Abstract
Quantitative mass spectrometry-based proteomics is a vital tool in modern life science research. In contrast to the popularity of approaches for relative protein quantitation, the widespread use of absolute quantitation has been hampered by inefficient and expensive production of labeled protein standards. To optimize production of isotopically labeled standards, we genetically modified a commonly employed protein expression Escherichia coli strain, BL21 (DE3), to construct an auxotroph for arginine and lysine. This bacterial strain allows low-cost, high-level expression of fully labeled proteins with no conversion of labeled arginine to proline. In combination with a fluorescence-based quantitation of standards and nontargeted LC–MS/MS analysis of unfractionated total cell lysates, this strain was used to determine the copy number of a post-translational modifier, small ubiquitin-like modifier (SUMO-2), in HeLa, human sperm, and chronic lymphocytic leukemia cells. By streamlining and improving the generation of labeled standards, this production system increases the breadth of absolute quantitation by mass spectrometry and will facilitate a far wider uptake of this important technique than previously possible.
Keywords: absolute SILAC, PSAQ, SUMO, quantitation, mass spectrometry, proteomics, biomarker, sperm cells, chronic lymphocytic leukemia
Short abstract
We developed a bacterial expression system optimized for the expression of isotope-labeled protein standards for absolute quantitation by mass spectrometry. This bacterial strain allows low-cost, high-level expression of fully labeled proteins with no conversion of labeled arginine to proline. These labeled proteins can be used to determine the copy number of cellular proteins by quantitative mass spectrometry.
Introduction
With rapid and ongoing developments in sample preparation, instrumentation and computational analysis, mass spectrometry-based quantitative proteomics is increasingly emerging as an approach of choice for addressing many aspects of biology.1−3 One of the most popular formats of relative quantitative proteomics,(4) stable-isotope labeling by amino acids in cell culture (SILAC),(5) relies on internal metabolic labeling of cellular proteomes under study. Conversely, absolute protein quantitation with stable isotopes depends on the external generation of isotopically labeled standards to accurately determine protein abundance in biological samples. This approach has been used for some time and has promising applications in systems biology and biomarker discovery. However, the popular employment of isotope-labeled peptides as standards, either synthesized chemically(6) or expressed in the form of a concatamer,(7) faces limitations compared to the more recently developed quantitation with full-length labeled proteins.(8) The important advantage of the latter approach, termed absolute SILAC(9) or PSAQ (protein standard absolute quantification),(8) is that standards are introduced earlier in the workflow and are subjected to the same fractionation and digestion conditions as the endogenous counterpart, thereby providing more reliable quantitation. Until now, a limited number of labeled standards have been produced for specific mass spectrometric measurements, resulting in a single proof-of-principle study with little further use of the material.7−15 For single studies, picomole quantities of standard are often sufficient; however, systems biology and biomarker discovery applications will require large-scale production of stringently quality controlled standards. Large-scale labeled protein production fulfills the current trend toward uncoupling the production of standards from the actual experiment by centrally producing labeled proteins and distributin g them for use by the scientific community. A notable example of this trend is the super SILAC approach,(16) in which libraries of labeled cell lines are centrally prepared and distributed to end-user laboratories. In this context, the widespread uptake of absolute SILAC depends strongly on how inexpensively and efficiently labeled proteins can be produced.
To address this need, we have generated a lysine and arginine auxotrophic form of the popular protein expression strain E. coli BL21 (DE3). The strain can be used in conjunction with commonly applied prokaryotic protein expression techniques to express relatively large quantities of proteins labeled 100% with “heavy” lysine and arginine residues, with no appreciable arginine to proline conversion. We demonstrate the application of the technology by expressing a 13C615N4-arginine and 13C615N2-lysine labeled form of the Small ubiquitin-like modifier (SUMO-2)(17) and use it to calculate the copy number of SUMO-2 in HeLa cells and two clinical samples.
Materials and Methods
Generation of an E. coli BL21 (DE3) Strain Auxotrophic for Arginine and Lysine
Auxotrophic mutants of E. coli BL21 (DE3)(18) were constructed by bacteriophage P1 transduction(19) to introduce in-frame deletions in lysA and argA genes from the Keio Collection of single-gene knockout library.(20) The deleted genes were replaced by a kanamycin resistance gene flanked by two FLP recombinase target sites, and the P1 lysates were prepared from lysA and argA mutants retrieved from the Keio Collection. P1 transduction was then carried out according to standard procedures as described previously(19) to introduce each mutation into the recipient E. coli BL21 (DE3). Following the selection of the transductants in the presence of kanamycin, the kanamycin resistance cassette was removed by the transient expression of FLP recombinase according to the procedure of Datsenko and Wanner.(21) This was repeated to obtain a strain with dual auxotrophy.
