Abstract
This work analyses the chitin-binding and catalytic domains of the human macrophage chitotriosidase and investigates the physiological role of this glycoside hydrolase in a complex mechanism such as the innate immune system, especially its antifungal activity. Accordingly, we first analyzed the ability of its chitin-binding domain to interact with chitin embedded in fungal cell walls using the β-lactamase activity reporter system described in our previous work. The data showed that the chitin-binding activity was related to the cell wall composition of the fungi strains and that their peptide-N-glycosidase/zymolyase treatments increased binding to fungal by increasing protein permeability. We also investigated the antifungal activity of the enzyme against Candida albicans. The antifungal properties of the complete chitotriosidase were analyzed and compared with those of the isolated chitin-binding and catalytic domains. The isolated catalytic domain but not the chitin-binding domain was sufficient to provide antifungal activity. Furthermore, to explain the lack of obvious pathologic phenotypes in humans homozygous for a widespread mutation that renders chitotriosidase inactive, we postulated that the absence of an active chitotriosidase might be compensated by the expression of another human hydrolytic enzyme such as lysozyme. The comparison of the antifungal properties of chitotriosidase and lysozyme indicated that surprisingly, both enzymes have similar in vitro antifungal properties. Furthermore, despite its more efficient hydrolytic activity on chitin, the observed antifungal activity of chitotriosidase was lower than that of lysozyme. Finally, this antifungal duality between chitotriosidase and lysozyme is discussed in the context of innate immunity.
Keywords: chitotriosidase, lysozyme, antifungal activity, Candida albicans
Introduction
In higher vertebrates, the innate immune system is the first line of defense against pathogens. It comprises cells and mechanisms that defend the host from infections by other organisms, in a nonspecific manner. It does not confer long-lasting or protective immunity to the host but plays an important role in triggering and optimizing the adaptative immune response. The first step in the activation of the innate immune system is the recognition of pathogen-associated molecular patterns (PAMPs) by C-type lectin receptors and Toll-like receptors (TLRs).1 TLRs are expressed predominantly in antigen-processing and -presentation cells such as macrophages, neutrophils, and dendritic cells, but their expression is not restricted to these cell types.
PAMPs are common molecules present on the cell surface of microbes including lipopolysaccharide (Gram-negative bacteria), peptidoglycan (PG), and lipoteichoic acid (Gram-positive bacteria). In the outer surface of the Candida albicans cell wall, N- and O-linked mannosyl groups of glycoproteins, phospholipomannan, and β-glucans have been described as PAMPs.2,3 Once the TLRs are attached to specific PAMPs, changes in the intracellular domain result in the activation of signaling events such as the translocation of transcription factors, cytokine modulation, and interferon-stimulated gene regulation leading to inflammatory responses and/or the release of antimicrobial agents. These antimicrobial agents could be at least classified into three types: (i) defensins, (ii) whey acidic protein motif containing proteins, and (iii) hydrolytic enzymes.3
Cell walls are essential for the survival of fungal cells. Digestion and inhibition of cell wall synthesis lead to cell lysis due to the high internal turgor pressure. Thus, substances that interfere with cell wall synthesis are considered as potential antifungal agents. In the most characterized yeast, Saccharomyces cerevisiae, the cell wall contains β(1 → 3)-glucans, β(1 → 6)-glucans, chitin, and mannoproteins that are linked together. Mannoproteins are attached to β(1 → 6)-glucans through a remnant of glysosylphosphatidylinositol anchors containing five α-linked mannosyl residues.4 The β(1 → 6)-glucans have some β(1 → 3)-linked branches, and it is to these branches that the reducing termini of chitin chains appear to be attached in a β(1 → 4) or β(1 → 2) linkage. The reducing ends of β(1 → 6)-glucans are connected to the nonreducing terminal glucoses of β(1 → 3)-glucans through a yet uncharacterized linkage. Because of their rigidity that result from the numerous crosslinks, the polysaccharides of fungal cell wall seem to have a major structural function and determine the shape of fungal cells, whereas the mannoproteins may act as “fillers” and are important determinants of the cell wall permeability.4
Chitinases catalyze the hydrolysis of chitin, which is a linear β(1 → 4)-linked polymer of N-acetylglucosamine units. Although chitin is not a component of human tissues, it is a major component of many pathogenic organisms' cell walls including not only fungi but also insects and nematodes. Chitotriosidase and acidic mammalian chitinase are two human chitinases that have been recently discovered. The former is expressed by macrophages and has received a lot of attention. In particular, it is a valuable diagnostic tool for monitoring the efficiency of the Gaucher's disease therapy.5–7 The latter is relatively abundant in the gastrointestinal tract and is characterized by an acidic isoelectric point and a distinct second pH optimum around 2.8 Both chitinases are synthesized as 50-kDa proteins each containing a 39-kDa N-terminal catalytic domain, a hinge region, and a C-terminal chitin-binding domain.
The production, storage, and secretion of chitotriosidase by macrophages and neutrophils, prime components of the innate immune system, suggest a role in innate immunity.9 Such a role has been described for many plant chitinases.10,11 Furthermore, van Eijk et al. showed that human chitotriosidase inhibited the growth of Cryptococcus neoformans and Mucor rouxii, and totally inhibited switching of a C. albicans isolate from yeast/single cell to hyphal growth.9,12 It was also demonstrated that the injection of recombinant human chitotriosidase improved survival in neutropenic mouse models of Candidiasis and Aspergillosis.12
Individuals homozygous for a 24-bp duplication within Exon 10 of the chitotriosidase gene have no functional chitotriosidase. According to Masoud et al.,13 the prevalence of homozygosity for this enzymatic activity deficiency among patients who had survived Candida sepsis suggests that chitotriosidase deficient persons are more susceptible to develop fungal infections. However, Hall et al.14 suggested that the immune system has several tiers of redundancy making sure that organisms remain viable despite defects in innate immunity genes.
In this study, we decided to compare the antifungal properties of the complete human macrophage chitotriosidase and its related domains, the chitin-binding and the catalytic domains, against C. albicans, which is one of the main causes of mortality in immunocompromised individuals. Contrary to what was observed for plant chitinases,15–17 we demonstrated that the catalytic domain is sufficient to present an antifungal activity and has the tendency to be even more efficient than the entire enzyme. In contrast, the isolated chitin-binding domain does not present any antifungal properties as it is the case for a few chitin-binding proteins.18–20 We have also asked whether the mutation that renders the human macrophage chitotriosidase inactive may be compensated by enzymes such as human lysozyme that has not only a documented action against bacteria but also a less-known antifungal activity.21,22
Results
Antifungal activity of the human macrophage chitotriosidase
The chitin-binding domain
Functionalization of the chitin-binding domains
To investigate the antifungal activity of the chitin-binding domain, we first analyzed its chitin-binding activity for chitin embedded in fungal cell walls. For that purpose, we used the β-lactamase reporter system described in our previous works.23,24 These works demonstrated the efficiency of the class A β-lactamase BlaP for the analysis of the properties of carbohydrate-binding domains. Using genetic engineering, we inserted the chitin-binding domains of the human macrophage chitotriosidase, the Acidic Mammalian Chitinase, and the one of the chitinase A1 from Bacillus circulans called ChBDI, ChBDII and ChBDIII, respectively, into a solvent-exposed loop of the carrier protein BlaP.23–26 Indeed, it was interesting to compare the chitin-binding properties of ChBDI with another human chitin-binding domain (ChBDII) and also a prokaryotic chitin-binding domain (CBDIII). The resulting genetic constructions encoding the chimeric proteins called BlaPChBDI, BlaPChBDII, and BlaPChBDIII were used to transform E. coli JM109 to achieve the overexpression of three soluble chimeric proteins in the periplasmic space. The proteins added with a polyhistidine tag were extracted from the periplasm by an osmotic cold shock and purified by chelation affinity and anion exchanger chromatographies.
