Summary
CsrA of Escherichia coli is an RNA binding protein that globally regulates gene expression by repressing translation and/or altering the stability of target transcripts. Here we explored mechanisms that control csrA expression. Four CsrA binding sites were predicted upstream of the csrA initiation codon, one of which overlapped its Shine-Dalgarno sequence. Results from gel shift, footprint, toeprint and in vitro translation experiments indicate that CsrA binds to these four sites and represses its own translation by directly competing with 30S ribosomal subunit binding. Experiments were also performed to examine transcription of csrA. Primer extension, in vitro transcription and in vivo expression studies identified two σ70-dependent (P2 and P5) and two σS-dependent (P1 and P3) promoters that drive transcription of csrA. Additional primer extension studies identified a fifth csrA promoter (P4). Transcription from P3, which is indirectly activated by CsrA, is primarily responsible for increased csrA expression as cells transition from exponential to stationary phase growth. Taken together, our results indicate that regulation of csrA expression occurs by a variety of mechanisms, including transcription from multiple promoters by two sigma factors, indirect activation of its own transcription, as well as direct repression of its own translation.
Introduction
Bacteria use a wide variety of mechanisms for sensing and responding to environmental changes. Elaborate global regulatory networks orchestrate sweeping changes in numerous cellular processes in response to physiological demands. Several adaptations in response to nutrient starvation and other stresses occur during the transition from exponential to stationary phase growth. Disrupting global regulatory genes that mediate these responses have profound consequences for bacterial growth and survival (Romeo, 1998; Marles-Wright and Lewis, 2007; Potrykus and Cashel, 2008; Hengge, 2009).
Csr (carbon storage regulator) is a global regulatory system in Escherichia coli that represses or activates expression of a variety of genes (Babitzke and Romeo, 2007; Edwards et al., 2011). The Csr system, also referred to as Rsm (repressor of secondary metabolites) in some organisms, is widely distributed in eubacteria and regulates virulence, quorum sensing, cell motility, biofilm development and carbon metabolism in various species (Romeo, 1998; Babitzke and Romeo, 2007; Timmermans and Van Melderen, 2010). The Csr system of E. coli consists of four major components, the RNA binding protein CsrA (Liu and Romeo, 1997), two non-coding sRNA antagonists of CsrA (CsrB and CsrC) (Liu et al., 1997; Weilbacher et al., 2003), and CsrD, a protein that is necessary for degradation of CsrB and CsrC by RNase E (Suzuki et al., 2006). CsrA is a homodimer with two identical RNA-binding surfaces that can simultaneously bind two sites within a transcript (Dubey et al., 2003; Mercante et al., 2006, 2009). Although considerable sequence variation exists among CsrA binding sites, GGA is a highly conserved motif that is often present in the loop of short RNA hairpins (Liu et al., 1997; Dubey et al., 2005). CsrB and CsrC contain several CsrA binding sites and function by sequestering CsrA, thereby preventing CsrA interaction with mRNA targets (Weilbacher et al., 2003; Babitzke and Romeo, 2007).
CsrA represses translation initiation of several genes by competing with 30S ribosomal subunit binding. CsrA typically binds to multiple sites in the leader region of target mRNAs, one of which overlaps the cognate Shine-Dalgarno (SD) sequence (Baker et al., 2002, 2007; Dubey et al., 2003; Wang et al., 2005; Lapouge et al., 2007; Yakhnin et al., 2007). Translational repression by CsrA often leads to more rapid mRNA decay. CsrA also activates expression of certain genes, although a detailed activation mechanism has not been reported for any organism. In the case of the E. coli flhDC operon, CsrA activates expression by binding to and stabilizing the mRNA encoding for a transcriptional activator of flagellum synthesis and chemotaxis (Wei et al., 2001; Wang et al., 2006; Smith and Hoover, 2009).
CsrA is an abundant protein in E. coli and its concentration increases from ~6 μM in exponential phase to ~17 μM as the cells enter the stationary phase of growth (Gudapaty et al., 2001). Although CsrA appears to regulate several hundred genes (Edwards et al., 2011), the regulation of csrA expression had not been investigated. Thus, we explored mechanisms that control csrA expression. We found that CsrA binds to four sites in its leader transcript, thereby repressing its own translation by competing with ribosome binding. This autoregulatory mechanism is similar to CsrA-mediated translation repression mechanisms that regulate expression of a variety of E. coli genes (Baker et al., 2002, 2007; Dubey et al., 2003; Wang et al., 2005). We also found that csrA is transcribed from five promoters, two of which are primarily transcribed by EσS and two that are transcribed by Eσ70. CsrA indirectly activates transcription from a strong EσS–dependent csrA promoter. We propose that antagonistic translational and transcriptional autoregulatory processes help to maintain optimal CsrA expression in E. coli.
Results
CsrA binds to four sites in its own leader transcript
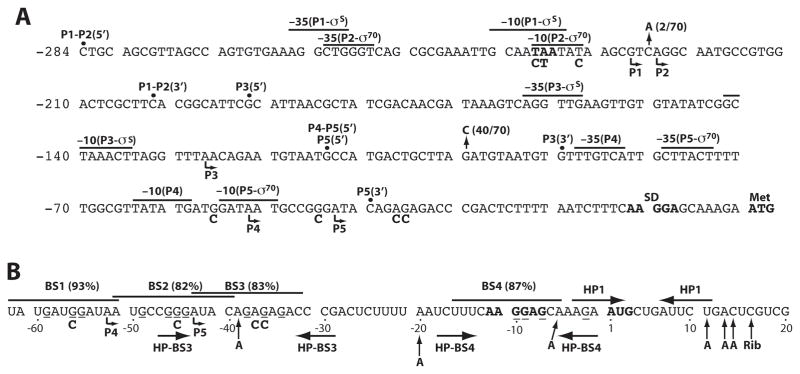
CsrA represses translation initiation of glgC, cstA, pgaA and hfq by binding as few as one (hfq) or as many as six (pgaA) sites in the untranslated leaders and/or translation initiation regions of these transcripts. In each case, one binding site overlaps the cognate SD sequence, such that bound CsrA blocks 30S ribosomal subunit binding (Baker et al., 2002, 2007; Dubey et al., 2003, Wang et al., 2005). It appears that CsrA represses translation of relA by a similar mechanism (Edwards et al., 2011). A position weighted matrix (pwm) search tool was previously developed to search for genes that might be regulated by CsrA. This program assigns values from 0 to 100% according to the minimum and maximum score calculated by the pwm (Baker et al., 2007). This program predicted the presence of four CsrA binding sites (BS1-BS4) in the csrA leader transcript with values ranging from 82% to 93% (Fig. 1B). Three of these binding sites (BS1, BS2 and BS4) contained a GGA motif, the most highly conserved component of CsrA binding sites (Dubey et al., 2005; Babitzke and Romeo, 2007). BS3 contained an AGA motif in place of the GGA, a situation that was previously observed for one of four CsrA binding sites in glgCAP leader RNA (Mercante et al., 2009). Furthermore, BS3 and BS4 had the potential to form short RNA hairpins with the GGA/AGA motif in the loop, a common feature of CsrA binding sites (Liu et al., 1997; Dubey et al., 2005; Babitzke and Romeo, 2007). Moreover, a pull down/RNA seq experiment independently identified csrA mRNA as an apparent target of CsrA binding (Edwards et al., 2011). Thus, experiments were performed to determine whether CsrA represses its own translation.
Fig. 1. Sequence of the csrA promoter and CsrA binding site regions.
A. csrA promoter region. Transcription start sites mapped by primer extension are marked by bent arrows. Putative -35 and -10 promoter elements for P1 through P5 are shown. Transcription start sites were verified for P1, P2, P3 and P5 by in vitro transcription. The csrA Shine-Dalgarno (SD) sequence and translation initiation codon (Met) are in bold. The alaS stop codon, which overlaps the P1 and P2 -10 promoter elements, is shown in boldface type. Vertical arrows mark the positions and frequency of two single nucleotide polymorphisms (SNPs) identified within the csrA promoter region of all sequenced E. coli strains, including pathogenic strains of E. coli. The 5′ endpoints for the P1-P5-csrA′-′lacZ, P3-P5-csrA′-′lacZ, P4-P5-csrA′-′lacZ and P5-csrA′-′lacZ translational fusions are shown. The common 3′ end point for all of the translational fusions is within the csrA coding sequence. The 5′ and 3′ endpoints are also shown for the P1-P2-csrA-lacZ, P3-csrA-lacZ, and P5-csrA-lacZ transcriptional fusions. The fusions containing only P5 were generated by deleting the -35 sequence element from P4. The identity of three point mutations in the overlapping -10 promoter elements for P1 and P2, as well as four point mutations in BS1-BS3, are shown below the wild type sequence. Numbering is with respect to the start of csrA translation.
