Summary
Analyses in mouse models have revealed crucial roles for MafA and MafB in islet β cells, with MafB required during development and MafA in adults. These two closely related transcription factors regulate many genes essential for glucose sensing and insulin secretion in a cooperative and sequential manner. Significantly, the switch from MafB to MafA expression also appears to be vital for functional maturation of the β cells produced by human embryonic stem (hES) cell differentiation. This review will summarize the discovery, distribution, and function of MafA and MafB in rodent pancreatic β cells, and describe some key questions regarding their importance to β cells.
β cell generation as a source of replacement therapy for diabetes mellitus
Pancreatic islet β cells are the only cell type in the body to secrete the insulin hormone in response to glucose. High levels of glucose stimulate insulin secretion from β cells. This hormone first acts on liver to increase glycogen synthesis and inhibit gluconeogenesis. Subsequently, insulin promotes glucose uptake and storage in skeletal muscle and adipose tissue. In addition to increased blood glucose concentrations, insulin is also secreted in response to amino acids, free fatty acids, potassium and neurotransmitters. The mass and functional capacity of β cells, in combination with peripheral insulin sensitivity, are determinants of glucose homeostasis.
Diabetes mellitus is a heterogeneous group of metabolic disorders characterized by chronic hyperglycemia, caused by absolute insulin deficiency due to autoimmune β cell destruction (i.e. type I diabetes) or a relative deficiency because the body cannot effectively use what is produced (type II diabetes). Since the discovery of insulin in the early 1920s, and its subsequent mass production, insulin therapy via administration of exogenous insulin has greatly improved the life expectancy of diabetics. However, patients eventually experience complications, including weight gain, kidney failure, retinopathy, nervous system damage and potentially life-threatening hypoglycemia, presumably due to the failure of exogenous insulin administration to regulate glucose levels as effectively as that provided by endogenous β cells. In 1999, a group of investigators in Canada transplanted human islets into seven type I diabetic patients, rendering them exogenous insulin-independent, with a normal or near normal glycohemoglobin levels1. Although most of these patients relapsed after three years and required external insulin treatment, this protocol provided encouragement and renewed interest for developing methods for generating sources of therapeutic β cells, as well as better protocols and reagents to prevent and minimize immunorejection.
Notably, the Canadian protocol utilized islets from two or more cadavers for each recipient, and revealed the very serious shortage of transplantable material. As a consequence, an enormous amount of effort has been focused on seeking alternative strategies to produce human islet β cells. Three major avenues have been actively pursued: 1) promoting replication of existing adult β cells; 2) directly differentiating stem cells to β cells; and 3) transdifferentiating non-β cells to a β cell fate2.
Under normal conditions, rodent β cells replicate at a low rate in adulthood2-4, but proliferation can be induced by glucose5, growth factors6, partial pancreatectomy7, 8, and insulin resistance8. However, similar conditions were unable to trigger substantial β cell replication in adult humans, including partial pancreatectomy and insulin resistance9, 10. The discrepancy between the capacity of human and rodent islet cells for replication has recently been attributed to inter-species differences in the distribution of G1/S phase cell cycle regulators11-15. Notably, in vitro overexpression of the G1/S regulators, Cdk6 and CyclinD3, in human islets induced robust β cell proliferation (15% upon Cdk6/CyclinD3 co-transduction versus 0.3% in control)15.
Progress has also been made in the making of insulin+ cells through in vitro differentiation of pluripotential stem cells as well as transdifferentiation of non-β cells in mouse models. The Baetge group used this information to establish a now well-recognized in vitro protocol to efficiently differentiate hES cells to definitive endoderm and subsequently to hormone-producing endocrine cells. Importantly, the insulin content of these cells was similar to mature human islet β cells and insulin was secreted upon administration of KCl and cAMP, agents that promote insulin release in the absence of stimulating high amounts of glucose. However, secretion was poorly responsive to stimulating glucose concentrations, the most important physiological effector16. Recently these investigators were able to produce β-like cells that release insulin in response to glucose by transplanting cells produced from an early in vitro differentiation stage under the epididymal fat pad of immunodeficient mice17. A notable feature of these glucose responsive cells was their ability to now produce MafA, a transcription factor necessary for adult β cell function in the rodents17. This resembles the natural developmental expression profile of β cell maturation in mice18-20, with the transition of MafB to MafA in insulin+ cells appearing to be critical for activity.
Interestingly, MafA, together with Pdx1 and Ngn3, were also the islet-enriched transcription factors capable of reprogramming adult mouse pancreatic acinar cells to β-like cells21. These insulin+ cells expressed many factors associated with β cell identity (e.g. Glut2/Slc2a2, Glucokinase, Nkx6.1, and Prohormone convertase 1/3), were ultrastructurally similar to islet β cells, secreted active insulin, and reduced high blood glucose levels upon destruction of endogenous islet β cells by streptozotocin administration, a commonly used strategy to mimic conditions found in type I diabetes21.
These results highlighted the necessity of producing MafA in biologically active insulin+ cells. Presently neither of these engineered β-like cells has been tested therapeutically in humans and our understanding of MafA and MafB function in islet cells in vivo results exclusively from rodent models. However, MafA is also enriched in human islet β cells22, suggesting that its regulatory properties are conserved. Here, we describe the discovery, distribution, and functions of MafA and MafB in rodent pancreatic β cells, and discuss pertinent issues regarding their significance in this context. A thorough understanding of how these two transcription factors impact β cells will likely provide perspectives valuable to generating functional insulin+ cells for therapeutic cell replacement therapy in diabetics.
MafA and MafB are principal large Maf transcription factors in pancreas
The large Maf protein family is composed of four distinct genes/proteins (see Box1), MafA, MafB, c-Maf and Nrl, which all contain N-terminal transactivation and C-terminal basic leucine-zipper DNA binding domains. The in vitro DNA binding properties of MafA, MafB, and c-Maf are indistinguishable23. c-Maf has been reported to be expressed in the pancreas20, 24-26, while pancreatic Nrl expression is undetectable27. Signficantly, pancreas development is unaffected in c-Maf null mice18.
Box 1. The Maf transcription factor family.
The Maf transcription factor family is a subgroup of the basic leucine-zipper (bZIP) family. The family members are named after v-Maf, the oncogenic component of the avian retrovirus AS42 originally isolated from a spontaneous musculoaponeurotic fibrosarcoma (Maf) in chicken94. Many maf-related genes have been cloned in human, mouse, rat, frog, quail, chick, zebrafish, and Drosophila by homology to the v-Maf bZIP domain95. In addition, Maf proteins show high conservation in the extended homology region (EHR) or ancillary DNA binding region, a small domain N-terminal to a basic amino acid rich region. The EHR in combination with this basic region imparts the DNA binding specificity to Maf proteins, which is 13-14 base pairs longer than the 8 base pair core binding site of other bZIP factors, like AP1 and CREB96-98. Maf proteins recognize several types of DNA-binding sequences, including palindromic Maf-recognition elements (MAREs, both T-MARE and C-MARE)97, 98 and AT-rich plus half MAREs99.