To check that the insertion of the three chitin-binding domains does not alter the β-lactamase activity of the protein carrier, kinetic parameters of nitrocefin hydrolysis were determined and compared with those of the parental β-lactamase (BlaP SmaI), harboring the engineered insertion loop. Data presented in Table I show that whatever the nature of the chitin-binding domain inserted into BlaP is the β-lactamase moiety remains active and exhibits kinetic parameters similar to those of the parental enzyme.
Table I.
Kinetic Studies of the Different Hybrid β-Lactamases
| BlaP (SmaI) | BlaPChBDI | BlaPChBDII | BlaPChBDIII | |
|---|---|---|---|---|
| KM (μM) | 40 ± 2 | 20 ± 1 | 41 ± 1 | 56 ± 2 |
| kcat (s−1) | 490 ± 20 | 195 ± 5 | 340 ± 10 | 415 ± 7 |
| kcat/KM (μM−1 s−1) | 12.2 ± 0.8 | 9.7 ± 0.5 | 8.3 ± 0.3 | 7.4 ± 0.3 |
The substrate was 100 μM nitrocefin. The values are compared with those reported for the parental enzyme without any insert (BlaP SmaI).27 Each kinetic has been performed twice in triplicates. These values represent the averages ± SD of the two independent experiments.
Next, to verify the biological functionality of the inserted chitin-binding domains, chitin-binding assays on insoluble purified β-chitin from crab shells were performed by monitoring the immobilized β-lactamase activity (Fig. 1). In these binding assays, we took into account the respective kinetic parameters of the different hybrid proteins to compare their binding activities. Although the immobilization of the enzyme onto a surface can affect its catalytic properties, we reasonably assume that this would affect the activity of the different chimeric proteins in the same way and thereby normalized the data using the kinetic parameters calculated for the proteins in solution. The results show that the chitin-binding activity of all the inserted domains is preserved and that the affinities for chitin increase in the following order ChBDII < ChBDI < ChBDIII. The carrier protein without any insert (BlaP SmaI) was used as a negative control. In these experiments, we took into account the respective kinetic parameters of the different hybrid proteins. The relative equilibrium association constant (Kr) of the three chimeric proteins for purified insoluble chitin was also determined by the method described by Gilkes et al.28 The calculated Kr values (Table II) were in the same value range compared with those observed for other classical carbohydrate-binding domains29 confirming that the chitin-binding domains were still functional after insertion into the β-lactamase BlaP. The Kr values increase in the following order ChBDI < ChBDII < ChBDIII. The prokaryotic chitin-binding domain thus has the highest affinity for purified insoluble chitin. The difference between ChBDII and ChBDI affinities observed when using the two different methods might be explained by the fact that the Kr values were determined in thermodynamic equilibrium conditions. In contrast, the immobilized enzymatic activities determined in the first experiment were measured after washings that disrupted this equilibrium.
Figure 1.
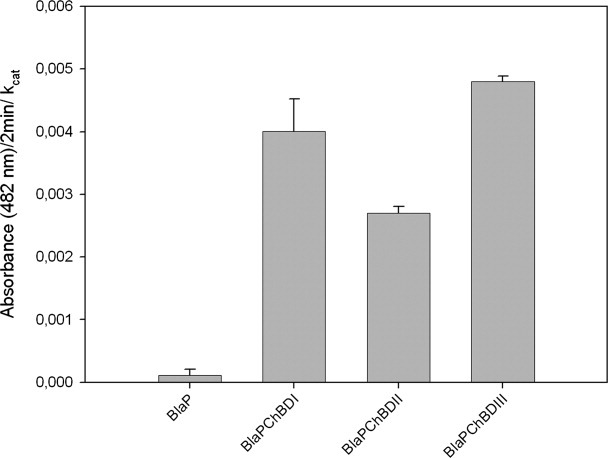
Interaction of the different hybrid proteins with purified insoluble chitin. Binding assay mixtures contained 10 mg (dry weight) of insoluble chitin and 25 pmol of protein in 500 μL of 50 mM sodium phosphate buffer, 150 mM NaCl (pH 7.4). Assay mixtures were incubated for 2 h at room temperature for binding. After removing the supernatant, the pellet was washed three times and incubated for 2 min with 1 mL of nitrocefin (100 μM). After filtration of the suspension, the absorbance of the filtrate was measured at 482 nm. This experiment has been repeated three times (each in duplicates). The bars represent the averages ± SD of these independent experiments. Error propagation was used to normalize the interaction of the different chimeric proteins with their respective kcat.
Table II.
Determination of the Relative Equilibrium Association Constant (Kr) for the Three Chimeric Proteins with Purified Insoluble Chitin, as Described By Gilkes et al.28
| BlaPChBDI | BlaPChBDII | BlaPChBDIII | |
|---|---|---|---|
| Kr (L g−1) | 5.4 ± 0.5 | 7.4 ± 0.7 | 10.1 ± 0.3 |
Each binding constant has been determined in two independent experiments (each performed in triplicates) using very low protein concentrations (see Materials and Methods section). The presented values correspond to the averages ± SD of these two independent experiments.
Interaction of the chitin-binding domains with fungal cell walls
To study the chitin-binding activity in the context of fungal cell walls, we compared the fungal binding properties of the three chimeric proteins with different pathogenic fungal species such as C. albicans, Candida tropicalis, C. neoformans, M. rouxii, and Trichoderma reesei. The immobilized β-lactamase activities of the cell wall obtained by incubating the hybrid proteins with the different fungal species are presented in Figure 2. As described earlier, the carrier protein without insert (BlaP SmaI) was used as a negative control and we took into account the respective kinetic parameters of the different hybrid proteins to compare their binding activities. In addition, the filamentous Gram-positive bacterium Streptomyces lividans TK24 was used as a negative control, because it does not contain any chitin. The data indicated that all the chimeric proteins recognized fungal cell wall chitin and that despite the fact that the prokaryotic chitin-binding domain had the higher affinity for purified chitin, the chitin-binding domain of the human macrophage chitotriosidase exhibits the strongest affinity for fungal cell walls in every tested fungal species. The observed binding properties also depend on the fungal species suggesting that the chitin content and/or accessibility differs in the different tested fungi.
Figure 2.
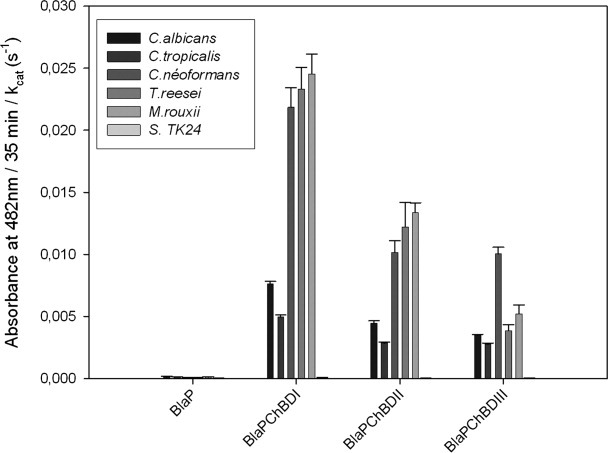
Interaction of the different chimeric proteins with fungal cell wall chitin. The cells were recovered by centrifugation of the culture and washed twice with 50 mM sodium phosphate buffer pH 7.5. The cell suspensions were incubated for 2 h with 25 pmol of each protein in 500 μL of 50 mM sodium phosphate buffer pH 7.5, 150 mM NaCl. After removing the supernatant, the cell pellets were washed three times and incubated for 35 min with 1 mL of nitrocefin (100 μM). After filtration of the suspension, the absorbance of the filtrate was measured at 482 nm. This experiment has been repeated three times (each in duplicates). The bars represent the averages ± SD of these independent experiments. Error propagation was used to normalize the interaction of the different chimeric proteins with their respective kcat.
