Abstract
Early studies suggested the existence of an hepatic receptor that is involved in the clearance of serpin:enzyme complexes. Subsequent work has identified this receptor as the LDL receptor-related protein 1 (LRP1). LRP1 is a multifunctional receptor that serves to transport numerous molecules into the cell via endocytosis and also serves as a signaling receptor. LRP1 plays diverse roles in biology, including roles in lipoprotein metabolism, regulation of protease activity, activation of lysosomal enzymes, and cellular entry of bacterial toxins and viruses. Deletion of the Lrp1 gene leads to lethality in mice, revealing a critical, but as of yet undefined, role in development. Its identification as a receptor for serpin:enzyme complexes confirms a major role for LRP1 in regulating protease activity.
Keywords: LRP1, LDL receptor family, Serpin:enzyme complexes, LDLa repeats, uPA:PAI-1 complexes, RAP
1. Introduction
The early work of Ohlsson et al. (1971), who investigated the clearance of trypsin–inhibitor complexes from the circulation, revealed the existence of a specific hepatic pathway responsible for removing complexes of proteases with their inhibitors. Subsequent work by Imber1 and Pizzo (1981) reinforced this concept by discovering that α2-macroglobulin (α2M) complexed with trypsin is rapidly removed by the liver, whereas the native form of the inhibitor is not removed from the circulation. In examining the specificity of these pathways, it was noted that various serpin:enzyme complexes could compete with one another for hepatic clearance (Fuchs et al., 1984) but did not seem to alter the hepatic uptake of modified α2M (Fuchs et al., 1982), raising the possibility of different pathways for these molecules. However, upon isolation of the hepatic receptor responsible for clearing α2M–protease complexes from the circulation (Ashcom et al., 1990; Moestrup and Gliemann, 1989) and confirming its identity to the LDL receptor-related protein (LRP, now called LRP1; Kristensen et al., 1990; Strickland et al., 1990), we now know that this receptor is responsible for clearing both α2M–protease complexes as well as serpin:enzyme complexes from the circulation (Kounnas et al., 1996).
2. Purification of LRP1
2.1. Isolation of full length LRP1 from tissue extracts
LRP1 has been purified from placental tissue (Ashcom et al., 1990) and from liver membrane extracts (Moestrup and Gliemann, 1989) by affinity chromatography over a Sepharose–α2M–methylamine column.
2.1.1. Purification of α2M and preparation of methylamine-reacted α2M–Sepharose
α2M is purified from human plasma employing the procedure detailed by Harpel (1976).
To prepare methylamine-activated α2M, incubate native α2M in 50 mM HEPES, 0.15 M NaCl, pH 8.0 with 100 mM methylamine at room temperature for 30 min.
Dialyze the α2M:Me extensively against 0.1 M NaHCO3, 0.5 M NaCl, pH 8.3.
After dialysis, couple to CNBr-activated Sepharose (Pharmacia Fine Chemical) as recommended by the manufacturer using 10 mg α2M:Me/ml resin.
Allow protein to couple to resin for 2 h at room temperature using gentle mixing (end-over-end).
Remove solution, and replace with 0.1 M Tris, pH 8.0 for an additional 2 h at room temperature.
Wash resin with 0.1 M sodium acetate, 0.5 M NaCl, pH 5.0 and then with 0.1 M sodium bicarbonate, 0.5 M NaCl, pH 8.3.
2.1.2. Purification of LRP1 from human placenta
All procedures, unless otherwise is indicated, are carried out at 4 °C.
Wash fresh placenta with cold TBS (50 mM Tris, 150 mM NaCI, pH 7.4) and 200 mM sucrose. Remove the fetal membranes and umbilical cord, and grind the placenta in a meat grinder. Use the tissue immediately or store at −80 °C until needed.
Suspend the tissue in an equal volume of TBS containing 0.005% digitonin, 1 mM each of MgCl2 and CaCl2, along with proteinase inhibitors: 1 mM PMSF, 0.02 mg/ml leupeptin, and 0.02 mg/ml d-Phe-Pro-Arg–Ch2Cl.
Stir for 15 min on ice, and then homogenize the mixture in a blender (three times for 30 s each). Following homogenization, centrifuge at 5000×g for 20 min.
Discard the supernatant, and suspend the pelleted tissue in an equal volume of extraction buffer (50 mM octyl-B-d-glucopyranoside in TBS containing 1 mM each of MgCl2 and CaCl2, 1 mM PMSF, 0.02 mg/ml leupeptin, 0.02 mg/ml d-Phe-Pro-Arg–Ch2Cl). Stir in extraction buffer for 1 h at 4 °C.
Centrifuge the suspension at 5000×g for 20 min.
Remove the supernatant, and subject to additional centrifugation at 11,000×g for 20 min.
Apply the resultant supernatant to a 120 ml Sepharose CL-4B column, and collect the unabsorbed material.
Mix this material with 40–60 ml of α2M:Me–Sepharose overnight at 4 °C using end-over-end mixing.
Wash the resin with eight column volumes of 25 mM octyl-β-d-glucopyranoside in TBS containing 1 mM CaCl2, 1 mM MgCl2, 1 mM PMSF.