The Maf protein family is subdivided into two major groups based on their molecular size, small (149-162 amino acids) and large (236-370 amino acids). Small Maf proteins (i.e. MafF, MafG and MafK) all lack a distinct transactivation domain. Small Maf homodimers can compete with large Maf proteins for cis-element binding and repress gene transcription100, 101. On the other hand, they can also form heterodimers with other bZIP factors such as the Cap-n-collar (CNC) family proteins and transactivate genes important to stress signaling, hematopoiesis, CNS function, inflammatory response and oncogenesis102. In contrast, the large Maf proteins (i.e. MafA, MafB, c-Maf, and Nrl) are characterized by the presence of a transactivation domain in the N-terminal region. These proteins are all recognized as key regulators of tissue specific gene expression and cellular differentiation in, for example, brain, retina, lens, kidney and pancreas 93, 103. In addition, the large Maf proteins can transform primary cells and serve as oncogenes in human cancers93, specifically c-Maf in angioimmunoblastic T-cell lymphomas104, and multiple myelomas105, 106 as well as MafB in multiple myelomas107.
MafA was first identified from a chicken lens cDNA expression library during a screen for proteins that bind to the lens-specific αCE2 enhancer element and activate lens crystallin gene transcription28, 29, and was initially termed L-Maf (i.e. Lens-specific Maf; the chicken orthologue to mammalian MafA). However, this protein has little, if any, impact on mammalian lens fiber differentiation30, 31. MafA was biochemically isolated in mammalian cells from the rodent β cell-line-specific RIPE3b1 electrophoretic mobility shift binding complex that is associated with the insulin C1 element activation23, 32. Human MAFA shares 93% of homology with the mouse protein ortholog and can also activate the insulin promoter driven reporter expression32. This factor complex is crucial for glucose-responsive insulin transcription in β cells (discussed in detail later). Notably, MafA also appears to be the principal islet-enriched transcription factor mediating insulin transcription in the thymus, which is crucial for the development of self-tolerant autoantibodies33. Two polymorphisms in the 5-flanking promoter region of human MAFA have been associated with type 1 diabetes, presumably because of decreased expression in the thymus33. Although transgenic Mafa promoter-driven activity was observed in many tissues outside the pancreas in mice34, 35, it is unclear if this represents endogenous expression.
Mammalian MafB was identified earlier than MafA as the protein product of the mutated gene in the X-ray induced Kreisler mutant, a hypomorphic allele caused by chromosomal inversion36, 37. These mice exhibit a number of abnormal behaviors, such as head tossing and running in circles, due to defects in hindbrain segmentation and otic development37. Later analysis of Mafb−/− mice showed that this factor controlled segmental identity in the hindbrain through activating the Hoxb3 gene expression in rhombomere(r)538. In addition, MafB plays critical roles in a variety of other cellular differentiation processes, including in kidney podocytes39, macrophages40-42, as well as islet α and β cells18. In the pancreas, MafB was originally characterized as the islet α cell-enriched25 activator binding to the glucagon G1 element (located between −77 and −51 base pair (bp) relative to the transcription starting site). Recent studies have revealed that MafB is produced in both insulin+ and glucagon+ cells during development and necessary in α and β cell differentiation (discussed in detail later). The human MAFB gene was recently cloned (GenBank: AL035665.32) and is 97% identical to the mouse MafB protein, although nothing is known about its distribution or significance in the human pancreas (see Box 1).
MafA and MafB have unusual expression patterns during islet cell development
MafA and MafB are expressed in a unique tempero-spatial regulated manner in relation to other islet-enriched factors in developing and postnatal murine islet cells. MafB is expressed earlier than MafA, with the initial production detected around E10.5 in the pancreatic epithelium20, 25. In contrast, MafA is first produced at E13.5 and only in insulin+ cells43. Their developmental expression pattern is also unusually late in comparison to all other islet-enriched transcription factors (e.g. Pdx1 (E8.5), Pax6 (E9.0), Ngn3 (E9.0), Isl1 (E9.5), Nkx2.2 (E9.5)). Moreover, these two large Maf transcription factors are mostly, if not exclusively (i.e. MafA) expressed in hormone-producing cells, a distinction from the broad distribution of other islet-enriched transcription factors in both early progenitor and committed islet cell populations. Except for a small portion of MafB+Ngn3+ cells, MafB is exclusively detected in glucagon+ cells and insulin+ cells during the primary and secondary transition20, 25 (Box2). However, MafB disappears from β cells within two weeks of birth and becomes an adult islet α cell-specific factor, while MafA expression persists in postnatal and adult β cells19.
Box 2. Primary and secondary transitions during pancreas development.
Hormone-expressing cells are produced during two sequential regulatory transitions108, often referred to as the primary and secondary transition. They differ in cell ultrastructure and hormone/enzyme content. The “primary transition” (prior to E13.5 in mice) is related to organ determination, that is when the foregut region is destined to form the pancreas. The first glucagon+ cells in mice are seen in the pancreatic epithelium at E9109, with a few insulin+ cells found one day later. During this early phase, only a small number of cells contain islet-like granules. The majority of pancreatic epithelial cells are at the “protodifferentiated” state, lacking secretory granules and producing low levels of terminal gene products (e.g. insulin and amylase)108. Although the physiological roles of these early cells are still unknown, it is generally believed that (at least) the insulin+ cells are dysfunctional. Notably, they do not express the principal glucose transporter of β cells, Glut2/Slc2a2109, 110.
The “secondary transition” is a relatively short period (between E13.5 and E15.5 in mice), characterized by an immense increase in pancreas-specific protein synthesis. The majority (if not all) islet β cells are formed during this transition109, 111. The protein concentrations of β cell-enriched products increase between 100- and 1,000-fold112. Similarly, exocrine cell differentiation is initiated at this time, accompanied by exponential increases in acinar enzyme gene expression, development of large amounts of rough endoplasmic reticulum, and formation of zymogen granules. It is also at this time that the structure of the adult organ begins to be recognizable. After the secondary transition, expansion of both the exocrine and endocrine pancreas occurs through proliferation of differentiated endocrine cell types. Endocrine cells migrate and aggregate to form islets, and become responsive to glucose after birth113-115.
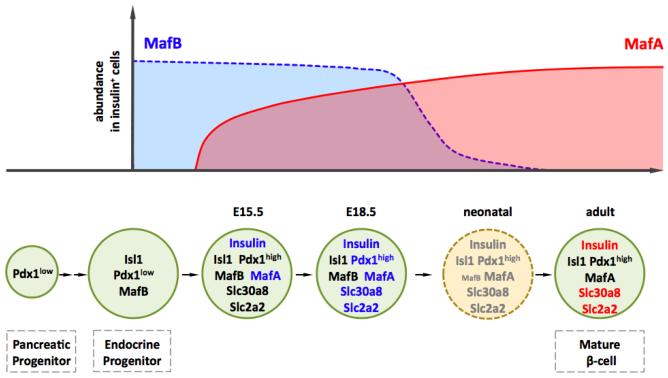
Quantitative immunohistochemical studies illustrated the dynamic nature of MafA and MafB production in the insulin+ cell population of the mouse fetal and neonatal pancreas (Figure 1). MafB was found in almost all insulin+ cells produced at E14.5 (91.7%) and E15.5 (89.3%), while only slightly reduced at the end of gestation (85%)19. The proportion of MafB+ insulin+ cells rapidly declined after birth, as very few co-expressing cells were detectable by postnatal day (P)14 (2.7%) and none by P2819. In contrast, a fraction of insulin+ cells co-express MafA at E14.5 (45%), which increases by E18.5 (75%) to that found in adult islets (80%). Mafa and Mafb mRNA expression is regulated in a similar dynamic manner44, 45. The steady-state Mafb messenger level is roughly 6-fold higher than that of Mafa in E15.5-E18.5 mouse insulin+ cells44, likely explaining why there is no effect on β cell development in Mafa mutant mice (discussed in detail later). Significantly, Mafa mRNA levels increase from neonates to adults by 10-fold in rats, consistent with its important role in adult islet β cell function45.