Expression, Purification and Quantitation of the Labeled Standard
The mature form of human SUMO-2 was subcloned into a pHisTEV vector and expressed in Escherichia coli BL21 (DE3) ΔlysA ΔargA cells as an N-terminally His-tagged protein. Expression was performed in M9 medium supplemented with 0.2% glucose, 50 μg/mL kanamycin, 1 mM MgSO4, 50 μg/mL 13C615N4-arginine and 50 μg/mL 13C615N2-lysine (Cambridge Isotope Laboratories, Andover, MA). After harvesting, recombinant, isotopically labeled SUMO-2 was purified by Ni-NTA (Ni2+-nitrilotriacetate)-affinity chromatography. After this first purification step, the His-tag was removed by Tobacco Etch Virus (TEV) Protease in 50 mM Tris/HCl pH 8.0, 5 mM β-mercaptoethanol and 50 mM NaCl and purified further by Ni-NTA-affinity chromatography and on a gel filtration Superdex-75 column (GE Healthcare, Piscataway, NJ).
6His-TEV-YFP-SUMO-2-FL-ECFP was expressed in E. coli B834 (DE3) and purified from bacterial lysates using Ni-NTA-affinity chromatography and anion-exchange chromatography as described.(22) Concentration of 6His-TEV-YFP-SUMO-2-FL-ECFP was calculated by measuring absorbance of ECFP at 433 nm and of YFP Venus(23) at 515 nm. The average of 20 measurements of absorbances was used and determined the concentration based on the Beer’s law and extinction coefficients for YFP and ECFP (92,200 and 28,750 M–1 cm–1, respectively).(22) Isotopically labeled SUMO-2 standard and 6His-TEV-YFP-SUMO-2-FL-ECFP were mixed, digested with the FASP method, as described below, and analyzed by mass spectrometry. The exact concentration of the labeled SUMO-2 standard was calculated from SILAC ratios based on the fluorescently determined concentration of 6His-TEV-YFP-SUMO-2-FL-ECFP.
Cell Preparation and Counting
Sperm Cells
Semen was collected from healthy normozoospermic (as defined by WHO (1999) guidelines) donors. Research donors were recruited at Ninewells Hospital, Dundee (HFEA Centre 004) in accordance with the Human Fertilisation and Embryology Authority Code of Practice Version 8 under local ethical approval (08/S1402/6) from Tayside Committee on Medical Research Ethics. Sperm cells were isolated using a 2 layer density gradient composed of 40% and 80% PureSperm(Nidacon Int AB, Mölndal, Sweden) buffered with noncapacitating buffer (NCB) [1.8 mM CaCl2, 5.4 mM KCl, 0.8 mM MgSO4.7H2O, 116.4 mM NaCl, 1.0 mM NaH2PO4, 5.6 mM d-glucose, 2.7 mM Na pyruvate, 41.8 mM Na lactate, 25 mM HEPES, pH 7.4]. Semen (maximum of 1 mL) was layered on top of each gradient and centrifuged at 300× g for 20 min in 15 mL conical centrifuge tubes. The 80% fractions were transferred to clean tubes by discarding the seminal fluid and the bulk of the density gradient then retrieving the sperm pellet from the bottom of the tube taking care to avoid contamination. Sperm cells were washed in NCB then centrifuged at 500 g for 5 min. The resulting pellets were resuspended in NCB. White blood cells were removed using CD45 Dynabeads (Invitrogen, Paisley, Scotland) in accordance to the manufacturer’s instructions. Ten fractions of each cell suspension were mixed with water (to dilute and immobilize) and cells were counted using an improved Neubauer hemocytometer (Weber Scientific International Ltd., Middlesex, U.K.).
Chronic Lymphocytic Leukemia (CLL) Cells
Studies involving isolation and analysis of human chronic lymphocytic leukemia cells (CLL) were approved by the Tayside Committee on Medical Research Ethics and Tayside Tissue Bank. Following informed consent, an enriched population of CLL cells was obtained by density gradient centrifugation (Ficoll-PaqueTM Plus, 1.077, GE Healthcare Bio-Sciences AS, Uppsala) from 20 mL of blood. Cells were counted with a Coulter counter (Beckman Coulter, Hialeah, FL).