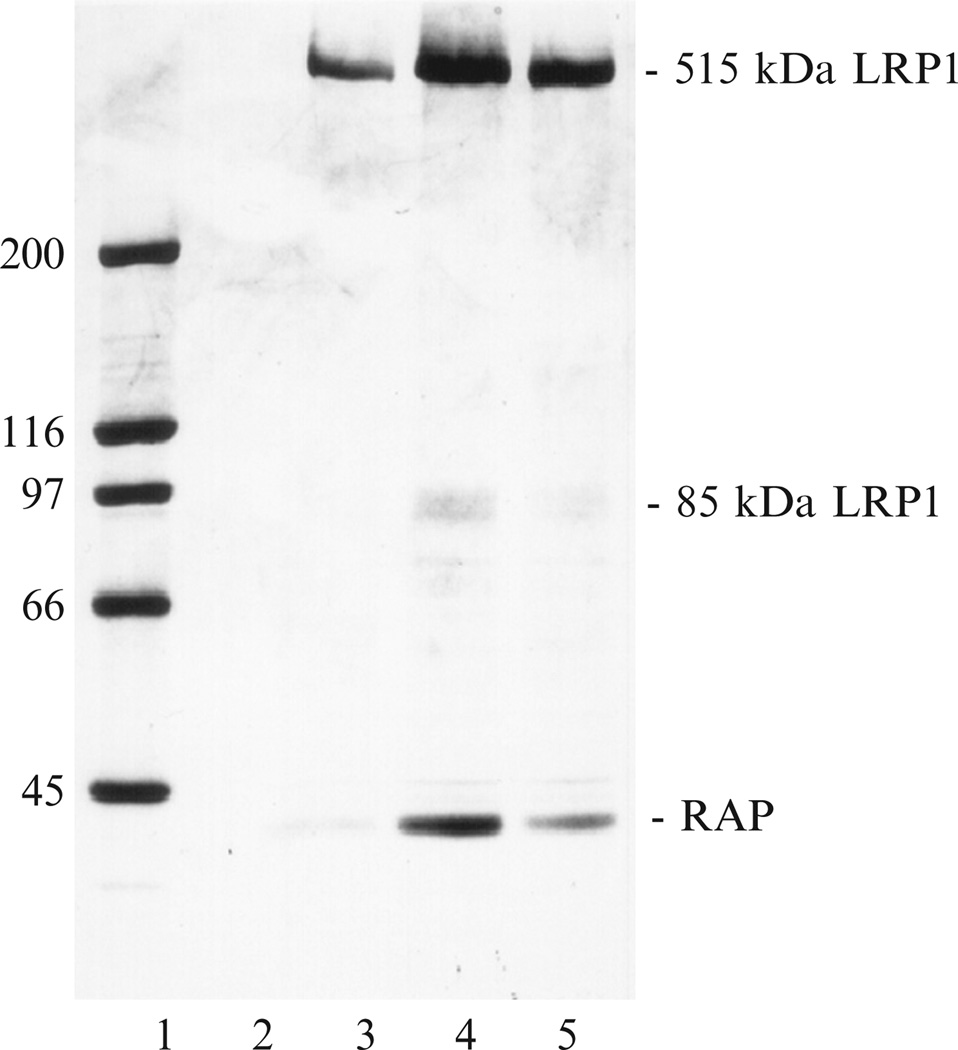
Elute LRP1 from the column with TBS containing 20 mM EDTA, 25 mM octyl-β-d-glucopyranoside. Collect fractions and measure the absorbance at 280 nm to monitor protein. Analyze each fraction by SDS-PAGE, and pool fractions containing LRP1 (see Fig. 2.1).
Apply LRP1 containing fractions to a Mono Q anion exchange column (Pharmacia Fine Chemicals) at room temperature, previously equilibrated with 20 mM octyl-β-d-glucopyranoside in 50 mM Tris, pH 8.2, at a flow rate of 0.5 ml/min.
After washing the column with equilibration buffer, elute LRP1 with a linear gradient from 0 to 1 M NaCl over 60 min at a flow rate of 0.5 ml/min. Protein is monitored at 280 nm, and 0.5 ml fractions are collected. LRP1 elutes at approximately 0.55 M NaCl.
Fractions containing LRP1 are pooled, analyzed by SDS-PAGE, and dialyzed into PBS containing 20 mM octyl-β-d-glucopyranoside, and stored frozen at −80 °C until used. The concentration of LRP1 is determined by absorbance measurements at 280 nm using an E1%280 nm of 13.5.
Figure 2.1.
Affinity chromatography of placental extract over Sepharose–α2M:Me. Fractions eluted from the α2M:Me–Sepharose affinity column were assessed by SDS-PAGE on a 5–15% gradient gel with a 4% stacking gel using the Laemmli buffer system. Lane 1, standards; lanes 2–5, fractions eluted from the affinity column. From Ashcom et al. (1990).
2.2. Isolation of soluble forms of LRP1 from plasma
Soluble forms of LRP1 circulate in the plasma as a consequence of shedding (Quinn et al., 1997), and Gaultier et al. (2008) have reported purification of soluble LRP1 using an affinity matrix in which GST-receptor-associated protein (RAP, a ligand of LRP1) was coupled to NHS-activated Sepharose 4 Fast Flow (GE Healthcare).
3. Expression of Receptor Fragments
3.1. Expression of individual LDLa repeats in Escherichia coli
LRP1 is composed of modules of β-propeller domains, EGF-repeats, and LDLa (also called complement-type repeats or ligand-binding repeats). For structural studies, a number of LDLa repeats have been expressed in E. coli BL21 (Blacklow and Kim, 1996; Dolmer et al., 1998) either as individual repeats or as fusion proteins with GST. In the case of the GST-fusion protein, GST is removed by thrombin digestion following purification. In all cases, the repeats need to be refolded. This can be accomplished as follows:
Dilute sample to approximately 0.2 mg/ml in 6 M guanidinium chloride, 50 mM Tris, 1 mM dithiothreitol, pH 8.5.