B. CsrA binding sites in the untranslated csrA leader transcript (BS1-BS4). Position weight matrix (pwm) scores are in parentheses. The conserved GGA motif for BS4 and the corresponding AGA motif for BS3 have the potential of being in the loop of short hairpins (HP-BS3 and HP-BS4). Another potential RNA hairpin (HP1) is shown. The P4 and P5 transcription start sites are shown (bent arrow). G residues that were protected by CsrA from RNase T1 cleavage are indicated (−) below the nucleotide. Positions of CsrA (A) and 30S ribosome toeprints (Rib) are marked with vertical arrows. The identity of four point mutations in BS1-BS3 is shown below the wild type sequence. Numbering is with respect to the start of csrA translation.
Quantitative gel mobility shift assays were performed to characterize the interaction of CsrA with csrA mRNA containing all four of the predicted CsrA binding sites (−105 to +42 relative to the start of csrA translation) (Fig. 1). A distinct band with some smearing was observed between 16 and 128 nM CsrA, indicating that CsrA formed a complex with this transcript with an apparent Kd value of 27 nM (Fig. 2A). As the concentration of CsrA was increased further, additional shifted species of higher molecular weights were observed. While the stoichiometry of the shifted complexes was not examined, the first shifted species likely contains one bound CsrA dimer per transcript, with the slower migrating species containing multiple CsrA molecules bound to each transcript.
Fig. 2. Gel mobility shift analysis of CsrA-csrA RNA interaction.
A. Determination of the apparent equilibrium binding constant (Kd) of CsrA-csrA RNA interaction. 5′-end-labeled csrA RNA (0.1 nM) was incubated with the concentration of CsrA shown at the bottom of each lane. Positions of bound (B) and free (F) RNA are shown. The simple binding curve for these data is shown at the right.
B. Competition assay. Labeled csrA RNA (0.1 nM) was incubated with CsrA in the absence or presence of specific (csrA and flhD) or non-specific (phoB) competitor RNA. Positions of bound (B) and free (F) RNA are shown.
The specificity of CsrA-csrA RNA interaction was investigated by performing competition experiments with specific (csrA and flhD) and non-specific (phoB) unlabeled RNA competitors (Fig. 2B). Unlabeled csrA was an effective competitor when it was in 500-fold molar excess over labeled csrA (0.1 nM labeled RNA vs. 50 nM unlabeled RNA). As the CsrA concentration was 250 nM, effective competition required a high concentration of unlabeled RNA to titrate out free CsrA before effective competition was observed. flhD RNA also competed for CsrA-csrA RNA interaction, although it required a higher concentration of unlabeled RNA. The finding that flhD did not compete as well as csrA may be explained by the fact that csrA and flhD RNAs contain four (see below) and two (unpublished data) CsrA binding sites, respectively. Importantly, the non-specific competitor RNA (phoB) did not compete with the CsrA-csrA RNA interaction. These results establish that CsrA binds specifically to its own transcript.
CsrA-csrA RNA footprint experiments were performed to determine whether CsrA binds to each of the predicted sites. Each reaction contained 4 nM labeled RNA. CsrA protected several G residues from RNase T1 cleavage at the lowest concentration of protein used (0.25 μM). With the exception of position −29, which lies between BS3 and BS4, CsrA protected all of the G residues extending from −59 to −2 (Fig. 1B and Fig. 3). Note that the G residues in BS2 were poorly cleaved even in the absence of CsrA. However, when the exposure intensity of this gel was increased it became evident that bound CsrA protected these residues from cleavage as well (Fig. 3B). While the footprinting results indicate that CsrA binds to all four of the predicted sites, BS3, which is the only binding site without the highly conserved GGA motif, was not protected as strongly as the other three sites.
Fig. 3. CsrA-csrA RNA footprint analysis.

A. 5′-end-labeled csrA RNA was treated with RNase T1 in the presence of the concentration of CsrA shown at the top of each lane. Partial alkaline hydrolysis (OH) and RNase T1 digestion (T1) ladders, as well as a control lane in the absence of RNase T1 treatment (C), are shown. The RNase T1 ladder was generated under denaturing conditions so that every G residue in the transcript could be visualized. Residues in which bound CsrA reduced RNase T1 cleavage are marked (−). Positions of the CsrA binding sites (BS1-BS4), the csrA Shine-Dalgarno (SD) sequence and the translation initiation codon (Met) are shown. Numbering is with respect to the start of csrA translation.
B. This gel fragment is a higher exposure of the BS2 region of the same gel shown in (A).
CsrA represses its own translation by competing with ribosome binding
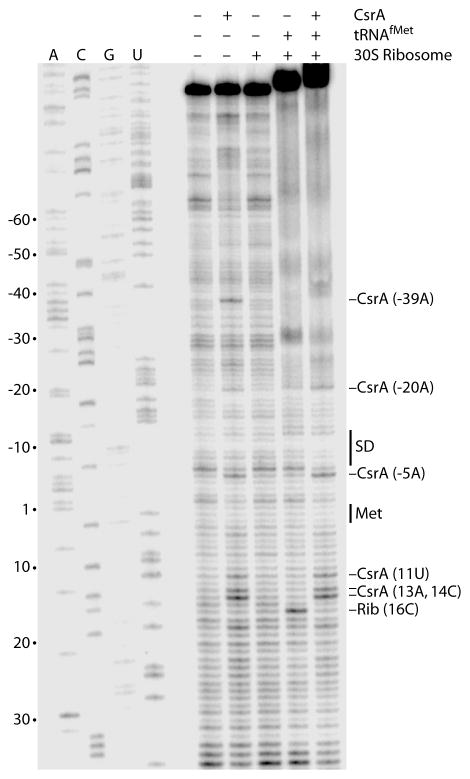
Since our footprint results demonstrated that CsrA binds to four sites in the csrA leader transcript including the site overlapping its SD sequence (BS4), toeprint experiments were performed to determine whether bound CsrA inhibited ribosome binding. CsrA-dependent toeprint bands were observed at several positions including −39A and −5A, which are at the 3′ edges of BS2 and BS4, respectively (Fig. 1B and Fig. 4, compare first and second lanes). CsrA-dependent stabilization of a short RNA hairpin (HP1) just downstream from BS4 could be responsible for the cluster of CsrA-dependent toeprints between 11U and 14C (Fig. 1B). However, we do not have an explanation for the CsrA-dependent toeprint at −20A. Toeprint experiments were also performed to identify the position of bound 30S ribosomal subunits. A tRNAfMet-dependent 30S ribosomal subunit toeprint was observed at position 16C, which is at the expected position 15 nt downstream from the A of the initiation codon (Fig. 4, compare third and fourth lanes). When CsrA was bound to the csrA transcript prior to the addition of 30S ribosomal subunits and tRNAfMet, all of the CsrA toeprints bands were observed except for that at −39A, whereas the ribosome toeprint was absent (Fig. 4, compare second, fourth and fifth lanes). These results demonstrate that CsrA competes with ribosome binding to its own transcript.
Fig. 4.
CsrA and 30S ribosomal subunit toeprint analysis of csrA RNA. The absence (−) or presence (+) of CsrA, tRNAfMet and/or 30S ribosomal subunits are shown at the top of each lane. CsrA was added prior to tRNAfMet and 30S ribosomal subunits when they were present in the same reaction. Positions of CsrA and 30S ribosomal subunit (Rib) toeprint bands are marked. Positions of the csrA Shine-Dalgarno (SD) sequence and the translation initiation codon (Met) are shown. Sequencing lanes to reveal A, C, G or U residues are labeled. Numbering is with respect to the start of csrA translation.