Figure 1. Roles of MafA and MafB in mouse β cells.
MafA and MafB are expressed in a dynamic temporal pattern (top panel)19, 20 and required at distinct stages in mouse β cells (bottom panels). MafB is critical to β cell terminal differentiation through regulation of key β genes (blue). Thus MafB binds and directly initiates the insulin and MafA transcription in developing β cells. In addition, it is required at E18.5 for expression of β cell factors involved in glucose-stimulated insulin secretion (e.g. pdx1, slc2a2, and slc30a8). MafA regulates most of these β cell genes (red) in adult when MafB in silenced.
The unique cellular and temporal expression pattern of MafA has been attributed to roughly 10kb of DNA sequences upstream from the transcription start site. This regulatory domain contains six highly conserved segments, termed Regions (R) 1 to 6, which drive Mafa-like expression in β cell lines and transgenic mice34. In particular, mouse R3 (−8118bp to −7750 bp (relative to transcription starting site)) is highly conserved between xenopus and humans, as well as the only region of identity retained in the chicken MAFA gene34. It alone is uniquely capable of conferring β-cell-line specific reporter expression34, while activity is lost in developing and adult β cells in vivo upon deletion of R3 from the R1-6 transgenic reporter34, 35. However, R3 was not necessary for transgenic activity of the MafaR1-6-driven reporter in many other non-pancreatic tissues34, 35. R3 activity is regulated in vitro by many distinct islet-enriched transcription factors, including Nkx2.2, Nkx6.1, NeuroD1, Foxa2, Pdx1, Pax6, MafB and Isl134, 46, all of whom are expressed earlier than MafA in the developing pancreas and are critical to β cell development and/or function47-49. Moreover, MafA is not present in the remaining insulin+ cells of these transcription factor knockout mice, the exception being NeuroD1 wherein another closely related basic helix-loop-helix factor probably compensates. Thus, β-cell-specific Mafa transcription is controlled by R3 through the actions of many key islet regulators34, 46.
Little is known about the factors influencing Mafb transcription, though ~8.2 kb of mouse upstream promoter sequences were capable of directing transgenic reporter expression to the pancreas50. The 5′-flanking region contains several potential regulatory elements, including two GC-boxes (5′-GGGCGG-3′), a palindromic sequence (5′-GTCAGCTGAC-3′), two half-Maf recognition sites (5′-GCTGAC-3′), and an E-box (5′-CAGCTG-3′; the underlined nucleotides are essential to activation). In addition, cell line based assays indicate that MafB positively regulates its own transcription51. However, the pancreatic transcription factors actually stimulating Mafb expression in developing α and β cells or silencing in postnatal β cells have not been determined.
The importance of MafA and MafB in β cell development and function
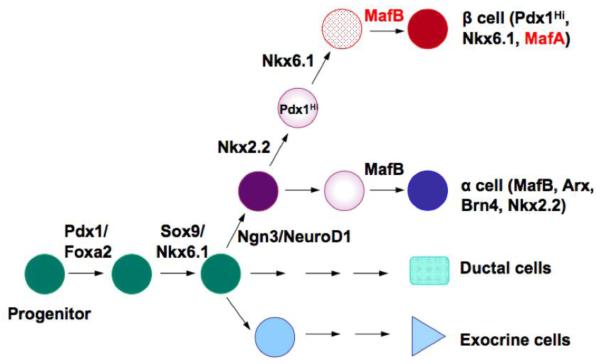
The pancreatic transcription factors play important in pancreas development (Figure 2). Almost all these transcription factor knockout mice result in either the loss in hormone+ cell numbers and/or respecification to another islet cell type47-49, 52. For instance, Pdx1 is critical for pancreas outgrowth during development, with loss leading to pancreas agenesis in mice and humans52-54. This condition results in death soon after birth unless treated for the loss of insulin producing β cells, and the digestive enzymes supplied by pancreatic acinar cells. In contrast, Ngn3 null mice are unable to produce any pancreatic islet cells during development due to the significance of this factor in the progenitor population, and die two to three days after birth from hyperglycemia55. Furthermore, β cells are completely lost and more grehlin+ ε cells are produced in mice lacking the NK homeodomain transcription factor, Nkx2.2, which is normally expressed in early pancreatic buds as well as later in Ngn3+ cells56. The pancreatic developmental phenotype of MafA and MafB null mice is distinct from essentially all other transcription factors, because MafB only impacts events required late in α and β cell maturation (Figure 1), while MafA does not affect β cell formation, despite being first detected during development in cells destined to populate the islet.
Figure 2. A cascade of transcription factors plays important roles in pancreas development.
The schematic diagram is a simplified model indicating transcription factors expressed at each stage of rodent pancreas development. The large Maf transcription factors MafB and MafA are located at the bottom of the cascade. MafB is required for the terminal differentiation of α and β cells while MafA is critical for β cell function in adult.
Only MafB is required during mouse β cell development
MafB is required for insulin and glucagon transcription in developing α and β cells, as illustrated by the reduction in hormone+ cell numbers throughout embryogenesis in Mafb−/− mice18, 57. In particular, MafB is essential for the first wave of insulin+ cells produced prior to E13.5 that are not completely functional, in addition to second wave β cells. Notably, the total number of endocrine cells is unchanged in mutant mice, and many proteins associated with β cell identity continue to be made. MafB binds to and is essential for of Insulin, Glucagon, and Mafa transcription during this period. Moreover, it is required for the maintenance of key β cell gene products (like Pdx1 and Slc2a2 (Glucose transporter; Glut2), which are made at normal levels until ~E15.5 and then cease to be produced in the Mafb−/− pancreas by E18.518, 57. Similar defects were also observed in KrENU mutant mice18, 57, a hypomorphic MafB basic DNA binding domain mutant produced by chemical mutagenesis with ethylnitrosourea (ENU)37.
