Abstract
To delineate temporal changes in the integrity and function of mitochondria/cardiomyocytes in hearts from mice exposed in utero to commonly used nucleoside analogs (NRTIs), CD-1 mice were exposed in utero to 80 mg AZT/kg, 40 mg 3TC/kg, 80 mg AZT/kg plus 40 mg 3TC/kg, or vehicle alone during days 12–18 of gestation and hearts from female mouse offspring were examined at 13 and 26 weeks postpartum. Alterations in cardiac mitochondrial DNA (mtDNA) content, oxidative phosphorylation (OXPHOS) enzyme activities, mtDNA mutations, and echocardiography of NRTI-exposed mice were assessed and compared with findings in vehicle-exposed control mice. A hybrid capture-chemiluminescence assay showed significant twofold increases in mtDNA levels in hearts from AZT- and AZT/3TC-exposed mice at 13 and 26 weeks postpartum, consistent with near doubling in mitochondrial numbers over time compared with vehicle-exposed mice. Echocardiographic measurements at 13 and 26 weeks postpartum indicated progressive thinning of the left ventricular posterior wall in NRTI-exposed mice, relative to controls, with differences becoming statistically significant by 26 weeks. Overall, progressive functional changes occurred in mouse mitochondria and cardiac tissue several months after in utero NRTI exposures; AZT and 3TC acted in concert to cause additive cardiotoxic effects of AZT/3TC compared with either drug alone.
Keywords: AZT, 3TC, Cardiotoxicity, Echocardiography Mitochondrial, Mitochondrial DNA content, Mitochondrial DNA mutation, Mitochondrial dysfunction, Mitochondrial toxicity, OXPHOS, Transplacental exposure
Introduction
Perinatal HIV-1 infection is a cause of HIV-1-associated child morbidity and mortality, with vertical transmission being the most common route of acquisition [1]. With nearly 50% of HIV-1-infected adults being women of childbearing age and 2 million HIV-1 infected women world-wide giving birth each year, thousands of children are at risk of becoming infected with HIV-1 [2]. Since the implementation of the Center for Disease Control and Prevention (CDC) guidelines in 1994, antiretroviral drug combinations based upon nucleoside reverse transcriptase inhibitors (NRTIs) such as zidovudine (3'-azido-2',3'-dideoxythymidine, AZT) have been successful in reducing mother-to-child transmission of HIV-1 from ~25 to <2% [3–5].
NRTIs are the backbone of highly active antiretroviral therapy (HAART), or combination drug therapies designed to inhibit viral replication in HIV-1-infected patients and to prevent vertical transmission of the virus in well-resourced settings [3, 4]. In HAART, one or more NRTIs such as AZT and lamivudine (2',3'-dideoxy-3'-thiacytidine, 3TC) have been given with either a protease inhibitor or a nonnucleoside reverse transcriptase inhibitor [4, 5]. Research conducted to determine the safety of NRTIs for the treatment of adult HIV-1 infection and for the prevention of mother-to-child transmission of the virus indicates that these compounds also may act as chemical mutagens and mitochondrial toxins [6–16].
AZT incorporation into fetal DNA has been demonstrated following transplacental exposure of mice, patas monkeys, rhesus monkeys, and newborn humans [9, 17–19]. Furthermore, incorporation of NRTIs into mitochondrial DNA (mtDNA) can lead to chain termination during mitochondrial DNA replication, changes in mtDNA levels, and mutations that can potentially persist and become pathogenic [17, 19, 20]. mtDNA depletion has been observed in cardiac and skeletal muscle of monkeys and leukocytes of human infants exposed in utero to AZT-based therapies, suggestive of the DNA chain termination properties of NRTIs [21–25].
Mutations in mtDNA caused by NRTIs can have a significant impact on the maintenance of normal mitochondrial enzymatic function [13]. For example, mtDNA encodes 13 polypeptides for oxidative phosphorylation (OXPHOS), with Complex I containing 7 mtDNA encoded subunits [26]. A mutation or lack of transcription of any of these components can affect the OXPHOS capability of the mitochondrion and its energy generating capacity, resulting in an increase in superoxide formation and further oxidative stress to mitochondria [11–13]. Small-scale mtDNA mutations have been found in HIV-1-infected adults given HAART [27–29] as well as in uninfected human infants and mouse pups given AZT or AZT/3TC in utero [20, 30].
Compared with the sizeable body of evidence showing a relationship between antiretroviral therapy, mitochondrial abnormalities, and cardiomyopathy in HIV-infected adults [11, 12, 14–16], few studies have focused on a potential link between perinatal or childhood exposure to NRTIs and mitochondrial dysfunction/cardiac disease in children [3–8, 22–25]. In HIV-1-infected adults given long-term NRTI-based therapy, mitochondrial dysfunction is considered a key component of the development of AIDS- or NRTI-related cardiomyopathy [11–15]. Although the precise role that viral infection and antiretrovirals play in the pathogenesis of cardiac disease in HIV-1-infected patients is unclear, symptoms related to mitochondrial damage often resolve once NRTI treatment is discontinued [12–15]. The degree to which NRTIs can induce mitochondrial damage and subsequently pose a risk for developing cardiac abnormalities in uninfected children who are exposed perinatally is still debated [3, 4, 7, 8, 16, 31, 32]. At the same time, experiments in mice and monkeys have shown that transplacental exposure to human-equivalent doses of AZT and other NRTIs, in the absence of HIV infection, results in persistent cardiac ultrastructural damage, dys-function, and genotoxicity [9, 17–21, 33–35]. Walker et al. [20] have hypothesized that a significant dose-related cardiac enlargement, found during the course of a 2-year transplacental carcinogenicity bioassay in mice, resulted from mitochondrial damage that persisted and led over time to delayed, progressive cardiotoxicity.
In the current work, the temporal relationships between transplacental NRTI exposures and mitochondrial and cardiac dysfunction were evaluated in hearts from female mice exposed in utero to AZT, 3TC, or a combination of AZT/3TC. The occurrence and persistence of cardiac changes were assessed by measuring alterations in mtDNA content, mtDNA mutations, OXPHOS enzyme activities, and echocardiography in NRTI-exposed and vehicle-exposed mice at 13 and 26 weeks postpartum. A companion report considers several limitations of the current study design, including the use of only female mice, and examines the relationships between in utero NRTI exposures and structural damage in mitochondria and cardiac tissue via light microscopic and ultrastructural studies of hearts from the same mouse offspring [35].
Materials and Methods
Animals, Exposures, and Tissue Disposition
Date-mated female CD-1 mice were treated with AZT or 3TC (obtained from Byron Chemical, Long Island City, NY) as described in a companion report [35]. Briefly, on gestation day 12, pregnant dams were randomly assigned to one of four groups (n = 3–7 pregnant mice/dose group) for treatments using vehicle (sterile PBS), 80 mg AZT/kg bw, 40 mg 3TC / kg bw, or 80 mg AZT/kg plus 40 mg 3TC/kg bw during gestation days 12 through 18. The pregnant dams were weighed and lightly anesthetized with isofluorane and O2 at 1 l/min, and then a NRTI(s) or vehicle was given once daily by gavage. Animals were housed and monitored under standard conditions [35]. All procedures using animals were approved by the Institutional Animal Care and Use Committee of the Lovelace Respiratory Research Institute.
Our previous studies demonstrated that female mouse offspring had greater sensitivity than males to mitochondrial damage and subsequent cardiac abnormalities occurring as a result of perinatal exposure to AZT and/or 3TC [20, 34]. Therefore, in this study, only female mice were randomly selected for euthanasia (by CO2 asphyxiation), necropsy, and collection of tissues at 13 and 26 weeks postpartum. At necropsy, hearts and other tissues were collected for evaluating mitochondrial integrity. Hearts were expunged of blood, weighed, sectioned, and stored or placed into appropriate fixatives for performing various assays (see Fig. 1). Data from microscopic studies of heart sections of the same mice are detailed in a companion report [35]. At each time point, hearts from four mice per treatment group were processed for isolation of mitochondria for measurement of OXPHOS enzyme activity, as described in detail below.
