same direction.263 This spatial feature of βgal and the above design resulted in some of expressed X-βgal tetramers being heterotetramers in which two flexible, adjacent N-terminal regions contained the N-terminal Arg and Leu residues.262 It was found that these “bivalent” Arg/Leu-βgal heterotetramers inhibited the degradation of reporter N-end rule substrates in S. cerevisiae much more efficaciously than either Arg-βgal alone or Leu-βgal alone.262 An inhibitor of this kind acts as a competitive inhibitor of the Type-1/2 sites of N-recognins and may also act, at the same time, as an allosteric activator of other substrate-binding sites of N-recognins, the ones that bind to internal (non-N-terminal) degrons of specific regulatory proteins. The latter inference (it remains to be verified for macromolecular-size inhibitors) stems from the known allosteric activation, by dipeptides with destabilizing N-terminal residues, of the autoinhibited third substrate-binding site of yeast Ubr1, the site that targets the internal degron of the Cup9 transcriptional repressor (Fig. 15 and Functions of the N-end rule pathway vis-á-vis their mechanism section). The Kwon laboratory has further advanced the bivalent-inhibitor approach by producing and characterizing small (<1 kDa) synthetic heterovalent inhibitors of the N-end rule pathway.264
Figure 15.
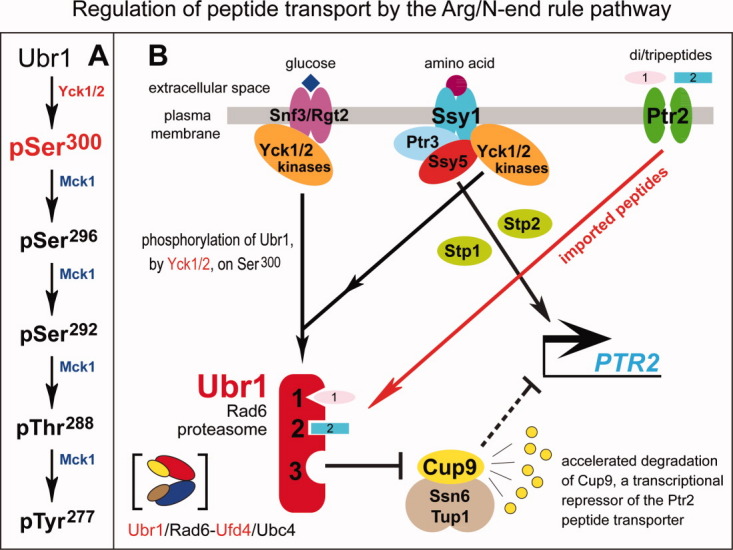
Regulation of peptide import by the Arg/N-end rule pathway in S. cerevisiae, and inputs by the amino acid-sensing SPS pathway. A: The “primed” cascade of Ubr1 phosphorylation in which the Yck1/Yck2-mediated phosphorylation on Ser300 of Ubr1 is essential for the normal regulation of peptide import39 [see also the legend to Fig. 7(A,B)]. B: Ubr1-mediated regulation of peptide import, and the involvement of the SPS pathway.27,29,138,295,296 Cup9 is a transcriptional repressor of the regulon that includes PTR2, which encodes the major importer of di/tripeptides. In the absence of Ubr1 (in ubr1Δ cells), Cup9 becomes long-lived, accumulates to high levels, and extinguishes expression of Ptr2. Therefore, ubr1Δ cells cannot import di/tripeptides. In wild-type (UBR1) cells growing in the absence of extracellular di/tripeptides, a relatively low but nonzero number of Ubr1 molecules have their third substrate-binding site “open” (not autoinhibited) and therefore can target Cup9 for degradation (t1/2 ∼ 5 min) via its internal degron, resulting in a low but significant steady-state concentration of Cup9 and, thus, a weak but significant expression of the Ptr2 transporter. In wild-type (UBR1) cells growing in the presence of extracellular di/tripeptides (some of which bear Type-1 and Type-2 destabilizing N-terminal residues), the imported peptides interact with the Type-1 and Type-2 binding sites of Ubr1. This binding allosterically increases the fraction of Ubr1 molecules whose third (Cup9-specific) site is “open” (active). The resulting decrease in the half-life of Cup9 (from ∼5 min to below 1 min) results in a low concentration of Cup9, and consequently to a strong induction of the Ptr2 transporter.27,29,138 Also shown is the amino acid-sensing SPS pathway (see Functions of the N-end rule pathway vis-á-vis their mechanisms section for details and additional references), which can influence the import of peptides at least in part through the Yck1/Yck2-mediated phosphorylation of Ubr1 on Ser.300 This phosphorylation is required (through a mechanism that remains to be determined) for normal levels of Ubr1 activity in the Ptr2-Cup9-Ubr1 circuit.39 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In contrast to S. cerevisiae, in which Ubr1 is the sole N-recognin of the Arg/N-end rule pathway, the genome of a multicellular eukaryote, for example, a mammal, encodes at least four UBR domain-containing E3 N-recognins, termed Ubr1, Ubr2, Ubr4, and Ubr5 (also called Edd)15,31,43 [Fig. 8(C)]. Sequelogs1 of the 570-kDa mouse Ubr4 N-recognin are known as PUSHOVER in D. melanogaster and as BIG in plants.15,31,265–269 Mammalian Ubr1 and Ubr2 are highly sequelogous N-recognins (they are also sequelogous to S. cerevisiae Ubr1), whereas the sequelogy between, for example, Ubr1 and either Ubr4 or Ubr5/Edd is largely confined to their ∼80-residue UBR domains. Mutational inactivation of human UBR1 causes JBS (Overview of the N-End Rule Pathway and Functions of the N-end rule pathway vis-á-vis their mechanism sections), and Ubr1−/− mice exhibit aspects of JBS as well.34 Mice lacking Ubr2 have a variety of defects, including genomic instability and impaired spermatogenesis (and infertility) in males.30,123 One function of mouse Ubr2 in meiotic spermatocytes is ubiquitylation of histone H2A, in a setting where this modification plays a role in transcriptional silencing.270 In contrast to viability of single-mutant Ubr1−/− and Ubr2−/− mice, double-mutant mouse strains that lacked both Ubr1 and Ubr2 died as ∼11-day (midgestation) embryos, with major defects in neurogenesis and cardiovascular development.271
Mammalian Ubr1, Ubr2, and Ubr4 N-recognins contain both Type-1 and Type-2 substrate-binding sites, whereas the Ubr5/Edd N-recognin lacks the Type-2 binding site31 [Fig. 8(C)]. Thus, in agreement with findings by the Bachmair laboratory about plant N-recognins,18,272,273 a mammalian N-recognin can contain both Type-1 and Type-2 substrate-binding sites or just one of them. The presence of a UBR domain in an E3 Ub ligase signifies its N-recognin property (i.e., its ability to bind to N-terminal Ndp residues of proteins or short peptides; Fig. 3) only in some cases, as the mouse Ubr3, Ubr6, and Ubr7 E3s are not N-recognins, despite the presence of a UBR domain43,274 (Figs. 7 and 8). The S. cerevisiae Ubr2 E3, which contains a UBR domain, is also not an N-recognin.43,275 Cognate in vivo ligands of UBR domains of non-N-recognin E3s remain to be identified. The identical names of mammalian Ubr2 and S. cerevisiae Ubr2 are functionally incongruent, in that only mammalian Ubr2 is an N-recognin.
Studies with mice, flies and plants indicated a broad range of physiological functions of Ubr4 and Ubr5/Edd (Refs. 15,31,265–269 and refs therein), but analyses of these E3s vis-á-vis their functions as N-recognins have yet to take place. Specific interactions of the Type-1/2 binding sites of the S. cerevisiae Ubr1 N-recognin with dipeptides bearing Ndp residues were shown to allosterically activate a third binding site of Ubr1, thereby accelerating the degradation of its target, Cup9, in a circuit that “senses” the presence of extracellular peptides and accelerates their uptake27,29,138 (Fig. 15). It would be, therefore, particularly informative to determine whether the analogous N-degron-binding sites of, for example, mammalian Ubr4 and Ubr5/Edd also function to regulate, allosterically, the activity of other substrate-binding sites of these E3s.
A recent advance276 opened up yet another dimension in studies of the mammalian N-end rule pathway. As in the rest of the Ub system, the “upstream” steps of polyubiquitylation of mammalian N-end rule substrates are mediated by the Ub-activating (E1) enzyme Uba1, which transfers the activated Ub moiety to cognate E2 enzymes such as Ube2A/Ube2B (the mammalian counterparts of the S. cerevisiae Rad6 E2). It was shown that Uba6-Use1, a different pair of E1-E2 enzymes, also functions in the Arg/N-end rule pathway, in that the Use1 E2 of this pair is a component of “alternative” E2-E3 Ub ligases that contain UBR-type N-recognins such as Ubr1 and Ubr2.276
N-terminal deamidation in the Arg/N-end rule pathway
In S. cerevisiae, the deamidation branch of the Arg/N-end rule pathway is mediated by the NTA1-encoded 52-kDa NtN,Q-amidase. Nta1 can deamidate either N-terminal Asn (N) or N-terminal Gln (Q), converting them into Asp or Glu, respectively, and thereby enabling the Nt-arginylation of resulting proteins by the Ate1 R-transferase23 [Fig. 2(A)]. Although there is enough of Nta1 in the yeast cytosol (and possibly in the nucleus as well) to mediate the deamidation branch of the Arg/N-end rule pathway,23 the bulk of Nta1 was shown to reside in mitochondria (http://yeastgfp.yeastgenome.org/), in agreement with our findings (H.-R. Wang and A. Varshavsky, unpublished data). NTA1 apparently encodes both forms of NtN,Q-amidase, as there are two alternative start (ATG) codons in the NTA1 open reading frame (ORF), with a putative mitochondrial presequence between them.23 Physiological substrates of the S. cerevisiae Nta1 NtN,Q-amidase are unknown. Apart from the inability of nta1Δ yeast cells (in contrast to wild-type cells) to degrade engineered cytosolic N-end rule substrates that bear N-terminal Asn or Gln, no defects could be detected, thus far, with nta1Δ cells, including the apparent absence of defects in their mitochondria (ref. 23; H.-R. Wang and A. Varshavsky, unpublished data). One possibility is that the mitochondrial pool of Nta1 may function in the recently discovered mitochondrial N-end rule pathway277,278 (Mitochondrial N-End Rule Pathway section).
In contrast to S. cerevisiae, where a single NtN,Q-amidase deamidates both N-terminal Asn and N-terminal Gln [Fig. 2(A)], the deamidation branch of the Arg/N-end rule pathway is bifurcated in multicellular eukaryotes. Specifically, the mammalian Ntan1-encoded 35-kDa NtN-amidase deamidates solely N-terminal Asn, whereas the Ntaq1-encoded 24-kDa NtQ-amidase deamidates solely N-terminal Gln25,42,116,117 (Fig. 3). Mouse Ntan1 and Ntaq1 are present largely in the cytosol and the nucleus,25,42 in contrast to the largely mitochondrial location of S. cerevisiae Nta1. Ntan1−/− mice, which lack NtN-amidase and cannot degrade N-end rule reporters bearing N-terminal Asn, are hypokinetic and have a significantly impaired spatial memory.25 The recently constructed Ntaq1−/− mouse strains, which lack NtQ-amidase and cannot degrade N-end rule reporters bearing N-terminal Gln, also exhibit behavioral abnormalities (K. Piatkov et al., unpublished data). Given their different N-terminal residues, Asn versus Gln, the physiological substrates of NtN-amidase (Ntan1) and NtQ-amidase (Ntaq1) are unlikely to overlap.
In D. melanogaster, the cleavage, by an initiator caspase, of the antiapoptotic E3 Ub ligase DIAP1 converts it into an N-end rule substrate (still active as a Ub ligase) that bears N-terminal Asn and is degraded by the deamidation branch of the Arg/N-end rule pathway140,141 (Fig. 10). Furthermore, an autocleavage, by the human USP1 deubiquitylase, produces a short-lived C-terminal fragment of USP1 that may be a physiological substrate of the Ntaq1 NtQ-amidase.279 A number of putative (unverified) substrates of NtN-amidase and NtQ-amidase are known to be produced by nonprocessive intracellular proteases, including caspases (Figs. 3 and 10, Substrates of the N-End Rule Pathway section, and Refs. 156–160).
Strong sequelogs1 of the mammalian Ntaq1 NtQ-amidase are present throughout the animal phylum (including fishes, insects and nematodes), in plants, and also in some fungi, such as the fission yeast S. pombe.42 Similarly, there are strong sequelogs of the mammalian Ntan1 NtN-amidase in other animals, and in some fungi as well. The S. cerevisiae Nta1 NtN,Q-amidase has sequelogs in many but not in all fungi. For example, S. pombe contains a sequelog of mammalian Ntaq1 but lacks a sequelog of S. cerevisiae Nta1. Remarkably, there is no significant sequelogy (sequence similarity1) among Nta1, Ntan1, and Ntaq1, although all three Nt-amidases catalyze identical or similar reactions.42 A parsimonious explanation is that a primordial Nta1 NtN,Q-amidase emerged after (not before) a split between lineages that gave rise to fungi and lineages that led to other eukaryotes. This otherwise plausible explanation is incomplete, given the presence of a sequelog of mammalian Ntaq1 in the yeast (fungus) S. pombe and the absence, from S. pombe, of a sequelog of S. cerevisiae Nta1.
Progress in biological research, including high-throughput structural studies, has produced a disposition all but improbable a decade ago: the mouse Ntaq1 NtQ-amidase, which was isolated, cloned and characterized in 2009, found itself surrounded by directly relevant genomic, proteomic and even crystallographic evidence.42 The earlier data, produced through studies of a specific gene and its encoded 24-kDa product of unknown biochemical nature, have become unified and informed by the revealed enzymatic identity of Ntaq1, its mechanistic similarity to transglutaminases, and its place in the Arg/N-end rule pathway42 (Figs. 3 and 14). Specifically, a mutant, termed tungus, in the D. melanogaster Cg8253 locus, which turned out to encode NtQ-amidase,42 has defective long-term memory.280 In addition, the expression of the nematode (C. elegans) counterpart of mouse Ntaq1 is regulated by neuron-specific transcription factors (ref. 42 and references therein). These findings are consistent with behavioral abnormalities of Ntaq1−/− mice and a major role of the Arg/N-end rule pathway in mammalian brain development.271
Figure 14.
Spalogy (spatial similarity1) between the Ntaq1 NtQ-amidase and Factor XIII transglutaminase. A and B: Crystal structures of the human Ntaq1 (C8orf32) NtQ-amidase340 (PDB: 3C9Q) and FXIII transglutaminase341 (PDB: 1FIE), respectively. C8orf32 is an initially uncharacterized human protein the structure of which was deposited in the Protein Data Bank (PDB: 3C9Q) by the Center for Eukaryotic Structural Genomics,340 and was later shown, by Wang et al.,42 to be the Ntaq1 NtQ-amidase. C and D: Structures around the active sites of Ntaq1 NtQ-amidase and Factor XIII transglutaminase, respectively. These regions are circled in A and B. C and D: The catalytic triad (Cys,314 His,373 and Asp396) of Factor XIII transglutaminase (D; ref. 341) and the corresponding residues (Cys,28 His,81 and Asp97) of human Ntaq1 (C) are indicated. Despite the striking spalogy between these regions of two enzymes (C and D), there is no significant sequelogy (sequence similarity) between them.42 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The rat Ntan1 gene, encoding NtN-amidase (Fig. 3), was found to be induced by approximately threefold after a 15-min exposure of rat hippocampal neurons to a static 100 mT magnetic field.281 A transient overexpression of Ntan1 in these cells (in the absence of magnetic field treatment) strongly down-regulated, through induced proteolysis, the level of Map2, a microtubule-associated protein.281 Map2 is also known to be downregulated by magnetic fields.281 Thus, Ntan1 might be involved in degradation of Map2, through an unknown processing step that would expose the Asn residue at the N-terminus of a cleaved Map2.