Dialyze against 50 mM Tris–HCl, pH 8.5, 10 mM CaCl2, 1 mM GSH, and 0.5 mM GSSG for 24 h at room temperature under oxygen-free conditions as described (Blacklow and Kim, 1996).
Purify refolded LDLa repeats by reverse-phase HPLC as described (Blacklow and Kim, 1996).
3.2. Expression of LRP1 fragments in cells
Soluble minireceptors representing each of the four ligand-binding domains of LRP1 have been expressed in human glioblastoma U87 cells (Bu and Rennke, 1996) as well as in COS-1 cells (Lee et al., 2006; Mikhailenko et al., 2001). In all cases, coexpression of RAP greatly increased the yield of soluble domains found in the media.
Plate COS-1 cells in 100-mm dishes and grow them to approximately 50% confluence.
Transfect the cells in serum-containing medium with 30 µg of pSec-TagB carrying cDNA for various LRP1 fragments using the FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis) according to the manufacturer’s protocol.
Twenty four hours after transfection, wash the cells, and change the medium to plain Dulbecco’s modified Eagle’s medium supplemented with 1% Nutridoma® -NS medium supplement (Roche Molecular Biochemicals).
Harvest this medium after 48 h of incubation, and detect soluble LRP1 fragments by immunoblot analysis using anti-myc antibody to detect recombinant proteins.
3.3. Expression of functional LRP1 “minireceptors”
Willnow et al. (1994) were the first to develop LRP1 “minireceptors” to identify regions on LRP1 that are involved in ligand binding. Since then, various “minireceptor” constructs covering all four clusters of ligand-binding repeats have been developed and used in functional studies to identify ligand-binding sites (Mikhailenko et al., 2001; Obermoeller-McCormick et al., 2001). These functional studies are best done in LRP1-deficient cells, such as the CHO 13-5-1 cell line (Fitzgerald et al., 1995). To express LRP1 minireceptors and investigate ligand uptake, the following protocol is used:
Plate CHO 13-5-1 cells in 6-well plates (5 × 104 cells/well) 24 h prior to the transfection.
Transfect cells with 2 µg of DNA/well in 1.5 ml of serum-containing medium using FuGENE 6 transfection reagent (Roche Molecular Biochemicals).
Thirty-six to forty hours following transfection, wash cells with phosphate-buffered saline (PBS) which are ready to be used in the ligand internalization experiments.
To measure ligand uptake, incubate transfected CHO 13-5-1 cells transiently transfected with mini-LRP1 constructs for 3 h at 37 °C with 125I-labeled ligands (5 nM).
After incubation, wash the cells with PBS and detach from plastic using 0.5 mg/ml trypsin, 0.5 mg/ml proteinase K, and 5 mM EDTA-containing buffer.
Internalized 125I-labeled ligand is defined as radioactivity associated with the cell pellet.
Nonspecific uptake of 125I-labeled ligand is determined by measuring 125I-labeled ligand uptake in the presence of excess unlabeled ligand and is subtracted from the total internalization.
The cell numbers for each experimental condition are measured in parallel wells that do not contain radioactivity.
4. Ligand Binding to LRP1
4.1. Determinants on LRP1 that bind to ligands
LRP1 binds numerous ligands, including proteases, protease inhibitor complexes, apoE-enriched lipoproteins, matrix proteins, and certain growth factors. By far, the largest class of structurally related ligands includes protease-inhibitor complexes, and the list of all serpin–protease complexes that have been reported to bind to LRP1 is listed in Table 2.1. Most of the ligands that are recognized by LRP1 appear to bind to one of the clusters of LDLa repeats (or ligand-binding repeats) that are present in the extracellular domain of LRP1. LRP1 contains four such clusters, termed I–IV. Cluster I contains two LDLa, cluster II contains eight LDLa repeats, cluster III contains 10 LDLa repeats, while cluster IV contains 11 LDLa repeats.
Table 2.1.
Serpin–enzyme complexes known to bind to LRP1
| Complex | References |
|---|---|
| uPA:plasminogen activator inhibitor-I (PAI-1) | Nykjær et al. (1994) |
| tPA:PAI-1 | Orth et al. (1994) |
| tPA:neuroserpin | Makarova et al. (2003) |
| Thrombin:antithrombin III | Kounnas et al. (1996) |
| Thrombin:heparin cofactor II | Kounnas et al. (1996) |
| Trypsin:α1-antitrypsin | Kounnas et al. (1996) |
| Elastase: α1-antitrypsin | Poller et al. (1995) |
| C1s:C1 inhibitor | Storm et al. (1997) |
| uPA:PAI-2 | Croucher et al. (2006) |
| Thrombin:protease nexin-1 | Knauer et al. (1997a,b) |
| Thrombin:protein C inhibitor | Kasza et al. (1997) |
| uPA:protein C inhibitor | Kasza et al. (1997) |
Recent structural studies examining the interaction of RAP D3 domain with two LDLa repeats from the LDL receptor have generated a model for how ligands may interact with LRP1 (Fisher et al., 2006). Prior work had established that the D3 domain of RAP contains a high-affinity LRP1 binding site, and random mutagenesis studies revealed a critical role for Lys270 and Lys256 in the binding interaction (Migliorini et al., 2003). Fisher et al. (2006) obtained a crystal structure of the RAP D3 domain in complex with two repeats from the LDL receptor. The results reveal that Lys270 and Lys256 each interact with a single repeat from the LDL receptor. Within an individual LDLa repeats, three conserved, calcium-coordinating acidic residues encircle the lysine side chain. This electrostatic interaction, combined with avidity effects resulting from the use of multiple sites, is thought to represent a model for ligand recognition by LRP1 and other LDL receptor family members.