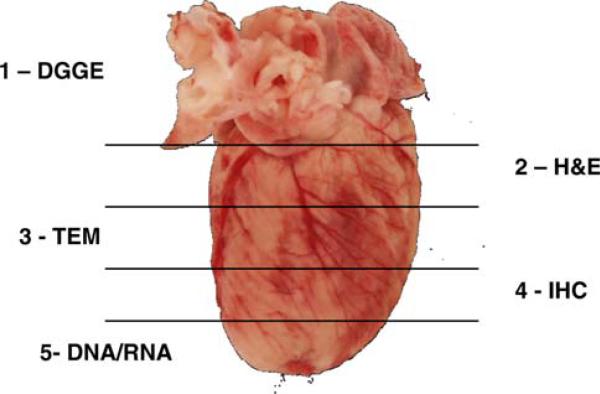
Fig. 1.
Diagram representing trimming of hearts from CD-1 female mice exposed transplacentally to AZT, 3TC, or AZT/3TC during the last 7 days of gestation. Each number designates the assay performed on the corresponding section of cardiac tissue. DGGE denaturing gradient gel electrophoresis, H&E hemotoxylin and eosin stain (as well as special stains), TEM transmission electron microscopy, IHC immunohistochemistry, DNA/RNA section preserved for DNA and/or RNA isolation for future studies
MtDNA Quantification by Hybrid Capture-Chemiluminescence Immunoassay (HC-CIA)
A HC-CIA technique was used to measure the amount of mtDNA in total genomic DNA isolated from minced hearts as an indicator of mtDNA integrity. At 13 weeks, hearts from 7 vehicle-exposed mice, 8 AZT-exposed mice, 4 3TC-exposed mice, and 8 AZT/3TC-exposed mice were examined; at 26 weeks, 10 vehicle-exposed, 8 AZT-exposed, 4 3TC-exposed, and 12 AZT/3TC-exposed hearts from mice were assessed. mtDNA levels were measured in cardiac tissue of mice from 5 of 6 vehicle-exposed litters, 6 of 6 AZT-exposed litters, 3 of 3 3TC-exposed litters, and 5 of 5 AZT/ 3TC-exposed litters. Sample DNA (0.2 lg in TE buffer, pH 7.4) was denatured at 95°C for 5 min, hybridized to 0.67 lM mouse ND-4 (NADH dehydrogenase-subunit 4) oligo (5'-amino ACCAGCCTAACACTTCTAGACAAACCG) in Expand High Fidelity PCR buffer without MgCl2 (Roche, Indianapolis, IN) in a total volume of 35 μ1 at 60°C for 30 min, and quickly cooled to 4°C. The hybridized sample was mixed with 2.0 μ1 of Biotin-ULS dye and 2.0 μ1 labeling solution (Kreatech Biotechnology, Amsterdam, Netherlands), made up to 40 μ1 with deionized water and incubated at 85°C for 30 min. Biotin-ULS-hybridized DNA, hereafter referred to as biotinylated-DNA, was briefly centrifuged at 3,000 rpm, and unreacted excess ULS dye was removed by filtration using YM-100 filters (Millipore Corp, Bedford, MA). The concentration of the biotinylated-DNA was measured using NanoDrop (Thermo Scientific, Waltham, MA) and diluted to a concentration of 20 ng/ml in 0.1M KH2PO4 buffer (pH 7.4) containing 1mM EDTA. A 100 μ1 aliquot of the diluted biotinylated-DNA was captured onto a white DNA-Bind (N-oxysuccinamide) 96-well plate in four replicate wells (Corning, Acton, MA), and incubated at 37°C for 1 h. A 500 bp amplicon of the human cystic fibrosis gene, prepared by PCR and modified with Biotin-ULS, was hybridized to a set of wells to serve as a non-specific blank. The wells were washed 3 times with wash solution (2× saline sodium citrate and 0.1% sodium dodecyl sulfate) and one time with TBST (10 mM TRIS, pH 8.0, 150 mM NaCl, 0.05% Tween 20). The plate was then incubated with 0.25% I-block (Casein, PE Applied Biosystems, Foster City, CA) in PBS for 1 h at 37°C and washed three times with TBST. An aliquot (100 μ1) of avidin-alkaline phosphatase (Applied Biosystems) diluted 1:2,000 in I-block buffer was added to the wells and incubated for 1 h at RT and rinsed three times with TBST. After incubation with AP substrate (CDP-Star with Emerald II, Applied Biosystems) for 30 min at RT, luminescence was measured using a TR 717 Luminometer (Applied Biosystems). The values for mtDNA quantity are expressed as luminescence units per ng total DNA.
OXPHOS Enzyme Activity Assays
OXPHOS enzyme assays were used to determine functional capacity of Complexes I and IV in cardiac mitochondria from mice exposed in utero to NRTIs compared to vehicle alone. At each time point, four mice from each treatment group (from 4 of 6 vehicle litters, 4 of 6 AZT litters, 3 of 3 3TC litters, and 4 of 5 AZT/3TC litters) were euthanized, and their hearts were processed for isolation of mitochondria for measurement of OXPHOS enzymatic activities. Mitochondrial isolation and quantification of specific activities of OXPHOS Complexes I and IV were performed on fresh tissue as previously described [36]. Briefly, hearts were individually minced using a scalpel blade and then homogenized in 1–3 ml of isolation buffer [210 mM mannitol, 70 mM sucrose, 1 mM EDTA, 20 mM HEPES (pH 7.4), 2 mM dithiothreitol, and 1 mM leupeptin] using a Glas-Col homogenizer (Terre Haute, IN) for 75 passes. Homogenized tissue was then centrifuged at 4°C at 1,000×g for 10 min to remove cellular debris and nuclei. The supernatant was transferred to a fresh tube and centrifuged at 4°C at 10,000×g for 20 min to collect the mitochondrial pellet. A small amount of cytosol remained on the mitochondrial pellet, and the pellet was snap frozen in liquid nitrogen and stored at -80°C until analyses. The Coomassie Brilliant Blue method, with bovine serum albumin as a standard, was used for quantifying total mitochondrial protein. Specific activities of Complexes I and IV were quantified using a Hewlett Packard diode array spectrophotometer [25]. Complex I NADH-ubiquinone oxidoreductase rotenone sensitive activity was measured for 1 min at a wavelength of 340 nm minus 425 nm by following the decrease in absorbance from the oxidation of NADH. For Complex IV, ferrocytochrome-c oxygen oxidoreductase activity was measured by following the oxidation of reduced cytochrome c for 30 s at a wavelength of 550 nm minus 540 nm.
PCR-Based Denaturing Gradient Gel Electrophoresis Analysis for mtDNA Mutations
A PCR-based vertical denaturing gradient gel electrophoresis (DGGE) technique was adapted to screen for sequence variants in the 22 tRNA genes and flanking regions of the mouse mitochondrial genome [20]. Sections of heart taken at necropsy for DGGE analysis were snap frozen in liquid nitrogen and stored at -80°C until processing. Total genomic DNA was isolated from minced heart sections of mice from 6 of 6 vehicle litters, 5 of 6 AZT litters, 3 of 3 3TC litters, and 5 of 5 AZT/3TC litters using the Wizard Genomic DNA purification Kit (Promega, Madison, WI). PCR was conducted using previously designed primers that amplify segments encoding the 22 tRNA genes and flanking regions [20]. The PCRs were performed in a 50 μ1 total volume that contained 1× reaction buffer (without MgCl2), 1.0 to 2.2 mM MgCl2, 0.2 mM of each deoxynucleotide triphosphate, 0.2 to 0.3 μM of each primer, 0.5 U GoTaq Flexi (Promega), and 100 ng of total DNA. A Perkin Elmer 9600 thermal cycler was used to perform PCR as follows: 94°C for 2 min, followed by 32–40 cycles of 94°C for 15 s, 59–61°C for 30 s, and 68°C for 1 min. A final extension at 72°C for 5 min completed the PCR. Once amplification was complete, the samples were heat-denatured at 99°C for 10 min, and then allowed to cool at the rate of 12°C per min to 37°C and held at 37°C for 30 min. Following reannealing, a psoralen–DNA crosslink was formed at one end of the individual PCR fragments by irradiating the samples on ice for 25 min using a 366-nm UV source (Ultra Violet Products Inc., San Gabriel, CA). An aliquot of the PCR products were run on an 8% polyacrylamide gel to verify PCR efficiency. Samples were then concentrated and resus-pended in 5 μ1 6× loading dye for loading onto vertical denaturing gels.