N-terminal arginylation in the Arg/N-end rule pathway
The Ate1 Arg-tRNA-protein transferase (R-transferase) catalyzes the conjugation of the carboxyl group of Arg to the α-amino group of a polypeptide's N-terminal residue [Figs. 2(A) and 3]. Strong sequelogs1 of the ∼60 kDa S. cerevisiae Ate1 R-transferase are present in all eukaryotes examined, including animals and plants.26,35,36,111,112,114 Both S. cerevisiae and mammalian genomes contain one ATE1 gene, whereas plants such as Arabidopsis thaliana contain two ATE1 genes, which encode highly sequelogous R-transferases.18,113 Mouse Ate1 pre-mRNAs are produced, in part, from the bidirectional DfaPAte1 promoter that contains a CpG island and also expresses, in the opposite direction, a gene termed Dfa (divergent from Ate1)35,282 [Fig. 9(A)]. Dfa pre-mRNAs are expressed from both DfaPAte1 and nearby promoters, are differentially spliced, and result in a complex set of mRNAs and proteins, largely though not exclusively in the testis. At least one isoform of mouse Dfa was shown to function, in a reporter-based in vivo assay, as a repressor of TATA-box transcriptional promoters.282 No functional or mechanistic connections between Dfa and R-transferase were detected so far, apart from the proximity of their head-to-head oriented genes.282
In addition to the DfaPAte1 promoter, mouse Ate1 pre-mRNAs are also produced from an upstream promoter that partially overlaps, in anti-sense direction, with ORFs encoding Dfa proteins.35,282 Six splicing-derived isoforms of the mouse Ate1 R-transferase have been identified26,35,112 [Fig. 9(B–D)]. The alternatively spliced exons of these isoforms are pairs of sequelogous exons, 1A versus 1B and 7A versus 7B [Fig. 9(D)]. Although the splicing-derived isoforms of R-transferase differ, in particular, in their substrate preferences, an unambiguously specific function of any Ate1 isoform (e.g., a distinct intracellular location or a unique in vivo substrate specificity) remains to be identified.
Both the mouse and S. cerevisiae Ate1 R-transferases are inhibited by low-μM levels of hemin (Fe3+-heme).36 Hemin binds to at least one of two conserved Heme Regulatory Motifs (HRMs) in R-transferase and inhibits its enzymatic activity through a redox mechanism that involves the formation of a disulfide bond between the vicinal Cys71 and Cys72 residues that are conserved among R-transferases in examined eukaryotes [Fig. 9(E,F)]. Remarkably, hemin also accelerates, in vivo, the degradation of mouse Ate1, thus, acting as both a “stoichiometric” and “catalytic” downregulator of R-transferase.36 Moreover, hemin also binds, at least in vitro, to both S. cerevisiae and mouse Ubr1 N-recognins. Although the binding of hemin to S. cerevisiae Ubr1 does not directly occlude any of the substrate-binding sites of Ubr1, hemin blocks the allosteric activation of the third binding site of Ubr1 that recognizes an internal degron of the Cup9 transcriptional repressor.36 [In the absence of hemin, this allosteric activation of the third binding site is caused by the occupancy of the Type-1/2 sites of Ubr1 that recognize destabilizing N-terminal residues (Fig. 15).] Thus, in addition to being a sensor of NO and oxygen,32,33 the Arg/N-end rule pathway is also a sensor of heme [hemin; Fig. 2(A) and 3]. Several hemoproteins, including cytochrome oxidase and NO synthases, contain hemin (Fe3+-heme) rather than Fe2+-heme as an essential prosthetic group.283 Because heme interacts with physiologically relevant gases such as O2, NO, and carbon monoxide (CO), there may be a physiological connection between the NO-sensing and heme-sensing properties of the Arg/N-end rule pathway [Fig. 2(A) and 3]. One function of this proteolytic circuit may be to coordinate the redox dynamics of heme, NO, oxygen, thiols, and other small effectors, by sensing them through components of the Arg/N-end rule pathway and by acting to alter their levels or spatiotemporal gradients, in part through conditional degradation of specific N-end rule substrates.
The inhibition of R-transferase by hemin is a property that is conserved from fungi to mammals.36 What selective pressures led to the retention of the heme-Ate1 connection over vast phylogenetic distances? Given the cardiovascular defects of Ate1−/− mouse embryos,26 their low levels of embryonic globin and heme, high levels of heme oxygenase,36 and their perturbed hematopoiesis (J. Sheng et al., unpublished data), the mammalian Arg/N-end rule pathway is likely to participate in the control of heme synthesis, transport, and/or catabolism. This might also be a role of Ate1-heme connections in S. cerevisiae. Physiological functions of Nt-arginylation in S. cerevisiae are still a terra incognita, as no defects could be detected, thus far, in ate1Δ yeast cells, apart from their inability to degrade N-end rule reporters with N-terminal Nds and Ndt residues.
The Rgs4, Rgs5, and Rgs16 regulators of G proteins bear N-terminal Cys and are targeted for regulated degradation by the NO/O2/Ate1-dependent branch of the mammalian Arg/N-end rule pathway26,32,33,41 (Overview of the N-End Rule Pathway and Functions of the N-end rule pathway vis-á-vis their mechanisms sections). Rgs4 is an inhibitor of tubulogenesis, a process that underlies the development and homeostasis of blood vessels and other tubular structures, such as those in the mammary gland, kidney and lung.284 Rgs4 and Rgs16 block signaling by the vascular endothelial growth factor (VEGF), and Rgs5 regulates vessel remodeling during neovascularization.284,285 As described above, hemin downregulates the Arg/N-end rule pathway in vivo, both through direct inhibition of the Ate1 R-transferase and through the induction of its degradation, and also (possibly) through effects of hemin on N-recognins.36 In particular, hemin upregulates Rgs4, Rgs5, and Rgs16, by inhibiting their degradation.36 Thus, it is likely (but remains to be verified) that the Arg/N-end rule pathway is a mediator of heme's effects on tubulogenesis. The sensor–effector link between the Arg/N-end rule pathway and heme may be relevant not only to normal conditions but also to perturbations of heme homeostasis, for example upon a “spontaneous” or wound-induced hemorrhage, or in a hemorrhage-prone setting of a growing tumor.
Heterozygous Ate1+/− mice appear indistinguishable from their wild-type counterparts, whereas Ate1−/− mice, which lack R-transferase and Nt-arginylation, die around embryonic day 15 (E15) with abnormalities that include cardiovascular defects.26 To bypass the embryonic lethality of the nonconditional absence of Nt-arginylation, the Cre-lox-tamoxifen technique was used to induce the loss of Ate1 in juvenile (∼1-month-old) Ate1+/− mice.41 The penetrance of Ate1+/−→Ate1−/− conversion ranged from ∼90% of cells in the brain and kidney to ∼60% of cells in the liver. Remarkably, the postnatal ablation of Ate1 caused a rapid decrease of body weight (in comparison to tamoxifen-treated control mice) and resulted in early death of ∼15% of Ate1-deficient mice, with surviving mice attaining only ∼70% of normal weight.41 This failure to thrive occurred despite higher than normal food intake by Ate1-deficient mice. These mice contained little or no visceral fat, exhibited an increased metabolic rate, a decreased fasting blood glucose level, and an increased intestinal import and retention of amino acids and/or peptides. Ate1-deficient mice were also resistant to diet-induced obesity, exhibited induction of the Ucp1 uncoupling protein in white adipose tissue (WAT), and expressed strongly reduced levels of mRNA encoding proopiomelanocortin (POMC), a pituitary-produced precursor of several neurohormones with systemic and brain functions. These functions include regulation of melanocytes, a process that is perturbed in Ate1-deficient mice.41 Because other roles of POMC-derived neurohormones include a downregulation of food intake, a strong reduction of POMC in Ate1-deficient mice is consistent with their hyperphagia.41 In addition, Ate1-deficient mice have an enhanced startle response and are strikingly hyperkinetic. They also suffer from premature kyphosis, that is, an excessive curvature of the upper back, and from frequent seizures.41 Ate1-deficient males are infertile, owing to defects in Ate1−/− meiotic spermatocytes.
The remarkably broad range of biological processes that are perturbed by the loss of Ate1 and Nt-arginylation during embryogenesis26 and postnatally41 remains to be understood in mechanistic, substrate-based terms. It is virtually certain that Rgs4, Rgs5, and Rgs16, a set of regulators of G proteins that are conditionally short-lived physiological Ate1 substrates,32,33,36 play a major role in many of specific phenotypes of Ate1−/− mice26,41 (Figs. 3 and 10, Overview of the N-End Rule Pathway and Functions of the N-end rule pathway vis-á-vis their mechanisms sections). But other Nt-arginylation substrates are likely to be involved as well.
Studies of plant R-transferases were initiated by the identification of the A. thaliana mutant delayed leaf senescence (dls1) as a null in the AtAte1 gene.125 Recent work, utilizing A. thaliana mutants lacking both AtAte1 and AtAte2 R-transferases and therefore completely lacking Nt-arginylation, has shown that this branch of the Arg/N-end rule pathway has a number of functions in plants, including seed germination (in part through the removal of sensitivity to the hormone abscisic acid), as well as various aspects of leaf and shoot development.18,113,126 In particular, N-terminal arginylation was shown to play a role in repressing the meristem-promoting BREVIPEDICELLUS (BP) gene in developing leaves.113 Most or all of these R-transferase functions require the activity of PROTEOLYSIS6 (PRT6), an E3 N-recognin Ub ligase of the plant Arg/N-end rule pathway that acts downstream of R-transferases.113,126,273
Results of recent studies by the Kashina laboratory suggested that Arg might be conjugated (presumably by the Ate1 R-transferase) to a number of different N-terminal residues of cellular proteins, that is, that the specificity of Nt-arginylation is not confined to N-terminal Asp, Glu and (oxidized) Cys, and is determined, instead, by currently unknown internal sequence motifs in protein substrates.127,286 While interesting if correct, this conjecture awaits verification, because it contradicts extensive previous evidence about the substrate specificity of R-transferase [Figs. 2(A) and 3]. A distinct kind of Arg conjugation was demonstrated to modify neurotensin, a 13-residue mammalian peptide that functions as a neurotransmitter and hormone. Arg is conjugated, via its α-amino group, to the γ-carboxyl group of an internal Glu residue of neurotensin, presumably during maturation of neurotensin precursors in the secretory pathway.287 In the resulting “branched” Arg-neurotensin conjugate, the orientation of the Arg moiety is opposite to that in linear, N-terminal Arg-protein conjugates that are produced by the Ate1 R-transferase. Given this difference in chemistry and “topology” of Arg conjugation,287 and the apparent absence of Ate1 from secretory compartments, it is likely (but remains to be determined) that neurotensin is arginylated by an enzyme distinct from Ate1.
Functions of the N-end rule pathway vis-á-vis their mechanisms
Subunit selectivity of the Arg/N-end rule pathway and degradation of a cohesin subunit
The subunit selectivity of processive proteolysis, first discovered in the context of the Arg/N-end rule pathway6,13 [Fig. 11(B,C)], underlies most functions of the Ub system, as it makes possible remodeling of protein complexes through subunit-selective degradation [Structure and Targeting of N-degrons section and Fig. 11(A–D)]. A cleaved subunit whose C-terminal fragment bears a destabilizing N-terminal residue would be a potential substrate for the degradation-mediated removal of this subunit from a protein complex.13,168 Because the Arg/N-end rule pathway can target both N-degrons and specific internal degrons [Figs. 2(A) and 3], this remodeling can involve not only a cleaved subunit that contains an N-degron but also a full-length subunit of a protein complex. The Arg/N-end rule pathway can use, in such settings, its capacity for subunit-specific proteolysis to reset the states of relevant circuits without destroying an entire oligomeric complex.
One example is the degradation of a separase-produced fragment of a cohesin subunit, a step that has been shown to be required for the high fidelity of chromosome cohesion/segregation.6,28 Although the identity of a residue at the (inferred) N-terminus of a separase-produced Scc1/Rad21 cohesin fragment varies among eukaryotes (for example, it is Arg in S. cerevisiae, Asn in S. pombe, Glu in mammals, and Cys in D. melanogaster),28,288 the N-terminal residue of a cohesin fragment is invariably destabilizing in the Arg/N-end rule pathway [Figs. 2(A,C) and 6]. This constraint on evolutionary alterations of the N-terminal residue is consistent with the necessity of removing, through degradation, a separase-produced cohesin fragment from the rest of cohesin, thereby “resetting” the cohesin complex for reconstitution through the addition of an uncleaved Scc1/Rad21 subunit.28,288
The C-terminal cohesin fragment can presumably dissociate (or be displaced) from the rest of cohesin complexes in vivo at a rate that allows the functioning of the cohesin machinery even in the absence of degradation of this fragment by the Arg/N-end rule pathway. But such a route is apparently inefficient enough to cause a greatly elevated frequency of chromosome loss in ubr1Δ S. cerevisiae cells and an increased genomic instability in ate1Δ mammalian cells. The separase-produced C-terminal fragments of Scc1/Rad21 cohesin are relatively stable proteins in both of these mutants, in contrast to their short half-lives in wild-type cells.6,28 It has also been shown that at least in yeast it is specifically the metabolic stabilization of the Scc1 cohesin fragment (as distinguished from other effects of the absence of the Arg/N-end rule pathway) that increases the frequency of chromosome loss.28 Another example of subunit-selective proteolysis is a circuit that regulates the import of peptides through the degradation of Cup9, a transcriptional repressor of the peptide transporter Ptr2. The S. cerevisiae Arg/N-end rule pathway targets Cup9, through its internal degron, for a subunit-selective, conditional degradation. Specifically, the short-lived Cup9 is a part of a repressor complex that also contains the more abundant and longer-lived Ssn6-Tup1 global repressor27,138 (Fig. 15).
Regulation of G proteins and the Arg/N-end rule pathway
As mentioned in Overview of the N-End Rule Pathway, Substrates of the N-End Rule Pathway, and N-terminal arginylation in the Arg/N-end rule pathway sections, the functions of the mammalian Arg/N-end rule pathway include the sensing of NO and oxygen, and the regulation of specific G proteins that are coupled to transmembrane receptors. These processes involve the NO/O2-dependent degradation of Rgs4, Rgs5, and Rgs16, a set of G-protein regulators32,33,36 (Figs. 3 and 10). The conditional Nt-arginylation and degradation of these RGS proteins by the Arg/N-end rule pathway alters the activity of cognate G-protein circuits.32,33 In particular, it has been shown that an induction of angiogenesis specifically in the heart results in a strong myocardial hypertrophy (through enlargement of cardiomyocytes), apparently because an increased density of the heart's vascular bed augments the levels of NO that is produced by endothelial cells.289 If so, this effect of increased NO on cardiomyocytes is likely to be mediated, at least in part, through an acceleration of the NO/O2-dependent degradation of Rgs4 (as well as Rgs5 and Rgs16) by the Arg/N-end rule pathway in NO-exposed cardiomyocytes. Indeed, the inhibition of Gq-protein signaling by Rgs4 is known to be a negative modulator of the myocardial hypertrophic response (ref. 289 and references therein). In sum, the Arg/N-end rule pathway is likely to be a major regulator of the heart. Recent studies indicated that the rate of degradation of Rgs4, largely by the Arg/N-end rule pathway (Fig. 3), influences other physiological and pathophysiological processes as well, including the invasiveness of breast cancer, responses of neurons to opiates, and responses of cells in culture to fluxes of calcium ions.290,291
The Cre recombinase-induced postnatal loss of the mouse Ate1 R-transferase and Nt-arginylation (N-terminal arginylation in the Arg/N-end rule pathway section) leads to a more than 10-fold increase in the level of the metabolically stabilized Rgs4 in Ate1-deficient mice, suggesting a major decrease in signaling by Rgs4-regulated G proteins.41 Although the loss of Nt-arginylation does result in a number of phenotypic alterations in Ate1-deficient mice (N-terminal arginylation in the Arg/N-end rule pathway section), it is remarkable that the strikingly strong effect of this loss on the levels of several RGS proteins32,41 is still compatible with most physiological functions. This robustness implies a compensatory feedback regulation, which remains to be explored.