It should be noted that in all these binding assays, the read-out was indirect and could not be confirmed by performing some competition type experiments using soluble chitin oligomers to compete off the different reporter proteins as we previously described that these chitin-binding domains only recognize insoluble chitin.23
Importance of fungal cell wall permeability on chitin accessibility
Fungal cell wall is composed of a chitin layer buried under a bulky β-1,3-glucan and a mannoprotein layer, which considerably limit chitin accessibility. To investigate the importance of chitin accessibility on chitin recognition by ChBDI and thus on the in vivo chitinolytic activity of the human macrophage chitotriosidase, we pretreated the different fungal strains with Type XIV protease, peptide-N-glycosidase F (PNGase F), or zymolyase before proceeding to binding assays with BlaPChBDI. Type XIV protease is a mixture of at least three proteolytic activities that target the peptide moieties of the mannoprotein layer. PNGase F is usually used to deglycosylate proteins, it cleaves the entire glycan moiety of mannoproteins. Zymolyase is a mixture of a β-1,3-glucan hydrolase and an alkaline protease, which may change the structure of the fungal cell wall to facilitate penetration of the glucan β-1,3-hydrolase. The data presented in Figure 3 indicate that for all the tested fungi, the different enzymatic treatments increase the binding of BlaPChBDI to chitin in the following order zymolyase > PNGase > Type XIV protease. This indicates that chitin accessibility is important in the recognition of fungi by chitin-binding proteins. Our data also indicate that removal of mannan from the cell wall is sufficient to improve the permeability to the hybrid protein. The slight amelioration of the binding observed when fungi are treated with the Type XIV protease suggests that glycosylation of proteins protect them against proteolysis.
Figure 3.
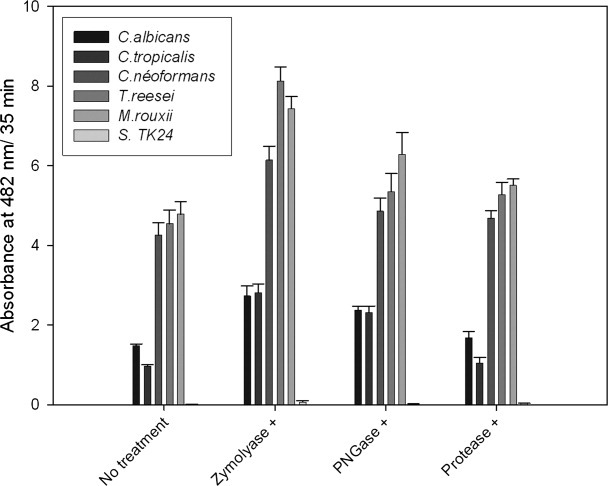
Interaction of BlaPChBDI with fungal chitin cell wall after treatment with zymolyase, PNGase, and Type XIV protease. In this experiment, the washed cells were incubated in the presence of the different hydrolytic enzymes before incubation with the chimeric proteins. After this treatment, cells were successively washed twice with 50 mM sodium phosphate buffer, incubated with the different hybrid proteins, washed again, and incubated with nitrocefin as described above. Finally, absorbance of the filtrate was measured at 482 nm. This experiment has been repeated three times (each in duplicates). The bars represent the averages ± SD of these independent experiments.
The catalytic domain
Functionality of the catalytic domain
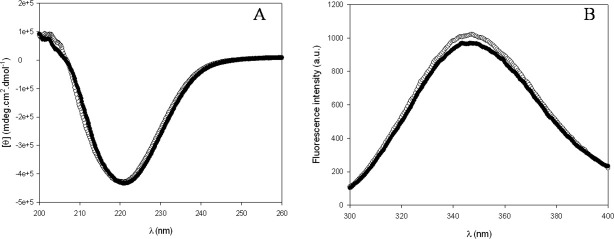
Besides the production of the isolated chitin-binding domain, we decided to express the catalytic domain to assess the contribution of both domains in the antifungal activity of the human macrophage chitotriosidase. The isolated catalytic domain, called Cat_chito, was successfully produced in a soluble form. The comparison of the far UV circular dichroism and fluorescence spectra of Cat_chito with those of the entire chitotriosidase [Fig. 4(a,b)] suggests that the isolated catalytic domain is correctly folded. Next, we investigated the ability of Cat_chito to hydrolyze different chitin forms such as soluble chitooligomers (4-MU-chitotriose), insoluble chitin [carboxymethyl (CM)-chitin-Remazol Brilliant Violet (RBV)], and crystalline chitin (chitin powder extracted from crab shells). Results are presented in Figure 5(a–c). The data show that the isolated catalytic domain hydrolyzes soluble chitooligomers with a catalytic efficiency (11,100 ± 200 μM/min−1) higher than that of the entire chitinase (9300 ± 200 μM/min−1) with a 380 nM protein concentration [Fig. 5(a)]. In contrast, if insoluble chitin hydrolysis is quite similar for both proteins [Fig. 5(b)], hydrolysis of crystalline chitin is severely affected when the chitin-binding domain is deleted [Fig. 5(c)]. Altogether these data suggest that the chitotriosidase catalytic domain folds correctly into an enzymatically active chitinase.
Figure 4.
Far-UV CD (A) and intrinsic fluorescence spectra (B) of the entire chitotriosidase (○) and its catalytic domain (Cat_chito) (•). Spectra were recorded in 50 mM sodium phosphate–citrate buffer (pH 5.6).
Figure 5.
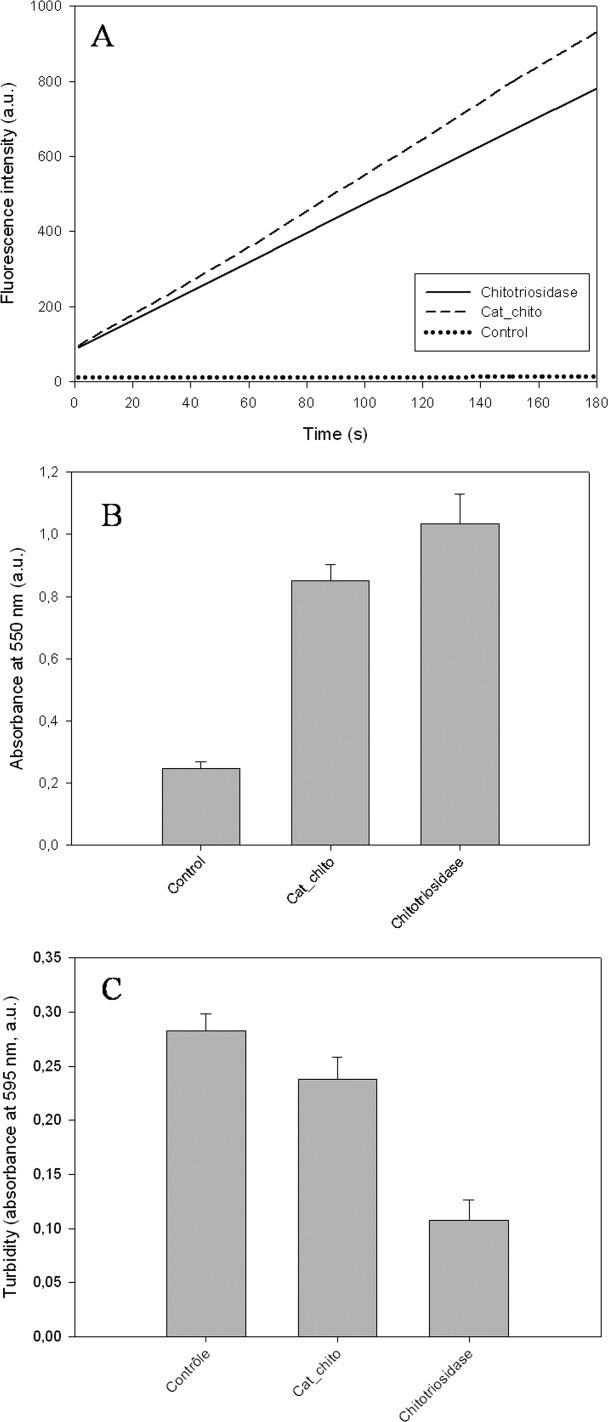
Comparison of the chitinolytic activities of the entire chitotriosidase and its catalytic domain (Cat_chito) on (A) soluble chitooligomer (4-MU-chitotriose), (B) insoluble chitin (CM-Chitin-RBV), and (C) crystalline chitin powder from crab shells.