In distinction to MafB, pancreas development was unaffected in Mafa−/− mice and pancreas-specific Mafa (MafaΔpanc) mutant mice19, 31. MafB probably acts in place of MafA in this context, since Mafb mRNA levels are normally 6-fold higher and elevated further (1.6-fold) in the E18.5 MafaΔpanc pancreas19, 44. Analysis of MafaΔpanc;Mafb−/− mice demonstrated that MafA does contribute during development to β cell gene transcription (e.g. insulin), but this effect was only prominent in the complete absence of MafB19. In contrast, all the other key islet insulin transcriptional activators play a critical role in pancreas development. Thus, α and β cell formation is impaired in Pax6−/− mice58-60, a severe and general reduction in islet endocrine cell numbers is found in the NeuroD1−/− mutant61, and β cell numbers are greatly decreased upon deletion of Pdx-1 by insulin-driven Cre recombinase in Pdx-1flox/flox mice62, 63. Hence, MafA and MafB are different from most other islet-enriched transcriptional regulators in not contributing in early cell lineage specification or terminal (β and α) cell differentiation, but only in islet cell maturation and/or function.
MafA and MafB actives genes associated with glucose-stimulated insulin secretion
MafA controls glucose-responsive transcription of insulin and other genes in islet β cells22, 23, 32, 64, 65. Elevated glucose concentrations acutely regulate MafA’s ability to bind and activate through the RIPE3b1/C1 DNA element66, a key cis-acting element of the mammalian insulin gene35, 67. MafA is a very potent activator of insulin transcription, with this factor independently capable of stimulating endogenous insulin production in αTC-6 cells (a glucagon+ MafB+ mouse islet α cell line) and chick embryonic endoderm in ovo43, 68. In contrast, MafB independently activates glucagon (and not insulin) production in βTC-3 cells (an insulin+ MafA+ islet β cell line), while an N-terminal MafB: C-terminal MafA chimera can stimulate insulin in early chick endoderm assays68 (see below).
Recent gene-profiling studies conducted with E18.5 wild type and Mafb−/− pancreata cast light on how MafB influences β cell formation19. Thus, gene ontology analysis revealed that MafB regulated many genes involved in aspects of mature β cell function, such as glucose sensing (e.g. Slc2a2), vesicle maturation (Slc30a8), Ca2+ signaling (Camk2b), and insulin secretion (Nnat). Strikingly, many of these genes were regulated in a similar way in adult MafaΔpanc islets, even though MafB was retained in a large fraction of the mutant insulin+ cell population (33.4 ± 5.6% of insulin+ cells expressed MafB). (MafaΔpanc mice were generated by crossing Mafafl/fl mice with transgenic mice producing Cre recombinase from the Pdx15.5 promoter fragment early in development and in a pancreas-wide pattern69.) Moreover, over-expression of a dominant negative form of MafA (DN-MafA) in the rat INS-1 β cell line also supported a significant role in glucose stimulated insulin secretion22, with this mutant inhibiting by sequester wild type MafA into a dimer complex incapable of cis-element binding. Thus, the mRNA levels of factors involved in metabolic-secretion coupling, proinsulin processing and glucagon-like peptide 1 receptor (GLP1R) signaling were reduced by DN-MafA (i.e. Gck, Glut2/Slc2a2, Pdx1, Nkx6.1, Glp1r, Pcsk1 and Pcx)22.
These results not only illustrated the functional interrelationship between these closely related large Maf transcription factors in the generation of islet β cells in vivo, but also provided evidence indicating that MafB and MafA do not act equivalently in these processes. Interestingly, MafA levels at P2 are only 7% of adult, and these rat islet β cells have poor glucose-stimulated insulin secretion properties. However, adenoviral-mediated over-expression of MafA in P2 islets greatly stimulated this activity, while Pdx1 infection had little effect45. These in vitro data further emphasize the significance of MafA to postnatal β cell maturation and function.
The phosphorylation status of MafA regulates DNA binding activity
Both MafA and MafB are heavily phosphorylated, a post-translational modification that impacts protein stability70-72, transactivation70, 71, 73, and oncogenic potential70. For example, a priming phosphorylation at serine (S)65 in MafA or S70 in MafB is necessary for ubiquitin-mediated degradation70, 71. In addition, S65 phosphorylation in MafA (and likely MafB) is required for priming Glycogen Synthase Kinase 3 (GSK3) for phosphorylation at S61, threonine (T)57, T53, and S49, which enhances transactivation and transformation potential70. Interestingly, this post-translational modification also differentially influences the activation properties of MafA and MafB. Thus, dephosphorylation by endogenous or exogenous phosphatases only inhibits the in vitro DNA-binding properties of MafA, and not MafB74.
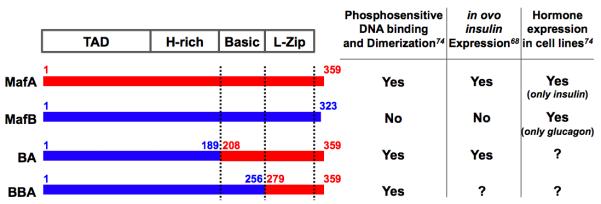
Analysis of MafA/B chimeras revealed that phosphatase sensitivity in these DNA binding assays was imparted by MafA sequences spanning the C-terminal dimerization region (amino acids (aa) 279-359), while the homologous MafB region (aa 257-323) enabled binding even after phosphatase treatment74. Mutational analysis showed that phosphorylation within the MafA (or MafB) N-terminal transactivation domain, and not the C-terminal basic leucine zipper (b-Zip) region, mediated phosphorylation-dependent MafA dimer formation and DNA binding74. Significantly, while only wild type MafA was able to stimulate endogenous insulin activation, the MafBB/A chimera containing the MafA C-terminal Zip region fused to the N-terminal MafB activation domain could impart this property onto MafB in the chick in ovo electroporation assay (Figure 3)68. It is possible that intramolecular interactions between the phosphoamino acid-rich transactivation and b-Zip domain not only control MafA DNA-binding activity, but also create distinctive interfaces for coregulatory proteins that are uniquely important in activation.
Figure 3. Summary of the differences between MafA and MafB in phosphatase-sensitive DNA binding and endogenous hormone gene activation assays.
The indicated MafB/A chimeras have been tested for phosphatase-sensitive DNA binding in gel shift assays, and for their ability to activate endogenous insulin and glucagon gene expression in both the chicken in ovo electroporation and cell line based assays. “?”, to be determined.
MafA is a positive indicator of β cell health and functionality
MafA levels appear to be a sensitive barometer of the activity status of islet β cells. Changing glucose levels can positively and negatively regulate MafA activity, processes influenced by transcriptional and/or post-transcriptional mechanisms34, 35, 67. Acutely elevating glucose concentration stimulates the hexosamine biosynthetic pathway and directly results in increased Mafa and insulin transcription35, 67. However, chronically high glucose levels have a rather specific detrimental effect on MafA activity in β cell lines and diabetic db/db mice75-79. Thus, supraphysiological (glucotoxic) glucose levels inhibit MafA and Pdx1 activity and in turn reduce insulin expression. For example, insulin gene transcription, insulin content and glucose-induced insulin secretion are progressively and drastically decreased in HIT-T15 β cells chronically grown for months at high, supraphysiological (11.1mM) versus a lower (0.8mM) glucose concentration80. The decrease in Pdx1 and MafA DNA binding activity is associated with the loss of insulin-driven reporter activity and insulin mRNA levels, with the change in MafA occurring earlier and correlated more closely with the loss in expression than Pdx180, 81. A similar response was observed in the βTC-6 cell line, except that MafA activity is principally affected77. Moreover, palmitate (a fatty acid mediator of β cell dysfunction under hyperglycemic conditions in vivo) decreases in MafA and Pdx-1 DNA activity in rat islets, causing reduced insulin expression82. The effect on Pdx1 activity under these conditions appears to be caused by a change in nuclear localization, while MafA mRNA processing80, stability75, and cellular localization83 have been connected with inhibition.