Regulation of apoptosis and the Arg/N-end rule pathway
As mentioned in N-terminal deamidation in the Arg/N-end rule pathway section, one example of involvement of the Arg/N-end rule pathway in the control of apoptosis is the conditional degradation of D. melanogaster DIAP1, a major antiapoptotic regulator140,141 (Fig. 10). A functional understanding of DIAP1 circuits vis-á-vis the Arg/N-end rule pathway remains to be attained. Some C-terminal fragments of other proteins that are cleaved by activated initiator and/or effector caspases are either previously demonstrated or putative (predicted) N-end rule substrates (Substrates of the N-End Rule Pathway section and Fig. 10). Such fragments bear destabilizing N-terminal residues and would be expected to be targeted for degradation by the Arg/N-end rule pathway (Refs. 6,140,141,146, and147 and references therein). If the cleavage of a protein by, for example, an initiator caspase produces a proapoptotic C-terminal fragment, a destabilizing N-terminal residue and (therefore) a short in vivo half-life of such a fragment would counteract the induction of apoptosis by spuriously activated caspases that are present even in unperturbed cells, given the level of noise in caspase-activation circuits. This antiapoptotic role of the Arg/N-end rule pathway was recently found to be significant not only in relatively unstressed cells but under proapoptotic conditions as well (K. Piatkov et al., unpublished data).
HIV-1 integrase and the Arg/N-end rule pathway
During infection by the HIV, the viral RNA genome is reverse-transcribed, and the resulting cDNA is inserted into the chromosomal DNA of infected mammalian cells. This process is mediated by HIV integrase, in association with viral and host proteins. The integrase is a member of the family of structurally related DNA transferases that includes phage transposases and mammalian VDG recombinases RAG1 and RAG2. The HIV integrase is initially a part of the HIV Gag-Pol polyprotein that resides in the virion. The integrase is excised from the polyprotein by the viral protease while still in the virion, and is released into the cytosol upon infection. At this stage, the excised integrase bears N-terminal Phe, an Ndp residue in the Arg/N-end rule pathway (Figs. 3 and 10), and has been shown to be targeted by this pathway for degradation132,133 (t1//2 < 30 min). In contrast, the otherwise identical integrase bearing, for example, N-terminal Met, was significantly longer-lived in vivo.132
Mutant HIV-1 viruses in which the N-terminal residue of (excised) integrase was changed from Phe to Met were viable but impaired (approximately fivefold or more) in replication and infectivity, in comparison to otherwise identical viruses containing Phe-integrase.133 This impairment included defects in early phases of infection, particularly reverse transcription and integration of the viral cDNA. Control experiments have shown that the impairment was not caused by a lower efficiency of Met-integrase excision from the polyprotein, in comparison to the excision of Phe-integrase.133 Specific mechanisms through which a metabolic stabilization (and thus higher steady-state levels) of integrase perturb both reverse transcription and integration remain to be understood. One possibility is that an excess of HIV-1 integrase might inhibit reverse transcription through the known physical interaction between the integrase and reverse transcriptase.133 The cited studies were carried out before the discovery of the Ac/N-end rule pathway.3 Therefore, although the replacement of N-terminal Phe of the HIV-1 integrase with N-terminal Met stabilized the resulting protein,132 it remains to be determined whether N-terminal Met, an Nds destabilizing residue in the Ac/N-end rule pathway [Fig. 2(B)], was Nt-acetylated in the posttranslationally formed Met-integrase (N-terminal sequence Met-Leu-Asp) after its release into the cytosol, and if so whether Met-integrase was targeted for degradation by the Ac/N-end rule pathway.
Many viruses encode polyproteins that yield, upon excision of individual proteins by viral proteases, either putative or confirmed N-end rule substrates. One example (in addition to HIV-1 integrase), is the polyprotein-derived nsP4, the RNA polymerase of the Sindbis virus. This enzyme has N-terminal Tyr (an Ndp residue in the Arg/N-end rule pathway; Fig. 3), is metabolically unstable, and has been shown to be partially stabilized by replacements of its N-terminal Tyr by the Met or Ala residues.131
Human birth defects and the N-end rule pathway
JBS is an autosomal recessive disorder caused by mutational inactivation of both copies of the UBR1 gene that encodes an N-recognin.34 JBS includes a congenital exocrine pancreatic insufficiency and inflammation, multiple malformations (such as nasal wing aplasia), and frequent mental retardation and deafness. The severity of JBS symptoms is decreased if at least one of the mutant copies of UBR1 encodes a partly functional missense hypomorph of the UBR1 E3 Ub ligase (C.-S. Hwang et al., unpublished data). Ubr1−/− mice292 exhibit milder versions of human JBS symptoms, including exocrine pancreatic insufficiency.34 The mechanistic cause(s) of JBS remains to be understood, in part because all other UBR-type N-recognins (The double-E3 design of the Arg/N-end rule pathway section), including UBR2, a strong sequelog of UBR1 that is expressed in exocrine pancreas as well, are retained in patients with JBS. Their cells, therefore, still contain the Arg/N-end rule pathway (Fig. 3). It is possible that UBR1, despite being 47% identical to UBR2,30,292 has a JBS-relevant physiological substrate(s) that is confined to UBR1. In addition, previous work has shown that S. cerevisiae Ubr1 is an activity-limiting component of the yeast Arg/N-end rule pathway.12 Thus, it is also possible that some JBS phenotypes may be caused by a suboptimal overall activity of the Arg/N-end rule pathway in patients with JBS, as distinguished from the loss of a function unique to UBR1.
Lyon and coworkers have recently identified the first human birth defect in the Ac/N-end rule pathway [Fig. 2(B)], an X-linked infantile lethal disorder caused by a hypomorphic missense mutation in NAA10 (hARD1), which encodes the catalytic subunit of the NatA Nt-acetylase (G. J. Lyon, a personal communication). This enzyme Nt-acetylates the N-terminal Ala, Ser, Thr, Val, Cys, and Gly residues of nascent proteins after the cotranslational removal of their N-terminal Met by Met-aminopeptidases.91–94,102 The early postnatal lethality of the human NAA10 syndrome is consistent with lethality of null NAA10 mutants in Trypanosoma brucei, C. elegans, and D. melanogaster (Refs. 293 and294 and references therein). Given the necessity of Nt-acetylation for the activity of AcN-degrons,3 an increased fraction of non-Nt-acetylated physiological substrates of NatA in human or other hypomorphic NAA10 mutants would be expected to (at least) partially stabilize these proteins against degradation. Detrimental effects of this stabilization (The Ac/N-End Rule Pathway section) may be one cause of the early postnatal lethality of the human NAA10 syndrome, despite a significant residual activity of the mutant NAA10 Nt-acetylase (G. J. Lyon, a personal communication).
Regulation of bacterial virulence and the Arg/N-end rule pathway
Listeria monocytogenes is a gram-positive bacterial pathogen of humans and other animals. Following internalization into mammalian cells, bacteria are initially contained within host vacuoles but rapidly escape into the cytosol (where they grow and divide), largely through the perforation of the phagosomal membrane by the pore-forming toxin called listeriolysin O (LLO). LLO that is secreted from bacterial cells into the cytosol of mammalian cells bears N-terminal Lys, an Ndp residue (Figs. 3 and 10). LLO is indeed a short-lived N-end rule substrate in mammalian cells.124 Moreover, replacement of N-terminal Lys of LLO with Val not only increased the in vivo half-life of LLO but also decreased the virulence of L. monocytogenes.124 Thus, the presence of strongly destabilizing residues at the N-termini of LLO toxins from L. monocytogenes and related bacteria124 is likely to have been selected, during evolution, to optimize, through downregulation, the levels of secreted LLO in the cytosol of infected mammalian cells.
Regulation of DNA repair and the Arg/N-end rule pathway
O6-methylguanine (O6meG) and related modifications of guanine in double-stranded DNA are lesions that can be produced by many alkylating agents, including N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), a potent carcinogen. O6meG is repaired through demethylation by the O6-methylguanine-DNA alkyltransferase (AGT). This protein is called MGMT (O6-methylguanine-DNA methyltransferase) in mammals and Mgt1 in S. cerevisiae. AGT proteins remove methyl and other alkyl groups from an alkylated O6 in guanine by transferring the adduct to an active-site Cys residue (ref. 40 and references therein). The resulting S-alkyl-Cys of AGT is not restored back to Cys, so repair proteins of this kind can act only once. Recent work has shown that S. cerevisiae Mgt1 (preferentially but not exclusively its S-alkylated form) is targeted for polyubiquitylation and degradation by the Arg/N-end rule pathway, specifically by its double-E3 Ubr1/Rad6-Ufd4/Ubc4 targeting complex40,44 [Fig. 2(A) and The double-E3 design of the Arg/N-end rule pathway section]. The Ubr1/Ufd4-dependent Arg/N-end rule pathway mediates not only the constitutive but also MNNG-accelerated degradation of Mgt1. Although the degradation signal of Mgt1 is near its N-terminus, it is not an N-degron.40,44
Because the Ubr1 and Ufd4 E3s physically interact, it is remarkable that the degron of Mgt1 can be recognized, independently, by either Ubr1 or Ufd4, possibly in a mutually exclusive manner, a previously undescribed mode of “alternative” degron recognition that remains to be understood both functionally and mechanistically (ref. 40; A. Shemorry et al., unpublished data). It is also unknown whether the targeting and polyubiquitylation of Mgt1 by the Ubr1/Rad6-Ufd4/Ubc4 complex can involve, in vivo, the DNA-bound Mgt1 protein or whether this targeting requires a dissociation of Mgt1 from DNA. A priori, it is likely (nothing is known about this at present) that Mgt1 functions as a part of a chromosome-associated protein complex. If so, degradation of Mgt1 may involve the subunit selectivity of the Arg/N-end rule pathway,13 an aspect of its activity that is also essential for selective degradation of the separase-produced C-terminal fragment of the Scc1 cohesin subunit that is bound to the rest of cohesin complex.28 In both cases, the alkylated (i.e., irreversibly inactivated) Mgt1 and the fragment of cohesin subunit are obligatory “dead-end” products of their respective circuits that are still parts of larger complexes. As discussed above in the context of cohesin, the Arg/N-end rule pathway operates, in these settings, as a homeostatic device that uses its capacity for subunit-selective proteolysis to reset the states of relevant circuits. It is likely (but remains to be determined) that MGMT, the mammalian counterpart of yeast Mgt1, is also regulated through degradation by the Arg/N-end rule pathway. The activity of MGMT in repairing alkylated DNA is of major relevance to the efficacy of DNA-alkylating anticancer drugs, a set of cytotoxic compounds extensively used in cancer therapy.255
Regulation of peptide transport and the Arg/N-end rule pathway
Peptides can serve as a source of amino acids and nitrogen in all organisms. The import of dipeptides and tripeptides (di/tripeptides) in S. cerevisiae is controlled by the Arg/N-end rule pathway295,296 through a specific design of the Ubr1 E3 Ub ligase. The Type-1/2 substrate-binding sites of Ubr1 bind to the Type-1 and Type-2 destabilizing N-terminal (Ndp) residues of proteins or short peptides, whereas the third binding site of Ubr1 recognizes an internal degron of Cup9, a homeodomain transcriptional regulator, largely a repressor, of more than 30 genes.38,138 The regulon of Cup9 includes PTR2, a gene encoding the main importer of dipeptide/tripeptide.297,298 The binding of imported di/tripeptides bearing destabilizing N-terminal residues to the Type-1/2 sites of Ubr1 allosterically activates the otherwise autoinhibited Cup9-binding site of Ubr1. The resulting increase in the fraction of Ubr1 molecules that can target Cup9 for polyubiquitylation leads to a faster degradation of Cup9 (t1/2 < 2 min) and thereby causes a derepression of the PTR2 gene. This positive-feedback circuit allows S. cerevisiae to detect the presence of extracellular peptides and to react by increasing the rate of their uptake27,29,138 (Fig. 15). Genes of the Cup9 regulon other than PTR2 that are also induced by di/tripeptides include additional components of the peptide transport and utilization system, for example, OPT2, which encodes an oligopeptide importer, and also peptidases that process imported peptides to amino acids.297,299
The properties of S. cerevisiae Ubr1 that underlie the regulation of peptide import through the conditional degradation of Cup9 were found to require phosphorylation of Ubr1 at Ser300 by the sequelogous type-I caseine kinases Yck1 and Yck2 [Figs. 7(A,B) and 15]. Interestingly, the phosphorylation of Ubr1 at Ser-300 by Yck1/2 initiates (primes) a specific phosphorylation cascade that results in phosphorylation of Ser296, Ser292, Thr288, and Tyr277 of Ubr1 by Mck1, a kinase of the glycogen synthase 3 (Gsk3) family39 [Fig. 7(A,B)]. In contrast to the functionally critical phosphorylation at Ser300 by Yck1/2, the subsequent phosphorylations of Ubr1 by the Mck1 kinase had at most minor effects on the known properties of Ubr1, including regulation of peptide import.39 A function of Ubr1 that requires the Mck1-mediated phosphorylation cascade [Fig. 7(A,B)] remains to be discovered.
The induction of the Ptr2 transporter by di/tripeptides, a process controlled by the Arg/N-end rule pathway (Fig. 15), is just one of regulatory inputs that couple Ptr2 expression to the availability and quality of nutrients. For example, Ptr2 expression is downregulated by certain nitrogen sources, including ammonia, but not by other nitrogen sources, such as urea or allantoin.300 Several systems, including the Arg/N-end rule pathway, ensure that a cell does not waste resources synthesizing large amounts of the Ptr2 transporter in the absence of extracellular peptides, or when a more efficacious nitrogen source, such as ammonia, is present. Ptr2 is induced not only by short peptides but also by extracellular amino acids, particularly Leu and Trp. This response would be adaptive in natural habitats, because amino acids that S. cerevisiae (a scavenging heterotroph) encounters outside a laboratory tend to be products of protein breakdown and therefore signify a likely presence of short peptides. It has also not been precluded that Ptr2 might import some amino acids, in addition to peptides.
Extracellular amino acids regulate S. cerevisiae Ptr2 through the SPS (SSY1-PTR3-SSY5) system.301–304 Ssy1 is an integral membrane protein and sensor of amino acids that does not function as a transporter. Ssy1 “measures” the concentration ratio of an amino acid across the plasma membrane and signals through the rest of the SPS pathway to induce a regulon that includes a number of amino acid transporters as well as the Ptr2 peptide transporter. This signaling uses a proteolytic activation of latent (conditionally cytosolic) transcriptional activators Stp1 and Stp2, leading to their import into the nucleus and the induction of the SPS regulon.301–304 Our recent work has shown that an extracellular amino acid such as Trp acts via the SPS system to induce the Ptr2-mediated import of di/tripeptides not only through activation of positive transcriptional regulators such as Stp1/2 but also through acceleration of the degradation of the Cup9 repressor by the Arg/N-end rule pathway138 (Fig. 15).
It is likely that the Arg/N-end rule pathway regulates peptide transport in multicellular eukaryotes as well. In contrast to S. cerevisiae, mammalian cells contain peptide transporters not only in the plasma membrane but also in the ER membrane. These ER proteins, called TAP transporters, mediate the import of peptides (including peptides produced by the proteasome-mediated degradation of cytosolic and nuclear proteins) from the cytosol into the lumen of ER, where these peptides are loaded onto MHC class I proteins en route to the cell surface, for presentation of MHC-peptide complexes to T lymphocytes.305 Might a mammalian peptide-sensing circuit based on the Arg/N-end rule pathway regulate the expression of TAP transporters as well? This and related questions remain to be addressed.