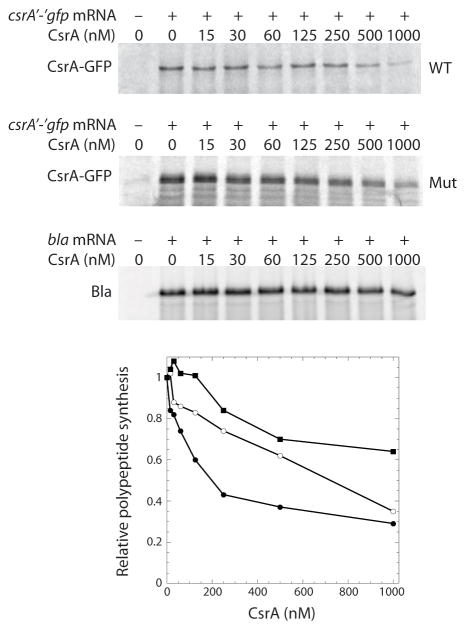
The in vitro binding studies demonstrated that bound CsrA blocks ribosome binding; therefore, in vitro translation experiments were performed with S-30 extracts produced from a CsrA-deficient strain to determine whether CsrA represses its own synthesis. Our initial attempt to examine the effect of purified CsrA on translation of csrA was problematic, as it appeared that the newly synthesized protein was rapidly degraded (not shown). Since we previously found that using gfp translational fusions circumvented similar problems in cell-free translation experiments (Yakhnin et al., 2006; Baker et al., 2007), a csrA′-′gfp translational fusion transcript (130 nM) was tested. The fusion transcript produced a stable CsrA-Gfp fusion protein. Importantly, addition of increasing concentrations of CsrA to the reaction led to a corresponding decrease in CsrA-Gfp synthesis. Translational repression is particularly evident at CsrA concentrations ≥ 250 nM. At the highest concentration of CsrA tested (1 μM), translation of the fusion protein was inhibited 70% (Fig. 5). Similar experiments were performed with the bla transcript as a negative control. Slight CsrA-dependent inhibition of bla translation was observed at the higher CsrA concentrations (Fig. 5).
Fig. 5.
Effect of CsrA on in vitro translation of csrA′-′gfp mRNA. The E. coli S-30 extract was prepared from a CsrA-deficient (csrA::kan) strain. Reactions were carried out with the concentration of CsrA indicated at the top of each lane in the absence (−) or presence (+) of wild type (WT) or binding site mutant (Mut) csrA′-′gfp or control (bla) transcripts. CsrA-GFP and Bla translation products were analyzed by SDS-PAGE. The relative levels of CsrA-GFP and Bla synthesis as a function of CsrA concentration is shown at the bottom. The level of polypeptide synthesis in the absence of CsrA was set to 1.0 for each transcript. Symbols are: ●, CsrA-GFP (WT); ○, CsrA-GFP (Mut); ν, Bla.
As a further test of CsrA-mediated autoregulation, a transcript containing a total of four point mutations in BS1, BS2 and BS3 was generated (Fig. 1). These mutations alter the highly conserved GGA motifs in BS1 and BS2, as well as the corresponding AGA motif in BS3. Note that BS4 was not altered because this binding site overlaps the csrA SD sequence (Fig. 1B). The mutant transcript was tested in gel shift and in vitro translation experiments. Gel mobility shift results indicated that the affinity for the mutant transcript was reduced almost 15-fold (not shown). As expected, the binding site mutations reduced CsrA-mediated translational repression, particularly between 125 and 500 nM CsrA (Fig. 5). Taken together, our biochemical results establish that CsrA represses its own synthesis by blocking ribosome access to its SD sequence.
Transcription of csrA is driven by five promoters
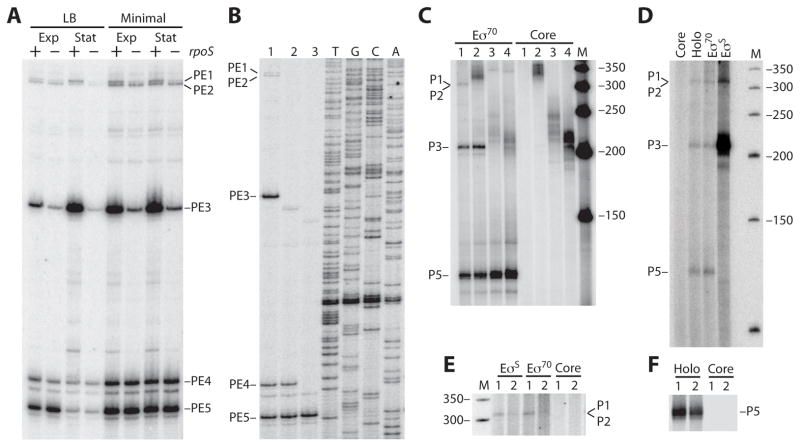
To gain a more thorough understanding of the mechanisms controlling csrA expression, primer extension studies were performed to identify potential csrA transcription start sites in vivo. Total cellular RNA was extracted in late exponential and early stationary phase growth from a strain containing pCSB68. This plasmid contained a csrA′-′lacZ translational fusion containing csrA sequences extending from −350 to +56 relative to the start of translation. Purified RNA was then subjected to primer extension analysis using a primer specific for the csrA′-′lacZ fusion junction. Several primer extension products (PE1-PE5) were detected when cells were grown in LB or minimal media (Fig. 6A). The relative abundance of PE3 increased in early stationary phase when cells were grown in LB and to a lesser extent in minimal media. A similar pattern was observed for PE1. In contrast, the level of PE5 decreased in stationary phase cultures grown in LB. Potential σ70-dependent promoters were identified that could give rise to each of these primer extension products, although P1, P2 and P3 had poor −35 sequence elements (Fig. 1A).
Fig. 6. Identification of five csrA promoters.
A. Primer extension mapping of the 5′ end of csrA transcripts. Wild type (+) and rpoS mutant (−) strains containing pCSB68 (csrA′-′lacZ) were grown in LB or minimal media. Total RNA was isolated from late exponential (Exp) and early stationary (Stat) phase cultures. RNA was hybridized to an end-labeled DNA primer and subsequently extended with reverse transcriptase. Sequencing reactions were performed using the same end-labeled primer (not shown). Reverse transcriptase products identified five potential transcription start sites (PE1-PE5). Potential promoter sequences (P1-P5) corresponding to PE1-PE5 are shown in figure 1A.
B. Primer extension analysis of P4. Total cellular RNA was isolated from strains grown in LB to late exponential phase. RNA was hybridized to an end-labeled DNA primer and subsequently extended with reverse transcriptase. Sequencing reactions were performed using the same end-labeled primer. Primer extension products (PE1-PE5) are marked. Lane 1, PLB1415 [pYH159 (P1-P5-csrA′-′lacZ)]; lane 2, PLB1418 [pYH162 (P4-P5-csrA′-′lacZ)]; lane 3, PLB1416 [pYH161 (P5-csrA′-′lacZ)].
C. In vitro transcription of csrA usingE σ70 or core RNA polymerase. Lanes 1, DNA template containing all 5 potential promoters (P1-P5). Lanes 2, DNA template containing P3-P5. Lanes 3, DNA template containing P4 and P5. Lanes 4, DNA template containing P5. Lane M, DNA size markers. Transcripts corresponding to P1 and/or P2, P3 and P5 are marked.
D. In vitro transcription using core, Eσ70 (Holo), reconstituted Eσ70 or reconstituted EσS. The DNA template contained all 5 promoters (P1-P5). Lane M, DNA size markers. Transcripts corresponding to P1 and/or P2, P3 and P5 are marked.
E. In vitro transcription using core, reconstituted Eσ70 or reconstituted EσS. The gel region corresponding to P1 and P2 is shown. Lanes 1, wild type DNA template containing all 5 potential promoters (P1-P5). Lanes 2, identical template as for lanes 1 except that it contained three point mutations in the overlapping -10 promoter sequences for P1 and P2 (Fig. 1A). Lane M, DNA size markers.
F. In vitro transcription using core or holo (Eσ70) RNA polymerase. The gel region corresponding to P5 is shown. Lanes 1, wild type DNA template. Lanes 2, identical template as for lanes 1 except that it contained four total point mutations in BS1–3 (Fig. 1).