HeLa Cells
HeLa cells were cultured in DMEM medium supplemented with 10% FBS and 100 units/ml penicillin and streptomycin. Cells were counted using the Cellometer Auto T4 Automatic cell counter (Nexcelom Bioscience LLC, Lawrence, MS).
Cell Lysis and Protein Digestion
Cell pellets were lysed in 100 mM Tris-HCl (pH 7.5) containing 4% SDS and processed by the FASP method.(24) The lysates were sonicated to reduce viscosity and incubated at 95 °C for 5 min. Following centrifugation for 20 min at 21 000× g, the supernatant was mixed with UA buffer (200 μL of 8 M urea in 100 mM Tris-HCl, pH 7.5), loaded onto Vivacon 500 μL ultrafiltration spin columns with nominal cutoff of 30 000 Da (Sartorius Stedim Biotech, Epsom, U.K.) and centrifuged at 14 000× g for 15 min. The samples were washed with 200 μL of UA buffer and centrifuged again to remove SDS. The concentrates were diluted with 100 μL of UA buffer containing 50 mM iodoacetamide and incubated in darkness for 30 min. After centrifugation the samples were washed three times with 200 μL of UA buffer and twice with 100 μL of 50 mM NaHCO3. Next, 50 μL of 50 mM NaHCO3 containing 2 μg of trypsin (Promega, Madison, WI) was added to the concentrate and the samples were incubated at room temperature overnight. The resulting digested peptides were eluted by centrifugation followed by two additional elutions with 50 μL of 50 mM NaHCO3 and were evaporated to dryness in a vacuum concentrator system (Concentrator Plus, Eppendorf, Hamburg, Germany) to remove NaHCO3. The dried residues were reconstituted in 40 μL of buffer A (0.5% acetic acid) and transferred to vials for MS analysis.
Liquid Chromatography–Mass Spectrometry and Data Analysis
Mass spectrometric analysis was performed by nanoscale LC–MS/MS using an LTQ-Orbitrap Velos instrument (Thermo Fisher Scientific, Bremen, Germany) coupled to an Ultimate U3000 (Dionex Corporation, Sunnyvale, CA) via a Proxeon nanoelectrospray source (Proxeon Biosystems, Denmark). Peptides were separated on a 75 μm PepMap C18 nano column (Dionex).
Data were acquired in data-dependent mode: full scan spectra were acquired with a resolution of 60 000 after accumulation of 1 000 000 ions. The “lock mass” option was used to improve the mass accuracy of precursor ions.(25) The 10 most intense ions were fragmented by collision-induced dissociation (CID) with normalized collision energy of 35% and recorded in the linear ion trap (target value of 5000) based on the survey scan and in parallel to the orbitrap detection of MS spectra.
Raw MS data were processed with MaxQuant, an integrated suite of software for quantitative proteomics (version 1.1.1.25).26,27 Enzyme specificity was set to trypsin, allowing for cleavage N-terminal to proline residues and up to two missed cleavages. Cysteine carbamidomethylation was considered as a fixed modification, and methionine oxidation and protein N-acetylation were set as variable modifications. MS/MS peak lists generated by MaxQuant were searched by the Andromeda database search engine(27) against the human International Protein Index (IPI) database (version 3.68).(28)
Results and Discussion
Generation of an Absolute SILAC-Compatible Bacterial Protein Expression Strain