Results of a recent study suggested that the mammalian Arg/N-end rule pathway plays a role, through the Ubr1 and Ubr2 N-recognins, in the control of protein synthesis by amino acids.306 Specifically, mammalian Ubr1 and Ubr2 appear to recognize (bind to) free Leu (ref. 306), as distinguished from the binding of these N-recognins to dipeptides bearing N-terminal Leu. Such dipeptides bind to, in particular, S. cerevisiae Ubr1 with Kd in a low-μM range38 (The double-E3 design of the Arg/N-end rule pathway section). In contrast, S. cerevisiae Ubr1 has a considerably (more than 10-fold) lower affinity for free amino acids.38 Therefore, it remains to be determined, using quantitative binding assays that involve free (nonimmobilized) Leu, whether its affinity for mammalian Ubr1 is in a range that would be likely to be relevant physiologically. The possibility of Ubr1/2 involvement in the control of protein synthesis was also suggested by the finding that overexpression of rat Ubr1 and Ubr2 in rat cells inhibited the activity of mTOR kinase, a key regulator of translation, and that the inhibition could be reversed by millimolar levels of Leu in the medium.306
The N-end rule pathway and degradation of misfolded proteins
Errors in specific steps of protein synthesis result, conservatively, in 5–20% of molecules of a 50 kDa protein containing at least one missense alteration that is not encoded by DNA.307,308 Premature termination of translation and frameshifts are additional sources of defective polypeptides. The misfolding and aggregation of mistranslated proteins and the resulting toxicity are significant even in the absence of environmental insults.307–311 Recent studies have shown that Ubr1, the E3 N-recognin of the S. cerevisiae Arg/N-end rule pathway, is a part of quality-control systems that function to selectively eliminate misfolded proteins in the nuclear and cytosolic compartments.119–121 In yeast, these systems include the import (assisted by chaperones of the Hsp70 family) of misfolded cytosolic proteins into the nucleus, followed by their degradation there by pathways that involve both the San1 E3 Ub ligase and the Ubr1-mediated Arg/N-end rule pathway.120,121,312–314 UBR-type N-recognins of the mammalian Arg/N-end rule pathway were also found to play a role in degradation of misfolded proteins.122
Does the S. cerevisiae Ubr1 N-recognin mediate the degradation of misfolded proteins as the double-E3 Ubr1–Ufd4 complex [Fig. 2(A)], or does it, perhaps, interact with other, “alternative” E3s as well? Does the targeting of misfolded proteins by Ubr1 involve N-degrons that would be produced through preliminary cleavages of misfolded proteins by (nonprocessive) proteases, or are such proteins targeted by the Arg/N-end rule pathway through their internal degrons, exposed as a result of misfolding? If so, are these degrons recognized by the substrate-binding sites of Ubr1 that target the internal degrons of the Cup9 repressor29,38 or the Mgt1 DNA repair protein,40 or does the targeting of misfolded proteins involve other, currently unknown binding sites of Ubr1? Does the in vivo degradation of some misfolded proteins also involve the Ac/N-end rule pathway and AcN-degrons3 [Fig. 2(B)]? These cotranslationally formed N-degrons are present in a majority of cellular proteins, and were suggested to mediate the selective elimination of unassembled, misfolded or otherwise aberrant proteins3 (The Ac/N-End Rule Pathway section). If AcN-degrons contribute to selective destruction of misfolded proteins, might some of the E3s involved that are presumed, at present, to function outside of the Ac/N-end rule pathway, have binding sites for AcN-degrons? As illustrated by these questions, our understanding of the Arg/N-end rule and Ac/N-end rule pathways as components of protein quality control is just beginning.
The roles of the N-end rule pathway described above are but a small subset of its currently known functions, summarized in Overview of the N-End Rule Pathway Section. Many of these functions were discovered through genetic approaches and therefore remain to be understood in mechanistic, substrate-based terms.
Prokaryotic N-End Rule Pathways
Although prokaryotes (bacteria and archaea) lack a bona fide Ub system, at least gram-negative bacteria contain a Ub-independent version of the N-end rule pathway, termed the Leu/N-end rule pathway (Overview of the N-End Rule Pathway and Substrates of the N-End Rule Pathway sections, Figs. 5 and 6). The Leu/N-end rule pathway was discovered in 1991 (ref. 14), and was characterized in gram-negative bacteria E. coli and Vibrio vulnificus.16,37 It comprises the following components.14,16,19,37,71–79,81,186,188,189,191,199,315–318 (1) ClpAP, a proteasome-like, ATP-dependent protease; (2) ClpS, the 12-kDa N-recognin of the Leu/N-end rule pathway that binds to N-terminal Leu, Phe, Trp or Tyr (the pathway's Ndp residues) and delivers N-end rule substrates to the ClpAP protease [Figs. 5 and 6(B–D)]; (3) Aat, an L/F-transferase that uses Leu-tRNA or Phe-tRNA as a cosubstrate to conjugate largely Leu (and occasionally Phe) to the N-termini of proteins bearing N-terminal Lys or Arg, the secondary destabilizing (Nds) residues of the Leu/N-end rule pathway [in contrast, Lys and Arg are Ndp residues in eukaryotes; Fig. 2(A); cf. Fig. 5]. Crystal structures of Aat suggested a catalytic mechanism of this enzyme316–318; (4) Bpt, an L-transferase that uses Leu-tRNA to conjugate Leu to N-terminal Asp, Glu, and (possibly) oxidized Cys [Fig. 5(B)].
V. vulnificus contains both the Aat and Bpt L-transferases, but E. coli contains only Aat. Therefore, N-terminal Arg, Lys, Asp and Glu are secondary destabilizing (Nds) residues in V. vulnificus, whereas in E. coli the N-terminal Asp and Glu are stabilizing (nondestabilizing) residues37 (Fig. 5). In V. vulnificus, the two L-transferases are encoded by the aat-bpt operon, but in several other gram-negative bacteria these genes are unlinked, and some gram-negatives lack one or the other of these L-transferases (e.g., E. coli lacks Bpt).37 Genomes of examined gram-negative bacteria encode ClpS, but archaea (archaebacteria) and gram-positive bacteria appear to lack significant sequelogs of Aat, Bpt, and ClpS, and therefore may lack the Leu/N-end rule pathway. This is a tentative conclusion, because, to the best of my knowledge, no biochemical tests, using N-end rule reporter proteins, for the presence of a Leu/N-end rule pathway or its analog have been carried out, thus far, with a gram-positive bacterium or an archaeon.
The ∼27-kDa bacterial Bpt L-tranferases are sequelogous to the ∼60-kDa eukaryotic Ate1 R-transferases [Fig. 6(A)], strongly suggesting their homology, that is, a common ancestry in evolution. In contrast to Ate1 R-transferases, Bpt L-transferases contain the CP....CXXCXXC motif [Fig. 6(A); X is any residue; in some Bpt enzymes, Pro is replaced by Ser or other residues]. This set of sequences is one of consensus motifs for binding a complex of nonheme iron and inorganic sulfide called a Fe-S cluster. Such complexes are present in many proteins and mediate a broad range of biological processes, including respiration, photosynthesis, redox catalysis, gene regulation, and a variety of enzymatic reactions.319,320 At least some cysteines of the CP....CXXCXXC motif are required for the activity of the V. vulnificus Bpt L-transferase. The presence of this motif [Fig. 6(A)] and spectroscopic properties of isolated Bpt are consistent with it being a Fe-S cluster protein (H. Wang and A. Varshavsky, unpublished data).
Specific mechanisms of the ClpS/ClpAP-mediated targeting and degradation of N-end rule substrates by the bacterial Leu/N-end rule pathway16,75,76,78,82 are described in Substrates of the N-End Rule Pathway section (Fig. 12). These mechanisms are understood in greater detail than the analogous but Ub-dependent targeting in the eukaryotic Arg/N-end rule pathway [Substrates of the N-End Rule Pathway section, Figs. 2(A), 3, and 11]. A sequelogy (sequence similarity) between the substrate-binding segment of the 12-kD bacterial ClpS N-recognin and the functionally analogous segment of a vastly larger E3 Ub ligase such as the yeast or mammalian Ubr1 N-recognin [Fig. 6(D)] suggests homology, that is, a common descent of bacterial and eukaryotic N-recognins (Evolution of the N-End Rule Pathway Section).
Two physiological substrates of the Leu/N-end rule pathway have recently been discovered.76,77 One of them is Dps, an 18-kDa DNA-binding protein that compacts the nucleoid of E. coli in starving cells, forming highly ordered, crystal-like structures. An unknown protease removes 5 residues from the initial N-terminus of Dps, producing N-terminal Leu (an Ndp residue) and thereby making Dps a short-lived N-end rule substrate. In a plausible model, a protease that produces the N-degron of Dps may do so in a regulated manner, at a time when the bulk of Dps should be removed from DNA through processive degradation by the Leu/N-end rule pathway. Remarkably, Dps can also be destroyed as the full-length (uncleaved) protein, by another processive protease, ClpXP, which targets the N-terminus-proximal segment of Dps. This segment is at least partially removed by a protease that produces the N-degron of Dps. An interplay between ClpS-dependent and ClpS-independent degradation of Dps, and physiological ramifications of this circuit remain to be understood.76,77
The other identified N-end rule substrate is the E. coli YgjG putrescine-aminotransferase (PATase). It is targeted for degradation by the Leu/N-end rule pathway through a route that requires Nt-leucylation by the Aat L/F-transferase76,77 [Fig. 5(A)]. The evidence by Ninnis et al.77 suggests that Aat can conjugate Leu or Phe to the (initially present) N-terminal Met residue of PATase, as distinguished from the previously characterized specificity of Aat, which conjugates (preferentially) Leu to N-terminal Arg or Lys of N-end rule substrates. It is possible that the active site of the Aat L/F-transferase might be able to accommodate the N-terminal Met of a substrate for its conjugation to Leu if the substrate's second residue is, for example, Asn, a small hydrophilic residue.77 It remains to be determined whether Aat can Nt-leucylate, in vivo, some N-terminal motifs that contain N-terminal Met. Physiological roles of the degradation of the putrescine-catabolizing PATase by the Leu/N-end rule pathway, for example, in controlling PATase levels as a function of in vivo putrescine levels,76,77 remain to be addressed as well.
Both gram-negative and gram-positive bacteria use the Nt-formylated Met residue (fMet) to initiate the synthesis of a polypeptide chain. The resulting N-terminal fMet of nascent bacterial proteins is cotranslationally deformylated by a ribosome-bound deformylase.321 In contrast to bacteria and similarly to eukaryotes, archaea initiate translation using unmodified Met, and in addition cotranslationally Nt-acetylate a large subset of their proteins.322 The protein α1 is one of two subunits that form the 20S core proteasome in the archaeal prokaryote Haloferax volcanii. Interestingly, the in vivo concentration of α1 was found to depend on the identity of a second-position residue that follows the N-terminal Met of the α1 protein.323 In particular, the levels of N-terminal mutants of the α1 subunit that were partially non-Nt-acetylated were strikingly higher than the levels of Nt-acetylated (wild-type) α1, in the absence of increased expression of α1 mRNA.323 Although the authors did not consider the possibility of AcN-degrons, their findings with H. volcanii α1 and its mutants may be accounted for by the presence of the Ac/N-end rule pathway in archaeal prokaryotes. This possibility remains to be examined.
Mitochondrial N-End Rule Pathway
Mitochondria of eukaryotic cells are descendants of gram-negative bacteria, which contain the Leu/N-end rule pathway [Fig. 3(B,C) and The Arg/N-End Rule Pathway section]. Might the matrix of mitochondria (a counterpart of bacterial cytosol) also contain an N-end rule pathway?5 Recent results by Pfanner and colleagues277,278,324 did reveal such a pathway. Specifically, most N-terminal presequences of nuclear-encoded mitochondrial matrix proteins are cleaved off by the mitochondrial processing protease (MPP) upon the import of these proteins from the cytosol into the matrix. The cleavage specificity of mitochondrial MPP often results in N-terminal residues of processed proteins that are destabilizing in both the bacterial Leu/N-end rule pathway and the eukaryotic (cytosolic/nuclear) Arg/N-end rule pathway [Figs. 2(A), 4(B), and 5]. Vögtle et al.277 identified an aminopeptidase, termed Icp55, in the mitochondrial matrix of S. cerevisiae that removes a single (bulky hydrophobic) destabilizing residue from the N-termini of matrix-imported, MPP-processed mitochondrial proteins. This processing step tends to yield proteins that bear unmodified N-terminal residues such as Ala, Ser or Thr. Remarkably, some of the normally long-lived mitochondrial matrix proteins become short-lived in icp55Δ cells, whose mitochondria lack the Icp55 protease.277
Thus, at least the bulky hydrophobic N-terminal residues, which are destabilizing in both the Arg/N-end rule pathway and the bacterial Leu/N-end rule pathway [Fig. 4(B)], are destabilizing in mitochondria as well, except that in wild-type cells these residues are cleaved off, by Icp55, from the N-termini of imported matrix proteins. One function of the Icp55 aminopeptidase is apparently to reduce or preclude the degradation of newly imported mitochondrial proteins by an N-end rule pathway in this organelle.277,278 Yet another mitochondrial processing peptidase, Oct1, also contributes, in addition to Icp55, to modifying the N-termini of specific proteins that are imported into the matrix, so that their mature N-terminal residues are not destabilizing in the bacterial-type N-end rule.324 In oct1Δ yeast cells, the substrates of the Oct1 protease retain destabilizing N-terminal residues [Figs. 4(B) and 5] and are short-lived in the matrix.324
Mitochondria presumably “inherited” their N-end rule pathway from their bacterial ancestors. It is likely that processing proteases such as Icp55 and Oct1 evolved as “suppressors” that prevent the degradation of otherwise short-lived mitochondrial N-end rule substrates that form upon the import of proteins from the cytosol into the matrix. If so, what exactly is the function of the N-end rule pathway in mitochondria? In particular, what mitochondrial proteins remain physiological N-end rule substrates in the presence of Icp55 and Oct1, which “rescue” specific proteins from becoming substrates of the N-end rule pathway? What is the identity of an N-recognin and a processive “downstream” protease that recognizes and degrades N-end rule substrates in mitochondria? These questions remain to be addressed. Because yeast Icp55 and Oct1 are evolutionarily conserved proteases, an N-end rule pathway is likely to be present in mitochondria of all eukaryotes. Remarkably, putative counterparts of mitochondrial Icp55 are also present at least in the cytosol.277 Thus, a previously unexplored possibility is that some cytosolic/nuclear substrates of the Arg/N-end rule pathway may be subject to N-terminal “editing” (and thus, a rescue from degradation) by specific aminopeptidases that would act, in such settings, similarly to the mitochondrial Icp55 and Oct1 proteases.
Chloroplasts of plant cells are descendants of ancient cyanobacteria, which may have also contained an N-end rule pathway. Indeed, chloroplasts (but apparently not mitochondria) contain a strong sequelog of bacterial ClpS N-recognins [Fig. 6(D)], suggesting a chloroplast Leu/N-end rule pathway. This possibility remains to be verified.