To determine which of these predicted promoters were recognized by σ70, we carried out in vitro transcription experiments on a variety of DNA templates with Eσ70 holoenzyme or with core RNA polymerase as a control. When the DNA template contained all five putative promoters, σ70-dependent transcripts corresponding to promoters P1/P2, P3 and P5 were observed (Fig. 6C, lanes 1). As PE1 and PE2 only differ in size by three nucleotides (Fig. 1A and 6A), this gel system was unable to resolve in vitro generated transcripts derived from P1 and P2. When the DNA template was truncated on the 5′ end such that only P3, P4 and P5 were present, σ70-dependent transcripts corresponding to P3 and P5 were observed (Fig. 6C, lanes 2). When a template containing P4 and P5 was used, a σ70-dependent transcript corresponding to P5, but not P4, was observed (Fig. 6C, lanes 3). The same P5-derived transcript was observed when the −35 promoter element for P4 was deleted from the P4-P5 template (Fig. 6C, lanes 4). Thus, we conclude that Eσ70 recognizes P1 and/or P2, P3, and P5 in vitro.
Although the housekeeping sigma factor (σ70) and the stationary phase and stress response sigma factor (σS) recognize similar promoter sequences, σS-dependent promoters have a highly conserved C residue immediately upstream of the −10 hexamer at the −13 position, as well as a less conserved G residue at the −14 position (Becker and Hengge-Aronis, 2001). Inspection of the csrA promoter region revealed this sequence arrangement for P1 and P3 (Fig. 1A). Therefore, we repeated the primer extension analysis in an rpoS mutant strain using the same growth conditions described above. The relative abundance of PE1 and PE3 were greatly reduced in the rpoS strain irrespective of the growth stage or media used, consistent with P1 and P3 functioning as σS-dependent promoters (Fig. 6A).
To verify that P1 and P3 were recognized byσS, in vitro transcription experiments were performed using a DNA template containing the entire csrA promoter region (P1-P5). Transcription reactions were carried out with EσS reconstituted from σS and core RNAP (Fig. 6D). Transcription reactions with core RNAP, Eσ70 holoenzyme and reconstituted Eσ70 served as controls. Our data indicate that P1 and P3 are authentic σS-dependent promoters (Fig. 6D). From our primer extension and in vitro transcription data we conclude that P1 is weakly transcribed by Eσ70 and EσS, that P2 is weakly transcribed by Eσ70, that P3 is a strongly transcribed by EσS and weakly by Eσ70, and that P5 is an intermediate strength promoter transcribed by Eσ70.
The lack of transcription from P4 in vitro was somewhat surprising as this promoter is similar to the σ70 consensus promoter and PE4 was a prominent band in our primer extension analysis (Fig. 1A and 6A). Thus, it was possible that PE4 represented transcription from P4 or a cleavage product from a transcript derived from P1, P2 or P3. To distinguish between these two possibilities, we carried out primer extension studies with strains containing plasmid-borne csrA′-′lacZ translational fusions containing all five promoters (P1-P5-csrA′-′lacZ), P4 and P5 (P4-P5-csrA′-′lacZ) or P5 only (P5-csrA′-′lacZ) in which the −35 sequence from P4 was deleted. If PE4 is derived from RNA cleavage we would only expect to observe this band from the strain containing the P1-P5-csrA′-′lacZ translational fusion. In contrast, if PE4 is derived from P4 we would expect to observe this band in strains containing P1-P5-csrA′-′lacZ or P4-P5-csrA′-′lacZ. As an equivalent amount of PE4 was observed whenever P4 was present in the fusion we conclude that P4 represents an authentic csrA promoter (Fig. 6B).
In vivo expression pattern of csrA from multiple promoters
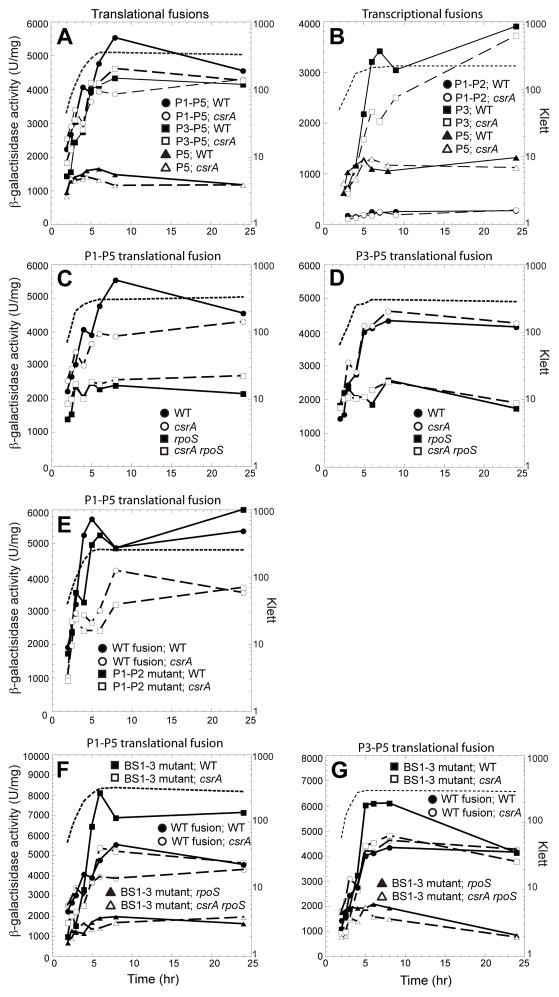
To further explore expression from the csrA promoter region, strains were constructed with chromosomally integrated csrA′-′lacZ translational fusions containing P1 through P5 (P1-P5-csrA′-′lacZ), P3 through P5 (P3-P5-csrA′-′lacZ), P4 and P5 (P4-P5-csrA′-′lacZ) or P5 only (P5-csrA′-′lacZ) (Fig. 1A). Note that the −35 promoter sequence for P4 was deleted from the latter fusion. Expression of the integrated P1-P5-csrA′-′lacZ and P3-P5-csrA′-′lacZ translational fusions increased several fold during the transition from exponential to stationary phase growth, whereas expression of the P4-P5-csrA′-′lacZ and P5-csrA′-′lacZ translational fusions exhibited modest increases during this transition (Fig. 7A). Furthermore, the latter two fusions exhibited nearly identical expression levels throughout growth. Thus it appears that transcription from P5 compensates for loss of transcription from P4 and/or that P5 effectively competes with the overlapping P4 promoter for RNA polymerase in vivo. This latter possibility is consistent with the absence of P4 transcription in vitro (Fig. 6C). Thus, we focused our attention on P1, P2, P3 and P5 in the expression studies described below.
Fig. 7. In vivo expression pattern of csrA from P1-P5 in wild type and rpoS strains.
Cells were grown in LB at 37 °C. Representative growth curves are shown in each panel (dashed line). Each experiment was performed at least three times. Results from representative experiments are shown.
A. Expression pattern of csrA′-′lacZ translational fusions. Symbols for β-galactosidase activity:●, P1-P5-csrA′-′lacZ (PLB1424);ν, P3-P5-csrA′-′lacZ (PLB1425); ○, P4-P5-csrA′-′lacZ (PLB1426);▲, P5-csrA′-′lacZ (PLB1427).
B. Expression pattern of csrA-lacZ transcriptional fusions. Symbols for β-galactosidase activity:●, P1-P2-csrA-lacZ (PLB1434); ν, P3-csrA-lacZ (PLB1429); ▲, P5-csrA-lacZ (PLB1707).
C. Effect of rpoS on expression of csrA-lacZ transcriptional fusions. Symbols for β-galactosidase activity:●, P1-P2-csrA-lacZ rpoS+ (PLB1434); ○, P1-P2-csrA-lacZ rpoS::Tn10 (PLB1457); ν, P3-csrA-lacZ rpoS+ (PLB1429);□, P3-csrA-lacZ rpoS::Tn10 (PLB1433);▲, P5-csrA-lacZ rpoS+ (PLB1707); △, P5-csrA-lacZ rpoS::Tn10 (PLB1709).
Expression studies were also carried out with P1-P2-csrA-lacZ, P3-csrA-lacZ and P5-csrA-lacZ transcriptional fusions (Fig. 7B). Expression from both P3 and P5 peaked during the transition from exponential to stationary phase growth, with expression from P3 being 3- to 4-fold higher than expression from P5. Expression from the P1-P2 fusion was considerably lower than from the other two fusions and was largely unaffected by the growth stage. These results generally reflect our primer extension results from cells grown in LB (Fig. 6A). Thus, we conclude that P3 is primarily responsible for the large increase of csrA expression during the transition from exponential to stationary phase growth.