4.2. Assays to measure the binding of ligands to LRP1
4.2.1. Quantitative measurements of ligand interaction using homologous ligand displacement experiments
Solid phase binding assays have been successful in measuring ligand association with LRP1 (Williams et al., 1992). Quantitative measurements can be readily made employing homologous ligand displacement experiments. In this assay, trace levels of 125I-labeled ligand is incubated with LRP1 immobilized in microtiter wells, and increasing concentrations of unlabeled ligand used to compete for binding. The data are analyzed by the program LIGAND (Munson and Rodbard, 1980), which has the advantage of fitting the nonspecific component as well.
Coat microtiter plates overnight at 4 °C with 100 µl of purified LRP1 (3–10 µg/ml) in 50 mM Tris, 150 mM NaCl, pH 7.4 (TBS) containing 5 mM Ca2+.
Block the wells with 10 mg/ml BSA in TBS, 5 mMCa2+ for 1 h at room temperature.
For high-affinity interactions such as the interaction of α2M* or RAP with LRP1, add 50–300 pMof 125I-labeled α2M* (23 pCi/pg) orRAP in TBS, 5 mM Ca2+, 30 mg/ml BSA to the wells in the presence of increasing concentrations (1–500 nM) of unlabeled ligands (α2M* or RAP).
Count aliquots of each stock solution to measure the total cpm added to each well.
Incubate overnight at 4 °C, and then wash the wells three times with TBS, 0.02% Tween 20.
Add 200 µ1 of 0.1 N NaOH to each well, and remove an aliquot (150 µl) for counting.
Analyze the data using the computer program LIGAND (Munson and Rodbard, 1980).
4.2.2. Using an ELISA to measure ligand binding to LRP1
Enzyme-linked immunosorbent assays have been extensively used to evaluate the binding of LRP1 to various proteins coated onto microtiter wells or to evaluate binding of proteins to LRP1 coated on microtiter wells.
For ELISAs, use 3 µg/ml of protein in 100 µl of coating buffer (50 mM Tris, pH 8.0, 150 mM NaCl (TBS), 5 mM CaCl2) to coat microtiter plate wells (Linbro/Titertek, Flow Laboratories, Inc., McLean, VA) for 4 h at 37 °C or 18 h at 4 °C.
Incubate with 5% BSA in 250 µl of coating buffer for 1 h at 37 °C to block unbound sites. Then, add purified ligands or LRP1 to the wells at threefold dilutions in concentrations ranging from 150 to 0.06 nM in TBS containing 5% BSA, 0.05% Tween 20, 5 mM CaCl2.
Following an 18-h incubation at 4 °C, wash the wells with TBS, 0.05% Tween, 5 mM CaC12 (TBS-Tween).
Detect bound ligand or receptor by incubating for 1 h at room temperature with antibodies (usually monoclonal) directed against the bound molecule. The antibodies are diluted in TBS-Tween.
After washing, add goat anti-mouse IgG conjugated to horseradish peroxidase and incubate for 1 h at room temperature.
After washing the wells, add the substrate 3,3′,5,5′-tetramethylbenzidine (Kirkegaard & Perry, Gaithersburg, MD), and measure the absorbance at 650 nm.
4.2.3. Surface Plasmon resonance measurements
Surface Plasmon resonance measurements have been useful for detecting binding of various ligands to LRP1 and other LDLR family members.
For these studies, activate the Biacore sensor chip (type CM5; Biacore AB) with a 1:1 mixture of 0.2 M N-ethyl-N_-(3-dimethylaminopropyl) carbodiimide and 0.05 M N-hydroxysuccinimide in water as described by the manufacturer.
Immobilize purified human LRP1 at the level of 3000 response units in a working solution of 10 µg/ml in 10 mM sodium acetate, pH 4.0. Flow over the chip at a rate of 5 µl/min.
Then block the remaining binding sites with 1 M ethanolamine, pH 8.5.
Wash out unbound protein with 0.5% SDS.
Use a second flow cell, similarly activated and blocked without immobilization of protein, as a negative control.
Use a flow cell with immobilized ovalbumin at the level of 500 response units as a control for nonspecific protein binding.
Perform all binding reactions in 10 mM HEPES, 0.15 M NaCl, pH 7.4 (HBS-P buffer; Biacore AB), containing 0.005% Tween 20.
Measure binding of ligands to LRP1 at 25 °C at a flow rate of 30 µl/min for 4 min, followed by 4 min of dissociation.
Subtract the bulk shift due to changes in refractive index measured on blank surfaces from the binding signal at each condition to correct for nonspecific signals.
Regenerate chip surfaces with subsequent 1-min pulses of 10 mM sodium acetate, pH 4.0, containing 1 M NaCl and 10 mM NaOH containing 1 M NaCl followed by 2 min of washing with running buffer to remove the high salt solution.
Binding of ligands is typically measured using twofold dilutions in HBS-P buffer over a range of concentrations (e.g., 0.6–50 nM).