Evolution of the N-End Rule Pathway
An N-terminal residue is present even in dipeptides. Thus, N-degrons of polypeptide substrates were available as targets of proteolytic systems from early stages of protein evolution, in agreement with the presence of N-end rule pathways in both prokaryotes and eukaryotes. Primordial polypeptides, produced by ribozymes that eventually became ribosomes,325,326 were at first (most likely) short, disordered and therefore prone to aggregation or other noncognate interactions. Thus, damage control pathways that involve chaperones and selective proteolysis182,327 were probably as vital at the dawn of protein-based life as they are in extant cells. Prior to emergence of quasi-modern proteins, primordial proteases and chaperones consisted, at least in part, of specific RNAs or other nucleic acids. Over ensuing eons, a succession of changes, driven by selection for cells that produced optimal amounts of fitness-increasing polypeptides, led to the emergence of the genetic code, ribosomes and the rest of the translation apparatus. This evolution also led to DNA-encoded polypeptides that began to resemble their modern counterparts, including functional properties of specific proteins and their ability to stay in solution. Fundamental similarities among the extant bacteria, archaea and eukaryotes indicate that the last universal common ancestor (LUCA) of modern organisms was a cell that expressed a broad range of proteins.326,328–330 LUCA also contained proteolytic systems, including, probably (for the reasons above), an N-end rule pathway.
The Nt-acetylation of cellular proteins apparently predated LUCA, as both bacteria, archaea and eukaryotes contain Nt-acetylases.91,92 Thus, a version of the Ac/N-end rule pathway3 [Fig. 2(B)] may have already been in place by the time of LUCA. As to the Leu/N-end rule and Arg/N-end rule pathways, their presence in Gram-negative bacteria and in eukaryotes, respectively [Figs. 2(A) and 5], suggests an emergence of a version of the Leu/N-end rule pathway in pre-LUCA cells. The N-terminal Leu, Phe, Tyr and Trp residues of N-end rule substrates are recognized both by the 12-kDa E. coli ClpS and by the Type-2 binding site of eukaryotic N-recognins such as the 200 kDa human UBR1 E3 Ub ligase [Fig. 6(B–D)]. The highly sequelogous segments in the substrate-binding sites of bacterial ClpS and eukaryotic Ubr1 suggests a common ancestry for these otherwise nonsequelogous proteins of vastly different sizes72,74,83–85,331 [Fig. 6(D)], presumably through the emergence, at first, of a ClpS-like protein in a lineage of cells that led to LUCA.
LUCA might have also contained early versions of Bpt and/or Aat L-transferases, which act upstream of ClpS in the Leu/N-end rule pathway (Fig. 5). After separation of prokaryotic and eukaryotic lineages, the expanding rule books of the N-end rule continued to be similar in all organisms [Fig. 4(B)], but the underlying machinery began to diverge. In particular, Ub ligases and deubiquitylases have emerged (or, alternatively, were retained) only in eukaryotes.37,332 Some bacteria express mediators or inhibitors of the Ub system that are injected into eukaryotic hosts during infection, but these proteins do not seem to have Ub-related functions in bacteria themselves.333–337 (Possible causes of the absence of a bona fide Ub system in extant prokaryotes are considered in Substrates of the N-End Rule Pathway section.) In addition, the problem of recognizing basic and acidic (as distinguished from bulky hydrophobic) N-terminal residues in later versions of the N-end rule pathway was solved quite differently in eukaryotes versus bacteria. For example, the largest known bacterial N-end rule, in the Gram-negative bacterium Vibrio vulnificus, is similar to the N-end rule of the eukaryotic Arg/N-end rule pathway37 [Figs. 2(A), 4(B), and 5]. Remarkably, however, this similarity results from different mechanisms in bacteria versus eukaryotes: the N-terminal Arg and Lys are Ndp (primary destabilizing) residues in the eukaryotic Arg/N-end rule pathway, in that these basic residues (and N-terminal His as well) are recognized directly by the UBR domains (Type-1 binding sites) of E3 N-recognins such as Ubr1 [Figs. 2(A), 3(C), 5, and 8(A,B)].83–85 In contrast, the N-terminal Arg and Lys are Nds (secondary destabilizing) residues in both E. coli and V. vulnificus, owing to the presence of the Aat L/F-transferase. This enzyme, which conjugates Leu (and occasionally Phe) to the N-terminal Arg or Lys residues14,71,316–318 (Fig. 5), is absent from examined eukaryotes. The Nt-leucylating bacterial Bpt L-transferases are sequelogous1 to Nt-arginylating Ate1 R-transferases of eukaryotes [Fig. 6(A) and ref. 37]. Strikingly, however, bacterial Bpt L-transferases are largely nonsequelogous to bacterial Aat L/F-transferases,37 despite the similarity of reactions catalyzed by Bpt and Aat (Fig. 5).
One difference between bacterial and eukaryotic N-end rule pathways is particularly remarkable in its consistency (no exceptions so far) and the absence of a robust explanation for its emergence on either functional or mechanistic grounds. Specifically, the bacterial Leu/N-end rule pathway uses Nt-leucylation of N-end rule substrates, whereas the otherwise similar eukaryotic Arg/N-end rule pathway uses Nt-arginylation [Figs. 2(A), 3, and 5]. The leucylation/arginylation dichotomy exists despite a significant sequelogy1 (sequence similarity) between the Nt-leucylating Bpt transferases of bacteria and the Nt-arginylating Ate1 R-transferases of eukaryotes [Fig. 6(A) and ref. 37]. This sequelogy suggests homology, that is, a common ancestry of Bpt and Ate1. Nevertheless, the Nt-arginylating and Nt-leucylating transferases are cleanly divided, in extant organisms, between Bpt L-transferases in bacteria and Ate1 R-transferases in eukaryotes. This dichotomy is even more remarkable than described above. Specifically, Plasmodium falciparum, an obligatory intracellular parasite and the cause of malaria in humans, is a eukaryote that contains a Plasmodium-specific R-transferase.37 However, this R-transferase, which is also present in other apicomplexans (a group of obligately parasitic unicellular eukaryotes that includes Plasmodium) but not in other examined eukaryotes, is a sequelog not of the bacterial Bpt L-transferase but of the bacterial Aat L/F-transferase.37 (This would not be expected a priori, given the sequelogy between bacterial Bpt and non-Plasmodium eukaryotic Ate1 R-transferases.)
Thus, irrespective of whether an Nt-arginylating R-transferase of a eukaryotic cell descended from a Bpt-type L-transferase or from a largely nonsequelogous Aat-type L/F-transferase (Fig. 5), the final result was, in all cases, the change of enzymatic specificity from that of L-transferase (Nt-leucylation) to that of R-transferase (Nt-arginylation). Why? There is no clear answer (to the best of my knowledge), in part because the replacement of Nt-leucylation in bacteria with Nt-arginylation in eukaryotes does not appear to have been necessary for “mechanistic” reasons. Specifically, most UBR-type N-recognins in the Arg/N-end rule pathway of eukaryotes contain a Type-2 substrate-binding site whose ability to recognize the N-terminal Leu, Phe, Tyr Trp or Ile residues of N-end rule substrates is essentially identical to the recognition specificity of ClpS, the bacterial N-recognin [Figs. 5, 6(B–D), and 7(A)]. Thus, the presumably ancestral Nt-leucylation should have sufficed in eukaryotes, without a change to Nt-arginylation. However, Nt-arginylation did emerge in eukaryotes and replaced Nt-leucylation not only completely but also early in eukaryotic evolution. Specifically, the S. cerevisiae and human Ate1 R-transferases are highly sequelogous enzymes. In addition, there is no Nt-leucylation in examined eukaryotes and there is no Nt-arginylation in examined prokaryotes.
For reasons suggested by Lynch210 (Structure and Targeting of N-degrons section), many aspects of molecular circuits in eukaryotic cells evolved through a genetic drift and occasional fixation of mildly deleterious mutations, as distinguished from adaptive Darwinian evolution based on positive selection. Although an initially nonadaptive evolution may be relevant to the emergence of Nt-arginylation in eukaryotes, it is most likely that a specific selection pressure favored, in primordial eukaryotes, a replacement of the (apparently) preceding Nt-leucylation by Nt-arginylation. What might be a reason for such a pressure? Discussions of LUCA and early evolution suffer from the problem that a less parsimonious, more convoluted scenario would be unlikely a priori (because of Occam's razor) but cannot be formally precluded. With this caveat, one difference between Arg and Leu that may have caused the ascendancy of Nt-arginylation in eukaryotes is high reactivity of the side chain of Arg, in comparison to Leu, and a multitude of metabolic transitions that involve Arg, often in the context of the Arg/N-end rule pathway. For example, Arg is a substrate of NO synthases and the immediate precursor of NO. Furthermore, NO is required for the in vivo oxidation of N-terminal Cys, a modification that allows Nt-arginylation of the resulting Cys-sulfinate or Cys-sulfonate by the Ate1 R-transferase of the Arg/N-end rule pathway32 (Fig. 3). Arg-tRNA is a cosubstrate of R-transferase and at the same time the source of Arg in proteins produced by the ribosomes, suggesting a competition between these uses of Arg. (But an analogous competition also exists for the consumption of Leu-tRNA by L-transferases vs. bacterial ribosomes.) Some Arg residues in proteins undergo enzymatic methylation or deimination, the latter a conversion of the positively charged Arg to the uncharged citrulline residue.338,339
Might the in vivo methylation or deimination of Arg in cellular proteins involve the N-terminal Arg residue as well? Might there be a regulatory connection, through Arg-tRNA and R-transferase, between the Arg/N-end rule pathway and translation by ribosomes? (Interestingly, the mouse Ate1 R-transferase binds to specific mRNAs and is associated with translationally active polysomes (R.-G. Hu et al., unpublished data; see also ref. 114.) These are some among a multitude of questions that can be asked about the Arg/N-end rule pathway. The answers at hand have revealed a strikingly multifunctional, universally present and extensively regulated proteolytic circuit [Figs. 2(A) and 3]. Explorations of bacterial and mitochondrial N-end rule pathways continue as well. The recent discovery of the Ac/N-end rule pathway [Figs. 2(B) and 4(A)] further expanded the already broad functional scope of the eukaryotic N-end rule pathway, and has also revealed the main physiological functions of Nt-acetylases and Met-aminopeptidases3. Many years after the initial discovery of the N-end rule4, this ancient system continues to be a fount of biological insights.
Acknowledgments
I thank T. Baker, D. Barford, C. Brower, D. Finley, M. Lynch, M. Maurizi, A. Shemorry, and B. Wadas for their helpful comments on the manuscript. I am particularly grateful to D. Finley for his detailed suggestions. I also thank D. Barford, H.K. Song, and K. Zeth for their assistance with and advice about figures.
References
- 1.Varshavsky A. Spalog and sequelog: neutral terms for spatial and sequence similarity. Curr Biol. 2004;14:R181–R183. doi: 10.1016/j.cub.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. Naming a targeting signal. Cell. 1991;64:13–15. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- 3.Hwang C-S, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 5.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varshavsky A. Discovery of cellular regulation by protein degradation. J Biol Chem. 2008;283:34469–34489. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A, Varshavsky A. The ubiquitin system. Nat Med. 2000;10:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 8.Varshavsky A. The early history of the ubiquitin field. Protein Sci. 2006;15:647–654. doi: 10.1110/ps.052012306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 10.Gonda DK, Bachmair A, Wünning I, Tobias JW, Lane WS, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 11.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 12.Bartel B, Wünning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson ES, Gonda DK, Varshavsky A. Cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature. 1990;346:287–291. doi: 10.1038/346287a0. [DOI] [PubMed] [Google Scholar]
- 14.Tobias JW, Shrader TE, Rocap G, Varshavsky A. The N-end rule in bacteria. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 15.Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Mogk A, Schmidt R, Bukau B. The N-end rule pathway of regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–689. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graciet E, Wellmer F. The plant N-end rule pathway: structure and functions. Trends Plant Sci. 2010;15:447–453. doi: 10.1016/j.tplants.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nature Biotechnol. 2000;18:494–496. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 21.Baker RT, Varshavsky A. Inhibition of the N-end rule pathway in living cells. Proc Natl Acad Sci USA. 1991;87:2374–2378. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Y, Varshavsky A. The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker RT, Varshavsky A. Yeast N-terminal amidase: a new enzyme and component of the N-end rule pathway. J Biol Chem. 1995;270:12065–12074. doi: 10.1074/jbc.270.20.12065. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki T, Varshavsky A. Degradation signals in the lysine-asparagine sequence space. EMBO J. 1999;18:6017–6026. doi: 10.1093/emboj/18.21.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon YT, Balogh SA, Davydov IV, Kashina AS, Yoon JK, Xie Y, Gaur A, Hyde L, Denenberg VH, Varshavsky A. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1 amidase and the asparagine branch of the N-end rule pathway. Mol Cell Biol. 2000;20:4135–4148. doi: 10.1128/mcb.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon YT, Kashina AS, Davydov IV, Hu R-G, An JY, Seo JW, Du F, Varshavsky A. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 27.Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 28.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–960. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 29.Du F, Navarro-Garcia F, Xia Z, Tasaki T, Varshavsky A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc Natl Acad Sci USA. 2002;99:14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon YT, Xia ZX, An JY, Tasaki T, Davydov IV, Seo JW, Xie Y, Varshavsky A. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tasaki T, Mulder LCF, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu R-G, Sheng J, Xin Q, Xu Z, Takahashi TT, Varshavsky A. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 33.Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, Davydov IV, Kwon YT. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zenker M, Mayerle J, Lerch MM, Tagariello A, Zerres K, Durie PR, Beier M, Hülskamp G, Guzman C, Rehder H, Beemer FA, Hamel B, Vanlieferinghen P, Gershoni-Baruch R, Vieira MW, Dumic M, Auslender R, Gil-da-Silva-Lopes VL, Steinlicht S, Rauh R, Shalev SA, Thiel C, Winterpacht A, Kwon YT, Varshavsky A, Reis A. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome) Nat Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 35.Hu R-G, Brower CS, Wang H, Davydov IV, Sheng J, Zhou J, Kwon YT, Varshavsky A. Arginyl-transferase, its specificity, putative substrates, bidirectional promoter, and splicing-derived isoforms. J Biol Chem. 2006;281:32559–32573. doi: 10.1074/jbc.M604355200. [DOI] [PubMed] [Google Scholar]
- 36.Hu R-G, Wang H, Xia Z, Varshavsky A. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci USA. 2008;105:76–81. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graciet E, Hu RG, Piatkov K, Rhee JH, Schwarz EM, Varshavsky A. Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen. Proc Natl Acad Sci USA. 2006;103:3078–3083. doi: 10.1073/pnas.0511224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z, Webster A, Du F, Piatkov K, Ghislain M, Varshavsky A. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J Biol Chem. 2008;283:24011–24028. doi: 10.1074/jbc.M802583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang C-S, Varshavsky A. Regulation of peptide import through phosphorylation of Ubr1, the ubiquitin ligase of the N-end rule pathway. Proc Natl Acad Sci USA. 2008;105:19188–19193. doi: 10.1073/pnas.0808891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang C-S, Shemorry A, Varshavsky A. Two proteolytic pathways regulate DNA repair by co-targeting the Mgt1 alkyguanine transferase. Proc Natl Acad Sci USA. 2009;106:2142–2147. doi: 10.1073/pnas.0812316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brower CS, Varshavsky A. Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS ONE. 2009;4:e7757. doi: 10.1371/journal.pone.0007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Piatkov KI, Brower CS, Varshavsky A. Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol Cell. 2009;34:686–695. doi: 10.1016/j.molcel.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tasaki T, Zakrzewska A, Dudgeon D, Jiang Y, Lazo JS, Kwon YT. The substrate recognition domains of the N-end rule pathway. J Biol Chem. 2009;284:1884–1895. doi: 10.1074/jbc.M803641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang C-S, Shemorry A, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol. 2010;12:1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Y. Structure, assembly and homeostatic regulation of the 26S proteasome. J Mol Cell Biol. 2011;2:308–317. doi: 10.1093/jmcb/mjq030. [DOI] [PubMed] [Google Scholar]