As the primer extension and in vitro transcription results indicated that P3 was transcribed by EσS (Fig. 6A and 6D), we examined expression of csrA in WT and rpoS (σS-deficient) strains. Expression of the P3-csrA-lacZ transcriptional fusion was 20-fold lower in the rpoS mutant strain (Fig. 7C), confirming that P3 is primarily transcribed by EσS in vivo. Expression levels from the P1-P2-csrA-lacZ and P5-csrA-lacZ transcriptional fusions were largely unaffected by rpoS (Fig. 7C). While the results for the P3 and P5 fusions are consistent with the primer extension and in vitro transcription results (Fig. 6), we expected to see an effect of rpoS on the P1-P2 fusion. Perhaps expression from P2 compensates for loss of σS-dependent expression from P1.
CsrA indirectly activates transcription from P3
As described above, expression from the integrated P1-P5-csrA′-′lacZ translational fusion increased several fold during the transition from exponential to stationary phase growth. Despite our previous finding that CsrA represses its own translation, we found that peak expression of the P1-P5-csrA′-′lacZ fusion was reduced ~40% in the csrA mutant strain, suggesting that CsrA indirectly activates its own transcription and that activation overshadows CsrA-mediated translational repression (Fig. 8A). The finding that expression from the P3-P5-csrA′-′lacZ and P5-csrA′-′lacZ translational fusions were unaffected by the csrA mutation, suggested that activation involves DNA sequences upstream from the 5′ endpoint used to generate the P3-P5 translational fusion (Fig. 1A and 8A).
Fig. 8. CsrA indirectly activates transcription from P3.
Cells were grown in LB at 37 °C. Representative growth curves are shown in each panel (dashed line). Each experiment was performed at least three times. Results from representative experiments are shown.
A. Effect of CsrA on expression of csrA′-′lacZ translational fusions. Symbols for β-galactosidase activity: ●, P1-P5-csrA′-′lacZ csrA+ (PLB1424); ○, P1-P5-csrA′-′lacZ csrA::kan (PLB1442); ν, P3-P5-csrA′-′lacZ csrA+ (PLB1425); □, P3-P5-csrA′-′lacZ csrA::kan (PLB1443);▲, P5-csrA′-′lacZ csrA+ (PLB1427); △, P5-csrA′-′lacZ csrA::kan (PLB1445).
B. Effect of csrA on expression of csrA-lacZ transcriptional fusions. Symbols for β-galactosidase activity: ●, P1-P2-csrA-lacZ csrA+ (PLB1434); ○, P1-P2-csrA-lacZ csrA::kan (PLB1455); ν, P3-csrA-lacZ csrA+ (PLB1429); □, P3-csrA-lacZ csrA::kan (PLB1453);▲, P5-csrA-lacZ csrA+ (PLB1707); △, P5-csrA-lacZ csrA::kan (PLB1708).
C. Effect of CsrA and RpoS on expression of the P1-P5-csrA′-′lacZ translational fusion. Symbols for β-galactosidase activity: ●, csrA+ rpoS+ (PLB1424); ○, rpoS+ csrA::kan (PLB1442);ν, csrA+ rpoS::Tn10 (PLB1712); □, csrA::kan rpoS::Tn10 (PLB1710).
D. Effect of CsrA and RpoS on expression of the P3-P5-csrA′-′lacZ translational fusion. Symbols for β-galactosidase activity: ●, csrA+ rpoS+ (PLB1425); ○, rpoS+ csrA::kan (PLB1443);ν, csrA+ rpoS::Tn10 (PLB1449); □, csrA::kan rpoS::Tn10 (PLB1716).
E. Effect of P1 and P2 promoter mutations on expression of the P1-P5-csrA′-′lacZ translational fusion. Symbols for β-galactosidase activity:●, WT fusion csrA+ (PLB1424); ○, WT fusion csrA::kan (PLB1442);ν, mutant fusion csrA+ (PLB1720); □, mutant fusion csrA::kan (PLB1721).
F. Effect of CsrA binding site mutations on expression of the P1-P5-csrA′-′lacZ translational fusion. Symbols for β-galactosidase activity: ●, WT fusion csrA+ rpoS+ (PLB1424); ○, WT fusion rpoS+ csrA::kan (PLB1442);ν, BS1-BS2-BS3 mutant fusion csrA+ rpoS+ (PLB1487); □, BS1-BS2-BS3 mutant fusion rpoS+ csrA::kan (PLB1489); ▲, BS1-BS2-BS3 mutant fusion csrA+ rpoS::Tn10 (PLB1714); △, BS1-BS2-BS3 mutant fusion csrA::kan rpoS::Tn10 (PLB1715).
G. Effect of CsrA binding site mutations on expression of the P3-P5-csrA′-′lacZ translational fusion. Symbols for β-galactosidase activity: ●, WT fusion csrA+ rpoS+ (PLB1425); ○, WT fusion rpoS+ csrA::kan (PLB1443);ν, BS1-BS2-BS3 mutant fusion csrA+ rpoS+ (PLB1495); □, BS1-BS2-BS3 mutant fusion rpoS+ csrA::kan (PLB1717); ▲, BS1-BS2-BS3 mutant fusion csrA+ rpoS::Tn10 (PLB1718); △, BS1-BS2-BS3 mutant fusion csrA::kan rpoS::Tn10 (PLB1719).
Similar expression studies were carried out with P1-P2-csrA-lacZ, P3-csrA-lacZ and P5-csrA-lacZ transcriptional fusions (Fig. 8B). The 5′ endpoints of the transcriptional fusions were identical to those for the corresponding translational fusions described above (Fig. 1A). Expression of the P1-P2-csrA-lacZ and P5-csrA-lacZ transcriptional fusions were unaffected by csrA, whereas expression from P3 was reduced by ~40% in the csrA mutant. Thus, it is apparent that CsrA-dependent activation can occur in the absence of sequences upstream from the common 5′ endpoint used to generate the P3-P5 translational and P3 transcriptional fusions.
To further examine CsrA-dependent activation, expression of the P1-P5 and P3-P5 translational fusions were tested in WT, csrA, rpoS and csrA rpoS strains (Fig. 8C and 8D). CsrA-dependent activation of the P1-P5 translational fusion was lost in the rpoS genetic background, consistent with CsrA-dependent activation functioning through P3 (Fig. 8C). To rule out the possibility that transcription from P1 or P2 is responsible for CsrA-dependent activation, expression studies were conducted with a fusion in which the overlapping −10 promoter sequences for these two promoters were mutated (Fig. 1A). Note that these mutations abolished transcription from P1 and P2 in vitro (6E). The finding that CsrA-dependent activation was retained with this mutant fusion confirms that CsrA indirectly activates transcription of P3 (Fig. 8E).
We next tested the effect of the mutations in the first three CsrA binding sites described above (Fig. 1). Consistent with the approximate 15-fold reduction in binding affinity and reduced translational repression in vitro (Fig. 5 and data not shown), expression of the mutant P1-P5-csrA′-′lacZ translational fusion was higher than expression from the WT fusion. Furthermore, CsrA-dependent activation of the mutant fusion was lost in an rpoS genetic background (Fig. 8F). Identical experiments were carried out with WT and binding site mutant P3-P5 translational fusions. As CsrA-dependent activation was observed for the mutant fusion in rpoS+ but not in rpoS mutant strain, it is apparent that loss of translational repression uncovered CsrA-dependent activation of P3 (Fig. 8G). Taken together, our results are consistent with a model in which CsrA represses its own translation by binding to BS1-BS4, while CsrA indirectly activates transcription of P3. Furthermore, CsrA-dependent activation of P3 involves sequences upstream and downstream of position −114 of the csrA promoter region (Fig. 1A).
Expression studies were also conducted with the P1-P5, P3-P5 and P5 translational fusions to compare the effects of CsrA deficiency and CsrA overproduction on csrA expression. Expression of the P1-P5 translational fusion was highest when the single chromosomal copy of csrA was present and lowest when the strain contained a plasmid that overproduced CsrA (Fig. 9A). Expression of the P3-P5 translational fusion was also lowest in strains that overproduced CsrA (Fig. 9B). In contrast, expression of the P5 translational fusion was unaffected by the level of CsrA in the cell (Fig. 9C). Taken together, these results suggest that a single copy of csrA indirectly increases expression of csrA, perhaps by activating an activator or repressing a repressor of P3 transcription. However, CsrA-dependent activation of csrA transcription is partly tempered by CsrA-mediated repression of its own translation. These results also suggest that overproduction of CsrA from a plasmid leads to stronger translational repression of transcripts originating from P1 or P3, thereby overshadowing the indirect affect of CsrA on increasing its own transcription.