A DGGE-4001 system (C.B.S. Scientific Company, Solana Beach, CA) was used to screen PCR products for formation of mutant/wild-type heteroduplexes and mutant homoduplexes. Optimized conditions used for gradient, voltage, and run time for final analysis of biological samples are reported here (Table 1). However, multiple experiments were necessary to obtain separation of mutant/ wild-type DNA heteroduplexes and homoduplexes. The resuspended PCR products were loaded onto a 1-mm thick, 8% polyacrylamide gel with varied linear denaturant gradients depending on the primer set used (Table 1). Denaturing gradient gels were submerged into the DGGE tank, with 1× TBE buffer that continuously circulated between the upper reservoir of the gel cassette and the buffer tank. For most of the primer sets, electrophoresis was conducted at 130 V and a constant temperature of 58°C for 17 h (Table 1, footnote c). The gels were stained for 15 min in ethidium bromide (1 μM/ml), destained for 10 min, and PCR products were visualized with a Spectroline UV transilluminator (Spectronics Corporation, Westbury, NY), and photographed using a Polaroid 667 with black and white film. Any band that contained presumptive resolved and unresolved mutant/wild-type heteroduplexes or mutant homoduplexes was excised carefully from the gel, crushed, and eluted overnight at 4°C in 200 μ1 of sterile H2O. A second round of PCR was performed on DNA obtained from eluted bands to purify the PCR product following the same conditions as described earlier. Samples were then run on a secondary DGGE gel using the same gradient appropriate for the primer set to confirm the identification of the heteroduplexes or mutant homoduplexes representing presumptive sequence variants. The resulting bands were excised carefully from the gel, crushed, and eluted overnight as described earlier to serve as template for an additional round of PCR, and the amplicon was used for determining the DNA sequencing. For DNA sequencing, the PCR amplicons were purified from salts, primers, and unincorporated dNTPs using the Millipore Montage PCR centrifugal filter devices (Bedford, MA). Millipore Micropure-EZ centrifugal filter devices were used in a second round of purification to remove polymerases. The purified PCR products were then sequenced using the Applied Biosystems 3,730×1 DNA Analyzer.
Table 1.
PCR and electrophoresis conditions for amplification and vertical DGGE analysis of the 22 mouse tRNA genes and flanking regions
| Target gene(s) and flanking region | Forward and reverse primersa | PCR conditions |
Gradient (%)b | ||
|---|---|---|---|---|---|
| MgCl2 (mM) | Annealing temperature (°C) | PCR cycles | |||
| tRNA 1 | MtRNA 1F/1R-PS | 2.0 | 59 | 32 | 28–54 |
| tRNA 2 | MtRNA 2F-PS/2R | 2.2 | 61 | 37 | 36–72 |
| tRNA 3 | MtRNA 3F/3R-PS | 1.0 | 63 | 37 | 28–54 |
| tRNA 4/5 | MtRNA 4F/4R-PS | 2.0 | 59 | 32 | 28–54 |
| tRNA 5/6 | MtRNA 5F/5R-PSc | 2.2 | 61 | 37 | 28–54 |
| tRNA 7/8/9 | MtRNA 6F-PS/6R | 1.0 | 61 | 40 | 25–54 |
| tRNA 10/11 | MtRNA 7F-PS/7R | 2.0 | 61 | 32 | 18–72 |
| tRNA 12/13 | MtRNA 8F-PS/8R | 2.2 | 61 | 32 | 25–54 |
| tRNA 14 | MtRNA 9F-PS/9R | 2.0 | 61 | 40 | 25–54 |
| tRNA 15 | MtRNA 10F/10R-PS | 2.0 | 61 | 32 | 18–72 |
| tRNA 16 | MtRNA 11F/11R-PSd | 2.2 | 61 | 40 | 25–54 |
| tRNA 17/18/19 | MtRNA 12F-PS/12R | 2.0 | 61 | 32 | 18–72 |
| tRNA 20 | MtRNA 13F/13R-PS | 1.0 | 61 | 32 | 25–54 |
| tRNA 21/22 | MtRNA 14F-PS/14R | 1.0 | 61 | 37 | 18–72 |
The oligonucleotide denoted with PS has a psoralen derivative linked to the 5′ end
Hundred percent is defined as 7 M urea plus 40% v/v formamide
Electrophoresis was conducted at 100 V for 17 h for primer set 5, and all others were run at 130 V for 17 h
PCR was carried out with 0.3 μM of each primer for primer set 11, and all other primers were amplified with 0.2 μM of each primer
As part of this work, mtDNA from a control mouse with no evidence of unique DGGE banding patterns was amplified with primer sets previously demonstrated to result in presumptive mutations in other samples. As recommended by Chan et al. [34], these PCR products were sequenced and used as reference sequences to identify sequence variations unique to this strain of mouse when compared to the published sequence data for Mus musculus domesticus.
Transthoracic Echocardiographic Measurements
The decision to obtain echocardiograms at the two selected time points in the current study was based on previous findings of echocardiographic alterations in 7-week-old B6C3F1 mice exposed in utero to AZT/3TC and the authors’ conclusion that older mice might provide more reliable data [20]. Therefore, 13- or 26-week-old female mice were lightly anesthetized with isofluorane and O2 at 1 l/min and an Acuson Seqouia 512 with a 15-MHz linear array transducer was used to record a transthoracic two-dimensional M-mode echocardiogram. Eight mice from each exposure group were examined at 13 weeks of age; 11 vehicle-exposed, 7 AZT-exposed, 8 3TC-exposed, and 11 AZT/3TC-exposed mice were examined at 26 week of age. The mice examined by echocardiography were from 5 of 6 vehicle-exposed litters, 5 of 6 AZT-exposed litters, 3 of 3 3TC-exposed litters, and 5 of 5 AZT/3TC-exposed litters. Software from the ultrasound instrument was used to obtain measurements of the thickness of the left ventricular posterior wall (LVPW), pulse wave measurements, and the internal diameter of the left ventricle in end-diastole (LVED) and end-systole (LVES), which was then used to calculate the percentage of fractional shortening using the formula [(LVED - LVES)/(LVED)] × 100.
Three separate measurements of each parameter (thickness of the LVPW, pulse wave measurements, and the internal diameter of the left ventricle in end-diastole and end-systole) were taken for each animal and averaged; averages for each mouse then were combined with those for animals in the same treatment group and time point to give an average for each measured end point in a given treatment group at 13 and 26 weeks of age.
Statistical Analyses
Two-way ANOVA was performed to determine whether differences existed in mtDNA levels, OXPHOS enzymatic activities, and echocardiographic measurements of NRTI-treated mice versus unexposed mice, or AZT/3TC-exposed versus AZT- or 3TC-exposed mice, at both time points. For assessing results of measurements of OXPHOS enzymatic activities, a balanced two-way ANOVA power and sample size analysis was performed using the SAS 9.2 GLM-POWER procedure (Copyright© 2002–2008 by SAS Institute Inc, Cary, NC). Multiple comparison analyses were performed when a difference was found between the groups using the Holm-Sidak method. A repeated measures one-way ANOVA was performed to determine whether differences existed in repeated echocardiographic measurements between NRTI-treated mice versus unexposed mice and AZT/3TC-exposed versus AZT- or 3TC-exposed mice. The null hypothesis for each test states that there is no difference between any of the treatment groups or time points. A P-value ≤ 0.05 was considered significant.
Since echocardiographic measurements were taken from mice at two time points, a repeated measures one-way ANOVA was utilized to identify possible differences within the exposure groups in relation to aging of the mice. Measurements at 26 weeks postpartum were subtracted from 13-week measurements in mice in which two measurements were obtained; the calculated values were then averaged across a group to give an average ± standard error for each group.
Results
Heart mtDNA Quantification
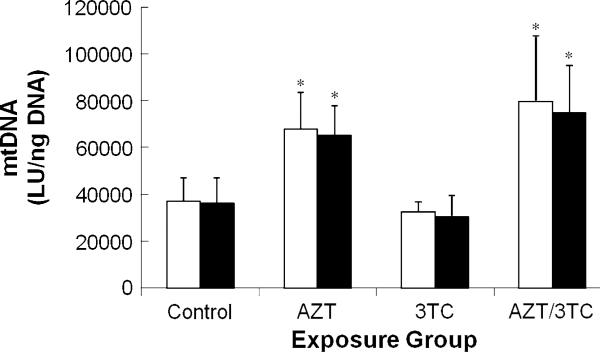
Table 2 summarizes the mtDNA levels found in hearts at 13 and 26 weeks after transplacental exposure of mice to AZT, 3TC, or AZT/3TC. At 13 weeks of age, significant twofold increases in mtDNA levels were seen in hearts from AZT- and AZT/3TC-exposed mice relative to both vehicle-exposed mice (P < 0.001) and 3TC-exposed mice (P ≤ 0.003) (Fig. 2). Mean mtDNA levels in 3TC-exposed mice at 13 weeks were lower than those of vehicle-exposed mice, but the difference was not significant. In each exposure group, mtDNA levels were similar at 13 and 26 week of age so that no significant age-related differences were observed.