It should be noted that in this assay, we used crab shell chitin as an insoluble chitin substrate that seems to be the most suitable and commonly used insoluble chitin, although it is unlikely to be a lysosomal component and therefore a common substrate for the chitotriosidase.
Antifungal activities of the chitin-binding and catalytic domains
The respective roles of the chitin-binding and catalytic domains in the chitotriosidase antifungal activity have not been investigated yet. To assess their contribution, we compared the effects of the entire chitotriosidase, Cat_chito and BlaPChBDI on C. albicans viability. In this aim, C. albicans viability assays were realized (Table III). In these assays, C. albicans yeast cells were exposed to the different tested proteins during 2 h. After adequate dilution, cells were plated on solid media to estimate the survival. The data indicate that at equal concentration, the susceptibility to the protein treatment was very similar for the complete chitinase compared with its catalytic domain, with a slightly stronger effect observed for the catalytic domain. This result is in contradiction with previous works15–17 describing that the catalytic domains of many plant chitinases did not present any antifungal activity and that the presence of the chitin-binding domain was essential for the antifungal properties. Our data also show that the chitin-binding domain does not present any antifungal activity contrary to what has been demonstrated for a few chitin-binding proteins18–20 that interfere with cell wall synthesis by binding to nascent chitin, by crosslinking the polymer or by interfering with growth polarity.
Table III.
BlaPChBDI (FBC), Cat_chito (FCat), and the Entire Chitotriosidase (FC) Fungicidal Effects on C. albicans
| Susceptibility to BlaPChBDI (FBC) | Susceptibility to Cat_chito (FCat) | Susceptibility to chitotriosidase (FC) |
|---|---|---|
| ≈0 | 0.52 ± 0.12 | 0.45 ± 0.11 |
The fungicidal activity F was calculated using the following formula F = (CFU of the control cell suspension – CFU of the tested cell suspension)/(CFU of the control cell suspension). This value is comprised between 0 and 1, the higher the F value, the higher the sensitivity of the yeast to the protein. Each protein was used at a concentration of 68 μM. Four independent experiments have been realized (each in triplicates). These values represent the averages ± SD of these independent experiments.
Human macrophage chitotriosidase versus human lysozyme
In addition to its well-documented antibacterial effects,30 it was demonstrated that in some in vitro conditions, exposure of C. albicans to the hen egg lysozyme causes a loss of viability.21,31,32 Thus, we have compared the antifungal activities of the human macrophage chitotriosidase to one of the human lysozyme against C. albicans with the idea that human lysozyme might compensate chitotriosidase deficiency in homozygous individuals for a mutation that inactivates the human chitinase. To do so, we realized three different experiments to investigate the effects of these two enzymes on (i) the inhibition of the switch between the yeast and hyphal forms of C. albicans, (ii) the fungus viability, and (iii) its biofilm formation inhibition.
Inhibition of C. albicans switch between the yeast and filamentous forms
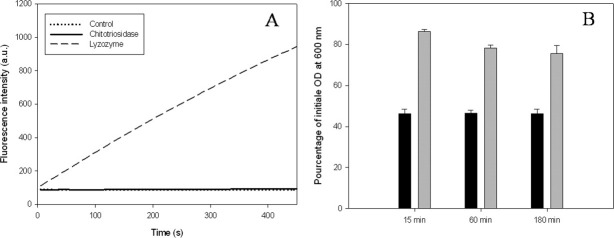
Microscopy data analysis (Fig. 6) shows that both human enzymes are able to inhibit C. albicans switch from the yeast to the hyphal forms. Furthermore, the minimal inhibitory concentration of lysozyme required to observe this effect is twice lower than the one required when using the chitotriosidase. The same result was obtained when the experiments were performed on solid media according to the procedure as described by van Eijk et al. (data not shown).
Figure 6.
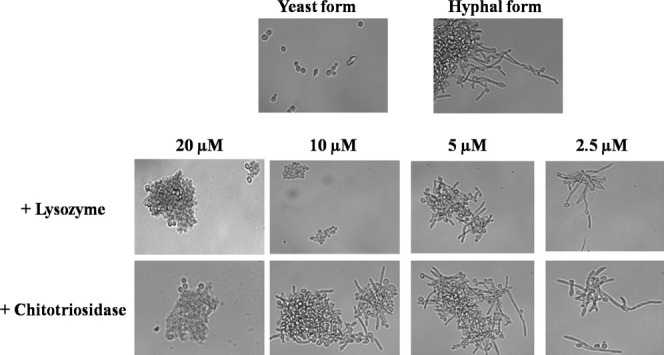
Analysis of the chitotriosidase and lysozyme antifungal activity on C. albicans switching from yeast to hyphal forms. Hyphae formation of C. albicans was induced by shifting the growth temperature from 28 to 37°C during 2 h. Lysozyme and chitotriosidase were added at 20, 10, 5, and 2.5 μM to assess their antifungal activities. This experiment has been repeated four times.
Effect on C. albicans viability
C. albicans viability assays have been achieved as described earlier. The data presented in Table IV indicate that, at equal concentration, the reduction of survival was twice more important for lysozyme than the one observed for chitotriosidase. Interestingly, no significant dose-dependent effect was observed for the two tested enzymes which might be explained by the fact that the cell wall permeability causes a bulking that would limit protein diffusion.
Table IV.
Fungicidal Effects of Various Lysozyme (FL) and Chitotriosidase (FC) Concentrations on C. albicans
| Protein concentration (μM) | Susceptibility to lysozyme (FL) | Susceptibility to chitotriosidase (FC) |
|---|---|---|
| 68 | 0.75 ± 0.12 | 0.38 ± 0.04 |
| 340 | 0.81 ± 0.19 | 0.47 ± 0.06 |
| 680 | 0.87 ± 0.21 | 0.52 ± 0.11 |
The fungicidal activity F was calculated as described in Table III. Four independent experiments have been realized (each in duplicates). These values represent the averages ± SD of these experiments.
Inhibition of C. albicans biofilm formation
Filamentation is a physiological state required for initiating the C. albicans biofilm formation. We examined the effect of chitotriosidase and lysozyme on biofilm formation using the method described by Ramage et al.33 Biofilm formation was monitored after 48 h of adherence in the presence of the two isolated enzymes or in combination. The results presented in Figure 7 indicate that as expected, the antifungal agent fluconazole (positive control) dramatically affects C. albicans biofilm formation.34 Treatments with chitotriosidase and lysozyme resulted in a lesser but significant decrease in the biofilm formation. The data also indicate that at equal protein concentration, lysozyme exhibits the stronger effect.
Figure 7.