5. Binding of Serpin–Enzyme Complexes to LRP1
5.1. Specificity of binding
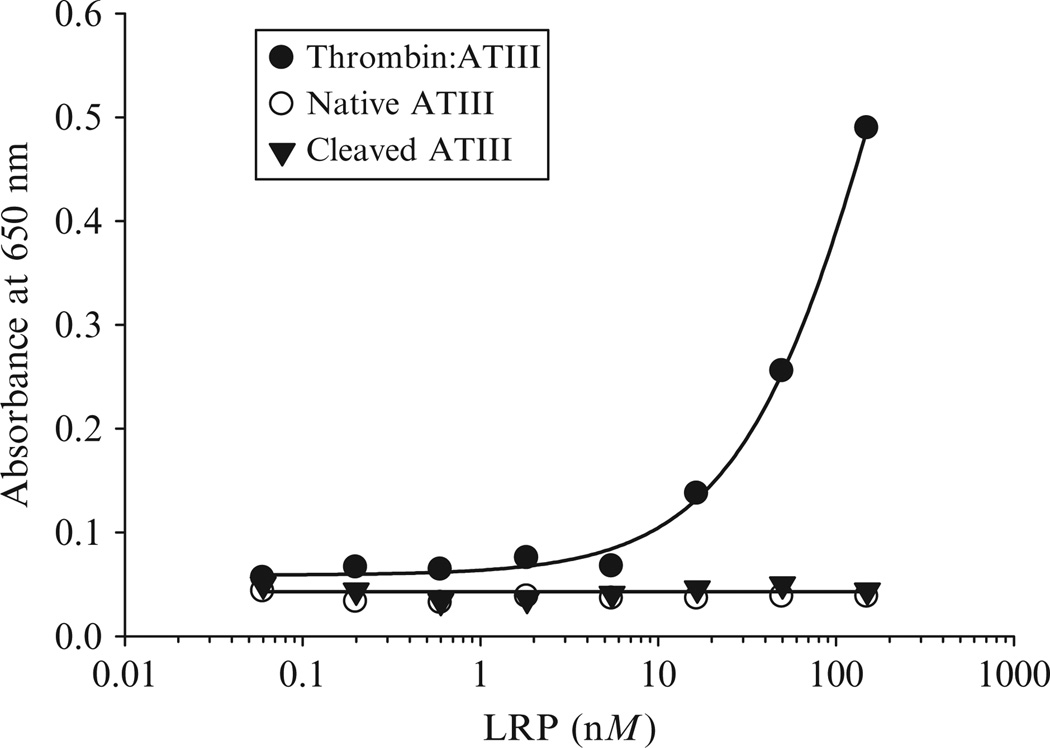
Serpins can exist in a variety of conformational states, including the native serpin, the proteolytically modified form in which the inhibitory capacity is abolished, and finally as a stable proteinase-complexed form. For most serpins, including antithrombin III, heparin cofactor II, α1-antitrypsin (Kounnas et al., 1996), and neuroserpin (Makarova et al., 2003), very little binding of the native or cleaved serpin to LRP1 occurs (Fig. 2.2). Thus, the only serpin form recognized by LRP1 is the stable proteinase-complexed form, consistent with the findings that only the proteinase-complexed forms of serpins are rapidly removed by the hepatic clearance pathway (Mast et al., 1991).
Figure 2.2.
LRP1 appears specific for the enzyme:serpin complexes. Increasing concentrations of LRP1 were incubated with microtiter wells coated with thrombin:ATIII (closed circles), native ATIII (open circles), or cleaved ATIII (closed triangles). Following incubation, bound LRP was detected with monoclonal antibody 8G1 (Adapted from Kounnas et al., 1996).
5.2. Determinants on serpins that are responsible for binding to LRP1
Proteinase cleavage of the exposed loop present in the serpin triggers a conformational change in the serpin and the formation of a covalent complex with the target proteinase. Studies investigating plasminogen activator inhibitor 1 (PAI-1) complexes with various proteinases have revealed that high-affinity LRP1 receptor binding is independent of the nature of the proteinase, since different PAI-1/proteinase complexes can cross-compete with one another, implying that the high-affinity receptor-binding epitope resides in the serpin alone (Stefansson et al., 1998). However, the specific regions of serpins involved in receptor recognition at the molecular level are not well defined and appear to be cryptic in nature. While studies investigating the clearance of serpin–enzyme complexes originally implicated a pentapeptide sequence located at the COOH-terminal fragment of α1-antitrypsin in receptor binding (Joslin et al., 1991), mutation of this region in heparin cofactor II failed to diminish the binding, internalization, or degradation of thrombin:heparin cofactor II complexes (Maekawa and Tollefsen, 1996).
Mutagenesis studies have revealed some serpin epitopes that contribute to serpin interactions with LRP1. Basic residues clustered to one face of PAI-1, composed of parts of β-sheet-A and α-helix-D, appear to be involved in mediating PAI-1-proteinase binding to LRP1 (Rodenburg et al., 1998; Skeldal et al., 2006; Stefansson et al., 1998). Alanine substitution of Lys-82 and Arg-120 reduced the ability of LRP1 to recognize PAI-1 complexed to urokinase plasminogen activator (uPA). Similarly, mutation of Arg-78 and Lys-124 to alanine also resulted in loss of binding of the complex to LRP1. Importantly, Stefansson et al. (1998) found that a PAI-1molecule with Arg-76 mutated to glutamic acid within the heparin-binding domain abolished binding to LRP1.