Table 2.
mtDNA levels in heart tissue from 13- and 26-week-old female CD-1 mice exposed transplacentally to AZT, 3TC, or AZT/3TCa
| Treatment group | Group sizes at 13 weeks (and 26 weeks)b | Mean mtDNA level ± SE at 13 weeks | Mean mtDNA level ± SE at 26 weeks | Comparisons of values at 13 versus 26 weeks of age (P-value) |
|---|---|---|---|---|
| Control | n = 7 (10) | 37,088 ± 3,720 | 36,084 ± 3,497 | P > 0.05 |
| 3TC | n = 4 (4) | 32,511 ± 2,167 | 30,448 ± 4,485 | P > 0.05 |
| AZT | n = 8 (8) | 67,916 ± 5,477 | 65,056 ± 4,505 | P > 0.05 |
| AZT/3TC | n = 8 (12) | 79,465 ± 9,983 | 74,895 ± 5,768 | P > 0.05 |
| Comparisons of NRTI treatment groups versus controls (P-values) | 3TC, P > 0.05 | 3TC, P > 0.05 | ||
| AZT, P < 0.001* | AZT, P < 0.001* | |||
| AZT/3TC, P < 0.001* | AZT/3TC, P < 0.001* | |||
| Comparisons of individual NRTI treatment groups (P-values) | 3TC vs. AZT, P = 0.003* | 3TC vs. AZT, P = 0.004* | ||
| 3TC vs. AZT/3TC, P < 0.001* | 3TC vs. AZT/3TC, P < 0.001* | |||
| AZT vs. AZT/3TC, P > 0.05 | AZT vs. AZT/3TC, P > 0.05 |
Values represent the average across all animals measured at specified time points expressed as luminescence units per ng total DNA ± SE; statistical analysis via two-way ANOVA with a Holm-Sidak post hoc test
Twenty-six-week group sizes are given in parentheses
Significant P-values
Fig. 2.
mtDNA levels (LU/ng DNA ± SE) in heart tissue from 13 week (white bars) and 26 week (black bars) female CD-1 mice exposed transplacentally to vehicle, AZT, 3TC, or AZT/3TC. Asterisk indicates average values that are significantly greater than the average control value and significantly greater than the average 3TC value within each time point. (13 weeks: control vs. AZT, P < 0.001; control vs. AZT/3TC P < 0.001, AZT vs. 3TC P = 0.003, AZT/3TC vs. 3TC P < 0.001; 26 weeks: control vs. AZT P < 0.001, control vs. AZT/3TC P < 0.001, AZT vs. 3TC P = 0.004, AZT/3TC vs. 3TC P < 0.001)
Cardiac OXPHOS Enzyme Specific Activities
Table 3 summarizes the specific activities of OXPHOS Complexes I and IV measured in hearts at 13 and 26 weeks after transplacental exposure of mice to AZT, 3TC, or AZT/3TC. Statistical analyses of the cardiac OXPHOS enzyme activities of NRTI-exposed mice relative to vehicle-exposed mice revealed no significant differences. Significant differences were also not found when comparisons were made among the NRTI-exposed groups and between the time points. However, there were several trends including non-significant increases in Complex I activity at 13 weeks in 3TC-exposed mice and at 26 weeks in AZT/3TC-exposed mice, as well as non-significant increases in Complex IV activity at both 13 and 26 weeks in 3TC- and AZT/3TC-exposed mice.
Table 3.
OXPHOS Complex I and IV enzyme activities in mitochondria from hearts of 13- and 26-week-old female CD-1 mice exposed transplacentally to AZT, 3TC, or AZT-3TC
| Treatment group | Group sizes at 13 weeks (and 26 weeks)a | Complex activity ± SE at 13 weeks | Complex activity ± SE at 26 weeks | Comparisons of values at 13 versus 26 weeks of age (P-value) |
|---|---|---|---|---|
| Complex I enzyme activity b | ||||
| Control | n = 4 (3) | 49.3 ± 9.6 | 32.9 ± 19.1 | P > 0.05 |
| 3TC | n = 4 (4) | 67.0 ± 19.4 | 41.9 ± 4.0 | P > 0.05 |
| AZT | n = 4 (4) | 36.5 ± 12.5 | 29.5 ± 9.6 | P > 0.05 |
| AZT/3TC | n = 4 (4) | 41.1 ± 14.9 | 115.2 ± 44.9 | P > 0.05 |
| Comparisons of NRTI treatment groups versus controls (P-values) | P > 0.05 for all NRTI group versus control comparisons | P > 0.05 for all NRTI group versus control comparisons | ||
| Comparisons of individual NRTI treatment groups (P-values) | P > 0.05 for all NRTI group comparisons | P > 0.05 for all NRTI group comparisons | ||
| Complex IV enzyme activity b | ||||
| Control | n = 4 (3) | 109.5 ± 8.9 | 64.7 ± 25.0 | P > 0.05 |
| 3TC | n = 4 (4) | 146.0 ± 11.3 | 128.9 ± 15.3 | P > 0.05 |
| AZT | n = 4 (4) | 104.6 ± 29.7 | 98.9 ± 34.1 | P > 0.05 |
| AZT/3TC | n = 4 (4) | 139.8 ± 42.7 | 156.0 ± 38.4 | P > 0.05 |
| Comparisons of NRTI treatment groups versus controls (P-values) | P > 0.05 for all NRTI group versus control comparisons | P > 0.05 for all NRTI group versus control comparisons | ||
| Comparisons of individual NRTI treatment groups (P-values) | P > 0.05 for all NRTI group comparisons | P > 0.05 for all NRTI group comparisons |
Twenty-six-week groups sizes are given in parentheses
Enzyme activity values (μmol/min/mg protein ± SE) represent averages across the number of animals indicated ± standard error. Statistically significant differences between exposure groups and time points were not found using two-way ANOVA
DGGE Analysis of Cardiac mtDNA
Among DNA samples from 61 mice across all groups, 9 sequence variants occurring in PCR amplicons for primer pairs 4 and 11 were detected as mutant homoduplexes on denaturing gradient gels and were confirmed by DNA sequencing. Confirmed sequence variants were found in 3 of 19 control mice, in 2 of 15 AZT-exposed mice, and 3 of 19 AZT/3TC-exposed mice (Table 4), with no significant differences in the spectrum of mutations observed among the groups. Among the three distinct sequence variants identified, one was a -T deletion at nucleotide 3,815 that was detected in one AZT- and one AZT/3TC-exposed mouse, but not in control mice. The other two were either a deletion or an insertion of a thymine base at nucleotide 9,821 and were found in both vehicle-exposed and AZT- or AZT/3TC-exposed mice. The wild-type sequences in the run of thymines beginning at base 9,821 in DNA samples from CD-1 mice in the current study proved to include 10 thymines as opposed to 9 reported for Mus musculus domesticus. In all cases, the +T insertion at base 9,821 was identified as a mutant homoduplex that did not migrate as far on primary and secondary DGGE gels as wild-type homoduplexes in samples without mutant sequences. In contrast, -T deletions at base 9,821 were identified as a mutant homoduplexes that migrated further than wild-type homoduplexes. For each of these single-base insertions or deletions, mutant homoduplexes were not accompanied by mutant/wild-type heteroduplexes or wild-type homoduplexes suggesting that all the sequences verified by DNA sequencing represent mouse polymorphisms.
Table 4.