Fig. 9. Effect of csrA overexpression on expression csrA′-′lacZ translational fusions. Cells were grown in LB at 37 °C. Representative growth curves are shown in each panel (dashed line). Each experiment was performed at least three times. Results from representative experiments are shown.
A. Effect of csrA overexpression on expression of the P1-P5-csrA′-′lacZ translational fusion. Symbols for β-galactosidase activity: ●, csrA+pYH172 (vector) (PLB1465); ○, csrA::kan pYH172 (vector) (PLB1467); ν, csrA+pYH171 (csrA+) (PLB1464); □, csrA::kan pYH171 (csrA+) (PLB1466).
B. Effect of csrA overexpression on expression of the P3-P5-csrA′-′lacZ translational fusion. Symbols for β-galactosidase activity: ●, csrA+pYH172 (vector) (PLB1471); ○, csrA::kan pYH172 (vector) (PLB1473); ν, csrA+pYH171 (csrA+) (PLB1470); □, csrA::kan pYH171 (csrA+) (PLB1472).
C. Effect of csrA overexpression on expression of the P5-csrA′-′lacZ translational fusion. Symbols for β-galactosidase activity: ●, csrA+pYH172 (vector) (PLB1479); ○, csrA::kan pYH172 (vector) (PLB1481); ν, csrA+pYH171 (csrA+) (PLB1478); □, csrA::kan pYH171 (csrA+) (PLB1480).
The results shown in figure 9C demonstrate that CsrA does not measurably repress translation of transcripts originating from P5 despite the presence of BS3 and BS4 in these transcripts (Fig. 1B). This finding suggested that the affinity of CsrA for transcripts containing only these two binding sites is insufficient to repress translation. As BS3 does not contain the highly conserved GGA motif (Fig. 1) and was least protected by CsrA in RNA footprints (Fig. 3), BS3 probably constitutes a low-affinity binding site. Consistent with this interpretation, gel mobility shift results indicated that the affinity of CsrA for a transcript containing only BS3 and BS4 was ~15-fold lower than for a transcript containing all four binding sites (data not shown). Thus, while BS4 is the binding site that overlaps the ribosome binding site, BS1 and BS2 are required to repress translation.
Discussion
CsrA is the key component of a global regulatory system in E. coli that represses or activates expression of a large number of genes. CsrA represses translation initiation of several genes by directly competing with 30S ribosomal subunit binding (Baker et al., 2002, 2007; Dubey et al., 2003; Wang et al., 2005; Edwards et al., 2011). The results presented here establish that CsrA regulates its own expression by essentially the same translational repression mechanism (Fig. 2, 3, 4 and 5). The CsrA-mediated translational repression mechanism identified in E. coli is assumed to function in several other organisms, although translational repression has only been verified for the hag gene of Bacillus subtilis (Babitzke and Romeo, 2007; Yakhnin et al., 2007; Timmermans and Van Melderen, 2010). In addition to direct competition for 30S ribosomal subunit binding, CsrA-mediated translational repression could occur by a variety of mechanisms. For example, it was recently shown that RsmA from Pseudomonas aeruginosa represses translation initiation of pslA by promoting formation of an RNA hairpin that sequesters the pslA SD sequence (Irie et al., 2010).
We also identified five promoters (P1 through P5) that drive transcription of csrA (Fig. 1A and 6). P1 is weak promoter that is recognized by σS and σ70 (Fig. 6 and 7). P2, which overlaps P1, is a weak σ70-dependent promoter (Fig. 6 and 7). P3 is a strong σS-dependent promoter that is also recognized by σ70, whereas P5 is a σ70-dependent promoter of intermediate strength (Fig. 6 and 7). Finally, an intermediate level of transcription was observed from P4 in vivo but not in vitro (Fig. 6). The lack of in vitro transcription from P4 could have occurred for several reasons. First, transcription might require a sigma factor other than σ70 or σS. Second, P4 might require an unidentified activator protein. Third, as the P4 and P5 promoter sequences overlap, P5 might compete with P4 transcription in vitro; however, we don’t understand how P4 would overcome P5 competition in vivo. Clearly, additional studies are required to understand P4 function.
Of particular interest, the large increase in csrA expression during the transition from exponential to stationary phase growth is largely due to σS-dependent transcription from P3 (Fig. 6 and 7). These results are consistent with previous studies showing that σS accumulates during the transition from exponential to stationary phase growth, thereby reprogramming the global gene expression pattern (Hengge, 2009).
Another interesting observation is that an intrinsic terminator is not found downstream from alaS, the gene immediately upstream of csrA. Thus, in addition to the csrA-specific promoters, transcriptional readthrough from alaS may contribute to csrA expression. As the alaS-csrA intergenic region, including the promoters and CsrA binding sites, only contains two single nucleotide polymorphisms (SNPs) among 70 sequenced E. coli strains (Fig. 1A), it is apparent that transcriptional and translational regulation of csrA expression is conserved in laboratory and pathogenic strains of E. coli.
In addition to directly repressing its own translation, CsrA indirectly activates its own transcription. Expression of the P1-P5-csrA′-′lacZ translational and P3-csrA-lacZ transcriptional fusions decreased in csrA mutant strains (Fig. 8A and 8B), suggesting that CsrA indirectly activates transcription of P3. This interpretation was confirmed by the finding that indirect activation of the P1-P5-csrA′-′lacZ translational fusion was retained when transcription from P1 and P2 was eliminated by mutating their overlapping −10 promoter sequences (Fig. 1A, 6E and 8E). Interestingly, CsrA-dependent activation was not observed for the wild type P3-P5-csrA′-′lacZ translational fusion; however, activation was observed when translational repression was removed by mutating the first three CsrA binding sites (Fig 8G), confirming the opposing effects of CsrA on its own transcription and translation. In addition, CsrA-dependent activation of the P1-P5-csrA′-′lacZ translational fusion was accentuated by mutations in these three CsrA binding sites (Fig. 8F). Thus, our results are consistent with a model in which indirect activation of P3 transcription by CsrA involves two distinct DNA sequences, one of which is located upstream from the 5′ endpoint of the P3-P5 translational fusion and one that is downstream from this fusion junction. Moreover, both of these regions appear to be required for maximal activation. As CsrA binds to the transcripts of 32 genes that encode known or putative transcriptional regulators (Edwards et al., 2011), it is likely that CsrA indirectly activates its transcription by activating an activator or repressing a repressor that directly regulates P3 transcription. While a definitive biological role for the opposing effects of CsrA on its own expression is lacking, its direct effect on translation would occur more rapidly than its indirect effect on transcription. Thus, in addition to contributing to the overall fine-tuning of CsrA activity in the cell, translational repression may provide a rapid mechanism to reduce CsrA synthesis when the concentration of free CsrA reaches a critical level.
The Csr regulatory circuitry in E. coli is also controlled by a variety of negative feedback loops (Fig. 10). In one case CsrA represses expression of CsrD, a protein that is required for degradation of CsrB and CsrC by RNase E. Thus, CsrA indirectly stabilizes its own sRNA antagonists (Suzuki et al., 2006). This regulatory loop is also present in Salmonella enterica (Jonas et al., 2010). In another regulatory loop CsrA indirectly stimulates transcription of csrB and csrC via the BarA-UvrY two-component signal transduction system (TCS) (Suzuki et al., 2002). This TCS responds to acetate and other short chain fatty acids (Chavez et al., 2010). Our discovery of CsrA-mediated translational autoregulation constitutes a third negative feedback loop. The circuitry surrounding CsrA implies that maintenance of optimal CsrA activity is a crucial process. As several promoters control transcription of csrA, perhaps each responding to specific signals, it is readily apparent that the control of csrA expression is complex. Perhaps complex regulation of csrA expression should not be surprising, as CsrA appears to directly control expression of hundreds of genes in E. coli (Edwards et al., 2011).
Fig. 10.

Model of the Csr regulatory circuitry. CsrA represses expression of target mRNAs, including its own transcript, by inhibiting translation initiation. CsrA is also capable of activating expression of mRNAs by unknown mechanisms. RpoS activates transcription of csrA. CsrA indirectly activates transcription of its RNA antagonists CsrB and CsrC via the BarA-UvrY two-component signal transduction system. Phosphorylated UvrY directly activates transcription of csrB and csrC. CsrB and CsrC contain several CsrA binding sites such that each sRNA is capable of sequestering several CsrA dimers. CsrA represses expression of CsrD, a protein that is required for degradation of CsrB and CsrC by RNase E. Finally, CsrA indirectly activates its own transcription by an unknown mechanism. Although not shown, ppGpp and DksA activate transcription of CsrB and CsrC via BarA-UvrY.