For protease nexin 1 (PN-1), a region which separates β-sheet-6B and α-helix-B, corresponding to Pro-47 through Ile-58, appears responsible for interacting with LRP1 (Knauer et al., 1997a,b). Thus a synthetic peptide representing this region (PHDNIVISPHGI) was shown to competitively inhibit the LRP1-dependent internalization of thrombin:PN1 complexes. An antibody prepared against this synthetic peptide inhibited PN1:thrombin complex degradation by 70%, but it had no effect on binding of the complex to cell surface heparins (Knauer et al., 1999). In addition, mutagenesis within the corresponding region of PN-1 (His-48Aand Asp-49A) reduced the catabolism rate of mutated PN-1 to 15% of wild type (Knauer et al., 1999).
5.3. Clearance of 125I-labeled serpin–enzyme complexes in mice
To measure the clearance of serpin–enzyme complexes from the circulation, the following protocol can be used:
Inject anesthetized mice with a bolus of 200 µl of serpin:enzyme complex (e.g., ATIII-125I-thrombin, 100 nM) in the presence or absence of competitor (e.g., RAP 110 µM) into the tail vein over a period of ~15 s.
Collect blood (40 µl) at selected time intervals following injection (1, 5, 10, and 20 min), by retro-orbital bleeding into 10 µl of 0.5 M EDTA.
Weigh the sample, and count for its 125Iodine content.
The initial time point, taken 1 min after injection, is considered to represent 100% radioactivity in the circulation.
Examine the clearance of each preparation in two mice and average the results.
6. Summary
Substantial evidence exists confirming the role of LRP1 as the hepatic receptor is responsible for clearing serpin–enzyme complexes from the circulation. It should be pointed out that other members of the LDL receptor family are also able to bind serpin–enzyme complexes. These include the VLDL receptor (Argraves et al., 1995; Kasza et al., 1997) and LRP2/gp330 (Stefansson et al., 1995). Since these receptors are not expressed in the liver, LRP1 is mainly responsible for hepatic clearance of serpin–enzyme complexes from the circulation. While structural studies are beginning to reveal the molecular mechanisms by which ligands interact with this receptor family, the exact mechanisms by which serpin–enzyme complexes are recognized by LRP1 still need to be delineated. It is clear that LRP1 only binds serpin–enzyme complexes and does not bind to the native or cleaved serpin. Current evidence suggests that determinants present on the serpin contribute to binding, but it is likely that determinants on the protease also contribute to LRP1 binding as well.
ACKNOWLEDGMENTS
This work was supported by grants HL054710, HL050784, HL072929 (DKS), and CA098369, HL084387, DK081376 (TMA). SCM is supported by T32HL007698.
REFERENCES
- Argraves KM, Battey FD, MacCalman CD, McCrae KR, Gåfvels M, Kozarsky KF, Chappell DA, Strauss JF, Strickland DK. The very low density lipoprotein receptor mediates the cellular catabolism of lipoprotein lipase and urokinase-plasminogen activator inhibitor type I complexes. J. Biol. Chem. 1995;270:26550–26557. doi: 10.1074/jbc.270.44.26550. [DOI] [PubMed] [Google Scholar]
- Ashcom JD, Tiller SE, Dickerson K, Cravens JL, Argraves WS, Strickland DK. The human α2-macroglobulin receptor: Identification of a 420-kD cell surface glycoprotein specific for the activated conformation of α2-macroglobulin. J. Cell Biol. 1990;110:1041–1048. doi: 10.1083/jcb.110.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklow SC, Kim PS. Protein folding and calcium binding defects arising from familial hypercholesterolemia mutations of the LDL receptor. Nat. Struct. Biol. 1996;3:758–762. doi: 10.1038/nsb0996-758. [DOI] [PubMed] [Google Scholar]
- Bu G, Rennke S. Receptor-associated protein is a folding chaperone for low density lipoprotein receptor-related protein. J. Biol. Chem. 1996;271:22218–22224. doi: 10.1074/jbc.271.36.22218. [DOI] [PubMed] [Google Scholar]
- Croucher D, Saunders DN, Ranson M. The urokinase/PAI-2 complex: A new high affinity ligand for the endocytosis receptor low density lipoprotein receptor-related protein. J. Biol. Chem. 2006;281:10206–10213. doi: 10.1074/jbc.M513645200. [DOI] [PubMed] [Google Scholar]
- Dolmer K, Huang W, Gettins PGW. Characterization of the calcium site in two complement-like domains from the low-density lipoprotein receptor-related protein (LRP) and comparison with a repeat from the low-density lipoprotein receptor. Biochemistry. 1998;37:17016–17023. doi: 10.1021/bi982022s. [DOI] [PubMed] [Google Scholar]
- Fisher C, Beglova N, Blacklow SC. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell. 2006;22:277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Fitzgerald DJ, Fryling CM, Zdanovsky A, Saelinger CB, Kounnas M, Winkles JA, Strickland D, Leppla S. Pseudomonas exotoxin-mediated selection yields cells with altered expression of low-density lipoprotein receptor-related protein. J. Cell Biol. 1995;129:1533–1541. doi: 10.1083/jcb.129.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs HE, Shifman MA, Pizzo SV. In vivo catabolism of α1-proteinase inhibitor-trypsin, antithrombin III-thrombin, and α2-macroglobulin-methylamine. Biochem. Biophys. Acta. 1982;716:151–157. doi: 10.1016/0304-4165(82)90263-x. [DOI] [PubMed] [Google Scholar]