Sequence variants identified by PCR-based DGGE and DNA sequence analysis of mtDNA from hearts of control and NRTI-exposed female CD-1 mice
| Exposure group, mouse numbera | Age (weeks) | Primer pair number | Location (base #) | Sequence variant |
|---|---|---|---|---|
| Unexposed controls (n = 3/19, 16%) | ||||
| D108 | 13 | 11 | 9,821 | –T base deletion |
| D126 | 26 | 11 | 9,821 | +T base insertion |
| D130 | 26 | 11 | 9,821 | +T base insertion |
| AZT-exposedb (n = 2/15, 13%) | ||||
| E002 | 26 | 11 | 9,821 | +T base insertion |
| E023 | 26 | 4 | (1) 3,815 | –T base deletion |
| 11 | (2) 9,821 | +T base insertion | ||
| AZT/3TC-exposedb (n = 3/19, 16%) | ||||
| F104 | 13 | 11 | 9,821 | –T base deletion |
| F105 | 26 | 11 | 9,821 | –T base deletion |
| F110 | 26 | 4 | 3,815 | –T base deletion |
Number of litters examined: controls = 6 of 6 l, AZT = 5 of 6 l, 3TC = 3 of 3 l, AZT/3TC = 5of5 l
All exposures occurred during days 12–18 of gestation, and dosing was based on total body weight of the pregnant dam: 80 mg AZT/kg bw, 40 mg 3TC/kg bw, or 80 mg AZT + 40 mg 3TC/kg bw
DNA sequencing was unable to confirm four additional presumptive sequence variants identified on denaturing gradient gels via formation of mutant/wild-type heteroduplexes; heteroduplexes were observed in a PCR amplicon for primer pair 11 in one AZT-exposed mouse, and heteroduplexes with the same migration patterns were found in PCR amplicons for primer pair 10 in three AZT/3TC-exposed mice. No heteroduplexes were detected in control mice.
Transthoracic Echocardiography
Two-way ANOVA analyses indicated no significant differences in fractional shortening or pulse wave measurements in NRTI treatment and control groups at 13 or 26 weeks of age; however, mice in all three NRTI exposure groups had a decrease in LVPW measurements at 13 weeks that became significantly different from the average control value in mice at 26 week of age (P ≤ 0.003) (Table 5). At 26 week of age, AZT/3TC coexposure resulted in LVPW measurements that were significantly less than those in the AZT-alone exposure group (P = 0.05) but not the 3TC-alone group. A significant increase in LVPW measurements also was found in vehicle-exposed mice as they aged from 13 to 26 weeks (P = 0.002). The failure to find a comparable significant increase in the thickness of the LVPW in all treated mouse groups from 13 to 26 weeks of age indicates a persistent effect of transplacental NRTIs on heart growth over time post exposure.
Table 5.
Summary of echocardiographic measurements from 13- and 26-week-old female CD-1 mice exposed transplacentally to AZT, 3TC, or AZT-3TCa,b
| Treatment group | Group sizes at 13 weeks and (26 weeks)b | Measurement ± SE at 13 weeks | Measurement ± SE at 26 weeks |
|---|---|---|---|
| F.S. %c | |||
| Control | n = 8 (11) | 40.3 ± 1.6 | 41.1 ± 1.8 |
| 3TC | n = 8 (8) | 42.6 ± 3.1 | 41.8 ± 2.0 |
| AZT | n = 8 (7) | 42.7 ± 4.1 | 43.1 ± 2.7 |
| AZT/3TC | n = 8 (11) | 37.3 ± 3.8 | 39.0 ± 2.0 |
| Comparisons of NRTI treatment groups versus controls (P-values) | P > 0.05 for all NRTI versus control comparisons | P > 0.05 for all NRTI versus control comparisons | |
| Comparisons of individual NRTI treatment groups (P-values) | P > 0.05 for all NRTI group comparisons | P > 0.05 for all NRTI group comparisons | |
| Pulse wave (m/s) | |||
| Control | n = 8 (11) | 0.71 ± 0.02 | 0.69 ± 0.03 |
| 3TC | n = 8 (8) | 0.78 ± 0.05 | 0.70 ± 0.04 |
| AZT | n = 8 (7) | 0.71 ± 0.10 | 0.77 ± 0.02 |
| AZT/3TC | n = 8 (11) | 0.75 ± 0.03 | 0.65 ± 0.04 |
| Comparisons of NRTI treatment groups versus controls (P-values) | P > 0.05 for all NRTI versus control comparisons | P > 0.05 for all NRTI versus control comparisons | |
| Comparisons of individual NRTI treatment groups (P-values) | P > 0.05 for all NRTI group comparisons | P > 0.05 for all NRTI group comparisons | |
| LVPW (cm)d,e | |||
| Control | n = 8 (11) | 0.090 ± 0.004 | 0.122 ± 0.010f |
| 3TC | n = 8 (8) | 0.081 ± 0.010 | 0.090 ± 0.003 |
| AZT | n = 8 (7) | 0.085 ± 0.005 | 0.093 ± 0.006 |
| AZT/3TC | n = 8 (11) | 0.089 ± 0.004 | 0.075 ± 0.003 |
| Comparisons of NRTI treatment groups versus controls (P-values) | P > 0.05 for all NRTI versus control comparisons | 3TC, P = 0.001* | |
| AZT, P = 0.003* | |||
| AZT/3TC, P < 0.001* | |||
| Comparisons of individual NRTI treatment groups (P-values) | P > 0.05 for all NRTI group comparisons | 3TC vs. AZT, P > 0.05 | |
| 3TC vs. AZT/3TC, P > 0.05 | |||
| AZT vs. AZT/3TC, P = 0.05* |
Values represent the average across all animals measured at specified time points ± SE; statistical analysis via two-way ANOVA with a Holm-Sidak post hoc test
Number of mice examined at 26 weeks in parentheses. Number of litters examined: control = 5 of 6 l, AZT = 5 of 6 l, 3TC = 3 of 3 l, AZT/3TC = 5of5 l
FS = Fractional shortening calculated as [(LVED – LVES)/(LVED)] × 100
For multiple comparisons between data for 13- and 26-week time points combined, mean values were significantly less than the mean control value for AZT (P = 0.006), 3TC (P = 0.001), and AZT/3TC (P < 0.001)
Three significant figures were retained to allow for reporting of standard errors
The mean vehicle-exposed mouse value at 26 weeks were significantly greater than the mean control value at 13 weeks (P = 0.002)
Significant P-values
Using a repeated measures one-way ANOVA, signifi-cant differences were only observed in measurements of the internal diameter of the left ventricle during end-diastole, the thickness of the LVPW, and fractional shortening percentages over time (Table 6). As the mice aged, a significant decrease in the diameter of LVED relative to temporal changes in control mice (0.01 ± 0.01 cm) and AZT- (0.01 ± 0.01 cm) was evident in 3TC- (-0.05 ± 0.02 cm; P-values = 0.003) and AZT/3TC-exposed mice (-0.04 ± 0.01 cm; P-values ≤ 0.029). The average fractional shortening percentage in AZT/3TC-exposed mice was significantly different from vehicle-exposed mice as they aged (P-value < 0.001). Significant differences in measurements of the internal diameter of the left ventricle during systole and pulse wave measurements were not found among the exposure groups relative to vehicle- exposed mice. Using the calculated differences between the 13 and 26 weeks time points, LVPW thickness measurements for the AZT- (0.01 ± 0.01 cm), 3TC- (0.01 ± 0.01 cm), and AZT/3TC-exposed (-0.01 ± 0.01 cm) animals (P-values ≤ 0.008) had averages that were significantly different from those for the unexposed control animals (0.04 ± 0.01). These findings (Table 6) confirmed the results of the initial data analyses (Table 5) when the exposure groups were compared across the two time points.
Table 6.