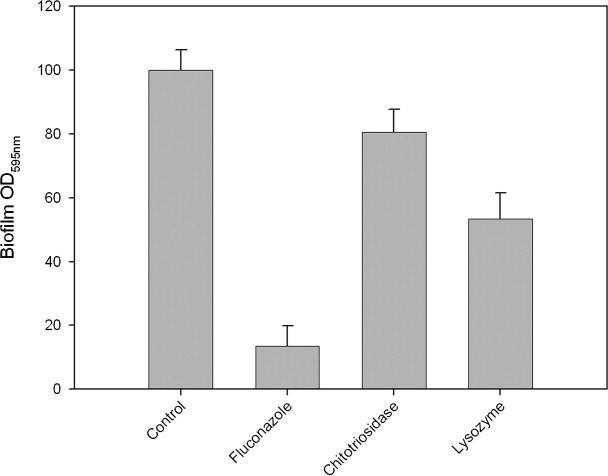
Analysis of C. albicans biofilm formation in the presence of chitotriosidase and lysozyme. All the absorbance values were standardized as a percentage; 100% corresponding to the absorbance value obtained for the control (cells incubated with buffer). The results are the means of two independent experiments each carried out with 20 replicates for each tested enzyme.
In all these assays, no significant increase in the antifungal activity was observed when both enzymes were combined (data not shown) suggesting that no in vitro synergy occurs between these two enzymes. This absence of synergy could be explained by the fact that the two enzymes target the same substrate.
Antibacterial effect
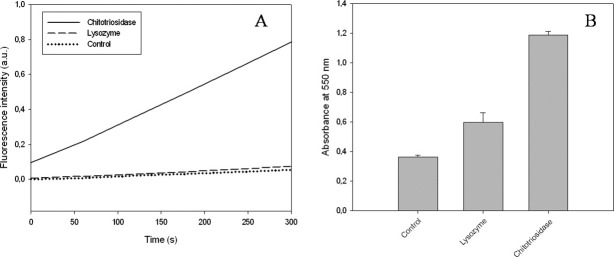
It is well established that lysozyme catalyzes the hydrolysis of β-1,4-linkages between N-acetylmuramic acid and N-acetyl-d-glucosamine in PG. However, we show here that this enzyme also has an antifungal effect. Thus, we investigated the possible hydrolysis of the β-1,4-linkages between N-acetyl-d-glucosamine units in chitin by lysozyme, and inversely, it was interesting to check whether chitotriosidase could also hydrolyze PG in addition to its chitin hydrolysis activity. The data presented in Figure 8(a,b) indicate that although lysozyme exhibits a stronger antifungal activity than chitotriosidase, it weakly hydrolyzes insoluble chitin (CM-chitin-RBV) and has no activity on soluble chitooligosaccharides (4-MU-chitotriose). In addition we investigated the hydrolytic activity of the chitotriosidase against peptidoglycan. Figure 9(a,b) shows that the enzyme is unable to hydrolyze purified PG but has a slight effect on Bacillus subtilis cell lysis. In contrast, as expected, lysozyme exhibits a strong activity against both purified PG and bacteria.
Figure 8.
Comparison of chitotriosidase and lysozyme activities on (A) a soluble chitooligomers (4-MU-chitotriose) and (B) insoluble chitin (CM-Chitin-RBV).
Figure 9.
Comparison of the activities of chitotriosidase and lysozyme on (A) purified fluorescein-labeled PG and (B) Bacillus subtilis cells. For the bacterial cell lysis experiment, the measured absorbance values were standardized as a percentage. 100% corresponding to the absorbance value obtained for the control (bacterial culture without added enzyme) at the same incubation time. The black and the gray bars represent the values obtained for lysozyme and chitotriosidase, respectively. This experiment has been repeated three times (each in duplicates). The bars represent the averages ± SD.
Discussion
The results of the present investigation are consistent with the following conclusions. The comparison of the fungal chitin interaction properties of ChBDI, ChBDII, and ChBDIII, which correspond to the chitin-binding domains of the human macrophage chitotriosidase, the human acidic mammalian chitinase,8 and chitinase chiA1 from B. circulans,29,35 respectively, has demonstrated that ChBDI exhibits the strongest affinity supporting the role of the human chitotriosidase in defense against chitin-containing pathogens. We also showed that the recognition of the chitin embedded in fugal cell wall was dependent of chitin accessibility as enzymatic treatments of the tested fungi result in a significant increase of ChBDI binding. This result emphasizes the importance of chitin accessibility in the recognition and antifungal activity of the chitotriosidase.
The analysis of the functional contribution of the chitin-binding (ChBDI) and catalytic (Cat_chito) domains in both chitin hydrolysis and the antifungal activity of the chitotriosidase demonstrated that addition of ChBDI to a C. albicans cell suspension did not resulted in a further decrease in survival. Surprisingly, the complete chitinase and the catalytic domain presented similar antifungal properties, the latter exhibited a slightly stronger effect.
As the chitotriosidase chitin-binding domain deletion did not result in a decrease of either the antifungal activity or the soluble and insoluble chitin hydrolysis, these data raise the question of the role of this domain in a human chitinase since this domain only seems to be important for crystalline chitin hydrolysis [Fig. 5(c)].
In this study, we also investigated the ability of lysozyme to complement a chitotriosidase deficiency. The results indicate that, in our in vitro conditions, no synergic effect occurred between these two enzymes in fungal growth or fungal biofilm formation inhibition experiments. But surprisingly, our data showed that, at the same concentration, lysozyme exhibited a stronger antifungal activity than chitotriosidase. We suggest that chitotriosidase and lysozyme are not synergistic in their antifungal activities because they share a common substrate since insoluble chitin both in vitro (on crab shell chitin) and in vivo (on fungal cell walls) is hydrolyzed by both enzymes. We believe that the higher antifungal activity observed for lysozyme is due to its smaller size compared to the one of the chitotriosidase. Indeed, the molecular masses of chitotriosidase and lysozyme are 50 and 14.7 kDa, respectively. As chitin accessibility is strongly decreased by the presence of a β-(1-3)-d-glucan layer and a mannoprotein layer that act as fillers,4 we postulated that lysozyme could access the fungal cell chitin much more easily than chitotriosidase. Indeed, an estimate of fungal wall pore size predicts that diffusion through the fungal wall could be difficult for proteins larger than 15 to 20 kDa.18 A slight increase in the antifungal activity of the catalytic domain (40 kDa) compared to what was observed for the complete chitinase supports this hypothesis.
The observed stronger antifungal activity of lysozyme suggests that in individuals lacking an active chitotriosidase, lysozyme could be expressed as an alternative as it was suggested by Hall et al.9 But we cannot exclude the expression of other enzymes such as proteases or other glycoside hydrolases that would facilitate the diffusion of chitotriosidase or lysozyme through the cell wall of pathogenic fungi by degrading the other components of the fungal cell wall.
In conclusion, we have particularly discussed the direct role of the chitotriosidase in immunity by inhibiting fungal growth. But we do not exclude that this enzyme could indirectly be involved in immunity by releasing some degradation compounds from the fungal cell walls that would activate the immune system by interacting with TLRs as postulated by Netea et al.2
In this study, we also demonstrated for the first time that human lysozyme that is usually associated with defense against bacteria also presents an antifungal activity that is even higher than that of chitotriosidase, the human chitinase reported to be an antifungal protein.