Chronic hyperglycemia causes β cell dysfunction in type 2 diabetes in part through the generation of excessive levels of reactive oxygen species (ROS), leading to cellular damage of macromolecules including proteins, lipids, carbohydrates, and nucleic acids. Exacerbating this problem, the low levels of antioxidant enzymes in the islet render β cells more sensitive to oxidative stress (e.g. compared to liver: superoxide dismutase, 50%; glutathione peroxidase, 1%; catalase, 1%)84, 85. MafA is translocated to the cytoplasm83 and p38 MAPK-mediated degradation is increased86 by oxidative stress. In contrast, nuclear Pdx1 levels appeared to be unaffected under these circumstances in db/db mice83. Importantly, reducing ROS by transgenic β cell over-expression of glutathione peroxidase-1 in db/db mice not only improved blood glucose levels to near normal, but also increased islet β cell volume, insulin granulation, and MafA nuclear content83. It was suggested that transgenic redox protein thioredoxin expression in db/db β cells also reduced cell failure in part by preventing MafA inactivation87. Notably, loss of MafA does not cause overt β cell dysfunction in vivo19, 31, suggesting that subsequent loss/inactivity of another factor(s) leads to this condition. Collectively, these results support MafA as a key regulator of glucose-stimulated insulin secretion and β cell function, perhaps representing an early indicator of cell stress in type 2 diabetes.
MafB expression is induced in maternal β cells during pregnancy
MafB is normally lost in β cells soon after birth (Figure 2). However, expression is induced in a fraction of maternal mouse insulin+ cells during pregnancy44. This contrasts with other transcription factors mediating changes in β function and/or proliferative capacity, whose expression is uniformly distributed in the islet cell population prior to pregnancy (e.g. Hnf4α88 and Menin89).
Due to fetal demands, the mother’s insulin sensitivity declines by one-third during pregnancy. Both β cell function and proliferative cell mass expansion occurs in maternal islets to adapt to the bodies increased requirements for insulin. The mother develops gestational diabetes if this does not occur, as found in 3 to 7 percent of pregnancies. Measurement of β cell BrdU incorporation illustrated that maternal MafB+ β cells were not undergoing cell replication44, different to other gestational diabetes mouse models associated with transcription factor dysfunction (Menin89, Hnf4α88, FoxM190). This suggests that MafB may have a unique role in β cell adaptation during pregnancy, possibly serving as a pre-mitotic factor to induce β cell proliferation and/or through involvement in functional enhancement during this physiological state of insulin resistance.
The potential role of MafA and MafB in islet β cell generation from non-β cells
Due to the significance of MafA in glucose-responsive transcription and adult β function, MafA has been utilized to induce β cell differentiation in both human and rodent stem cells and differentiated cell types (Table 1). This protein alone or in combination with other pancreatic transcription factors was able to induce the expression of insulin and other key β cell markers. In most cases, the generated insulin+ cells improved blood glucose levels in streptozotocin-induced diabetic animals, a mouse model of type 1 diabetes. Significantly, MafA was 1 of only 3 islet factors identified in a large transcription factor screen that together effectively reprogrammed adult pancreatic acinar cells to islet β-like cells (i.e. Pdx1, Ngn3, MafA). Notably, the resultant insulin+ cells not only produced many β cell identity markers but also functionally and ultrastructurally resembled endogenous β cells21.
Table 1.
Differentiation or transdifferentiation experiments that use MafA to generate β cells.
| Model | Original cell type |
Other TF* administrated |
Delivery method |
Conversion efficiency** |
β-cell or endocrine markers induced |
Other β cell characteristics |
Glucose-stimulated insulin secretion/c- peptide production |
Improve hyperglycemia in animals / transplanted animals |
Refs |
|---|---|---|---|---|---|---|---|---|---|
|
In vitro differentiatied Mouse ES cell line A2lox |
stem cell line | none | doxycycline inducible transgene |
n.a. | Insulin, Slc2a2, Gck, | n.a. | yes | n.a. | 116 |
| Placenta- derived multipotent stem cells |
stem cell | none | lentivirus | n.a. | Insulin, Slc2a2, Nkx2.2 |
n.a. | yes | yes | 117 |
| Live mouse | exocrine cell | Pdx1, Ngn3 | adenovirus injection |
>20% | Insulin, Slc2a2, Gck, Pcsk1, Nkx6.1, NeuroD, Nkx2.2 |
cell size, ultrastructure |
n.a. | yes | 21 |
| AR42J-B13 cells |
pancreatic progenitor- like cell line |
Ngn3, Nkx6.1 | adenovirus | n.a. | Insulin, Slc2a2, Pdx1, Nkx2.2 |
n.a. | n.a. | n.a. | 118 |
| Live mouse | hepatocyte | Ngn3, NeuroD2 | adenovirus injection |
0.5-1% | Insulin | n.a. | n.a. | yes | 119 |
| Live rat | intestinal epithelial cell |
none | oral administration of adenovirus |
n.a. | Insulin, Pdx1, Pcsk1, Kir6.2 |
n.a. | n.a. | yes | 120 |
| Chicken embryo |
gut endoderm cells |
alone or with Ngn3 |
electroporation | n.a. | Insulin | n.a. | n.a. | n.a. | 68 |
TF, transcription factors.
Conversion efficiency is determined as percentage of insulin+ cells over the infected/electroporated cells.
The ability of MafB to collaborate with other factors to generate insulin+ cells has only been tested in chick gut endoderm, wherein MafB alone or together with Ngn3 was unable to induce insulin production68. Significantly, MafB has only been associated with functionally immature insulin+ cell populations produced during the most recognized hES differentiation procedure16, 17. However, these cells remained dysfunctional with regard to glucose-responsive insulin secretion until MafA production was observed, a process that presently requires transplantation in vivo17. The induction of MafA is also observed during rodent pancreas development and postnatally in vivo, and indicates that the MafB to MafA transition may serve as a functional read out of the β-like cells produced from stem cells and/or through transdifferentiation approaches.
Hence, MafA appears to be a potent regulator essential for the generation of β-like cells. Moreover, its actions are required over MafB in specific and distinct steps of β cell maturation. Strikingly, MafA activity is inhibited by proinflammatory cytokines implicated in the pathogenesis of diabetes mellitus, like interleukin-1β and tumor necrosis factor-α. These cytokines have been detected in the pancreas of type 1 diabetic patients as well as mouse models of disease and cause MafA inactivation, β cell dysfunction and apoptosis91, 92. Therefore, efforts will also need to be focused on developing inhibitors for such proinflammatory cytokines to protect newly generated therapeutic cells from inactivation.
Concluding remarks
Studies on MafA and MafB using mouse models have revealed crucial roles in islet β cell formation and function, with MafB required during development and MafA in adults. Gene expression analyses disclosed that the two closely related transcription factors regulate key β cell genes in a cooperative and sequential manner. Research in mouse models as well as in vitro hES cell differentiation showed that the switch from MafB+ to MafA+ appears vital to functional maturation of β cells.