- 46.Förster F, Lasker K, Nickell S, Sali A, Baumeister W. Toward an integrated structural model of the 26S proteasome. Mol Cell Proteomics. 2010;9:1666–1677. doi: 10.1074/mcp.R000002-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–693. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 49.Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Liu F, Walters KJ. Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci. 2010;35:352–360. doi: 10.1016/j.tibs.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 52.Stolz A, Wolf DH. Endoplasmic reticulum-associated protein degradation: a chaperone-assisted journey to hell. Biochim Biophys Acta. 2010;1803:694–705. doi: 10.1016/j.bbamcr.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev. 2009;109:1561–1574. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isaacson MK, Ploegh HL. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5:559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 57.Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 58.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–958. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 61.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daulni A, Tansey WP. Damage control: DNA repair, transcription, and the ubiquitin-proteasome system. DNA Repair. 2009;8:444–448. doi: 10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 63.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 64.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 65.Wolf DH, Hilt W. The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochim Biophys Acta. 2004;1695:19–31. doi: 10.1016/j.bbamcr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 66.Darwin KH. Prokaryotic ubiquitin-like protein, proteasomes, and pathogenesis. Nat Rev Microbiol. 2009;7:485–491. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burns KE, Liu W-T, Boshoff HIM, Dorrestein PC, Barry CE., III Proteasomal protein degradation in mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cerda-Maira FA, Pearce MJ, Fuortes M, Bishai WR, Hubbard SR, Darwin KH. Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis. Mol Microbiol. 2010;77:1123–1135. doi: 10.1111/j.1365-2958.2010.07276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sutter M, Damberger FF, Imkamp F, Allain FHT, Weber-Ban E. Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J Am Chem Soc. 2010;132:5610–5612. doi: 10.1021/ja910546x. [DOI] [PubMed] [Google Scholar]
- 70.Humbard MA, Miranda HV, Lim J-M, Krause DJ, Pritz JR, Zhou G, Chen SH, Wells L, Maupin-Furlow JA. Ubiquitin-like small archaeal modifier proteins (SAMPS) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shrader TE, Tobias JW, Varshavsky A. The N-end rule in Escherichia coli: cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J Bact. 1993;175:4364–4374. doi: 10.1128/jb.175.14.4364-4374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 73.Wang KH, Roman-Hernandez G, Grant RA, Sauer TT, Baker TA. The molecular basis of N-end rule recognition. Mol Cell. 2008;32:406–414. doi: 10.1016/j.molcel.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schuenemann VJ, Kralik SM, Albrecht R, Spall SK, Truscott KN, Dougan DA, Zeth K. Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS. EMBO Rep. 2009;10:508–514. doi: 10.1038/embor.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Román-Hernández G, Grant RA, Sauer RT, Baker TA. Molecular basis of substrate selection by the N-end rule adaptor protein ClpS. Proc Natl Acad Sci USA. 2009;106:8888–8893. doi: 10.1073/pnas.0903614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmidt R, Zahn R, Bukau B, Mogk A. ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol Microbiol. 2009;72:506–517. doi: 10.1111/j.1365-2958.2009.06666.x. [DOI] [PubMed] [Google Scholar]
- 77.Ninnis RL, Spall SK, Talbo GH, Truscott KN, Dougan DA. Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. EMBO J. 2009;28:1732–1744. doi: 10.1038/emboj.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Donatis GM, Singh SK, Viswanathan S, Maurizi MR. A single ClpS monomer is sufficient to direct the activity of the ClpA hexamer. J Biol Chem. 2010;285:8771–8781. doi: 10.1074/jbc.M109.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hou JY, Sauer RT, Baker TA. Distinct structural elements of the adaptor ClpS are required for regulating degradation by ClpAP. Nat Struct Mol Biol. 2008;15:288–294. doi: 10.1038/nsmb.1392. [DOI] [PubMed] [Google Scholar]
- 80.Varshavsky A. The N-end rule at atomic resolution. Nat Struct Mol Biol. 2008;15:1238–1240. doi: 10.1038/nsmb1208-1238. [DOI] [PubMed] [Google Scholar]
- 81.Dougan DA, Truscott KN, Zeth K. The bacterial N-end rule pathway: expect the unexpected. Mol Microbiol. 2010;76:545–558. doi: 10.1111/j.1365-2958.2010.07120.x. [DOI] [PubMed] [Google Scholar]
- 82.Román-Hernández G, Hou JY, Grant RA, Sauer RT, Baker TA. The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.06.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi WS, Jeong B-C, Joo YJ, Lee M-R, Kim J, Eck MJ, Song HK. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat Struct Mol Biol. 2010;17:1175–1181. doi: 10.1038/nsmb.1907. [DOI] [PubMed] [Google Scholar]
- 84.Matta-Camacho E, Kozlov G, Li FF, Gehring K. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat Struct Mol Biol. 2010;17:1182–1188. doi: 10.1038/nsmb.1894. [DOI] [PubMed] [Google Scholar]
- 85.Sriram SM, Kwon YT. The structural basis of N-end rule recognition. Nat Struct Mol Biol. 2010;17:1164–1165. doi: 10.1038/nsmb1010-1164. [DOI] [PubMed] [Google Scholar]
- 86.Jörnvall H. Acetylation of protein N-terminal amino groups: structural observations on alpha-amino acetylated proteins. J Theor Biol. 1975;55:1–12. doi: 10.1016/s0022-5193(75)80105-6. [DOI] [PubMed] [Google Scholar]
- 87.Mullen JR, Kayne PS, Moerschell RP, Tsunasawa S, Gribskov M, Colavito-Shepanski M, Grunstein M, Sherman F, Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park EC, Szostak JW. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 1992;11:2087–2093. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gautschi M, Just S, Mun A, Ross S, Rücknagel P, Dubaquié Y, Ehrenhofer-Murray A, Rospert S. The yeast N-alpha-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol Cell Biol. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song O-K, Wang X, Waterborg JH, Sternglanz R. An N-alpha-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. J Biol Chem. 2003;278:38109–38112. doi: 10.1074/jbc.C300355200. [DOI] [PubMed] [Google Scholar]
- 91.Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, Gevaert K. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast to humans. Proc Natl Acad Sci USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 93.Goetze S, Qeli E, Mosimann C, Staes A, Gerrits B, Roschitzki B, Mohanty S, Niederer EM, Laczko E, Timmerman E, Lange V, Hafen E, Aebersold R, Vandekerckhove J, Basler K, Ahrens CH, Gevaert K, Brunner E. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 2009;7:e1000236. doi: 10.1371/journal.pbio.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Starheim KK, Gromyko D, Evjens E, Ryningen A, Varhaug JE, Lillehaug JR, Arnesen T. Knockdown of human N-alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol Cell Biol. 2009;29:3569–3581. doi: 10.1128/MCB.01909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Polevoda B, Brown S, Cardillo TS, Rigby S, Sherman F. Yeast N(alpha)-terminal acetyltransferases are associated with ribosomes. J Cell Biochem. 2008;103:492–508. doi: 10.1002/jcb.21418. [DOI] [PubMed] [Google Scholar]
- 96.Addlagatta A, Hu X, Liu JO, Matthews BW. Structural basis for the functional differences between Type I and Type II human methionine aminopeptidases. Biochemistry. 2005;44:14741–14749. doi: 10.1021/bi051691k. [DOI] [PubMed] [Google Scholar]
- 97.Lowther WT, Matthews BW. Metalloaminopeptidases: common functional themes in disparate structural surroundings. Chem Rev. 2002;102:4581–4608. doi: 10.1021/cr0101757. [DOI] [PubMed] [Google Scholar]
- 98.Moerschell RP, Hosokawa Y, Tsunasawa S, Sherman F. The specificities of yeast methionine aminopeptidase and acetylation of amino-terminal methionine in vivo. Processing of altered iso-1-cytochromes created by oligonucleotide transformation. J Biol Chem. 1990;265:19638–19643. [PubMed] [Google Scholar]
- 99.Li X, Chang Y-H. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc Natl Acad Sci USA. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bradshaw RA, Brickey WW, Walker KW. N-terminal processing: the methionine aminopeptidase and N-alpha-acetyl transferase families. Trends Biochem Sci. 1998;23:263–267. doi: 10.1016/s0968-0004(98)01227-4. [DOI] [PubMed] [Google Scholar]
- 101.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Helbig AO, Gauci S, Raijmakers R, van Breukelen B, Slijper M, Mohammed S, Heck AJR. Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol Cell Proteom. 2010;9:928–939. doi: 10.1074/mcp.M900463-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- 104.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 105.Behnia R, Panic B, Whyte JRC, Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- 106.Setty SRG, Strochlic TI, Tong AHY, Boone C, Burd CG. Golgi targeting of ARF-like GTPase Arl3p requires its N-alpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- 107.Jackson CL. N-terminal acetylation targets GTPases to membranes. Nat Cell Biol. 2004;6:379–380. doi: 10.1038/ncb0504-379. [DOI] [PubMed] [Google Scholar]
- 108.Hofmann I, Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and effects their motility. J Cell Sci. 2006;119:1494–1503. doi: 10.1242/jcs.02958. [DOI] [PubMed] [Google Scholar]
- 109.Graham TR. Membrane targeting: getting Arl to the Golgi. Curr Biol. 2004;14:R483–R485. doi: 10.1016/j.cub.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 110.Coulton AT, East DA, Galinska-Rakoczy A, Lehman W, Mulvihill DP. The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. J Cell Sci. 2010;123:3235–3243. doi: 10.1242/jcs.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Balzi E, Choder M, Chen W, Varshavsky A, Goffeau A. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem. 1990;265:7464–7471. [PubMed] [Google Scholar]
- 112.Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Graciet E, Walter E, Maoiléidigh DÓ, Pollmann S, Meyerowitz EM, Varshavsky A, Wellmer F. The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc Natl Acad Sci USA. 2009;106:13618–13623. doi: 10.1073/pnas.0906404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang J, Han X, Saha S, Xu T, Rai R, Zhang F, Wolf YI, Wolfson A, Yates JR, III, Kashina A. Arginyltransferase is an ATP-independent self-regulating enzyme that forms distinct functional complexes in vivo. Chem Biol. 2011;18:121–130. doi: 10.1016/j.chembiol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stewart AE, Arfin SM, Bradshaw RA. The sequence of porcine protein N-terminal asparagine amidohydrolase. A new component of the N-end rule pathway. J Biol Chem. 1995;270:25–28. doi: 10.1074/jbc.270.1.25. [DOI] [PubMed] [Google Scholar]
- 116.Grigoryev S, Stewart AE, Kwon YT, Arfin SM, Bradshaw RA, Jenkins NA, Copeland NG, Varshavsky A. A mouse amidase specific for N-terminal asparagine. The gene, the enzyme, and their function in the N-end rule pathway. J Biol Chem. 1996;271:28521–28532. doi: 10.1074/jbc.271.45.28521. [DOI] [PubMed] [Google Scholar]
- 117.Cantor JR, Stone EM, Georgiou G. Expression and biochemical characterization of the human enzyme N-terminal asparagine amidohydrolase. Biochemistry. 2011;50:3025–3033. doi: 10.1021/bi101832w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Davydov IV, Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J Biol Chem. 2000;275:22931–22941. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- 119.Eisele F, Wolf DH. Degradation of misfolded proteins in the cytoplasm by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 120.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prasad R, Kawaguchi S, Ng DTW. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, Wu K, Johnson J, Cyr DM, Caplan AJ. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell. 2010;21:2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ouyang Y, Kwon YT, An JY, Eller D, Tsai S-C, Diaz-Perez S, Trke J, Teitell MA, Marahrens Y. Loss of Ubr2, an E3 ubiquitin ligase, leads to chromosome fragility and impaired homologous recombinational repair. Mut Res. 2006;596:64–75. doi: 10.1016/j.mrfmmm.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 124.Schnupf P, Zhou J, Varshavsky A, Portnoy DA. Listeriolysin O secreted by Listeria monocytogenes into the host cell cytosol is degraded by the N-end rule pathway. Inflammation Immunity. 2007;75:5135–5147. doi: 10.1128/IAI.00164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yoshida S, Ito M, Gallis J, Nishida I, Watanabe A. A delayed leaf senescence mutant is defective in arginyl-tRNA-protein arginyl-transferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 2002;32:129–137. doi: 10.1046/j.1365-313x.2002.01407.x. [DOI] [PubMed] [Google Scholar]
- 126.Holman TJ, Jones PD, Russell L, Medhurst A, Ubeda TS, Talloji P, Marquez J, Schmuths H, Tung SA, Taylor I, Footitt S, Bachmair A, Theodoulou FL, Holdsworth MJ. The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:4549–4554. doi: 10.1073/pnas.0810280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rai R, Wong CC, Xu T, Leu NA, Dong DW, Guo C, McLaughlin KJ, Yates JR, III, Kashina A. Arginyltransferase regulates alpha cardiac actin function, myofibril formation and contractility during heart development. Development. 2008;135:3881–3889. doi: 10.1242/dev.022723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Saha S, Mundia MM, Zhang F, Demers RW, Korobova F, Svitkina T, Perieteanu AA, Dawson JF, Kashina A. Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol Biol Cell. 2010;21:1350–1361. doi: 10.1091/mbc.E09-09-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kurosaka S, Leu NA, Zhang F, Bunte R, Saha S, Wang J, Guo C, He W, Kashina A. Arginylation-dependent neural crest cell migration is essential for mouse development. PLoS Genet. 2010;6:e1000878. doi: 10.1371/journal.pgen.1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang F, Saha S, Shabalina SA, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329:1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.de Groot RJ, Rümenapf T, Kuhn RJ, Strauss JH. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc Natl Acad Sci USA. 1991;88:8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mulder LCF, Muesing MA. Degradation of HIV-1 integrase by the N-end rule pathway. J Biol Chem. 2000;275:29749–29753. doi: 10.1074/jbc.M004670200. [DOI] [PubMed] [Google Scholar]
- 133.Lloyd AG, Ng YS, Muesing MA, Simon V, Mulder LC. Characterization of HIV-1 integrase N-terminal mutant viruses. Virology. 2007;360:129–135. doi: 10.1016/j.virol.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lecker SH, Solomon V, Price SR, Kwon YT, Mitch WE, Goldberg AL. Ubiquitin conjugation by the N-end rule pathway and mRNAs for its components increase in muscles of diabetic rats. J Clin Invest. 1999;104:1411–1420. doi: 10.1172/JCI7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR, III, Mogilner A, Zebroski H, Kashina A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- 136.Caprio MA, Sambrooks CL, Durand ES, Hallak M. The arginylation-dependent association of calreticulin with stress granules is regulated by calcium. Biochem J. 2010;429:63–72. doi: 10.1042/BJ20091953. [DOI] [PubMed] [Google Scholar]
- 137.Yang F, Cheng Y, An JY, Kwon YT, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. The ubiquitin ligase Ubr2, a recognition E3 component of the N-end rule pathway, stabilizes Tex.19.1 during spermatogenesis. PLoS One. 2010;5:e14017. doi: 10.1371/journal.pone.0014017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xia Z, Turner GC, Hwang C-S, Byrd C, Varshavsky A. Amino acids induce peptide uptake via accelerated degradation of CUP9, the transcriptional repressor of the PTR2 peptide transporter. J Biol Chem. 2008;283:28958–28968. doi: 10.1074/jbc.M803980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Turner GC, Varshavsky A. Detecting and measuring cotranslational protein degradation in vivo. Science. 2000;289:2117–2120. doi: 10.1126/science.289.5487.2117. [DOI] [PubMed] [Google Scholar]