CsrA (RsmA) homologs have been identified in a large number of bacterial species. Depending on the particular organism, Csr controls a variety of cellular processes and behaviors, such as quorum sensing, biofilm development, motility and chemotaxis, central carbon flux and pathogenesis (Babitzke and Romeo, 2007; Timmermans and Van Melderen, 2010). It is particularly exciting that the E. coli Csr circuitry is interconnected with the σS global regulatory network, resulting in a concerted response to cellular stresses, such as nutrient limitation. Thus, it is tempting to speculate that CsrA activity in the cell is fine tuned in response to a wide variety of signals, ultimately governing cellular physiology and behavior on a scale that extends well beyond the genes and processes known to be under direct control of the Csr system.
Experimental Procedures
Bacterial strains and plasmids
E. coli strains used in this study are listed in table 1. Detailed descriptions of the strains and plasmids used in this study are in Supporting Information.
Table 1.
E. coli strains used in this study
| Strain | Descriptiona | Source |
|---|---|---|
| CAG45133 | λ[rpoHp3::lacZ] rpoS::Tn10 Tcr | Costanzo et al., 2006 |
| CF7789 | ΔlacI-lacZ (MluI) | M. Cashel |
| PLB940 | CF7789/pCSB68 Apr | This study |
| PLB941 | CF7789/csrA::kan pCSB68 Apr | This study |
| PLB986 | CF7789/rpoS::Tn10 Tcr pCSB68 Apr | This study |
| PLB1415 | S17-1 λpir/pYH159 Apr | This study |
| PLB1416 | S17-1 λpir/pYH161 Apr | This study |
| PLB1418 | S17-1 λpir/pYH162 Apr | This study |
| PLB1424 | CF7789/P1-P5-csrA′-′lacZ | This study |
| PLB1425 | CF7789/P3-P5-csrA′-′lacZ | This study |
| PLB1426 | CF7789/P4-P5-csrA′-′lacZ | This study |
| PLB1427 | CF7789/P5-csrA′-′lacZ | This study |
| PLB1429 | CF7789/P3-csrA-lacZ | This study |
| PLB1433 | CF7789/P3-csrA-lacZ rpoS::Tn10 Tcr | This study |
| PLB1434 | CF7789/P1-P2-csrA-lacZ | This study |
| PLB1442 | CF7789/P1-P5-csrA′-′lacZ csrA::kan | This study |
| PLB1443 | CF7789/P3-P5-csrA′-′lacZ csrA::kan | This study |
| PLB1445 | CF7789/P5-csrA′-′lacZ csrA::kan | This study |
| PLB1449 | CF7789/P3-P5-csrA′-′lacZ rpoS::Tn10 Tcr | This study |
| PLB1453 | CF7789/P3-csrA-lacZ csrA::kan | This study |
| PLB1455 | CF7789/P1-P2-csrA-lacZ csrA::kan | This study |
| PLB1457 | CF7789/P1-P2-csrA-lacZ rpoS::Tn10 Tcr | This study |
| PLB1464 | CF7789/P1-P5-csrA′-′lacZ pYH171 (csrA+) | This study |
| PLB1465 | CF7789/P1-P5-csrA′-′lacZ pYH172 (vector) | This study |
| PLB1466 | CF7789/P1-P5-csrA′-′lacZ csrA::kan pYH171 (csrA+) | This study |
| PLB1467 | CF7789/P1-P5-csrA′-′lacZ csrA::kan pYH171 (vector) | This study |
| PLB1470 | CF7789/P3-P5-csrA′-′lacZ pYH171 (csrA+) | This study |
| PLB1471 | CF7789/P3-P5-csrA′-′lacZ pYH172 (vector) | This study |
| PLB1472 | CF7789/P3-P5-csrA′-′lacZ csrA::kan pYH171 (csrA+) | This study |
| PLB1473 | CF7789/P3-P5-csrA′-′lacZ csrA::kan pYH172 (vector) | This study |
| PLB1478 | CF7789/P5-csrA′-′lacZ pYH171 (csrA+) | This study |
| PLB1479 | CF7789/P5-csrA′-′lacZ pYH172 (vector) | This study |
| PLB1480 | CF7789/P5-csrA′-′lacZ csrA::kan pYH171 (csrA+) | This study |
| PLB1481 | CF7789/P5-csrA′-′lacZ csrA::kan pYH172 (vector) | This study |
| PLB1487 | CF7789/P1-P5-csrA′-′lacZ BS1-BS3 mutationsb | This study |
| PLB1489 | CF7789/P1-P5-csrA′-′lacZ BS1-BS3 mutations csrA::kan | This study |
| PLB1707 | CF7789/P5-csrA-lacZ | This study |
| PLB1708 | CF7789/P5-csrA-lacZ csrA::kan | This study |
| PLB1709 | CF7789/P5-csrA-lacZ rpoS::Tn10 Tcr | This study |
| PLB1710 | CF7789/P1-P5-csrA′-′lacZ csrA::kan rpoS::Tn10 Tcr | This study |
| PLB1712 | CF7789/P1-P5-csrA′-′lacZ rpoS::Tn10 Tcr | This study |
| PLB1714 | CF7789/P1-P5-csrA′-′lacZ BS1-BS3 mutations rpoS::Tn10 Tcr | This study |
| PLB1715 | CF7789/P1-P5-csrA′-′lacZ BS1-BS3 mutations csrA::kan rpoS::Tn10 Tcr | This study |
| PLB1716 | CF7789/P3-P5-csrA′-′lacZ csrA::kan rpoS::Tn10 Tcr | This study |
| PLB1718 | CF7789/P1-P3-csrA′-′lacZ BS1-BS3 mutations rpoS::Tn10 Tcr | This study |
| PLB1719 | CF7789/P1-P3-csrA′-′lacZ BS1-BS3 mutations csrA::kan rpoS::Tn10 Tcr | This study |
| PLB1720 | CF7789/P1-P5-csrA′-′lacZ P1-P2 promoter mutationsc | This study |
| PLB1721 | CF7789/P1-P5-csrA′-′lacZ P1-P2 promoter mutations csrA::kan | This study |
| S17-1 λpir | recA thi pro hsdR?M+ RP4-2 Tc::Mu Km::Tn7 λpir+ | de Lorenzo et al., 1990 |
| TRCF7789 | CF7789/csrA::kan | Romeo et al., 1993 |
All csrA fusions were integrated into the λ att site via the CRIM system (Haldimann and Wanner, 2001). The integratedP1-P5-csrA′-′lacZ, P3-P5-csrA′-′lacZ, P4-P5-csrA′-′lacZ, and P5-csrA′-′lacZ translational fusions contain −284 to +56, −192 to +56, −114 to +56, and −114 to −90 plus −82 to +56 relative to the start of csrA translation, respectively. TheP1-P2-csrA-lacZ, P3-csrA-lacZ and P5-csrA-lacZ transcriptional fusions contain −284 to −201, −192 to −89 and −114 to −90 plus −82 to −40 relative to the start of csrA translation, respectively (Fig. 1A).
CsrA binding site mutations: BS1, G(−56)C; BS2 G(−45)C; BS3 A(−37)C and G(−36)C.
Mutations in the overlapping P1 and P2 −10 promoter sequences: T(−237)C, A(−236)T, T(−232)C.
β-galactosidase assay
Bacterial cultures were grown in LB medium at 37 °C. Cells were harvested at various times during growth, washed with 10 mM Tris-HCl (pH 7.5) and frozen as cell pellets at −20 °C. Cell extracts were prepared by suspending frozen cell pellets in 0.5 ml of BugBuster (Novagen). After 30 min of incubation at 37°C, 0.3 ml of Z buffer containing 0.2 mg/ml lysozyme was added, incubation was then continued for 30 min at 37 °C. Following removal of cell debris, protein concentrations were determined by the Bio-Rad protein assay. β-galactosidase assays were performed using the cell extracts as described previously (Baker et al., 2002).