Materials and Methods
Construction, expression, and purification of the hybrid proteins
The three hybrid proteins were constructed, expressed, and purified as described previously.23,24
Kinetic studies of the hybrid β-lactamases
The kinetic parameters of the purified bifunctional hybrid β-lactamases were determined using a 150 μM nitrocefin substrate solution prepared in 50 mM sodium phosphate buffer at pH 7. The initial rates of hydrolysis were monitored at 482 nm. The Km and kcat values were calculated by fitting the data to the Henri-Michaelis equation and its linearized form according to Hanes.36
Binding assay of the hybrid β-lactamases to purified insoluble chitin
Identical amounts (50 nM) of each chimeric protein and of the parental β-lactamase were incubated for 2 h with the same amount (10 mg) of insoluble chitin (Sigma). The mixtures were centrifuged at 13,000g during 10 min to separate the supernatants containing the unbound proteins from the pellet containing chitin with the captured protein. The pellets were washed three times with 50 mM sodium phosphate buffer, 500 mM NaCl and then incubated in the presence of 150 μM nitrocefin for 1 min at 25°C. The suspension was rapidly filtered, and the absorbance of the hydrolysis product was measured at 482 nm.
Determination of Kr
Binding assays were conducted as follows: various concentrations of the chimeric protein (25 nM–3 μM) were incubated in the presence of 10 mg of chitin in a final volume of 500 μL of 50 mM sodium phosphate buffer, 150 mM NaCl, pH 7.5 at 22°C with continual mixing. The mixtures were centrifuged at 13,000g for 15 min at 4°C and the supernatant containing the free proteins was collected. The free protein concentration was determined with the help of the reporter β-lactamase activity. The amount of bound protein was calculated as the difference between the initial protein concentration and the free protein concentration after binding. The relative equilibrium association constants (Kr) were determined as described by Gilkes et al.28 using the following equation:
where [B] is the concentration of bound ligand (mol g chitin–1), [F] the concentration of free ligand (molar), [N0] the concentration of binding sites in the absence of ligand, a the number of lattice units occupied by a single ligand molecule, and Ka the equilibrium association constant (L mol−1). The Ka value cannot be isolated from this equation but the relative equilibrium association constant, Kr (L g chitin−1) is defined as:
Expression of the human chitotriosidase and its catalytic domain in Pichia pastoris
Recombinant human chitotriosidase and its catalytic domain were produced using the pPIC9K P. pastoris expression vector (Invitrogen) as described by Gratepanche et al.37 Briefly, the genes encoding the catalytic domain of the chitotriosidase and the complete enzyme (GenBank, gi:4502808) followed by a His6-tag on the C-terminal ends were amplified from the plasmid pReceiver-YAD (GeneCopoeia™) and cloned into the pPIC9K vector (Invitrogen) by standard methods. The pPIC9K plasmid contains the α-factor secretion signal that directs the recombinant protein into the secretory pathway. The constructs were digested with Sal I and used to transform P. pastoris strain SMD168 by electroporation. This resulted in insertion of the constructs at the AOX1 locus of P. pastoris, generating a His+ Mut+ phenotype. Transformants were selected for the His+ phenotype on 2% agar containing regeneration dextrose biotin (1M sorbitol, 2% dextrose, 1.34% yeast nitrogen base, 4 × 10−5% biotin, and 0.005% of l-glutamic acid, l-methionine, l-lysine, l-leucine, and l-isoleucine) medium and then further selected for high copy number by their ability to grow on 2% agar containing 1% yeast extract, 2% peptone, 2% dextrose medium, and the G418 antibiotic at various concentrations (0.5–4 mg/mL; Invitrogen). The proteins were expressed in a shaker flask and harvested 72 h after induction by methanol.
Purification of the recombinant chitotriosidase and its catalytic domain
The proteins were purified by using a cation exchange column (P-Sepharose, Pharmacia) equilibrated in the first purification step with a 50 mM acetate buffer pH 5 (Buffer A). The P. pastoris supernatants dialyzed against the same buffer were applied onto the column. Elution was performed by a linear NaCl (0–1M) gradient. The active fractions eluted at 600 mM NaCl were pooled and dialyzed against Buffer B (50 mM sodium phosphate buffer, 150 mM NaCl, pH 7.5) for the second purification step on an Ni-NTA-Sepharose column (Novagen, USA) equilibrated with Buffer B. The column was washed with seven column volumes of the same buffer, three column volumes of Buffer B + 2M NaCl, and three column volumes of Buffer B + 10 mM imidazole. Elution was performed with an increasing linear (10–500 mM) imidazole gradient in Buffer A. Active fractions eluted at 200 mM imidazole and yielded a single band on sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis. Proteins integrity was verified by both N-terminal sequencing using the Edman degradation procedure and electrospray ionization mass spectrometry. For both proteins, we observed a difference (400 Da) between the theoretical and experimental molecular weights, this difference was explained by the analysis of the N-terminal sequence of the two proteins. Results indicated that both sequences contained four residues of the α-factor signal peptide (Glu–Ala–Glu–Ala) incorrectly cleaved by P.pastoris endoproteases kex2. Furthermore, a weak heterogeneity is also observed for both proteins with the presence of species that differ from each other by increments of 162 kDa, this has been attributed to a fortuitous glycosylation of the proteins.
Detection of fungal chitin
A colony of the tested fungi (C. albicans [the American Type Culture Collection (ATCC) 10231-IHEM3731], C. tropicalis (IHEM6246), C. neoformans (ATCC 32265-IHEM3969), T. reesei (ATCC 26921-IHEM4122) provided by the Belgian Co-ordinated Collections of Microorganisms and M. rouxii (ATCC 44260) provided by ATCC) was inoculated into 30 mL of YPD medium containing 2% dextrose and grown at 28°C for 20 h. To detect chitin, 4.8 mL of this culture (DO600 nm = 0.5) was pelleted by centrifugation and washed twice with 50 mM sodium phosphate buffer pH 7.5. The pellet was then resuspended in 500 μL of phosphate buffer containing 5 pmol of each chimeric protein. Incubation was performed at 4°C during 2 h under continuous mixing. The cell suspensions were centrifuged at 8000g during 10 min to separate the supernatant containing the unbound protein from the pellet containing chitin with the captured protein. The pellets were washed three times with 50 mM sodium phosphate buffer, 0.2% Tween and then incubated in the presence of 1 mL of 100 μM nitrocefin for 35 min at 25°C. The suspension was rapidly filtered and the absorbance of the filtrate was measured at 482 nm. Two negative controls were made, the first one using the parental β-lactamase BlaP without inserted chitin-binding domain and the second using the filamentous bacterium S. lividans TK24.
Detection of fungal chitin in the presence of zymolyase, PNGase, and protease
Three different enzymes were used in these assays: zymolyase (or lyticase 20T; ICN Immuno Biologicals, Costa Mesa, CA); PNGase F (from Elizabethkingia meningoseptica; Sigma), and the protease Type XIV from Streptomyces griseus (Sigma).
The procedure applied in this experiment was exactly the same as described above excepted that the washed cells were incubated in the presence of the different hydrolytic enzymes before the incubation with the chimeric proteins. Briefly, cell pellets were incubated with 100 units of zymolyase 20T, 100 units of PNGase F, or 4 mg of protease in 1 mL of 50 mM sodium phosphate buffer pH 7.4. For the samples incubated with the zymolyase, the buffer was supplemented with 1.2M sorbitol and 27 mM β-mercaptoethanol. After this treatment, cells were washed twice with 50 mM sodium phosphate buffer and then incubated with the different chimeric proteins. The experiment was completed exactly as described earlier.
Antifungal activity of the chitotriosidase and the lysozyme
The C. albicans hyphae formation inhibition assays in solid medium were carried out according to the procedure described by van Eijk et al.12 Briefly, a disc, removed from an actively growing fungal culture in YPD medium (2% dextrose, 100 μg/mL of ampicillin), was added to wells containing a fresh YPD agar medium (2% dextrose, 100 μg/mL of ampicillin, 2% agar). After 48 h of incubation at 37°C, 20 μL of each tested enzyme was placed on a sterile paper disc placed in the center of the well. The plate was then incubated for an additional 24 h at 37°C. The enzyme concentration was 100 μM for each tested protein. In liquid medium, filament formation was induced by switching the culture temperature of an actively growing fungal culture in YPD medium (2% dextrose, 100 μg/mL of ampicillin) from 28 to 37°C during 2 h. The lysozyme and chitotriosidase concentrations used in these tests to inhibit hyphae formation were 20, 10, 5, and 2.5 μM.