Widespread use of absolute SILAC and its application in biomarker discovery and systems biology, which is the ultimate goal of absolute quantitation, rely on efficient and inexpensive production of protein standards. “Heavy” arginine and lysine are the most frequently used stable isotope-labeled amino acids in SILAC-based studies; however, these amino acids are nonessential to commonly available E. coli expression strains. Hence, the endogenous production of lysine and arginine in a regular expression strain dramatically compromises the incorporation efficiency.(10) Conversely, the use of strains auxotrophic for lysine and arginine, but not optimized for protein expression, leads to nonoptimal expression.(9) Cell-free synthesis, an alternative approach that in absence of a dedicated absolute SILAC-compatible strain has in some cases been used to produce limited amounts of a standard,8,9,11 suffers from higher costs, relatively low levels of protein expression, difficulties in protein purification and occasionally incomplete labeling.(11)
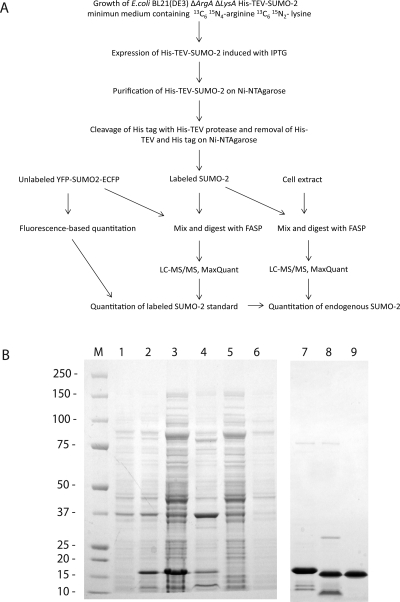
To obtain both the full incorporation of SILAC amino acids and optimal protein yields, the E. coli BL21 (DE3) strain(18) was modified to be auxotrophic for lysine and arginine. BL21 (DE3) is a widely used E. coli strain for high-level protein expression that employs the T7 RNA Polymerase-based system, carries additional tRNAs to overcome the problem of codon bias and is protease compromised to minimize post-translational processing. To abolish the synthesis of lysine and arginine we deleted the diaminopimelate decarboxylase (lysA) and N-acetylglutamate synthase (argA) genes by using the Keio collection of mutants,(20) which allows in-frame single-gene deletion of target genes (Figure 1A and Materials and Methods). An advantage of the system is that the same cosmid and resistance marker can be employed to make multiple knockouts due to the elimination of the resistance marker by FLP recombinase. Therefore, this auxotrophic strain does not have any antibiotic resistance and is amenable to generation of any protein that can be expressed recombinantly. Arginine and lysine auxotrophy was confirmed by the absence of growth in the minimal medium without added arginine or lysine (Figure 1B). Importantly, although in many cases proteins expressed in E. coli are insoluble, whether or not the protein of interest is soluble is inconsequential for this method as insoluble aggregates are easily and completely purified by differential centrifugation and on metal affinity resins in presence of urea. The recently developed FASP digestion(24) allows complete solubilization of proteins under highly denaturing conditions before buffer exchange on the filter and subsequent proteolytic digestion. Our strategy for the production of labeled standards and their incorporation into the workflow of quantitative mass spectrometry are outlined in Figure 2A.
Figure 1.
Bacterial strain for the production of SILAC-labeled SUMO-2. (A) Genetic modification of the E. coli BL21 (DE3) strain. (B) SILAC-compatible strain auxotroph for lysine and arginine. Growth on M9 minimal agar plates with and without lysine and arginine is shown.
Figure 2.
Experimental workflow. (A) Labeled SUMO-2 is recombinantly expressed in the E. coli BL21 (DE3) strain auxotroph for arginine and lysine and purified by nickel-affinity column chromatography and gel filtration. Concentration of heavy SUMO-2 was calculated by mass spectrometry using a fluorescently labeled SUMO-2 as a reference. Isotopically labeled SUMO-2 was then spiked in cell extracts to determine the copy number of endogenous SUMO-2. (B) Purification of the SUMO-2 standard labeled with heavy lysine and arginine: lane M, molecular mass marker; lane 1, total cell lysate from uninduced cells; lane 2, total bacterial extract from induced cells; lane 3, supernatant; lane 4, pellet; lane 5, flow-through from the Ni-NTA column; lane 6, wash; lane 7, 6His-TEV-SUMO-2 purified from Ni-NTA-affinity column; lane 8, cleavage by TEV protease; lane 9, purified SUMO-2.