However, many unanswered questions remain to be addressed to completely understand how MafA and MafB work in β cells. For example, do MafA and MafB act equivalently, and thus, do differences in expression levels explain the Mafb and Mafa knockout phenotypes. If so, what is the threshold level required for these factors to successfully activate β cell gene transcription? If MafA and MafB have distinct capabilities, as suggested by biochemical and cell line-based studies, how are the differences implemented? One potential mechanism is recruitment of discrete binding co-regulatory partners. Ironically there are hundreds of coregulatory factors (http://www.nursa.org), yet only a hand full have been linked to β cells, presently limited to p300/CBP-associated factor in regards to MafA70. In addition, it is also vital to understand the driving force (e.g. signaling pathways) and executors (e.g. transcription factors and epigenetic factors) that silence MafB and drive high MafA expression in postnatal β cells, and how these elements can be employed in the development of β cells for therapeutic purposes. Last, but certainly not least, our information on human MAFA and MAFB function is presently limited to their actions as oncogenes in cancer (see Box1, multiple myelomas and angioimmunoblastic T-cell lymphomas)93. Thus, it is crucial to determine if the highly conserved human orthologs of MafA (i.e. 93% to rodent) and MafB (97%) have similar functional properties in the pancreas, since clearly the ultimate goal is to translate the knowledge obtained from model organisms to humans.
Abbreviation
- Pdx1
pancreatic duodenal homeobox1
- Ngn3
neurogenin 3
- MafA
musculoaponeurotic fibrosarcoma oncogene family, A
- MafB
musculoaponeurotic fibrosarcoma oncogene family, B
- Pax6
paired box gene 6
- Isl1
Islet1 transcription factor, LIM/homeodomain
- Nkx2.2
NK2 transcription factor related, locus 2
- MAPK
mitogen-activated protein kinases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Shapiro AM, et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343:230–238. doi: 10.1056/NEJM200007273430401. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann AM, Gannon M. Molecular regulation of pancreatic beta-cell mass development, maintenance, and expansion. J Mol Endocrinol. 2007;38:193–206. doi: 10.1677/JME-06-0053. [DOI] [PubMed] [Google Scholar]
- 3.Finegood DT, et al. Dynamics of beta-cell mass in the growing rat pancreas. Estimation with a simple mathematical model. Diabetes. 1995;44:249–256. doi: 10.2337/diab.44.3.249. [DOI] [PubMed] [Google Scholar]
- 4.Teta M, et al. Very slow turnover of beta-cells in aged adult mice. Diabetes. 2005;54:2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- 5.Alonso LC, et al. Glucose infusion in mice: a new model to induce beta-cell replication. Diabetes. 2007;56:1792–1801. doi: 10.2337/db06-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miettinen PJ, et al. Downregulation of EGF receptor signaling in pancreatic islets causes diabetes due to impaired postnatal beta-cell growth. Diabetes. 2006;55:3299–3308. doi: 10.2337/db06-0413. [DOI] [PubMed] [Google Scholar]
- 7.Dor Y, et al. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 8.Flier SN, et al. Evidence for a circulating islet cell growth factor in insulin-resistant states. Proc Natl Acad Sci U S A. 2001;98:7475–7480. doi: 10.1073/pnas.131192998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler AE, et al. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 10.Menge BA, et al. Partial pancreatectomy in adult humans does not provoke beta-cell regeneration. Diabetes. 2008;57:142–149. doi: 10.2337/db07-1294. [DOI] [PubMed] [Google Scholar]
- 11.Georgia S, Bhushan A. Beta cell replication is the primary mechanism for maintaining postnatal beta cell mass. J Clin Invest. 2004;114:963–968. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kushner JA, et al. Cyclins D2 and D1 are essential for postnatal pancreatic beta-cell growth. Mol Cell Biol. 2005;25:3752–3762. doi: 10.1128/MCB.25.9.3752-3762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiaschi-Taesch N, et al. Survey of the human pancreatic beta-cell G1/S proteome reveals a potential therapeutic role for cdk-6 and cyclin D1 in enhancing human beta-cell replication and function in vivo. Diabetes. 2009;58:882–893. doi: 10.2337/db08-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavine JA, et al. Contamination with E1A-positive wild-type adenovirus accounts for species-specific stimulation of islet cell proliferation by CCK: a cautionary note. Mol Endocrinol. 2010;24:464–467. doi: 10.1210/me.2009-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiaschi-Taesch NM, et al. Induction of human beta-cell proliferation and engraftment using a single G1/S regulatory molecule, cdk6. Diabetes. 2010;59:1926–1936. doi: 10.2337/db09-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Amour KA, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 17.Kroon E, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 18.Artner I, et al. MafB is required for islet beta cell maturation. Proc Natl Acad Sci U S A. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Artner I, et al. MafA and MafB Regulate Genes Critical to {beta} Cells in a Unique Temporal Manner. Diabetes. 2010 doi: 10.2337/db10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura W, et al. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Q, et al. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, et al. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia. 2007;50:348–358. doi: 10.1007/s00125-006-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka TA, et al. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kataoka K, et al. Differentially expressed Maf family transcription factors, c-Maf and MafA, activate glucagon and insulin gene expression in pancreatic islet alpha- and beta-cells. J Mol Endocrinol. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- 25.Artner I, et al. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- 26.Gosmain Y, et al. Pax-6 and c-Maf functionally interact with the alpha-cell-specific DNA element G1 in vivo to promote glucagon gene expression. J Biol Chem. 2007;282:35024–35034. doi: 10.1074/jbc.M702795200. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q, et al. Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene. 1996;12:207–211. [PubMed] [Google Scholar]
- 28.Ogino H, Yasuda K. Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- 29.Reza HM, et al. L-Maf, a downstream target of Pax6, is essential for chick lens development. Mech Dev. 2002;116:61–73. doi: 10.1016/s0925-4773(02)00137-5. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi T, et al. Neither MafA/L-Maf nor MafB is essential for lens development in mice. Genes Cells. 2009;14:941–947. doi: 10.1111/j.1365-2443.2009.01321.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C, et al. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olbrot M, et al. Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci U S A. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noso S, et al. Insulin transactivator MafA regulates intrathymic expression of insulin and affects susceptibility to type 1 diabetes. Diabetes. 2010;59:2579–2587. doi: 10.2337/db10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raum JC, et al. FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol Cell Biol. 2006;26:5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raum JC, et al. Islet beta-cell-specific MafA transcription requires the 5′-flanking conserved region 3 control domain. Mol Cell Biol. 2010;30:4234–4244. doi: 10.1128/MCB.01396-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deol MS. The Abnormalities of the Inner Ear in Kreisler Mice. J Embryol Exp Morphol. 1964;12:475–490. [PubMed] [Google Scholar]