The C. albicans viability assays were performed using a procedure modified from Samaranayake et al.31 C. albicans yeast cells were cultured overnight at 28°C in YPD medium. Cells were harvested and washed twice before being resuspended in a 50 mM sodium phosphate buffer; 50 mM NaCl, pH 7 to yield a final concentration of 7 × 106 cells/mL (OD520 nm= 0.65). The cell suspension was diluted 10-fold with a 100 mM phosphate buffer, pH 7.5 containing 68, 340, and 680 μM of lysozyme and chitotriosidase. Samples were incubated for 2 h at 37°C and continually mixed. After appropriate dilutions, the samples were plated on YPD solid media and incubated at 37°C overnight. The results are the means of at least three independent experiments, each carried out in duplicates or triplicates.
Biofilm formation of C. albicans in the presence of the chitotriosidase and the lysozyme
C. albicans biofilm formation was monitored as described by Jin et al.38 and Al-Fattani and Douglas.34 C. albicans was grown in YPD medium (2% dextrose) at 28°C overnight. Cells were harvested and washed twice in 50 mM phosphate buffered saline (PBS), pH 7.4. Before biofilm formation experiments, all washed cell suspensions were adjusted to an OD520 nm value of 0.38 in a yeast nitrogen base medium. One hundred microliter of this standardized cell suspension was added to the 96-well polystyrene plates in the presence of the tested enzyme. The plate was incubated at 37°C for 48 h to allow biofilm formation. The growth medium was removed from each well and nonadherent cells were removed by washing six times with 200 μL of sterile 50 mM sodium phosphate buffer, pH 7.4. Finally, biofilms were stained with 110 μL of a 0.4% crystal violet aqueous solution for 45 min at room temperature. The plate was then washed twice with sterile distilled water and destained with 200 μL of 95% ethanol. After 45 min of destaining, 100 μL of the destaining solution was transferred to a new well and the amount of the crystal violet stain in the destaining solution was measured at 595 nm. The final enzyme concentrations used in this experiment were 25 μM of each isolated enzyme or in combination. Fluconazole was used as a negative control since Al-Fattani and Douglas34 demonstrated that this antifungal agent is able to inhibit C. albicans biofilm formation.
Chitinolityc activity measurements
For soluble chitooligosaccharides hydrolysis, the enzyme preparations were incubated at 37°C with 22 μM 4-methylumbelliferone-chitotriose (4-MU-chitotriose; Sigma) in 50 mM sodium phosphate buffer, pH 7.5. The formed 4-MU was detected fluorometrically (excitation at 366 nm and emission at 445 nm). The final concentration of chitotriosidase was 3 nM, for lysozyme, assays were also performed with a 10-fold higher concentration. The correlation between the measured fluorescence intensity and the molarity of released methylumbelliferone was realized using a calibration curve obtained with isolated 4-MU.
For insoluble chitin hydrolysis, enzymes incubated in 400 μL of 50 mM sodium phosphate buffer pH 7.5 were mixed with 100 μL of 2 mg/mL CM-Chitin-RBV. The reaction was carried out at 37°C for 2 h and stopped by adding 100 μL of 2M HCl and incubating on ice for 15 min. The absorbance of the mixtures was measured at 520 nm after centrifugation at 12,000g for 5 min. Sodium phosphate buffer incubated with the substrate was used as the blank.
Crystalline chitin hydrolysis was performed with chitin powder extracted from crab shells. One milligram of chitin (dry weight) was incubated with 5 μM enzyme in a final volume of 1 mL of 50 mM sodium phosphate buffer pH 7.5. After 24 h of incubation at 25°C, turbidity of the samples was measured at 595 nm.
Lysozyme activity measurements
Detection of lysozyme activity was performed using the EnzChek® Lysozyme Assay Kit (E22013) (Molecular Probes). This assay measures lysozyme activity on fluorescein-labelled Micrococcus lysodeikticus cell walls, so that the fluorescence of the label is quenched. Lysozyme hydrolytic action relieves this quenching. For each assay, 50 μL of the DQ lysozyme substrate (50 μg/mL) is incubated in the presence of 50 μL of the kit reaction buffer and 5 μL of the tested enzyme. Final protein concentrations in these assays were 350 nM for each enzyme. The fluorescence was measured using excitation/emission wavelengths of 494 and 518 nm, respectively. For chitotriosidase, the assays were also performed with a fourfold higher concentration.
Glossary
Abbreviations:
- ChBDI
human macrophage chitotriosidase chitin-binding domain
- ChBDII
acidic mammalian chitinase chitin binding domain
- ChBDIII
Bacillus circulans chitinase A1 chitin-binding domain
- Cat_chito
human macrophage chitotriosidase catalytic domain
- PAMP
pathogen-associated molecular pattern
- TLR
toll-like receptor
References
- 1.Hazeki K, Nigorikawa K, Hazeki O. Role of phosphoinositide 3-kinase in innate immunity. Biol Pharm Bull. 2007;30:1617–1623. doi: 10.1248/bpb.30.1617. [DOI] [PubMed] [Google Scholar]
- 2.Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, Hobson RP, Bertram G, Hughes HB, Jansen T, Jacobs L, Buurman ET, Gijzen K, Williams DL, Torensma R, McKinnon A, MacCallum DM, Odds FC, Van der Meer JW, Brown AJ, Kullberg BJ. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulain D, Jouault T. Candida albicans cell wall glycans, host receptors and responses: elements for a decisive crosstalk. Curr Opin Microbiol. 2004;7:342–349. doi: 10.1016/j.mib.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Kollar R, Petrakova E, Ashwell G, Robbins PW, Cabib E. Architecture of the yeast cell wall. The linkage between chitin and beta(1 → 3)-glucan. J Biol Chem. 1995;270:1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- 5.Bussink AP, van Eijk M, Renkema GH, Aerts JM, Boot RG. The biology of the Gaucher cell: the cradle of human chitinases. Int Rev Cytol. 2006;252:71–128. doi: 10.1016/S0074-7696(06)52001-7. [DOI] [PubMed] [Google Scholar]
- 6.Fusetti F, von Moeller H, Houston D, Rozeboom HJ, Dijkstra BW, Boot RG, Aerts JM, van Aalten DM. Structure of human chitotriosidase. Implications for specific inhibitor design and function of mammalian chitinase-like lectins. J Biol Chem. 2002;277:25537–25544. doi: 10.1074/jbc.M201636200. [DOI] [PubMed] [Google Scholar]
- 7.Malaguarnera L. Chitotriosidase: the yin and yang. Cell Mol Life Sci. 2006;63:3018–3029. doi: 10.1007/s00018-006-6269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, Place A, Aerts JM. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J Biol Chem. 2001;276:6770–6778. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 9.Hall AJ, Morroll S, Tighe P, Gotz F, Falcone FH. Human chitotriosidase is expressed in the eye and lacrimal gland and has an antimicrobial spectrum different from lysozyme. Microbes Infect. 2008;10:69–78. doi: 10.1016/j.micinf.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Abeles FB, Bosshart RP, Forrence LE, Habig WH. Preparation and purification of glucanase and chitinase from bean leaves. Plant Physiol. 1971;47:129–134. doi: 10.1104/pp.47.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verburg JG, Huynh QK. Purification and characterization of an antifungal chitinase from Arabidopsis thaliana. Plant Physiol. 1991;95:450–455. doi: 10.1104/pp.95.2.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Eijk M, van Roomen CP, Renkema GH, Bussink AP, Andrews L, Blommaart EF, Sugar A, Verhoeven AJ, Boot RG, Aerts JM. Characterization of human phagocyte-derived chitotriosidase, a component of innate immunity. Int Immunol. 2005;17:1505–1512. doi: 10.1093/intimm/dxh328. [DOI] [PubMed] [Google Scholar]