Expression of 13C615N4-Arginine and 13C615N2-Lysine Labeled SUMO-2
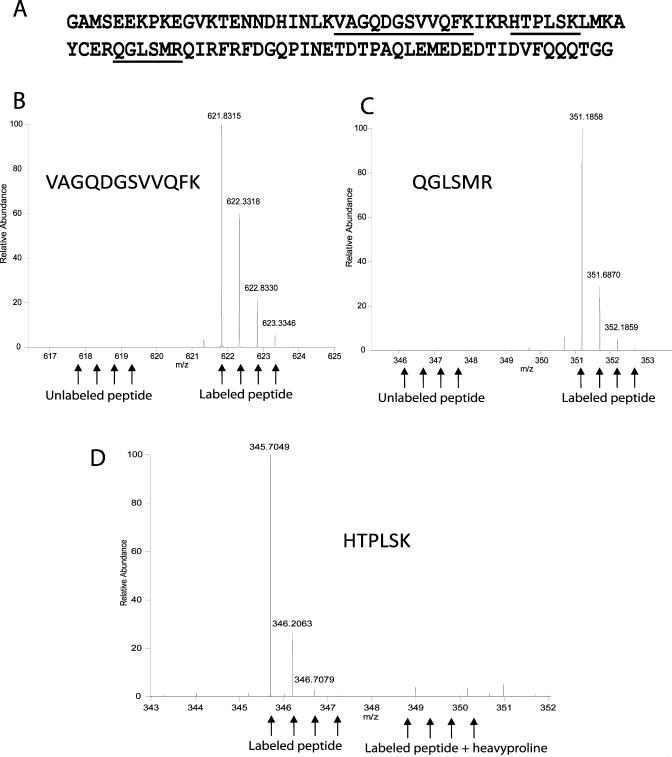
After optimization of growth conditions (see Materials and Methods), we expressed and purified 30 mg of labeled SUMO-2 from 0.5 L of modified medium. Considering materials alone, this equates to less than 4 US$ per mg protein by current materials costs. His6-tagged SUMO-2 was purified by nickel affinity chromatography. The His6-tag was cleaved away from SUMO-2 by TEV protease and untagged SUMO-2 was further purified by nickel affinity chromatography and gel filtration (Figure 2B). Crucially, we achieved complete incorporation of isotopically labeled arginine and lysine and observed no arginine to proline conversion (Figure 3). This greater than 99.9% incorporation level of isotope-labeled proteins compares favorably with those generated in a regular BL21(DE3) strain (i.e., not auxotrophic for arginine and lysine), which was only approximately 70%.(10) Incomplete labeling of the standard, especially in case of labeling efficiency below 95%, constitutes a serious problem in the quantitation of low abundance proteins that will be masked by the contaminating “light” peptides. Similarly, metabolic conversion of heavy arginine to heavy proline, reported in certain microrganisms29,30 and cell lines(31) but not here (Figure 3D), compromises quantitation accuracy by creating multiple peaks for every heavy proline-containing peptide.
Figure 3.
Complete incorporation of heavy isotopes and absence of arginine-to-proline conversion. Recombinant isotopically labeled SUMO-2 produced in BL21 (DE3) ΔlysA ΔargA was digested in solution with trypsin and analyzed by LC–MS/MS. (A) Amino acid sequence of the SUMO-2 standard. Highlighted text indicates the peptides whose MS spectra are reported in the lower panels. The arrows show the isotopic distribution of the heavy peptides and the calculated positions of the light counterparts (B and C) or of the heavy proline-containing peptide (D). Incorporation efficiency for both lysine (B) and arginine (C) containing peptides is greater than 99.9%. (D) Labeled arginine is not converted to heavy proline in BL21 (DE3) ΔlysA ΔargA.
Quantitation of the SUMO-2 Standard
The accuracy of quantitation of a protein of interest relies on the knowledge of the concentration of the corresponding standard. The commonly used methods for protein quantitation, such as amino acids analysis and measurement of absorbance at 280 nm,(9) require the standard to be highly purified. Otherwise, the presence of contaminating proteins would lead to an overestimation of the concentration of the standard. However, it is often desirable to avoid an extensive purification that is laborious, involves multiples steps and leads to loss of material. Here we reasoned that fluorescence spectroscopy combined with quantitative mass spectrometry(11) can be used to determine the concentration of an absolute SILAC standard even in complex mixtures. Contaminating proteins do not absorb light at absorbance wavelengths of fluorescent proteins and are therefore invisible to fluorescence measurements. Thus, we adapted a previously developed fluorescence-based strategy(11) to quantify the protein concentration of the labeled SUMO-2 standard. First, we purified a recombinant unlabeled YFP-ECFP fusion of SUMO-2(22) and calculated its concentration from the absorbance measurements of YFP and ECFP. In the second step, the labeled SUMO-2 standard was mixed with YFP-SUMO-2-ECFP, digested with trypsin and quantified by mass spectrometry (Figure 2A; Materials and Methods). The SILAC ratio between the isotope labeled, nonfluorescent SUMO-2 and its fluorescent analogue allows the calculation of the concentration of the absolute SILAC SUMO-2 standard.
It is theoretically possible that the accuracy of the fluorescence strategy is compromised by the incorrect folding of fluorophores. Although the use of two fluorophores might help to compensate in case the activity of only one fluorescent protein is compromised, the incorrect folding of the whole fusion protein could affect both fluorophores. However, YFP-SUMO2-ECFP is completely cleaved by the SUMO-2 protease,(22) and as protease cleavage is dependent on the folded structure of SUMO, it is likely that the protein is correctly folded and that therefore the two fluorophores are active.