Summary of changes between repeated echocardiographic measurements at 13 and 26 weeks of age in female CD-1 mice exposed transplacentally to AZT 3TC or AZT-3TCa,b
| Treatment group | Group size | LVED (cm)c | LVES (cm) | FS% | LVPW (cm) | Pulse wave (m/s) |
|---|---|---|---|---|---|---|
| Control | n = 7 | 0.01 ± 0.01 | 0.01 ± 0.01 | –1.02 ± 2.86 | 0.04 ± 0.01 | –0.07 ± 0.02 |
| 3TC | n = 8 | –0.05 ± 0.02 | –0.03 ± 0.02 | –0.81 ± 4.34 | 0.01 ± 0.01 | –0.10 ± 0.07 |
| AZT | n = 7 | 0.00 ± 0.01 | 0.01 ± 0.02 | –0.91 ± 3.02 | 0.01 ± 0.01 | 0.01 ± 0.02 |
| AZT/3TC | n = 6 | –0.04 ± 0.01 | –0.03 ± 0.02 | 2.62 ± 4.54 | –0.01 ± 0.01 | –0.06 ± 0.12 |
| Comparisons of NRTI treatment groups versus controls (P-values) | 3TC, P = 0.003* | P > 0.05 for all treated groups vs. controls | 3TC, P > 0.05 | 3TC, P = 0.002* | P > 0.05 for all treated groups vs. controls | |
| AZT, P > 0.05 | AZT, P = 0.008* | |||||
| AZT, P > 0.05 | AZT/3TC, P < 0.001* | AZT/3TC, P = 0.003* | ||||
| AZT/3TC, P = 0.029* | ||||||
| Comparisons of individual NRTI treatment groups (P-values) | 3TC vs. AZT, P = 0.003* | P > 0.05 between all treated groups | P > 0.05 between all treated groups | P > 0.05 between all treated groups | P > 0.05 between all treated groups | |
| 3TC vs. AZT/3TC, P > 0.05 | ||||||
| AZT vs. AZT/3TC, P = 0.028* |
Differences calculated as the average measurement at 26 weeks minus the average measurement at 13 weeks ± SE, with a positive value indicating an increase in the average value from 13 to 26 weeks, and a negative value indicating a decrease in the average value from 13 to 26 weeks. Repeated measures one-way ANOVA test
Data presented is only from mice in which measurements at both time points were obtained, and significant findings are suggestive of an effect involving treatment and aging
Significant findings in this parameter were only found when aging was taken into account; thus, without chronicity, no effect was seen
Significant P-values
Discussion
The current study was designed to delineate temporal changes in cardiac or mitochondrial integrity and function following in utero exposure of female CD-1 mice to AZT, 3TC, or AZT/3TC. Transplacental exposure of mice resulted in significant increases in mtDNA content for AZT or AZT/3TC (Table 2), non-significant trends toward increases in electron transport chain Complexs I and IV activities for 3TC or AZT/3TC (Table 3), and significant differences in select echocardiographic measurements for AZT, 3TC, or AZT/3TC (Tables 5 and 6). Differences in specific markers in AZT- and 3TC-exposed mice suggest that mechanisms for the induction of cardiac damage by these two NRTIs may be dissimilar, and their combined effects may explain the occurrence and progression of greater alterations in echocardiographic measurements in AZT/3TC-exposed mice. The work reported here also broadened the findings from earlier studies of mitochondrial dysfunction and mutations in mice exposed in utero to AZT and/or 3TC [20, 34] by assessing alterations at interim time points in the life of the mouse.
The twofold increases in mtDNA content without corresponding increases in OXPHOS enzyme activities in hearts of AZT- and AZT/3TC-exposed mice at first appears inconsistent, but may reflect successful adaptation to the cardiotoxic effects of these treatments [35]. These findings are similar to the elevations in mtDNA, with normal OXPHOS activity values, observed in hearts of 1-year-old E. patas monkeys exposed transplacentally to NRTIs compared to concurrent controls [33]. This increase in mtDNA quantity may reflect a compensatory mechanism of the heart to produce more mitochondria over time in the face of damaged incompetent mitochondria in order to meet the energy demands of the tissue. Although the numbers of mitochondria in some cardiomyocytes and the overall levels of mtDNA were significantly elevated in hearts of AZT and AZT/3TC-exposed mice, the presence of mitochondria with substantial morphologic damage (likely accompanied by depletion of mtDNA and enzyme function) in large numbers of cardiomyocytes [35] may have masked significant increases in OXPHOS and Complex IV activities in a subset of cells when the tissue as a whole was examined. Also, to determine whether the small sample size of whole hearts examined in the current study may have contributed to the negative findings for significant alterations in OXPHOS activities in NRTI-treated mice, power and sample size analysis was performed based upon data in Table 3. To achieve a power of 80% in detecting a treatment effect, a difference between the two time points, and the interaction effect between the treatment and time point a total sample size across all treatment groups and time points of 72, 1,224, and 56, respectively, are needed for enzymatic activity measurement in Complex I at the significance level of α = 0.05. Similarly, for Complex IV, a sample size of at least 64 is required to achieve 80% power to detect a treatment effect, 312 to detect a difference between the two time points, and 152 to detect an interaction between the treatment and time point. Interestingly, the sample size requirements in both Complex I and IV necessary to achieve 80% power in detection of a difference between the two time points is considerably larger than the requirements for detecting effects from either treatment alone or the interaction between treatment and time point. This observation suggests that the interval between the time points utilized in this study may not have been long enough to allow for the accumulation of measurable changes.
Notably, the latter postulate is supported by findings in older mice and monkeys in which NRTI-associated increases in cytochrome c oxidase activity were observed following NRTI exposure [9, 19, 20] and have been suggested to arise from mutations in genes associated with electron transfer and/or genes encoding Complex IV subunits, which could lead to reduced enzyme function and a subsequent compensatory feedback mechanism of overproduction [19]. Studies in animal models have shown clearly that mitochondrial damage and enzymatic dys-function induced by NRTIs can lead to neuropathy, hepatic disease, and skeletal and/or cardiac myopathy [12, 14, 20]. Divi and colleagues [37] also exposed human cells long-term to AZT and measured mtDNA levels and OXPHOS gene expression at interim time points; their findings of increased mtDNA levels with decreased OXPHOS gene expression during early passages and mtDNA depletion with increased OXPHOS gene expression during later passages illustrates the progression of drug-induced damage and the range in cellular responses.
Studies of the occurrence of small-scale mtDNA mutations in experimental animals or humans treated with NRTIs are relatively few [20, 27–29, 34]. A study of 18-month-old female mice exposed for the last 7 days of gestation to daily doses of 0 or 480 mg/kg AZT found significant elevations in the frequency of sequence variants in hearts that correlated positively with the proportion of atypical cardiac mitochondria in exposed mice compared with controls [20], suggesting that AZT may induce various mtDNA mutations, some of which may be lethal or persistent and potentially pathogenic [20]. Chan et al. [34] performed the same assay for mtDNA mutations in mice following treatment with human-equivalent doses of AZT, 3TC, or AZT/3TC throughout gestation and up to postnatal day 28, and found that sequence variants were elevated in 10-week-old females treated with AZT or AZT/3TC. In these earlier studies, wild-type reference DNA was added with each PCR amplicon ‘to force’ heteroduplex formation during the melting–annealing reaction prior to analysis by DGGE, allowing for the detection of heteroplasmic sequence variants in cases where mutant homoduplexes do not resolve from wild-type homoduplexes [20, 34]. This step was omitted in current work, complicating comparisons to the earlier mouse studies and perhaps underestimating the number of mtDNA sequence variants occurring at 13 and 26 after in utero exposure to AZT and/or 3TC.
Few point mutations have been reported in HIV-1-infected adults receiving NRTIs [27–29], but DGGE analysis of umbilical cord tissue from newborn infants showed that mtDNA sequence variants were significantly elevated by threefold in HIV-1-uninfected infants receiving prepartum AZT/3TC compared with unexposed controls (P < 0.001), with 24 changes observed in 19/52 (37%) treated newborns versus only 8 changes found in 7/55 (13%) control newborns [30]. Variants in unexposed newborns were predominately synonymous and homo-plasmic, representing reported polymorphisms, while AZT/3TC-exposed infants had a shift in mutations driven by increases in non-synonymous (miscoding) heteroplasmic sequence variants at novel and polymorphic sites. Thus, further research is needed to clarify mutagenic responses across studies involving different NRTI treatment regimens, species, and age groups.
Echocardiographic measurements at 13 and 26 weeks postpartum suggested progressive changes in hearts of NRTI-exposed mice. First, AZT- and 3TC-exposed mice had significantly less growth of the LVPW relative to control mice, while AZT/3TC-exposed mice had even greater loss of wall thickness as the mice aged. In contrast, vehicle-exposed mice appeared to exhibit normal cardiac growth that was shown by an increase in the measurement of the LVPW over time. Whether the NRTI-related thinning of the posterior wall of the left ventricle is a reflection of dilation or an overall loss of cardiomyocytes remains to be determined. Such ventricular changes would be conducive to delayed onset of cardiomyopathy as was seen in female mice that developed a significant dose-related cardiomegaly (not otherwise specified) following in utero AZT exposure [20]. Second, when differences between the repeated measures at 13 and 26 weeks of age were calculated and compared across groups, 3TC- and AZT/3TC-exposed mice exhibited a significant decrease in LVED measurements from 13 to 26 weeks relative to controls, suggesting a drug-induced effect on these parameters with age (Table 6). These assessments show differences that were not apparent when averages for each group were compared across two time points, and illustrate the potential effect that in utero exposure to NRTIs may have on development of cardiac tissue.