- 13.Masoud M, Rudensky B, Elstein D, Zimran A. Chitotriosidase deficiency in survivors of Candida sepsis. Blood Cells Mol Dis. 2002;29:116–118. doi: 10.1006/bcmd.2002.0547. [DOI] [PubMed] [Google Scholar]
- 14.Hall AJ, Quinnell RJ, Raiko A, Lagog M, Siba P, Morroll S, Falcone FH. Chitotriosidase deficiency is not associated with human hookworm infection in a Papua New Guinean population. Infect Genet Evol. 2007;7:743–747. doi: 10.1016/j.meegid.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan Y, Chye ML. A Brassica juncea chitinase with two-chitin binding domains show anti-microbial properties against phytopathogens and Gram-negative bacteria. Plant Signal Behav. 2008;3:1103–1105. doi: 10.4161/psb.3.12.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan Y, Ramalingam S, Nagegowda D, Taylor PW, Chye ML. Brassica juncea chitinase BjCHI1 inhibits growth of fungal phytopathogens and agglutinates Gram-negative bacteria. J Exp Bot. 2008;59:3475–3484. doi: 10.1093/jxb/ern197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh Y, Kawase T, Nikaidou N, Fukada H, Mitsutomi M, Watanabe T, Itoh Y. Functional analysis of the chitin-binding domain of a family 19 chitinase from Streptomyces griseus HUT6037: substrate-binding affinity and cis-dominant increase of antifungal function. Biosci Biotechnol Biochem. 2002;66:1084–1092. doi: 10.1271/bbb.66.1084. [DOI] [PubMed] [Google Scholar]
- 18.Bormann C, Baier D, Horr I, Raps C, Berger J, Jung G, Schwarz H. Characterization of a novel, antifungal, chitin-binding protein from Streptomyces tendae Tu901 that interferes with growth polarity. J Bacteriol. 1999;181:7421–7429. doi: 10.1128/jb.181.24.7421-7429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, Xie W, Gong Z. Characteristics and antifungal activity of a chitin binding protein from Ginkgo biloba. FEBS Lett. 2000;478:123–126. doi: 10.1016/s0014-5793(00)01834-2. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen KK, Nielsen JE, Madrid SM, Mikkelsen JD. Characterization of a new antifungal chitin-binding peptide from sugar beet leaves. Plant Physiol. 1997;113:83–91. doi: 10.1104/pp.113.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamaya T. Lytic action of lysozyme on Candida albicans. Mycopathol Mycol Appl. 1970;42:197–207. doi: 10.1007/BF02051947. [DOI] [PubMed] [Google Scholar]
- 22.Marquis G, Montplaisir S, Garzon S, Strykowski H, Auger P. Fungitoxicity of muramidase. Ultrastructural damage to Candida albicans. Lab Invest. 1982;46:627–636. [PubMed] [Google Scholar]
- 23.Vandevenne M, Filee P, Scarafone N, Cloes B, Gaspard G, Yilmaz N, Dumoulin M, Francois JM, Frere JM, Galleni M. The Bacillus licheniformis BlaP beta-lactamase as a model protein scaffold to study the insertion of protein fragments. Protein Sci. 2007;16:2260–2271. doi: 10.1110/ps.072912407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandevenne M, Gaspard G, Yilmaz N, Giannotta F, Frere JM, Galleni M, Filee P. Rapid and easy development of versatile tools to study protein/ligand interactions. Protein Eng Des Sel. 2008;21:443–451. doi: 10.1093/protein/gzn021. [DOI] [PubMed] [Google Scholar]
- 25.Chevigne A, Yilmaz N, Gaspard G, Giannotta F, Francois JM, Frere JM, Galleni M, Filee P. Use of bifunctional hybrid beta-lactamases for epitope mapping and immunoassay development. J Immunol Methods. 2007;320:81–93. doi: 10.1016/j.jim.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Ruth N, Quinting B, Mainil J, Hallet B, Frere JM, Huygen K, Galleni M. Creating hybrid proteins by insertion of exogenous peptides into permissive sites of a class A beta-lactamase. FEBS J. 2008;275:5150–5160. doi: 10.1111/j.1742-4658.2008.06646.x. [DOI] [PubMed] [Google Scholar]
- 27.Matagne A, Misselyn-Bauduin AM, Joris B, Erpicum T, Granier B, Frere JM. The diversity of the catalytic properties of class A beta-lactamases. Biochem J. 1990;265:131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilkes NR, Jervis E, Henrissat B, Tekant B, Miller RC, Jr, Warren RA, Kilburn DG. The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J Biol Chem. 1992;267:6743–6749. [PubMed] [Google Scholar]
- 29.Hashimoto M, Ikegami T, Seino S, Ohuchi N, Fukada H, Sugiyama J, Shirakawa M, Watanabe T. Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12. J Bacteriol. 2000;182:3045–3054. doi: 10.1128/jb.182.11.3045-3054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laible NJ, Germaine GR. Bactericidal activity of human lysozyme, muramidase-inactive lysozyme, and cationic polypeptides against Streptococcus sanguis and Streptococcus faecalis: inhibition by chitin oligosaccharides. Infect Immun. 1985;48:720–728. doi: 10.1128/iai.48.3.720-728.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samaranayake YH, Samaranayake LP, Pow EH, Beena VT, Yeung KW. Antifungal effects of lysozyme and lactoferrin against genetically similar, sequential Candida albicans isolates from a human immunodeficiency virus-infected southern Chinese cohort. J Clin Microbiol. 2001;39:3296–3302. doi: 10.1128/JCM.39.9.3296-3302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu T, Samaranayake LP, Leung WK, Sullivan PA. Inhibition of growth and secreted aspartyl proteinase production in Candida albicans by lysozyme. J Med Microbiol. 1999;48:721–730. doi: 10.1099/00222615-48-8-721. [DOI] [PubMed] [Google Scholar]
- 33.Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol. 2002;68:5459–5463. doi: 10.1128/AEM.68.11.5459-5463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Fattani MA, Douglas LJ. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol. 2006;55:999–1008. doi: 10.1099/jmm.0.46569-0. [DOI] [PubMed] [Google Scholar]
- 35.Ferrandon S, Sterzenbach T, Mersha FB, Xu MQ. A single surface tryptophan in the chitin-binding domain from Bacillus circulans chitinase A1 plays a pivotal role in binding chitin and can be modified to create an elutable affinity tag. Biochim Biophys Acta. 2003;1621:31–40. doi: 10.1016/s0304-4165(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 36.Frere JM. Enzymic mechanisms involving concomitant transfer and hydrolysis reactions. Biochem J. 1973;135:469–481. doi: 10.1042/bj1350469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gratepanche S, Gamain B, Smith JD, Robinson BA, Saul A, Miller LH. Induction of crossreactive antibodies against the Plasmodium falciparum variant protein. Proc Natl Acad Sci USA. 2003;22:13007–13012. doi: 10.1073/pnas.2235588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin Y, Yip HK, Samaranayake YH, Yau JY, Samaranayake LP. Biofilm-forming ability of Candida albicans is unlikely to contribute to high levels of oral yeast carriage in cases of human immunodeficiency virus infection. J Clin Microbiol. 2003;41:2961–2967. doi: 10.1128/JCM.41.7.2961-2967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]