- Fuchs HE, Michalopoulos GK, Pizzo SV. Hepatocyte uptake of alpha 1-proteinase inhibitor-trypsin complexes in vitro: Evidence for a shared uptake mechanism for proteinase complexes of alpha 1-proteinase inhibitor and antithrombin III. J. Cell. Biochem. 1984;25:231–243. doi: 10.1002/jcb.240250405. [DOI] [PubMed] [Google Scholar]
- Gaultier A, Arandjelovic S, Li X, Janes J, Dragojlovic N, Zhou GP, Dolkas J, Myers RR, Gonias SL, Campana WM. A shed form of LDL receptor-related protein-1 regulates peripheral nerve injury and neuropathic pain in rodents. J. Clin. Invest. 2008;118:161–172. doi: 10.1172/JCI32371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpel PC. Human alpha2-macroglobulin. Methods Enzymol. 1976;45:639–652. doi: 10.1016/s0076-6879(76)45055-3. [DOI] [PubMed] [Google Scholar]
- Imber MJ, Pizzo SV. Clearance and binding of two electrophoretic “fast” forms of human alpha 2-macroglobulin. J. Biol. Chem. 1981;256:8134–8139. [PubMed] [Google Scholar]
- Joslin G, Fallon RJ, Bullock J, Adams SP, Perlmutter DH. The SEC receptor recognizes a pentapeptide neodomain of alpha 1- antitrypsin-protease complexes. J. Biol. Chem. 1991;266:11282–11288. [PubMed] [Google Scholar]
- Kasza A, Petersen HH, Heegaard CW, Oka K, Christensen A, Dubin A, Chan L, Andreasen PA. Specificity of serine proteinase/serpin complex binding to very- low-density lipoprotein receptor and alpha2-macroglobulin receptor/low-density-lipoprotein-receptor-related protein. Eur. J. Biochem. 1997;248:270–281. doi: 10.1111/j.1432-1033.1997.00270.x. [DOI] [PubMed] [Google Scholar]
- Knauer MF, Hawley SB, Knauer DJ. Identification of a binding site in protease nexin I (PN1) required for the receptor mediated internalization of PN1-thrombin complexes. J. Biol. Chem. 1997a;272:12261–12264. doi: 10.1074/jbc.272.19.12261. [DOI] [PubMed] [Google Scholar]
- Knauer MF, Kridel SJ, Hawley SB, Knauer DJ. The efficient catabolism of thrombin-protease nexin 1 complexes is a synergistic mechanism that requires both the LDL receptor- related protein and cell surface heparins. J. Biol. Chem. 1997b;272:29039–29045. doi: 10.1074/jbc.272.46.29039. [DOI] [PubMed] [Google Scholar]
- Knauer MF, Crisp RJ, Kridel SJ, Knauer DJ. Analysis of a structural determinant in thrombin-protease nexin 1 complexes that mediates clearance by the low density lipoprotein receptor-related protein. J. Biol. Chem. 1999;274:275–281. doi: 10.1074/jbc.274.1.275. [DOI] [PubMed] [Google Scholar]
- Kounnas MZ, Church FC, Argraves WS, Strickland DK. Cellular internalization and degradation of antithrombin III-thrombin, heparin cofactor II-thrombin, and α1-antitrypsin- trypsin complexes is mediated by the low density lipoprotein receptor-related protein. J. Biol. Chem. 1996;271:6523–6529. doi: 10.1074/jbc.271.11.6523. [DOI] [PubMed] [Google Scholar]
- Kristensen T, Moestrup SK, Gliemann J, Bendtsen L, Sand O, Sottrup-Jensen L. Evidence that the newly cloned LRP is the α2M receptor. FEBS Lett. 1990;276:151–155. doi: 10.1016/0014-5793(90)80530-v. [DOI] [PubMed] [Google Scholar]
- Lee D, Walsh JD, Mikhailenko I, Yu P, Migliorini M, Wu Y, Krueger S, Curtis JE, Harris B, Lockett S, Blacklow SC, Strickland DK, et al. RAP uses a histidine switch to regulate its interaction with LRP in the ER and golgi. Mol. Cell. 2006;22:423–430. doi: 10.1016/j.molcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Maekawa H, Tollefsen DM. Role of the proposed serpin-enzyme complex receptor recognition site in binding and internalization of thrombin-heparin cofactor II complexes by hepatocytes. J. Biol. Chem. 1996;271:18604–18610. doi: 10.1074/jbc.271.31.18604. [DOI] [PubMed] [Google Scholar]
- Makarova A, Mikhailenko I, Bugge TH, List K, Lawrence DA, Strickland DK. The LDL receptor-related protein modulates protease activity in the brain by mediating the cellular internalization of both neuroserpin and neuroserpin: tPA complexes. J. Biol. Chem. 2003 doi: 10.1074/jbc.M309150200. M309150200. [DOI] [PubMed] [Google Scholar]
- Mast AE, Enghild JJ, Pizzo SV, Salvesen G. Analysis of the plasma elimination kinetics and conformational stabilities of native, proteinase-complexed, and reactive site cleaved serpins: Comparison of α1-proteinase inhibitor, α1-antichymotrypsin, antithrombin III, α2-antiplasmin, angiotensinogen, and ovalbumin. Biochemistry. 1991;30:1723–1730. doi: 10.1021/bi00220a039. [DOI] [PubMed] [Google Scholar]
- Migliorini MM, Behre EH, Brew S, Ingham KC, Strickland DK. Allosteric modulation of ligand binding to low density lipoprotein receptor-related protein by the receptor-associated protein requires critical lysine residues within its carboxyl-terminal domain. J. Biol. Chem. 2003;278:17986. doi: 10.1074/jbc.M212592200. [DOI] [PubMed] [Google Scholar]