- 140.Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, Meier P. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 141.Varshavsky A. The N-end rule and regulation of apoptosis. Nat Cell Biol. 2003;5:373–376. doi: 10.1038/ncb0503-373. [DOI] [PubMed] [Google Scholar]
- 142.O'Neill GM, Golemis EA. Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics. Mol Cell Biol. 2001;21:5094–5108. doi: 10.1128/MCB.21.15.5094-5108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Law SF, O'Neill GM, Fashena SJ, Einarson MB, Golemis EA. The docking protein HEF1 is an apoptotic mediator at focal adhesion sites. Mol Cell Biol. 2000;20:5194–5195. doi: 10.1128/mcb.20.14.5184-5195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Foveau B, Leroy C, Ancot F, Deheuninck J, Li Z, Fafeur V, Tulasne D. Amplification of apoptosis through sequential caspase cleavage the Met tyrosine kinase receptor. Cell Death Diff. 2007;14:752–764. doi: 10.1038/sj.cdd.4402080. [DOI] [PubMed] [Google Scholar]
- 145.Tulasne D, Deheuninck J, Lourenco FC, Lamballe F, Ji Z, Leroy C, Puchois E, Moumen A, Maina F, Mehlen P, Fafeur V. Proapoptotic function of the MET tyrosine kinase receptor through caspase cleavage. Mol Cell Biol. 2004;24:10328–10339. doi: 10.1128/MCB.24.23.10328-10339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goldschneider D, Mehlen P. Dependence receptors: a new paradigm in cell signaling and cancer. Oncogene. 2010;29:1865–1882. doi: 10.1038/onc.2010.13. [DOI] [PubMed] [Google Scholar]
- 147.Ancot F, Foveau B, Lefebvre J, Leroy C, Tulasne D. Proteolytic cleavages give receptor tyrosine kinases the gift of ubiquity. Oncogene. 2009;28:2185–2195. doi: 10.1038/onc.2009.88. [DOI] [PubMed] [Google Scholar]
- 148.Conacci-Sorrell M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–493. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu YM, Huang CL, Kung HJ, Huang CYF. Proteolytic activation of Etk/Bmx tyrosine kinase by caspases. J Biol Chem. 2001;276:17672–17678. doi: 10.1074/jbc.M010964200. [DOI] [PubMed] [Google Scholar]
- 150.Furne C, Ricard J, Cabrera JR, Pays L, Bethea JR, Mehlen P, Liebl DJ. EphrinB3 is anti-apoptotic ligand that inhibits the dependence receptor functions of EphA4 receptors during adult neurogenesis. Biochim Biohys Acta. 2009;1793:231–238. doi: 10.1016/j.bbamcr.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tu S, Cerione RA. Cdc42 is a substrate for caspase and influences Fas-induced apoptosis. J Biol Chem. 2001;276:19656–19663. doi: 10.1074/jbc.M009838200. [DOI] [PubMed] [Google Scholar]
- 152.Chen L, Marechal V, Moreau J, Levine AJ, Chen J. Proteolytic cleavage of the mdm2 oncoprotein during apoptosis. J Biol Chem. 1997;272:22966–22973. doi: 10.1074/jbc.272.36.22966. [DOI] [PubMed] [Google Scholar]
- 153.Leverrier S, Vallentin A, Joubert D. Positive feedback of protein kinase C proteolytic activation during apoptosis. Biochem J. 2002;368:905–913. doi: 10.1042/BJ20021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Emoto Y, Kisaki H, Manome Y, Kharbanda S, Kufe D. Activation of protein kinase C-delta in human myeloid leukemia cells treated with 1-beta-d-arbabinofuranosylcytosine. Blood. 1996;87:1990–1996. [PubMed] [Google Scholar]
- 155.Datta R, Kojima H, Yoshida K, Kufe D. Caspase-3-mediated cleavage of protein kinase C-theta in induction of apoptosis. J Biol Chem. 1997;272:20317–20320. doi: 10.1074/jbc.272.33.20317. [DOI] [PubMed] [Google Scholar]
- 156.Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Diff. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 157.Fischer U, Jänicke RU, Schultze-Osthoff K. Many cuts to rua comprehensive update of caspase substrates. Cell Death Diff. 2003;10:786–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Agard NJ, Maltby D, Wells JA. Inflammatory stimuli regulate caspase substrate profiles. Mol Cell Proteom. 2010;9.5:880–893. doi: 10.1074/mcp.M900528-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cathelin S, Rébé C, Haddaoui L, Simioni N, Verdier F, Fontenay M, Launay S, Mayeux P, Solary E. Identification of proteins cleaved downstream of caspase activation in monocytes undergoing macrophage differentiation. J Biol Chem. 2006;281:17779–17788. doi: 10.1074/jbc.M600537200. [DOI] [PubMed] [Google Scholar]
- 160.Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J Biol Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nat Rev Cancer. 2011;11:364–374. doi: 10.1038/nrc3050. [DOI] [PubMed] [Google Scholar]
- 162.Uhlmann F. The mechanism of sister chromatid cohesion. Exp Cell Res. 2004;296:80–85. doi: 10.1016/j.yexcr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 163.Hsieh J-D, Cheng EH-Y, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 164.Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas E-MM, Dierlamm J, Chan WC, Staudt LM, Thome M. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, Baki L, Wen P, Efthimiopoulos S, Shao Z, Wisniewski T, Robakis NK. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;21:1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Witko-Sarsat V, Canteloup S, Durant S, Desdouets C, Chabernaud R, Lemarchand P, Descamps-Latscha B. Cleavage of p21waf1 by proteinase-3, a myeloid-specific serine protease, potentiates cell proliferation. J Biol Chem. 2002;277:47338–47347. doi: 10.1074/jbc.M202789200. [DOI] [PubMed] [Google Scholar]
- 167.Hamilton MH, Cook LA, McRackan TR, Schey KL, Hildebrandt JD. Gamma-2 subunit of G protein heterotrimer is an N-end rule ubiquitylation substrate. Proc Natl Acad Sci USA. 2003;100:5081–5086. doi: 10.1073/pnas.0831228100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Prakash S, Inobe T, Hatch AJ, Matouschek A. Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol. 2009;5:29–36. doi: 10.1038/nchembio.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun. 2011;2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Verhoef LGGC, Heinen C, Selivanova A, Halff EF, Salomons FA, Dantuma NP. Minimal length requirement for proteasomal degradation of ubiquitin-dependent substrates. FASEB J. 2009;23:123–133. doi: 10.1096/fj.08-115055. [DOI] [PubMed] [Google Scholar]
- 172.Heinen C, Ács K, Hoogstraten D, Dantuma NP. C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat Commun. 2011;2:191. doi: 10.1038/ncomms1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 174.Petroski MD, Deshaies RJ. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol Cell. 2003;11:1435–1444. doi: 10.1016/s1097-2765(03)00221-1. [DOI] [PubMed] [Google Scholar]
- 175.Piwko W, Jentsch S. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat Struct Mol Biol. 2006;13:691–697. doi: 10.1038/nsmb1122. [DOI] [PubMed] [Google Scholar]
- 176.Takeuchi J, Chen H, Coffino P. Proteasome substrate degradation requires association plus extended peptide. EMBO J. 2007;26:123–131. doi: 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 178.Jentsch S, Rumpf S. Cdc48 (p97): a "molecular gearbox" in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 179.Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xie Y, Varshavsky A. Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci USA. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xie Y, Varshavsky A. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat Cell Biol. 2002;4:1003–1007. doi: 10.1038/ncb889. [DOI] [PubMed] [Google Scholar]
- 182.Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2010;2:a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 184.Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yewdell JW, Lascina JR, Rechsteiner MC, Nicchitta CV. Out with the old, in with the new? Comparing methods for measuring protein degradation. Cell Biol Int. 2011;35:457–462. doi: 10.1042/CBI20110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Guo F, Esser L, Singh SK, Maurizi MR, Xia D. Crystal structure of the heterodimeric complex of the adaptor, ClpS, with the N-domain of the AAA+ chaperone, ClpA. J Biol Chem. 2002;22:46753–46762. doi: 10.1074/jbc.M208104200. [DOI] [PubMed] [Google Scholar]
- 187.Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- 188.Zeth K, Ravelli RB, Paal K, Cusack S, Bukau B, Dougan DA. Structural analysis of the adaptor protein ClpS with the N-terminal domain of ClpA. Nat Struct Biol. 2002;9:906–911. doi: 10.1038/nsb869. [DOI] [PubMed] [Google Scholar]
- 189.Wang KH, Oakes ESC, Sauer RT, Baker TA. Tuning the strength of a bacterial N-end rule signal. J Biol Chem. 2008;283:24600–24607. doi: 10.1074/jbc.M802213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- 191.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein folding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 192.Xia D, Esser L, Singh SK, Guo F, Maurizi MR. Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone. J Struct Biol. 2004;146:166–179. doi: 10.1016/j.jsb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 193.Aubin-Tam M-E, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Maillard RA, Gheorghe C, Sen M, Righini M, Tan J, Kaiser CM, Hodges C, Martin A, Bustamante C. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang KH, Sauer RT, Baker TA. ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev. 2007;15:403–408. doi: 10.1101/gad.1511907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SsB adaptor to the AAA+ ClpXP protease. Mol Cell. 2004;16:343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 198.Hwang W, Lang MJ. Mechanical design of of translocating motor proteins. Cell Biochem Biophys. 2009;54:11–22. doi: 10.1007/s12013-009-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Striebel LC, Kress W, Weber-Ban E. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr Opt Struct Biol. 2009;19:209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 200.Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 202.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis. PLoS Biol. 2010;5:e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45:11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Miranda HV, Nembharda N, Sub D, Hepowita N, Krausea DJ, Pritza JR, Phillipsa C, Söll D, Maupin-Furlowa JA. E1- and ubiquitin-like proteins provide a direct linkb between protein conjugation and sulfur transfer in archaea. Proc Natl Acad Sci USA. 2011;108:4417–4422. doi: 10.1073/pnas.1018151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, Takai K, Takami H. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast MATalpha2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 209.Xie W, Ngo DT. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol. 2010;21:533–539. doi: 10.1016/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 210.Lynch M. The Origins of Genome Architecture. 2007. Sinauer Associates, Inc: Sinauer Associates, Inc.
- 211.Baltimore D. Discovering Nf-kappaB. Cold Spring Harb Perspect Biol. 2009;1:a000026. doi: 10.1101/cshperspect.a000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–1079. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- 213.Varshavsky A. Ubiquitin fusion technique and related methods. Meth Enzymol. 2005;399:777–799. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 214.Ecker DJ, Stadel JM, Butt TR, Marsh JA, Monia BP, Powers DA, Gorman JA, Clark PE, Warren F, Shatzman A. Increasing gene expression in yeast by fusion to ubiquitin. J Biol Chem. 1989;264:7715–7719. [PubMed] [Google Scholar]
- 215.Park EC, Finley D, Szostak JW. A strategy for the generation of conditional mutations by protein destabilization. Proc Natl Acad Sci USA. 1992;89:1249–1252. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dohmen RJ, Varshavsky A. Heat-inducible degron and the making of conditional mutants. Meth Enzymol. 2005;399:799–822. doi: 10.1016/S0076-6879(05)99052-6. [DOI] [PubMed] [Google Scholar]
- 217.Johnsson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across cell membranes. EMBO J. 1994;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lévy F, Johnsson N, Rümenapf T, Varshavsky A. Using ubiquitin to follow the metabolic fate of a protein. Proc Natl Acad Sci USA. 1996;93:4907–4912. doi: 10.1073/pnas.93.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 221.Müller J, Johnsson N. Split-ubiquitin and the split-protein sensors: chessman for the endgame. Chembiochem. 2008;9:2029–2038. doi: 10.1002/cbic.200800190. [DOI] [PubMed] [Google Scholar]
- 222.Tarassov K, Messier V, Landry CR, Radinovic S, Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 223.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 224.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 226.Varshavsky A. The N-end rule. Cold Spring Harb Symp Quant Biol. 1996;60:461–478. doi: 10.1101/sqb.1995.060.01.051. [DOI] [PubMed] [Google Scholar]
- 227.Falnes PO, Olsnes S. Modulation of the intracellular stability and toxicity of diphtheria toxin through degradation by the N-end rule pathway. EMBO J. 1998;17:615–625. doi: 10.1093/emboj/17.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Falnes PO, Welker R, Krausslich HG, Olsnes S. Toxins are activated by HIV-type-1 protease through removal of signal for degradation by the N-end rule pathway. Biochemical J. 1999;343:199–207. [PMC free article] [PubMed] [Google Scholar]
- 229.Shapira A, Gal-Tanamy M, Nahary L, Litvak-Greenfeld D, Zemel R, Tur-Kaspa R, Benhar I. Engineered toxins "zymoxins" are activated by the HCV NS3 protease by removal of an inhibitory protein domain. PLoS One. 2011;14:e15916. doi: 10.1371/journal.pone.0015916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jucovic M, Walters FS, Warren GW, Palekar NV, Chen JS. From enzyme to zymogen: engineering Vip2, an ADP-ribosyltransferase from Bacillus cereus, for conditional toxicity. Protein Eng Des. 2008;21:631–638. doi: 10.1093/protein/gzn038. [DOI] [PubMed] [Google Scholar]
- 231.Law SK, Wang RR, Mak AN, Wong KB, Zheng YT, Shaw PC. A switch-on mechanism to activate maize ribosome-inactivating protein for targeting HIV-infected cells. Nucleic Acids Res. 2010;38:6803–6812. doi: 10.1093/nar/gkq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Johnson RJ, Lin SR, Raines RT. A ribonuclease zymogen activated by the NS3 protease of the hepatitis C virus. FEBS J. 2006;273:5457–5465. doi: 10.1111/j.1742-4658.2006.05536.x. [DOI] [PubMed] [Google Scholar]
- 233.Turcotte RF, Raines RT. Design and characterization of an HIV-specific ribonuclease zymogen. AIDS Res Hum Retroviruses. 2008;24:1357–1363. doi: 10.1089/aid.2008.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pratt MR, Schwartz EC, Muir TW. Small-molecule-mediated rescue of protein function by an inducible proteolytic shunt. Proc Natl Acad Sci USA. 2007;104:11209–11214. doi: 10.1073/pnas.0700816104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Johnston JA, Johnson ES, Waller PR, Varshavsky A. Methotrexate inhibits proteolysis of dihydrofolate reductase by the N-end rule pathway. J Biol Chem. 1995;270:8172–8178. doi: 10.1074/jbc.270.14.8172. [DOI] [PubMed] [Google Scholar]
- 236.Lévy F, Johnston JA, Varshavsky A. Analysis of a conditional degradation signal in yeast and mammalian cells. Eur J Biochem. 1999;259:244–252. doi: 10.1046/j.1432-1327.1999.00024.x. [DOI] [PubMed] [Google Scholar]
- 237.Iwamoto M, Bjöklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Taxis C, Stier G, Spadaccini R, Knop M. Efficient protein depletion by genetically controlled deprotection of a dormant N-degron. Mol Systems Biol. 2009;5:267. doi: 10.1038/msb.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and MATalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- 241.Kreft SG, Hochstrasser M. An unusual transmembrane helix in the Doa10 ERAD ubiquitin ligase modulates degradation of its cognate E2. J Biol Chem. 2011;286:20163–20174. doi: 10.1074/jbc.M110.196360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Mogk A, Bukau B. When the beginning marks the end. Science. 2010;327:966–967. doi: 10.1126/science.1187274. [DOI] [PubMed] [Google Scholar]