- 37.Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 38.Manzanares M, et al. Segmental regulation of Hoxb-3 by kreisler. Nature. 1997;387:191–195. doi: 10.1038/387191a0. [DOI] [PubMed] [Google Scholar]
- 39.Sadl V, et al. The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev Biol. 2002;249:16–29. doi: 10.1006/dbio.2002.0751. [DOI] [PubMed] [Google Scholar]
- 40.Aziz A, et al. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326:867–871. doi: 10.1126/science.1176056. [DOI] [PubMed] [Google Scholar]
- 41.Aziz A, et al. Development of macrophages with altered actin organization in the absence of MafB. Mol Cell Biol. 2006;26:6808–6818. doi: 10.1128/MCB.00245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarrazin S, et al. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell. 2009;138:300–313. doi: 10.1016/j.cell.2009.04.057. [DOI] [PubMed] [Google Scholar]
- 43.Matsuoka TA, et al. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci U S A. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pechhold S, et al. Transcriptional analysis of intracytoplasmically stained, FACS-purified cells by high-throughput, quantitative nuclease protection. Nat Biotechnol. 2009;27:1038–1042. doi: 10.1038/nbt.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguayo-Mazzucato C, et al. Mafa expression enhances glucose-responsive insulin secretion in neonatal rat beta cells. Diabetologia. 2011;54:583–593. doi: 10.1007/s00125-010-2026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du A, et al. Islet-1 is required for the maturation, proliferation, and survival of the endocrine pancreas. Diabetes. 2009;58:2059–2069. doi: 10.2337/db08-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gradwohl G. Development of the endocrine pancreas. Diabetes Metab. 2006;32:532–533. doi: 10.1016/s1262-3636(06)72807-5. [DOI] [PubMed] [Google Scholar]
- 48.Kim SK, MacDonald RJ. Signaling and transcriptional control of pancreatic organogenesis. Curr Opin Genet Dev. 2002;12:540–547. doi: 10.1016/s0959-437x(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 49.Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamada M, et al. The mouse mafB 5′-upstream fragment directs gene expression in myelomonocytic cells, differentiated macrophages and the ventral spinal cord in transgenic mice. J Biochem. 2003;134:203–210. doi: 10.1093/jb/mvg130. [DOI] [PubMed] [Google Scholar]
- 51.Huang K, et al. Molecular cloning and functional characterization of the mouse mafB gene. Gene. 2000;242:419–426. [PubMed] [Google Scholar]
- 52.Offield MF, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 53.Ahlgren U, et al. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 54.Jonsson J, et al. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 55.Gradwohl G, et al. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prado CL, et al. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishimura W, et al. Preferential reduction of beta cells derived from Pax6-MafB pathway in MafB deficient mice. Dev Biol. 2008;314:443–456. doi: 10.1016/j.ydbio.2007.12.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashery-Padan R, et al. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol. 2004;269:479–488. doi: 10.1016/j.ydbio.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 59.Sander M, et al. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 60.St-Onge L, et al. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 61.Naya FJ, et al. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahlgren U, et al. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gannon M, et al. pdx-1 function is specifically required in embryonic beta cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev Biol. 2008;314:406–417. doi: 10.1016/j.ydbio.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kataoka K, et al. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J Biol Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- 65.Shieh SY, Tsai MJ. Cell-specific and ubiquitous factors are responsible for the enhancer activity of the rat insulin II gene. J Biol Chem. 1991;266:16708–16714. [PubMed] [Google Scholar]
- 66.Sharma A, Stein R. Glucose-induced transcription of the insulin gene is mediated by factors required for beta-cell-type-specific expression. Mol Cell Biol. 1994;14:871–879. doi: 10.1128/mcb.14.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vanderford NL, et al. Glucose induces MafA expression in pancreatic beta cell lines via the hexosamine biosynthetic pathway. J Biol Chem. 2007;282:1577–1584. doi: 10.1074/jbc.M605064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Artner I, et al. MafA is a dedicated activator of the insulin gene in vivo. J Endocrinol. 2008;198:271–279. doi: 10.1677/JOE-08-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu G, et al. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 70.Rocques N, et al. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol Cell. 2007;28:584–597. doi: 10.1016/j.molcel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 71.Guo S, et al. The stability and transactivation potential of the mammalian MafA transcription factor are regulated by serine 65 phosphorylation. J Biol Chem. 2009;284:759–765. doi: 10.1074/jbc.M806314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han SI, et al. MafA stability in pancreatic beta cells is regulated by glucose and is dependent on its constitutive phosphorylation at multiple sites by glycogen synthase kinase 3. Mol Cell Biol. 2007;27:6593–6605. doi: 10.1128/MCB.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao L, et al. The RIPE3b1 activator of the insulin gene is composed of a protein(s) of approximately 43 kDa, whose DNA binding activity is inhibited by protein phosphatase treatment. J Biol Chem. 2000;275:10532–10537. doi: 10.1074/jbc.275.14.10532. [DOI] [PubMed] [Google Scholar]
- 74.Guo S, et al. Phosphorylation within the MafA N terminus regulates C-terminal dimerization and DNA binding. J Biol Chem. 2010;285:12655–12661. doi: 10.1074/jbc.M110.105759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harmon JS, et al. Oxidative stress-mediated, post-translational loss of MafA protein as a contributing mechanism to loss of insulin gene expression in glucotoxic beta cells. J Biol Chem. 2005;280:11107–11113. doi: 10.1074/jbc.M410345200. [DOI] [PubMed] [Google Scholar]
- 76.Kitamura YI, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 77.Poitout V, et al. Chronic exposure of betaTC-6 cells to supraphysiologic concentrations of glucose decreases binding of the RIPE3b1 insulin gene transcription activator. J Clin Invest. 1996;97:1041–1046. doi: 10.1172/JCI118496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sharma A, et al. The reduction of insulin gene transcription in HIT-T15 beta cells chronically exposed to high glucose concentration is associated with the loss of RIPE3b1 and STF-1 transcription factor expression. Mol Endocrinol. 1995;9:1127–1134. doi: 10.1210/mend.9.9.7491105. [DOI] [PubMed] [Google Scholar]
- 79.Matsuoka TA, et al. Regulation of MafA expression in pancreatic beta-cells in db/db mice with diabetes. Diabetes. 2010;59:1709–1720. doi: 10.2337/db08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olson LK, et al. Reduction of insulin gene transcription in HIT-T15 beta cells chronically exposed to a supraphysiologic glucose concentration is associated with loss of STF-1 transcription factor expression. Proc Natl Acad Sci U S A. 1995;92:9127–9131. doi: 10.1073/pnas.92.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harmon JS, et al. Reconstitution of glucotoxic HIT-T15 cells with somatostatin transcription factor-1 partially restores insulin promoter activity. Diabetes. 1998;47:900–904. doi: 10.2337/diabetes.47.6.900. [DOI] [PubMed] [Google Scholar]