Absolute Quantitation of SUMO-2 in Clinical Samples
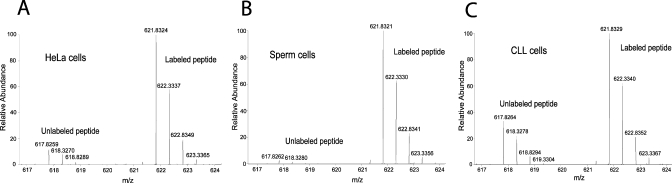
Because of its important role in both physiological and pathological states, such as cancer,(32) and its nature as a protein modifier, we reasoned that absolute SILAC would be the ideal technique for the investigation of SUMO-2 as a biomarker. Several studies have already successfully adapted SILAC-based proteomics to the investigation of sumoylation dynamics.33,34 Here we extend the scope of quantitative proteomic analysis of sumoylation to include absolute SILAC. As a proof of principle, we decided to determine the absolute abundance of SUMO-2 in two clinically relevant human samples, namely sperm and CLL cells, for which no evidence of SUMO-2 modification has been reported so far. Notably, this was achieved with no fractionation of the sample and without any targeted proteomics approach. CLL cells were isolated from peripheral blood of a patient with chronic lymphocytic leukemia,(35) which is the most common type of adult leukemia and involves uncontrolled proliferation of B-cells, whereas sperm cells(36) were collected from healthy donors. Cell lysis was performed under highly denaturing conditions to inactivate proteases and ensure complete solubilization of proteins. One of the main advantages of absolute SILAC compared to peptide-based absolute quantitation methods is that the standard can be added immediately after lysis and before any further manipulation of the sample, thus minimizing variations in sample preparation between endogenous protein and the standard. A known amount of the heavy SUMO standard was spiked into the sample and tryptic digestion was performed on a centrifugal filter unit with nominal cutoff of 30 kDa according to the FASP protocol.(24) Note that the 30 kDa cutoff was determined by the manufacturer with folded rather than denatured proteins(24) and that this filter efficiently retains detergent-denatured SUMO-2 (data not shown). Following digestion, peptides were eluted and subjected to LC–MS/MS analysis and the ratios were determined by the MaxQuant data processing software(26) from 4 technical replicates. CLL (chronic lymphocytic leukemia) cells had a copy number comparable to HeLa cells (3.63 ± 0.003 × 106 and 3.80 ± 0.05 × 106 molecules per cell, respectively) whereas sperm cells contained significantly fewer copies of SUMO-2 (1.11 ± 0.05 × 105 molecules per cell; Figure 4, Table 1).
Figure 4.
Copy number of SUMO-2. MS spectra of the doubly charged peptide VAGQDGSVVQFK from (A) HeLa, (B) sperm and (C) CLL cells.
Table 1. Copy Number of SUMO-2 for HeLa, Sperm and CLL (Chronic Lymphocytic Leukemia) Cells.
| sample | number of cells | ratio H/L (mean ± SDa) | pmol endogenous SUMO-2 | molecules per cell (mean ± SDa) |
|---|---|---|---|---|
| HeLa cells | 5.00 × 105 | 8.44 ± 0.10 | 1.90 | 3.80 ± 0.05 × 106 |
| Sperm cells | 3.95 × 106 | 36.42 ± 1.67 | 0.44 | 1.11 ± 0.05 × 105 |
| CLL cells | 3.00 × 106 | 2.95 ± 0.03 | 10.89 | 3.63 ± 0.003 × 106 |
Standard deviation (SD) is calculated from technical replicates.
Physiochemical properties of SUMO conjugated proteins are different among themselves and as compared to unconjugated SUMO and the labeled SUMO internal standard. Thus, no fractionation should be performed at the protein level, as this would compromise the accuracy of quantitation, but rather proteins should be digested with an approach that does not discriminate between different subsets of SUMO. Here we have employed the FASP approach, which allows complete solubilization of proteins under highly denaturing conditions before subsequent proteolytic digestion. After digestion, peptides with the same sequence derived from the heavy SUMO-2 standard have, apart from the higher mass, the same physiochemical properties of those derived from endogenous free and conjugated SUMO-2. Although in general a fractionation step at the peptide level would not affect the quantitation, it was not necessary in this study as SUMO-2 was detectable and quantifiable in unfractionated complex mixtures.