RNA directed in vitro translation
His-tagged CsrA (CsrA-H6) was purified as described previously (Dubey et al., 2005). RNA templates for in vitro translation were synthesized using the MEGAscript kit (Ambion). CsrA-deficient E. coli S-30 extract was prepared from TRCF7789 (csrA::kan). In vitro translation reactions followed previously published procedures (Baker et al., 2007). Reaction mixtures contained buffer, S-30 extract, RNA, protease inhibitors, an energy regeneration system, [35S]methionine, the other 19 amino acids and various concentration of CsrA. A detailed procedure is in Supporting Information.
In vitro transcription assay
Multi-round in vitro transcription assays followed a published procedure (Babitzke et al., 2003). DNA templates contained promoters P1 through P5 (P1-P5), P3 through P5 (P3-P5), P4 and P5 (P4-P5), or P5 only. Additional templates contained mutations in the overlapping −10 promoter sequences for P1 and P2, or mutations in CsrA binding sites BS1, BS2 and BS3 (Fig. 1). Reaction mixtures contained buffer, template DNA, the four NTPs, [α-32P]-UTP, and E. coli RNA polymerase holoenzyme (Eσ70), core enzyme, or reconstituted Eσ70 or EσS. A detailed procedure is in Supporting Information.
Gel mobility shift assay
Various in vitro generated RNAs were 5′-end labeled. Binding reactions contained buffer, yeast RNA, 0.1 nM labeled csrA RNA and various concentrations of CsrA. Competition assay mixtures also contained unlabeled competitor RNA. Apparent equilibrium binding constants (Kd) of CsrA-csrA RNA interaction were calculated as described previously (Yakhnin et al., 2000). A detailed procedure is in Supporting Information.
RNA footprint assay
Binding reactions (10 μl) containing various concentrations of CsrA and 4 nM csrA RNA were otherwise identical to those described for the gel shift assay. After the initial binding reaction, 0.075 U RNase T1 (Roche) was added to the reaction mixtures, and incubation was continued for 15 min at 37 °C. Reactions were terminated by the addition of 10 μl of gel loading buffer and placed on ice. Partial alkaline hydrolysis and RNase T1 digestion ladders of each transcript were prepared as described previously (Bevilacqua and Bevilacqua, 1998). Samples were fractionated through standard 6% polyacrylamide sequencing gels. Radioactive bands were visualized with a phosphorimager.
Toeprint assay
Toeprint assays were performed by modifying a published procedure (Yakhnin et al., 2007). E. coli 30S ribosomal subunits were purified as described previously (Baker et al., 2007). Toeprint reactions contained buffer, csrA RNA, a 5′ end-labeled DNA primer complementary to the 3′ end of the transcript, 4 μM CsrA and/or 260 nM 30S ribosomal subunits ± 5 μM tRNAfMet and dNTPs. Mixtures containing CsrA were incubated for 30 min at 37 °C to allow CsrA-mRNA complex formation prior to the addition of 30S ribosomal subunits. The reaction mixture was incubated for 15 min at 37 °C following the addition of 0.25 U of AMV reverse transcriptase (Roche). A detailed procedure is in Supporting Information.
Primer extension assay
Strains PLB940 (CF7789/pCSB68) and PLB986 (CF7789/rpoS::Tn10 pCSB68) were grown in LB or M9 minimal medium supplemented with 0.2% glucose at 37 °C in a shaking water bath. Strains PLB1415 (S17-1 λpir/pYH159), PLB1416 (S17-1 λpir/pYH161) and PLB1418 (S17-1 λpir/pYH162) were grown in LB under the same conditions. Ampicillin (100 μg/ml) was included in all cultures to maintain plasmid selection. Ten ml aliquots were harvested at late exponential and early stationary phase growth and added to an equal volume of frozen killing buffer (10 mM Tris-HCl, pH 7.2, 5 mM MgCl2, 25 mM sodium azide, 12.5% ethanol, and 500 μg/ml chloramphenicol). Cells pellets were suspended in 1 ml of a 2:1 mixture of RNA Protect bacterial reagent (Qiagen):TE buffer, placed on ice for 10 min, and centrifuged. Cell pellets were frozen at −80 °C. Total RNA was isolated using the RNeasy bacterial protocol (Qiagen) and traces of genomic DNA were removed with 2 U of Turbo DNase (Ambion). RNA samples were extracted with phenol and precipitated. Oligonucleotide primers complementary to the csrA′-′lacZ fusion junctions were 5′-end labeled with [γ-32P]-ATP. 2.5 μg of RNA was annealed to 0.4 pmol of the labeled primer in TE buffer by heating to 80 °C and then slowly cooling to room temperature. Reverse transcription reactions (4 μl) contained the hybridization mixture and 1x Superscript III reverse transcriptase buffer, 0.5 mM dNTP, 0.8 U RNasin, 20 μg/ml acetylated BSA, 1 mM DTT and 5 U of Superscript III reverse transcriptase (Invitrogen). Reactions were incubated for 30 min at 42 °C and then quenched by adding 3 μl of stop buffer (20 mM EDTA, 95% formamide, 0.1% SDS, 0.05% xylene cyanol, 0.05% bromphenol blue) and placed on ice. Samples were heated for 5 min at 95 °C prior to fractionating through standard 6% sequencing gels. Sequencing reactions were performed using the same end-labeled primer and pCSB68 as the template. Radioactive bands were visualized using a phosphorimager.
Supplementary Material
Acknowledgments
We thank Katsushiko Murakami for σ70 and Richard Burgess for σS. We also thank Adrianne Edwards and Christopher Vakulskas for helpful suggestions. This work was supported by National Institutes of Health Grant GM059969.
References
- Babitzke P, Romeo T. CsrB sRNA Family: Sequestration of RNA-binding Regulatory Proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Schaak J, Yakhnin AV, Bevilacqua PC. Role of RNA structure in transcription attenuation in Bacillus subtilis: the trpEDCFBA operon as a model system. Methods Enzymol. 2003;371:392–404. doi: 10.1016/S0076-6879(03)71030-1. [DOI] [PubMed] [Google Scholar]
- Baker CS, Eöry LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol. 2007;189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- Becker G, Hengge-Aronis R. What makes an Escherichia coli promoter σS dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 ofσS. Mol Microbiol. 2001;39:1153–1165. doi: 10.1111/j.1365-2958.2001.02313.x. [DOI] [PubMed] [Google Scholar]
- Bevilacqua JM, Bevilacqua PC. Thermodynamic analysis of an RNA combinatorial library contained in a short hairpin. Biochemistry. 1998;37:15877–15884. doi: 10.1021/bi981732v. [DOI] [PubMed] [Google Scholar]
- Chavez RG, Alvarez AF, Romeo T, Georgellis D. The physiological stimulus for the BarA sensor kinase. J Bacteriol. 2010;192:2009–2012. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, σE, by guanosine 3′,5′-bispyrophosphate (ppGpp) J Bacteriol. 2006;188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, Babitzke P. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol. 2003;185:4450–4460. doi: 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07663.x. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. Regulatory interactions of Csr components: The RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol. 2001;183:6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Proteolysis of σS (RpoS) and the general stress response in Escherichia coli. Res Microbiol. 2009;160:667–76. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Ahmad I, Romeo T, Römling U, Melefors O. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol. 2010;12:524–540. doi: 10.1111/j.1462-2920.2009.02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P, Haas D. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol Microbiol. 2007;66:341–356. doi: 10.1111/j.1365-2958.2007.05909.x. [DOI] [PubMed] [Google Scholar]
- Liu MY, Romeo T. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J Bacteriol. 1997;179:4639–4642. doi: 10.1128/jb.179.14.4639-4642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Gui G, Wei B, Preston JF, III, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- Marles-Wright J, Lewis RJ. Stress responses of bacteria. Curr Opin Struct Biol. 2007;17:755–760. doi: 10.1016/j.sbi.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol. 2009;392:511–528. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem. 2006;281:31832–31842. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Smith TG, Hoover TR. Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv Appl Microbiol. 2009;67:257–295. doi: 10.1016/S0065-2164(08)01008-3. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67:2897–908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Prüß BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Yakhnin AV, Trimble JJ, Chiaro CR, Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 2000;275:4519–4524. doi: 10.1074/jbc.275.6.4519. [DOI] [PubMed] [Google Scholar]
- Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol Microbiol. 2007;64:1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- Yakhnin H, Yakhnin AV, Babitzke P. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol Microbiol. 2006;61:1252–1266. doi: 10.1111/j.1365-2958.2006.05278.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.