- 243.Hassink G, Kikkert M, van Voorden S, Lee S-J, Spaapen R, van Laar T, Cloleman CS, Bartee E, Früh K, Chau V, Wiertz E. TEB4 is a C4HC3 RING-finger containing ubiquitin ligase of the endoplasmic reticulum. Biochem J. 2005;388:647–655. doi: 10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Singh RK, Kabbaj M-HM, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homodimer interface. EMBO J. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou M-M. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Dohmen RJ, Madura K, Bartel B, Varshavsky A. The N-end rule is mediated by the UBC2 (RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci USA. 1991;88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 251.Ju D, Wang X, Xu H, Xie Y. The armadillo repeats of the Ufd4 ubiquitin ligase recognize ubiquitin-fusion proteins. FEBS Lett. 2007;581:265–270. doi: 10.1016/j.febslet.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 252.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 253.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 254.Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun. 2007;354:329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Scheffner M, Staub O. HECT E3s and human disease. BMC Biochem. 2007;8(Suppl. I):S6. doi: 10.1186/1471-2091-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Johnson ES, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 258.Park Y, Yoon SK, Yoon JB. The HECT domain of TRIP12 ubiquitinates substrates of the ubiquitin fusion degradation pathway. J Biol Chem. 2009;284:1540–1549. doi: 10.1074/jbc.M807554200. [DOI] [PubMed] [Google Scholar]
- 259.Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn't fit all. Trends Biochem Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 260.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 261.Scott DC, Monda JK, Grace CRR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. A dual mechanism for Rub1 ligation to Cdc53. Mol Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kwon YT, Levy F, Varshavsky A. Bivalent inhibitor of the N-end rule pathway. J Biol Chem. 1999;274:18135–18139. doi: 10.1074/jbc.274.25.18135. [DOI] [PubMed] [Google Scholar]
- 263.Jacobson RH, Zhang XJ, DuBose RF, Matthews BW. Three-dimensional structure of beta-galactosidase from E. coli. Nature. 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 264.Lee MJ, Pal K, Tasaki T, Roy S, Jiang S, An JY, Banerjee R, Kwon YT. Synthetic heterovalent inhibitors targeting recognition E3 components of the N-end rule pathway. Proc Natl Acad Sci USA. 2008;105:100–105. doi: 10.1073/pnas.0708465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Shim SY, Wang J, Asada N, Neumayer G, Tran HC, Ishiguro K, Sanada K, Nakatani Y, Nguyen MD. Protein 600 is a microtubule/endoplasmic reticulum-associated protein in CNS neurons. J Neurosci. 2008;28:3604–3614. doi: 10.1523/JNEUROSCI.5278-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Nakatani Y, Konishi H, Vassilev A, Kurooka H, Ishiguro K, Sawada J, Ikura T, Korsmeyer SJ, Qin J, Herlitz AM. p600, a unique protein required for membrane morphogenesis and cell survival. Proc Natl Acad Sci USA. 2005;102:15093–15098. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Cojocaru M, Bouchard A, Cloutier P, Cooper JJ, Varzavand K, Price DH, Coulombe B. Transcription factor IIS cooperates with the E3 ligase UBR5 to ubiquitinate the CDK9 subunit of the positive transcription elongation factor B. J Biol Chem. 2011;286:5012–5022. doi: 10.1074/jbc.M110.176628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hu G, Wang X, Saunders DN, Henderson M, Russell AJ, Herring BPZ, Zhou J. Modulation of myocardin function by the ubiquitin E3 ligase UBR5. J Biol Chem. 2010;285:11800–11809. doi: 10.1074/jbc.M109.079384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Tomaić V, Pim D, Thomas M, Massimi P, Myers MP, Banks L. Regulation of the human papillomavirus type 18 E6/E6AP ubiquitin ligase complex by the HECT domain-containing protein EDD. J Virol. 2011;85:3120–3127. doi: 10.1128/JVI.02004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.An JY, Kim E-A, Jiang Y, Zakrzewska A, Kim DE, Lee MJ, Mook-Jung I, Zhang Y, Kwon YT. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci USA. 2010;107:1912–1917. doi: 10.1073/pnas.0910267107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.An JY, Seo JW, Tasaki T, Lee MJ, Varshavsky A, Kwon YT. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci USA. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Stary S, Yin X-J, Potuschak T, Schlögelhofer P, Potuschak T, Nizhynska V, Bachmair A. PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N-end rule pathway with specificity for aromatic amino-terminal residues. Plant Physiol. 2003;133:1360–1366. doi: 10.1104/pp.103.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Garzón M, Eifler K, Faust A, Scheel H, Hofmann K, Koncz C, Yephremov A, Bachmair A. PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 2007;581:3189–3196. doi: 10.1016/j.febslet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 274.Tasaki T, Sohr R, Xia Z, Hellweg R, Hörtnagl H, Varshavsky A, Kwon YT. Biochemical and genetic studies of UBR3, a ubiquitin ligase with a function in olfactory and other sensory systems. J Biol Chem. 2007;282:18510–18520. doi: 10.1074/jbc.M701894200. [DOI] [PubMed] [Google Scholar]
- 275.Wang L, Mao X, Ju D, Xie Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J Biol Chem. 2004;279:55218–55223. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- 276.Lee P, Sowa ME, Gygi SP, Harper JW. Alternative ubiquitin activation/conjugation cascades interact with N-recognin ubiquitin ligases of the N-end rule pathway to control degradation of RGS proteins. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.05.034. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 278.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 279.Huang TT, Nijman SMB, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HP, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 280.Dubnau J, Chiang A-S, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 281.Hirai T, Taniura H, Goto Y, Ogura M, Sng JCG, Yoneda Y. Stimulation of the ubiquitin-proteasome pathway through expression of amidohydrolase for N-terminal asparagine (Ntan1) in cultured rat hippocampal neurons exposed to static magnetism. J Neurochem. 2006;96:1519–1530. doi: 10.1111/j.1471-4159.2006.03655.x. [DOI] [PubMed] [Google Scholar]
- 282.Brower CS, Veiga L, Jones RH, Varshavsky A. Mouse Dfa is a repressor of TATA-Box promoters and interacts with the Abt1 activator of basal transcription. J Biol Chem. 2010;285:17218–17234. doi: 10.1074/jbc.M110.118638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 284.Albig AR, Schiemann WP. Identification and characterization of regulator of G protein signaling 4 (RGS4) as a novel inhibitor of tubulogenesis: RGS4 inhibits mitogen-activated protein kinases and vascular endothelial growth factor signaling. Mol Biol Cell. 2005;16:609–625. doi: 10.1091/mbc.E04-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Berger M, Bergers G, Arnold B, Hämmerling GJ, Ganss R. Regulator of G-protein signaling-5 induction in pericytes coincides with active vessel remodeling during neovascularization. Blood. 2005;105:1094–1101. doi: 10.1182/blood-2004-06-2315. [DOI] [PubMed] [Google Scholar]
- 286.Wong CCL, Xu T, Rai R, Bailey AO, Yates JR, III, Wolf YI, Zebroski H, Kashina A. Global analysis of posttranslational protein arginylation. PLoS Biol. 2007;5:e258. doi: 10.1371/journal.pbio.0050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Eriste E, Norberg Å, Nepomuceno D, Kuei C, Kamme F, Tran D-T, Strupat K, Jörnvall H, Liu C, Lovenberg TW, Sillard R. A novel form of neurotensin post-translationally modified by arginylation. J Biol Chem. 2005;280:35089–35097. doi: 10.1074/jbc.M502567200. [DOI] [PubMed] [Google Scholar]
- 288.Hauf S, Waizenegger IC, Peters J-M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 289.Tirziu D, Chorianopoulos E, Moodie KL, Palac RT, Zhuang ZW, Tjwa M, Roncal C, Eriksson U, Fu Q, Elfenbein A, Hall AE, Carmeliet P, Moons L, Simons M. Myocardial hypertrophy in the absence of external stimuli is induced by angiogenesis in mice. J Clin Invest. 2007;117:3188–3197. doi: 10.1172/JCI32024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Xie Y, Wolff DW, Wei T, Wang B, Deng C, Kirui JK, Jiang H, Qin H, Abel PW, Tu Y. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 2009;69:5743–5751. doi: 10.1158/0008-5472.CAN-08-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Sjögren B, Neubig RR. Thinking outside of the "RGS box": new approaches to therapeutic targeting of regulators of G protein signaling. Mol Pharm. 2010;78:550–557. doi: 10.1124/mol.110.065219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3-alpha) of the N-end rule pathway. Mol Cell Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wang Y, Mijares M, Gall MD, Turan T, Javier A, Bornemann DJ, Manage K, Warrior R. Drosophila variable nurse cells encodes arrest defective 1 (ARD1), the catalytic subunit of the major N-terminal acetyltransferase complex. Dev Dyn. 2010;239:2813–2827. doi: 10.1002/dvdy.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ingram AK, Cross GA, Horn D. Genetic manipulation indicates that ARD1 is an essential N(alpha)-acetyltransferase in Trypanosoma brucei. Mol Biochem Parasitol. 2000;111:309–317. doi: 10.1016/s0166-6851(00)00322-4. [DOI] [PubMed] [Google Scholar]
- 295.Alagramam K, Naider F, Becker JM. A recognition component of the ubiquitin system is required for peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:225–234. doi: 10.1111/j.1365-2958.1995.tb02237.x. [DOI] [PubMed] [Google Scholar]
- 296.Byrd C, Turner GC, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Cai H, Kauffman S, Naider F, Becker JM. Genomewide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics. 2006;172:1459–1476. doi: 10.1534/genetics.105.053041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Cai H, Hauser M, Naider F, Becker JM. Differential regulation and substrate preferences in two peptide transporters of Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:1805–1813. doi: 10.1128/EC.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Wiles AM, Cai H, Naider F, Becker JM. Nutrient regulation of oligopeptide transport in Saccharomyces cerevisiae. Microbiology. 2006;152:3133–3145. doi: 10.1099/mic.0.29055-0. [DOI] [PubMed] [Google Scholar]
- 300.Godard P, Urrestarazu A, Vissers S, Kontos K, Bontempi G, van Helden J, André B. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3065–3086. doi: 10.1128/MCB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Andréasson C, Heessen S, Ljungdahl PO. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 2006;20:1563–1568. doi: 10.1101/gad.374206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wu B, Ottow K, Poulsen P, Gaber RF, Albers E, Kielland-Brandt MC. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J Cell Biol. 2006;173:327–331. doi: 10.1083/jcb.200602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Boles E, André B. Role of transporter-like sensors in glucose and amino acid signalling in yeast. Top Curr Genet. 2004;9:121–153. [Google Scholar]
- 304.Liu Z, Thornton J, Spirek M, Butow RA. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol Cell Biol. 2008;28:551–563. doi: 10.1128/MCB.00929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Hergeta M, Baldaufa C, Schölza C, Parceja D, Wiesmüllerb K-H, Tampéa R, Abelea R, Bordignonc E. Conformation of peptides bound to the transporter associated with antigen processing (TAP) Proc Natl Acad Sci USA. 2011;108:1349–1354. doi: 10.1073/pnas.1012355108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kume K, Iizumi Y, Shimada M, Ito Y, Kishi T, Yamaguchi Y, Handa H. Role of N-end rule ligases UBR1 and UBR2 in regulating the leucine-mTOR signaling pathway. Genes Cells. 2010;15:339–349. doi: 10.1111/j.1365-2443.2010.01385.x. [DOI] [PubMed] [Google Scholar]
- 307.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 311.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 312.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac T, Gottschling DE, Gardner RG. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Hampton RY. San1-mediated quality control: substrate recognition "sans" chaperones. Mol Cell. 2011;41:2–3. doi: 10.1016/j.molcel.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 314.Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 315.Abramochkin G, Shrader TE. Aminoacyl-tRNA recognition by the leucyl/phenylalanyl-tRNA protein transferase. J Biol Chem. 1996;271:22901–22907. doi: 10.1074/jbc.271.37.22901. [DOI] [PubMed] [Google Scholar]
- 316.Suto K, Shimizu Y, Watanabe K, Ueda T, Fukai S, Nureki O, Tomita K. Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J. 2006;25:5942–5950. doi: 10.1038/sj.emboj.7601433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Watanabe K, Toh Y, Suto K, Shimizu Y, Oka N, Wada T, Tomita K. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature. 2007;449:867–871. doi: 10.1038/nature06167. [DOI] [PubMed] [Google Scholar]
- 318.Dong X, Kato-Murayama M, Muramatsu T, Mori H, Shirouzu M, Bessho Y, Yokoyama S. The crystal structure of leucyl/phenylalanyl-tRNA-protein transferase from Escherichia coli. Prot Sci. 2007;16:528–534. doi: 10.1110/ps.062616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Cheek J, Broderick JB. Adenosylmethionine-dependent iron-sulfur enzymes: versatile clsters in a radical new role. J Biol Inorg Chem. 2001;6:209–226. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- 320.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, functon and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 321.Bingel-Erlenmeyer R, Kohler R, Kramer G, Sandikci A, Antolic S, Maier T, Schaffitzel C, Wiedmann B, Bukau B, Ban N. A peptide deformylase–ribosome complex reveals mechanism of nascent chain processing. Nature. 2008;452:108–111. doi: 10.1038/nature06683. [DOI] [PubMed] [Google Scholar]
- 322.Falb M, Aivaliotis M, Garcia-Rizo C, Bisle B, Tebbe A, Klein C, Konstantinidis K, Siedler F, Pfeiffer F, Oesterhelt D. Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: a large-scale proteomics survey. J Mol Biol. 2006;362:915–924. doi: 10.1016/j.jmb.2006.07.086. [DOI] [PubMed] [Google Scholar]
- 323.Humbard MA, Zhou G, Maupin-Furlow JA. The N-terminal penultimate residue of 20S proteasome alpha1 influences its N-alpha acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J Bact. 2009;191:3794–3803. doi: 10.1128/JB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Vögtle F-N, Prinz C, Kellermann J, Lottspeich F, Pfanner N, Meisinger C. Mitochondrial protein turnover: role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-02-0169. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Schrum JP, Zhu TF, Szostak JW. The origins of cellular life. Cold Spring Harb Perspect Biol. 2010;2:a002212. doi: 10.1101/cshperspect.a002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Fox GE. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2010;2:a003483. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Åkerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Ranea JAG, Sillero A, Thornton JM, Orengo CA. Protein superfamily evolution and the Last Universal Common Ancestor (LUCA) J Mol Evol. 2006;63:513–525. doi: 10.1007/s00239-005-0289-7. [DOI] [PubMed] [Google Scholar]
- 329.Koonin EV, Novozhilov AS. Origin and evolution of the genetic code: the universal enigma. IUBMB Life. 2009;61:99–111. doi: 10.1002/iub.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Spirin A. The ribosome as a conveying thermal ratchet machine. J Biol Chem. 2009;284:21103–22119. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Lupas AN, Koretke KK. Bioinformatic analysis of ClpS, a protein module involved in prokaryotic and eukaryotic protein degradation. J Struct Biol. 2003;141:77–83. doi: 10.1016/s1047-8477(02)00582-8. [DOI] [PubMed] [Google Scholar]
- 332.Catic A, Sun ZY, Ratner DM, Misaghi S, Spooner E, Samuelson J, Wagner G, Ploegh HL. Sequence and structure evolved separately in a ribosomal ubiquitin variant. EMBO J. 2007;26:3474–3483. doi: 10.1038/sj.emboj.7601772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKγ to dampen the host NF-κB-mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Munro P, Flatau G, Lemichez E. Bacteria and the ubiquitin pathway. Curr Opt Microbiol. 2007;10:39–46. doi: 10.1016/j.mib.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 335.Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: a re-emerging field. Cell. 2010;143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Hicks SW, Galán JE. Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opt Microbiol. 2010;13:41–46. doi: 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Randow F, Lehner PJ. Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol. 2009;11:527–534. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 338.Bedford MT, Richard S. Arginine methylation: an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 339.Vossenaar ER, Zendman AJW, van Venrooij WJ, Pruijn GJM. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 340.Bitto E, Bingman CA, McCoy JG, Wesenberg GE, Phillips GN. Crystal structure of the uncharacterized human protein C8orf32 with bound peptide. Protein Data Bank. 2008 PDB 3CQ9. [Google Scholar]
- 341.Pedersen LC, Yee VC, Bishop PD, Le Trong I, Teller DC, Stenkamp RE. Transglutaminase factor XIII uses proteinase-like catalytic triad to crosslink macromolecules. Protein Sci. 1994;3:1131–1135. doi: 10.1002/pro.5560030720. [DOI] [PMC free article] [PubMed] [Google Scholar]