Conclusions
The bacterial strain described here allows high-yield and inexpensive production of full-length SILAC labeled protein standards with complete incorporation of labeled amino acids and no proline to arginine conversion. BL21 (DE3) ΔlysA ΔargA could either be employed directly by individual laboratories or one large batch of rigorously quality controlled protein standards could be produced centrally and distributed to final users. Low cost production of large amounts of labeled proteins will also make feasible the production of more sophisticated second-generation SILAC standards, such as the creation of different types of ubiquitin and SUMO polymers and phosphorylated proteins by postproduction in vitro reactions. In addition, this system is not limited to the production of proteins, but it can also generate multiple labeled peptides as part of artificial proteins (QconCATs).7,15 We envisage that our strain will be widely used and that it will dramatically improve the generation of labeled standards for absolute quantitation using mass spectrometry.
Acknowledgments
We thank the Tayside Tissue Bank, Dundee (http://www.tissuebank.dundee.ac.uk) for B-CLL samples. We also thank Michael H. Tatham for comments on the manuscript and Katherine Hands for help with the B-CLL samples. This work was supported by the European Union (RUBICON network of excellence) and by Cancer Research UK (grant number C434/A7794). I.M. is a Sir Henry Wellcome Postdoctoral Fellow.
Glossary
Abbreviations:
- SILAC
stable isotope labeling by amino acids in cell culture
- PTM
post-translational modification
- Ubl
ubiquitin-like protein
- SUMO
small ubiquitin-like modifier
- CLL
chronic lymphocytic leukemia
- LC–MS/MS
liquid chromatography–tandem mass spectrometry
- QconCAT
quantitative concatamer
- PSAQ
protein standard absolute quantification
- Lys
lysine
- Arg
arginine
- LTQ
linear quadrupole ion trap
- ppm
parts per million
- SD
standard deviation
References
- Aebersold R.; Mann M. Mass spectrometry-based proteomics. Nature 2003, 422 (6928), 198–207. [DOI] [PubMed] [Google Scholar]
- Cravatt B. F.; Simon G. M.; Yates J. R. 3rd The biological impact of mass-spectrometry-based proteomics. Nature 2007, 450 (7172), 991–1000. [DOI] [PubMed] [Google Scholar]
- Domon B.; Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010, 28 (7), 710–21. [DOI] [PubMed] [Google Scholar]
- Ong S. E.; Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005, 1 (5), 252–62. [DOI] [PubMed] [Google Scholar]
- Ong S. E.; Blagoev B.; Kratchmarova I.; Kristensen D. B.; Steen H.; Pandey A.; Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1 (5), 376–86. [DOI] [PubMed] [Google Scholar]
- Gerber S. A.; Rush J.; Stemman O.; Kirschner M. W.; Gygi S. P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 2003, 100 (12), 6940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon R. J.; Doherty M. K.; Pratt J. M.; Gaskell S. J. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2005, 2 (8), 587–9. [DOI] [PubMed] [Google Scholar]
- Brun V.; Dupuis A.; Adrait A.; Marcellin M.; Thomas D.; Court M.; Vandenesch F.; Garin J. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol. Cell. Proteomics 2007, 6 (12), 2139–49. [DOI] [PubMed] [Google Scholar]
- Hanke S.; Besir H.; Oesterhelt D.; Mann M. Absolute SILAC for accurate quantitation of proteins in complex mixtures down to the attomole level. J. Proteome Res. 2008, 7 (3), 1118–30. [DOI] [PubMed] [Google Scholar]
- Janecki D. J.; Bemis K. G.; Tegeler T. J.; Sanghani P. C.; Zhai L.; Hurley T. D.; Bosron W. F.; Wang M. A multiple reaction monitoring method for absolute quantification of the human liver alcohol dehydrogenase ADH1C1 isoenzyme. Anal. Biochem. 2007, 369 (1), 18–26. [DOI] [PubMed] [Google Scholar]
- Nanavati D.; Gucek M.; Milne J. L.; Subramaniam S.; Markey S. P. Stoichiometry and absolute quantification of proteins with mass spectrometry using fluorescent and isotope-labeled concatenated peptide standards. Mol. Cell. Proteomics 2008, 7 (2), 442–7. [DOI] [PubMed] [Google Scholar]
- Singh S.; Springer M.; Steen J.; Kirschner M. W.; Steen H. FLEXIQuant: a novel tool for the absolute quantification of proteins, and the simultaneous identification and quantification of potentially modified peptides. J. Proteome Res. 2009, 8 (5), 2201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L.; Hunter C. L. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 2006, 5 (4), 573–88. [DOI] [PubMed] [Google Scholar]