- 82.Hagman DK, et al. Palmitate inhibits insulin gene expression by altering PDX-1 nuclear localization and reducing MafA expression in isolated rat islets of Langerhans. J Biol Chem. 2005;280:32413–32418. doi: 10.1074/jbc.M506000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harmon JS, et al. beta-Cell-specific overexpression of glutathione peroxidase preserves intranuclear MafA and reverses diabetes in db/db mice. Endocrinology. 2009;150:4855–4862. doi: 10.1210/en.2009-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lenzen S, et al. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radic Biol Med. 1996;20:463–466. doi: 10.1016/0891-5849(96)02051-5. [DOI] [PubMed] [Google Scholar]
- 85.Tiedge M, et al. Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes. 1997;46:1733–1742. doi: 10.2337/diab.46.11.1733. [DOI] [PubMed] [Google Scholar]
- 86.Kondo T, et al. p38 MAPK is a major regulator of MafA protein stability under oxidative stress. Mol Endocrinol. 2009;23:1281–1290. doi: 10.1210/me.2008-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamoto M, et al. Transgenic expression of antioxidant protein thioredoxin in pancreatic beta cells prevents progression of type 2 diabetes mellitus. Antioxid Redox Signal. 2008;10:43–49. doi: 10.1089/ars.2007.1586. [DOI] [PubMed] [Google Scholar]
- 88.Gupta RK, et al. Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4alpha. Genes Dev. 2007;21:756–769. doi: 10.1101/gad.1535507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karnik SK, et al. Menin controls growth of pancreatic beta-cells in pregnant mice and promotes gestational diabetes mellitus. Science. 2007;318:806–809. doi: 10.1126/science.1146812. [DOI] [PubMed] [Google Scholar]
- 90.Zhang H, et al. Gestational diabetes mellitus resulting from impaired beta-cell compensation in the absence of FoxM1, a novel downstream effector of placental lactogen. Diabetes. 2010;59:143–152. doi: 10.2337/db09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oetjen E, et al. Inhibition of MafA transcriptional activity and human insulin gene transcription by interleukin-1beta and mitogen-activated protein kinase kinase kinase in pancreatic islet beta cells. Diabetologia. 2007;50:1678–1687. doi: 10.1007/s00125-007-0712-2. [DOI] [PubMed] [Google Scholar]
- 92.Ortis F, et al. Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells. Diabetes. 59:358–374. doi: 10.2337/db09-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eychene A, et al. A new MAFia in cancer. Nat Rev Cancer. 2008;8:683–693. doi: 10.1038/nrc2460. [DOI] [PubMed] [Google Scholar]
- 94.Nishizawa M, et al. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci U S A. 1989;86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Coolen M, et al. Phylogenomic analysis and expression patterns of large Maf genes in Xenopus tropicalis provide new insights into the functional evolution of the gene family in osteichthyans. Dev Genes Evol. 2005;215:327–339. doi: 10.1007/s00427-005-0476-y. [DOI] [PubMed] [Google Scholar]
- 96.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 97.Kataoka K, et al. Structure-function analysis of the maf oncogene product, a member of the b-Zip protein family. J Virol. 1993;67:2133–2141. doi: 10.1128/jvi.67.4.2133-2141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kerppola TK, Curran T. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene. 1994;9:3149–3158. [PubMed] [Google Scholar]
- 99.Yoshida T, et al. The 5′-AT-rich half-site of Maf recognition element: a functional target for bZIP transcription factor Maf. Nucleic Acids Res. 2005;33:3465–3478. doi: 10.1093/nar/gki653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Motohashi H, et al. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 101.Kataoka K, et al. Transactivation activity of Maf nuclear oncoprotein is modulated by Jun, Fos and small Maf proteins. Oncogene. 1996;12:53–62. [PubMed] [Google Scholar]
- 102.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 103.Kataoka K. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J Biochem. 2007;141:775–781. doi: 10.1093/jb/mvm105. [DOI] [PubMed] [Google Scholar]
- 104.Murakami YI, et al. c-Maf expression in angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. 2007;31:1695–1702. doi: 10.1097/PAS.0b013e318054dbcf. [DOI] [PubMed] [Google Scholar]
- 105.Chang H, et al. c-Maf nuclear oncoprotein is frequently expressed in multiple myeloma. Leukemia. 2007;21:1572–1574. doi: 10.1038/sj.leu.2404669. [DOI] [PubMed] [Google Scholar]
- 106.Hurt EM, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–199. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 107.van Stralen E, et al. Identification of primary MAFB target genes in multiple myeloma. Exp Hematol. 2009;37:78–86. doi: 10.1016/j.exphem.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 108.Rutter WJ, et al. Regulation of specific protein synthesis in cytodifferentiation. J Cell Physiol. 1968;72(Suppl 1):1–18. doi: 10.1002/jcp.1040720403. [DOI] [PubMed] [Google Scholar]
- 109.Pictet RL, et al. An ultrastructural analysis of the developing embryonic pancreas. Dev Biol. 1972;29:436–467. doi: 10.1016/0012-1606(72)90083-8. [DOI] [PubMed] [Google Scholar]
- 110.Pang K, et al. Beta cells arise from glucose transporter type 2 (Glut2)-expressing epithelial cells of the developing rat pancreas. Proc Natl Acad Sci U S A. 1994;91:9559–9563. doi: 10.1073/pnas.91.20.9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Slack JM. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 112.Han JH, et al. Selective expression of rat pancreatic genes during embryonic development. Proc Natl Acad Sci U S A. 1986;83:110–114. doi: 10.1073/pnas.83.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de los M. Espinosa, et al. Insulin release from isolated human fetal pancreatic islets. Science. 1970;168:1111–1112. doi: 10.1126/science.168.3935.1111. [DOI] [PubMed] [Google Scholar]
- 114.Heinze E, Steinke J. Insulin secretion during development: response of isolated pancreatic islets of fetal, newborn and adult rats to theophylline and arginine. Horm Metab Res. 1972;4:234–236. doi: 10.1055/s-0028-1094056. [DOI] [PubMed] [Google Scholar]
- 115.Norlin S, et al. Nuclear factor-{kappa}B activity in {beta}-cells is required for glucose-stimulated insulin secretion. Diabetes. 2005;54:125–132. doi: 10.2337/diabetes.54.1.125. [DOI] [PubMed] [Google Scholar]
- 116.Chen C, et al. Characterization of an In Vitro Differentiation Assay for Pancreatic-Like Cell Development from Murine Embryonic Stem Cells: Detailed Gene Expression Analysis. Assay Drug Dev Technol. 2011 doi: 10.1089/adt.2010.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chiou SH, et al. MafA promotes the reprogramming of placenta-derived multipotent stem cells into pancreatic islets-like and insulin-positve cells. J Cell Mol Med. 2010 doi: 10.1111/j.1582-4934.2010.01034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ogihara T, et al. Combined expression of transcription factors induces AR42J-B13 cells to differentiate into insulin-producing cells. Endocr J. 2008;55:691–698. doi: 10.1507/endocrj.k07e-169. [DOI] [PubMed] [Google Scholar]
- 119.Song YD, et al. Islet cell differentiation in liver by combinatorial expression of transcription factors neurogenin-3, BETA2, and RIPE3b1. Biochem Biophys Res Commun. 2007;354:334–339. doi: 10.1016/j.bbrc.2006.12.216. [DOI] [PubMed] [Google Scholar]
- 120.Nomura S, et al. MafA differentiates rat intestinal cells into insulin-producing cells. Biochem Biophys Res Commun. 2006;349:136–143. doi: 10.1016/j.bbrc.2006.08.032. [DOI] [PubMed] [Google Scholar]