The echocardiographic alterations in young mice exposed in utero to NRTIs are consistent with findings from uninfected infants born to HIV-1-infected mothers who received HAART during pregnancy. Initial reports from Lipshultz et al. [38, 39] indicated that perinatal exposure to AZT monotherapy was not significantly associated with cardiac abnormalities in HIV-1-uninfected children. A follow-up study of HAART and cardiovascular status among larger numbers of uninfected children who received peri-natal exposure to combination antiretroviral drug therapies suggested significant occurrence of cardiac dysfunction (abnormal left ventricular shortening and thinning of the ventricular septum) that progressed through at least 2 years after birth [40]. Notably, echocardiographic changes are more pronounced in females of both species, suggesting that unknown gender-related factors affect the outcome.
The findings from this study and a companion report [35] provide further evidence for the induction and progression of functional and structural changes in mitochondria and cardiac tissue months after in utero exposure of an animal model to NRTIs and indicate that AZT and 3TC act in concert to cause additive cardiotoxic effects of AZT/3TC. Since in utero NRTI-induced mitochondrial damage may predispose to cardiovascular symptoms that may develop later in life, the need for long-term follow-up of NRTI-exposed children and the development of avenues to minimize the apparent toxicities of this class of anti-retrovirals is imperative [16].
Acknowledgments
We wish to thank Mr. Steve Randock, Ms. Cynthia Herrera, and Ms. Wendy Piper (LRRI) for technical assistance in preparing figures and the manuscript; Dr. Beth Goens (University of New Mexico) for assistance with the echocardiography. This work was supported, in part, by NIH grant R01 HL 72727 to VEW and 1 F31 HL081928 to SMT from the National Heart, Lung, and Blood Institute and in part by the intramural program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- AZT
Zidovudine or 3'-azido-2',3'-dideoxythymidine
- DGGE
Denaturing gradient gel electrophoresis
- FS
Fractional shortening
- HAART
Highly active antiretroviral therapy
- HC-CIA
Hybrid capture-chemiluminescence immunoassay
- LVED
Left ventricle in end-diastole
- LVES
Left ventricle in end-systole
- LVPW
Left ventricular posterior wall
- mtDNA
Mitochondrial DNA
- NRTIs
Nucleoside reverse transcriptase inhibitors
- OXPHOS
Oxidative phosphorylation
- 3TC
Lamivudine or 2',3'-dideoxy-3'-thiacytidine
Contributor Information
Salina M. Torres, College of Pharmacy, University of New Mexico, Albuquerque, NM 87131, USA Lovelace Respiratory Research Institute, Albuquerque, NM 87108, USA.
Rao L. Divi, Center for Research, National Cancer Institute, NIH, Bethesda, MD 20982, USA
Dale M. Walker, SKS Consulting Services, Siler City, NC 27344, USA
Consuelo L. McCash, Lovelace Respiratory Research Institute, Albuquerque, NM 87108, USA
Meghan M. Carter, Lovelace Respiratory Research Institute, Albuquerque, NM 87108, USA
Matthew J. Campen, Lovelace Respiratory Research Institute, Albuquerque, NM 87108, USA
Tracey L. Einem, Center for Research, National Cancer Institute, NIH, Bethesda, MD 20982, USA
Yvonne Chu, Center for Research, National Cancer Institute, NIH, Bethesda, MD 20982, USA.
Steven K. Seilkop, SKS Consulting Services, Siler City, NC 27344, USA
Huining Kang, Department of Internal Medicine, Division of Epidemiology and Biostatistics, University of New Mexico, Albuquerque, NM 87106, USA.
Miriam C. Poirier, Center for Research, National Cancer Institute, NIH, Bethesda, MD 20982, USA
Vernon E. Walker, College of Pharmacy, University of New Mexico, Albuquerque, NM 87131, USA Lovelace Respiratory Research Institute, Albuquerque, NM 87108, USA; BioMosaics, Inc., Burlington, VT 05405, USA; Department of Pathology, University of Vermont, Burlington, VT 05405, USA; Genetic Toxicology Laboratory, University of Vermont, 665 Spear St., Building C, Burlington, VT 05405, USA.
References
- 1.Cooper ER, Charurat M, Mofenson L, Hanson IC, Pitt J, Diaz C, et al. Combination antiretroviral strategies for the treatment of pregnant HIV-1-infected women and prevention of perinatal HIV-1 transmission. Journal of Acquired Immune Deficiency Syndromes. 2002;29:484–494. doi: 10.1097/00126334-200204150-00009. [DOI] [PubMed] [Google Scholar]
- 2.UNAIDS The Joint United Nations Programme on HIV/AIDS. http://www.unaids.org/en/ [PubMed]
- 3.Watts DH. Treating HIV during pregnancy. An update on safety issues. Drug Safety. 2006;29:467–490. doi: 10.2165/00002018-200629060-00002. [DOI] [PubMed] [Google Scholar]
- 4.Thorne C, Newell ML. Safety of agents used to prevent mother-to-child transmission of HIV: Is there any cause for concern? Drug Safety. 2007;30:203–213. doi: 10.2165/00002018-200730030-00004. [DOI] [PubMed] [Google Scholar]
- 5.European Collaborative Study Mother to child transmission of HIV infection in the era of highly active antiretroviral therapy. Clinical Infectious Diseases. 2005;40:458–465. doi: 10.1086/427287. [DOI] [PubMed] [Google Scholar]
- 6.Blanche S, Tardieu M, Rustin P, Slama A, Barret B, Firtion G, et al. Persistent mitochondrial dysfunction and perinatal exposure to antiretroviral nucleoside analogues. Lancet. 1999;354:1084–1089. doi: 10.1016/S0140-6736(99)07219-0. [DOI] [PubMed] [Google Scholar]
- 7.Blanche S, Tardieu M, Benhammou V, Warszawski J, Rustin P. Mitochondrial dysfunction following perinatal exposure to nucleoside analogues. AIDS. 2006;20:1685–1690. doi: 10.1097/01.aids.0000242814.42344.77. [DOI] [PubMed] [Google Scholar]
- 8.Walker VE, Poirier MC. Foreword: Special issue on health risks of perinatal exposure to nucleoside reverse transcriptase inhibitors. Environmental and Molecular Mutagenesis. 2007;48:159–165. doi: 10.1002/em.20296. [DOI] [PubMed] [Google Scholar]
- 9.Olivero OA, Fernandez JJ, Antiochos BB, Wagner JLST, Claire ME, Poirier MC. Transplacental genotoxicity of combined antiretroviral nucleoside analogue therapy in Erythrocebus patas monkeys. Journal of Acquired Immune Deficiency Syndromes. 2002;29:323–329. doi: 10.1097/00126334-200204010-00001. [DOI] [PubMed] [Google Scholar]
- 10.Torres SM, Walker DM, Carter MM, Cook DL, Jr., McCash CL, Cordova EM, et al. Mutagenicity of zidovudine, lamivudine, and abacavir following exposure of human lymphoblastoid cells or in utero exposure of CD-1 mice to single agents or drug combinations. Environmental and Molecular Mutagenesis. 2007;48:224–238. doi: 10.1002/em.20264. [DOI] [PubMed] [Google Scholar]
- 11.Lewis W. Cardiomyopathy, nucleoside reverse transcriptase inhibitors and mitochondria are linked through AIDS and its therapy. Mitochondrion. 2004;4:141–152. doi: 10.1016/j.mito.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Kohler JJ, Lewis W. A brief overview of mechanisms of mitochondrial toxicity from NRTIs. Environmental and Molecular Mutagenesis. 2007;48:166–172. doi: 10.1002/em.20223. [DOI] [PubMed] [Google Scholar]
- 13.Lim SE, Copeland WC. Differential incorporation and removal of antiviral deoxynucleotides by human DNA polymerase gamma. The Journal of Biological Chemistry. 2001;276:23616–23623. doi: 10.1074/jbc.M101114200. [DOI] [PubMed] [Google Scholar]
- 14.Dagan T, Sable C, Bray J, Gerschenson M. Mitochondrial dysfunction and antiretroviral nucleoside analog toxicities: What is the evidence? Mitochondrion. 2002;1:397–412. doi: 10.1016/s1567-7249(02)00003-x. [DOI] [PubMed] [Google Scholar]