- Zinn N.; Winter D.; Lehmann W. D. Recombinant isotope labeled and selenium quantified proteins for absolute protein quantification. Anal. Chem. 2010, 82 (6), 2334–40. [DOI] [PubMed] [Google Scholar]
- Rivers J.; Simpson D. M.; Robertson D. H.; Gaskell S. J.; Beynon R. J. Absolute multiplexed quantitative analysis of protein expression during muscle development using QconCAT. Mol. Cell. Proteomics 2007, 6 (8), 1416–27. [DOI] [PubMed] [Google Scholar]
- Geiger T.; Cox J.; Ostasiewicz P.; Wisniewski J. R.; Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 2010, 7 (5), 383–5. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R.; Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007, 8 (12), 947–56. [DOI] [PubMed] [Google Scholar]
- Studier F. W.; Rosenberg A. H.; Dunn J. J.; Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990, 185, 60–89. [DOI] [PubMed] [Google Scholar]
- Miller J. H.A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, N.Y., 1992. [Google Scholar]
- Baba T.; Ara T.; Hasegawa M.; Takai Y.; Okumura Y.; Baba M.; Datsenko K. A.; Tomita M.; Wanner B. L.; Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006, 2, 2006–0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A.; Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000, 97 (12), 6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. F.; Hattersley N.; Samuel I. D.; Hay R. T.; Tatham M. H. A fluorescence-resonance-energy-transfer-based protease activity assay and its use to monitor paralog-specific small ubiquitin-like modifier processing. Anal. Biochem. 2007, 363 (1), 83–90. [DOI] [PubMed] [Google Scholar]
- Nagai T.; Ibata K.; Park E. S.; Kubota M.; Mikoshiba K.; Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20 (1), 87–90. [DOI] [PubMed] [Google Scholar]
- Wisniewski J. R.; Zougman A.; Nagaraj N.; Mann M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6 (5), 359–62. [DOI] [PubMed] [Google Scholar]
- Olsen J. V.; de Godoy L. M.; Li G.; Macek B.; Mortensen P.; Pesch R.; Makarov A.; Lange O.; Horning S.; Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 2005, 4 (12), 2010–21. [DOI] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26 (12), 1367–72. [DOI] [PubMed] [Google Scholar]
- Cox J.; Neuhauser N.; Michalski A.; Scheltema R. A.; Olsen J. V.; Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10 (4), 1794–805. [DOI] [PubMed] [Google Scholar]
- Kersey P. J.; Duarte J.; Williams A.; Karavidopoulou Y.; Birney E.; Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics 2004, 4 (7), 1985–8. [DOI] [PubMed] [Google Scholar]
- Soufi B.; Kumar C.; Gnad F.; Mann M.; Mijakovic I.; Macek B. Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis. J. Proteome Res. 2010, 9 (7), 3638–46. [DOI] [PubMed] [Google Scholar]
- Bicho C. C.; de Lima Alves F.; Chen Z. A.; Rappsilber J.; Sawin K. E. A genetic engineering solution to the “arginine conversion problem” in stable isotope labeling by amino acids in cell culture (SILAC). Mol. Cell. Proteomics 2010, 9 (7), 1567–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D.; Pinkse M. W.; Oostwaard D. W.; Mummery C. L.; Heck A. J.; Krijgsveld J. An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat. Methods 2007, 4 (9), 677–8. [DOI] [PubMed] [Google Scholar]
- Tatham M. H.; Geoffroy M. C.; Shen L.; Plechanovova A.; Hattersley N.; Jaffray E. G.; Palvimo J. J.; Hay R. T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008, 10 (5), 538–46. [DOI] [PubMed] [Google Scholar]
- Golebiowski F.; Matic I.; Tatham M. H.; Cole C.; Yin Y.; Nakamura A.; Cox J.; Barton G. J.; Mann M.; Hay R. T. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2009, 2 (72), ra24. [DOI] [PubMed] [Google Scholar]
- Schimmel J.; Larsen K. M.; Matic I.; van Hagen M.; Cox J.; Mann M.; Andersen J. S.; Vertegaal A. C. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell. Proteomics 2008, 7 (11), 2107–22. [DOI] [PubMed] [Google Scholar]
- Oscier D.; Fegan C.; Hillmen P.; Illidge T.; Johnson S.; Maguire P.; Matutes E.; Milligan D. Guidelines on the diagnosis and management of chronic lymphocytic leukaemia. Br. J. Hamaetol. 2004, 125 (3), 294–317. [DOI] [PubMed] [Google Scholar]
- Barratt C. L.; Kay V.; Oxenham S. K. The human spermatozoon - a stripped down but refined machine. J. Biol. 2009, 8 (7), 63. [DOI] [PMC free article] [PubMed] [Google Scholar]