- Mikhailenko I, Battey FD, Migliorini M, Ruiz JF, Argraves K, Moayeri M, Strickland DK. Recognition of alpha 2-macroglobulin by the low density lipoprotein receptor-related protein requires the cooperation of two ligand binding cluster regions. J. Biol. Chem. 2001;276:39484–39491. doi: 10.1074/jbc.M104382200. [DOI] [PubMed] [Google Scholar]
- Moestrup SK, Gliemann J. Purification of the rat hepatic α2-macroglobulin receptor as an approximately 440 kDa single chain polypeptide. J. Biol. Chem. 1989;264:15574–15577. [PubMed] [Google Scholar]
- Munson PJ, Rodbard D. LIGAND: A versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- Nykjær A, Kjoller L, Cohen RL, Lawrence DA, Gliemann J, Andreasen PA. Both pro-uPA and uPA:PAI-1 complex bind to the α2-macroglobulin receptor/LDL receptor-related protein: Evidence for multiple independent contacts between the ligands and receptor. Ann. NY Acad. Sci. 1994;737:483–485. doi: 10.1111/j.1749-6632.1994.tb44346.x. [DOI] [PubMed] [Google Scholar]
- Obermoeller-McCormick LM, Li Y, Osaka H, Fitzgerald DJ, Schwartz AL, Bu G. Dissection of receptor folding and ligand-binding property with functional minireceptors of LDL receptor-related protein. J. Cell Sci. 2001;114:899–908. doi: 10.1242/jcs.114.5.899. [DOI] [PubMed] [Google Scholar]
- Ohlsson K, Ganrot PO, Laurell CB. In vivo interaction beween trypsin and some plasma proteins in relation to tolerance to intravenous infusion of trypsin in dog. Acta Chir. Scand. 1971;137:113–121. [PubMed] [Google Scholar]
- Orth K, Willnow T, Herz J, Gething MJ, Sambrook J. Low density lipoprotein receptor-related protein is necessary for the internalization of both tissue-type plasminogen activator-inhibitor complexes and free tissue-type plasminogen activator. J. Biol. Chem. 1994;269:21117–21122. [PubMed] [Google Scholar]
- Poller W, Willnow TE, Hilpert J, Herz J. Differential recognition of α1-antitrypsin-elastase and α1-antichymotrypsin-cathepsin G complexes by the low density lipoprotein receptor-related protein. J. Biol. Chem. 1995;270:2841–2845. doi: 10.1074/jbc.270.6.2841. [DOI] [PubMed] [Google Scholar]
- Quinn KA, Grimsley PG, Dai YP, Tapner M, Chesterman CN, Owensby DA. Soluble low density lipoprotein receptor-related protein (LRP) circulates in human plasma. J. Biol. Chem. 1997;272:23946–23951. doi: 10.1074/jbc.272.38.23946. [DOI] [PubMed] [Google Scholar]
- Rodenburg KW, Kjoller L, Petersen HH, Andreasen PA. Binding of urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex to the endocytosis receptors alpha2-macroglobulin receptor/low-density lipoprotein receptor-related protein and very-low-density lipoprotein receptor involves basic residues in the inhibitor. Biochem. J. 1998;329:55–63. doi: 10.1042/bj3290055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeldal S, Larsen JV, Pedersen KE, Petersen HH, Egelund R, Christensen A, Jensen JK, Gliemann J, Andreasen PA. Binding areas of urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex for endocytosis receptors of the low-density lipoprotein receptor family, determined by site-directed mutagenesis. FEBS J. 2006;273:5143–5159. doi: 10.1111/j.1742-4658.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- Stefansson S, Kounnas MZ, Henkin J, Mallampalli RK, Chappell DA, Strickland DK, Argraves WS. gp330 on type II pneumocytes mediates endocytosis leading to degradation of pro-urokinase, plasminogen activator inhibitor- 1 and urokinase-plasminogen activator inhibitor-1 complex. J. Cell Sci. 1995;108:2361–2368. doi: 10.1242/jcs.108.6.2361. [DOI] [PubMed] [Google Scholar]
- Stefansson S, Muhammad S, Cheng XF, Battey FD, Strickland DK, Lawrence DA. Plasminogen activator inhibitor-1 contains a cryptic high affinity binding site for the low density lipoprotein receptor-related protein. J. Biol. Chem. 1998;273:6358–6366. doi: 10.1074/jbc.273.11.6358. [DOI] [PubMed] [Google Scholar]
- Storm D, Herz J, Trinder P, Loos M. C1 inhibitor-C1s complexes are internalized and degraded by the low density lipoprotein receptor-related protein. J. Biol. Chem. 1997;272:31043–31050. doi: 10.1074/jbc.272.49.31043. [DOI] [PubMed] [Google Scholar]
- Strickland DK, Ashcom JD, Williams S, Burgess WH, Migliorini M, Argraves WS. Sequence identity between the α2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J. Biol. Chem. 1990;265:17401–17404. [PubMed] [Google Scholar]
- Williams SE, Ashcom JD, Argraves WS, Strickland DK. A novel mechanism for controlling the activity of α2-macroglobulin receptor/low density lipoprotein receptor-related protein. Multiple regulatory sites for 39-kDa receptor-associated protein. J. Biol. Chem. 1992;267:9035–9040. [PubMed] [Google Scholar]
- Willnow TE, Orth K, Herz J. Molecular dissection of ligand binding sites on the low density lipoprotein receptor-related protein. J. Biol. Chem. 1994;269:15827–15832. [PubMed] [Google Scholar]