- 15.Brinkman K, ter Hofstede JM, Burger DM, Smeitink JAM, Koopmans PP. Adverse effects of reverse transcriptase inhibitors: Mitochondrial toxicity as common pathway. AIDS. 1998;12:1735–1744. doi: 10.1097/00002030-199814000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Foster C, Lyall H. HIV and mitochondrial toxicity in children. Journal of Antimicrobial Chemotherapy. 2008;61:8–12. doi: 10.1093/jac/dkm411. [DOI] [PubMed] [Google Scholar]
- 17.Olivero OA, Anderson LM, Diwan BA, Haines DC, Harbaugh SW, Moskal TJ, et al. Transplacental effects of 3′-azido-2′,3′-dideoxythymidine (AZT): Tumorigenicity in mice and genotoxicity in mice and monkeys. Journal of the National Cancer Institute. 1997;89:1602–1608. doi: 10.1093/jnci/89.21.1602. [DOI] [PubMed] [Google Scholar]
- 18.Olivero OA, Shearer GM, Chougnet CA, Kovacs AA, Landay AL, Baker R, et al. Incorporation of zidovudine into leukocyte DNA from HIV-1-positive adults and pregnant women, and cord blood from infants exposed in utero. AIDS. 1999;13:919–925. doi: 10.1097/00002030-199905280-00007. [DOI] [PubMed] [Google Scholar]
- 19.Poirier MC, Patterson TA, Slikker W, Jr., Olivero OA. Incorporation of 3′-azido-3′-deoxythymidine (AZT) into fetal DNA, and fetal tissue distribution of drug, after infusion of pregnant late-term rhesus macaques with a human-equivalent AZT dose. Journal of Acquired Immune Deficiency Syndromes. 1999;22:477–483. doi: 10.1097/00126334-199912150-00008. [DOI] [PubMed] [Google Scholar]
- 20.Walker DM, Poirier MC, Campen MJ, Cook DL, Jr., Divi RL, Nagashima K, et al. Persistence of mitochondrial toxicity in hearts of female B6C3F1 mice exposed in utero to 3′-azido-3′-deoxythymidine. Cardiovascular Toxicology. 2004;4:133–153. doi: 10.1385/ct:4:2:133. [DOI] [PubMed] [Google Scholar]
- 21.Gerschenson M, Erhart SW, Paik CY, St Claire MC, Nagashima K, Skopets B, et al. Fetal mitochondrial heart and skeletal muscle damage in Erythrocebus patas monkeys exposed in utero to 3′-azido-3′-deoxythymidine. AIDS Research and Human Retroviruses. 2000;16:635–644. doi: 10.1089/088922200308864. [DOI] [PubMed] [Google Scholar]
- 22.Poirier MC, Divi RL, Al-Harthi L, Olivero OA, Nguyen V, Walker B, et al. Long-term mitochondrial toxicity in HIV-uninfected infants born to HIV-infected mothers. Journal of Acquired Immune Deficiency Syndromes. 2003;33:175–183. doi: 10.1097/00126334-200306010-00010. [DOI] [PubMed] [Google Scholar]
- 23.Shiramizu B, Shikuma KM, Kamemoto L, Gerschenson M, Erdem G, Pinti M, et al. Placenta and cord blood mitochondrial DNA toxicity in HIV-infected women receiving nucleoside reverse transcriptase inhibitors during pregnancy. Journal of Acquired Immune Deficiency Syndromes. 2003;32:370–374. doi: 10.1097/00126334-200304010-00004. [DOI] [PubMed] [Google Scholar]
- 24.Divi RL, Walker VE, Wade NA, Nagashima K, Seilkop SK, Adams ME, et al. Mitochondrial damage and DNA depletion in cord blood and umbilical cord from infants exposed in utero to Combivir. AIDS. 2004;18:1013–1021. doi: 10.1097/00002030-200404300-00009. [DOI] [PubMed] [Google Scholar]
- 25.Divi RL, Leonard SL, Kuo MM, Nagashima K, Thamire C, St. Claire MC, et al. Transplacentally exposed human and monkey newborn infants show similar evidence of nucleoside reverse transcriptase inhibitor-induced mitochondrial toxicity. Environmental and Molecular Mutagenesis. 2007;47:201–209. doi: 10.1002/em.20201. [DOI] [PubMed] [Google Scholar]
- 26.Schon EA. Complements of the house. Journal of Clinical Investigation. 2004;114:760–762. doi: 10.1172/JCI22942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McComsey G, Tan DJ, Lederman M, Wilson E, Wong LJ. Analysis of the mitochondrial DNA genome in the peripheral blood leukocytes of HIV-infected patients with or without lipoatrophy. AIDS. 2002;16:513–518. doi: 10.1097/00002030-200203080-00001. [DOI] [PubMed] [Google Scholar]
- 28.McComsey G, Bai RK, Maa JF, Seekins D, Wong LJ. Extensive investigations of mitochondrial DNA genome in treated HIV-infected subjects: Beyond mitochondrial DNA depletion. Journal of Acquired Immune Deficiency Syndromes. 2005;39:181–188. [PubMed] [Google Scholar]
- 29.Martin AM, Hammond E, Nolan D, Pace C, Den Boer M, Taylor L, et al. Accumulation of mitochondrial DNA mutations in human immunodeficiency virus-infected patients treated with nucleoside-analogue reverse-transcriptase inhibitors. American Journal of Human Genetics. 2003;72:549–560. doi: 10.1086/367849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres SM, Walker DM, McCash CL, Ming J, Cordova EM, Pons RM, et al. Mutational analysis of the mitochondrial tRNA genes and flanking regions in umbilical cord tissue from uninfected infants receiving prepartum AZT-based therapies for prophylaxis of HIV-1. Environmental and Molecular Mutagenesis. 2009;50:10–26. doi: 10.1002/em.20433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alimenti A, Burdgec DR, Ogilvie GS, Money DM, Forbes JC. Lactic acidemia in human immunodeficiency virus-uninfected infants exposed to perinatal antiretroviral therapy. Pediatric Infectious Disease Journal. 2003;22:782–788. doi: 10.1097/01.inf.0000086400.93257.74. [DOI] [PubMed] [Google Scholar]
- 32.Spector SA, Saitoh A. Mitochondrial dysfunction: Prevention of HIV-1 mother-to-infant transmission outweighs fear. AIDS. 2006;20:1777–1778. doi: 10.1097/01.aids.0000242825.97495.a7. [DOI] [PubMed] [Google Scholar]
- 33.Divi RL, Leonard SL, Kuo MM, Walker BL, Orozco CC, St Claire MC, et al. Cardiac mitochondrial compromise in 1-yr-old Erythrocebus patas monkeys perinatally-exposed to nucleoside reverse transcriptase inhibitors. Cardiovascular Toxicology. 2005;5:333–346. doi: 10.1385/ct:5:3:333. [DOI] [PubMed] [Google Scholar]
- 34.Chan SS, Santos JH, Meyer JN, Mandavilli BS, Cook DL, Jr., McCash CL, et al. Mitochondrial toxicity in hearts of CD-1 mice following perinatal exposure to AZT, 3TC, or AZT/3TC in combination. Environmental and Molecular Mutagenesis. 2007;48:190–200. doi: 10.1002/em.20191. [DOI] [PubMed] [Google Scholar]
- 35.Torres SM, March TH, Carter MM, McCash CL, Seilkop SK, Walker DM, Walker VE. In utero exposure of female CD-1 mice to AZT and/or 3TC: I. Persistence of microscopic lesions in cardiac tissue. Cardiovascular Toxicology. 2008 doi: 10.1007/s12012-010-9061-3. 2010 Jan 27. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods in Enzymology. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 37.Divi RL, Kaverkos KJ, Humsi JA, Shockley ME, Thamire C, Nagashima K, et al. Morphological and molecular course of mitochondrial pathology in cultured human cells exposed long-term to zidovudine. Environmental and Molecular Mutagenesis. 2007;48:179–189. doi: 10.1002/em.20245. [DOI] [PubMed] [Google Scholar]
- 38.Lipshultz SE, Easley KA, Orav J, Kaplan S, Starc TJ, Bricker JT, et al. Absence of cardiac toxicity of Zidovudine in infants. New England Journal of Medicine. 2000;43:759–766. doi: 10.1056/NEJM200009143431102. [DOI] [PubMed] [Google Scholar]
- 39.Lipshultz SE, Easley KA, Orav EJ, Kaplan S, Starc TJ, Bricker JT, et al. Cardiovascular status of infants and children of women infected with HIV-1 (P2C2 HIV): A cohort study. The Lancet. 2002;360:353–368. doi: 10.1016/S0140-6736(02)09607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipshultz SE, Shearer WT, Rich K, Thompson B, Cheng I, Harmon T, et al. Antiretroviral therapy (ART)–associated cardiotoxicity in uninfected but ART-exposed infants born to HIV-infected Women: the prospective NHLBI CHA-ART-1 Study. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2005;28:9. [Google Scholar]