Abstract
Cellular therapy is under intense basic science and clinical investigation as a therapeutic intervention. One of the challenges lies in tracking these cells in vivo. While there are many ways to label and track cells—each with strengths and weaknesses—the green fluorescent protein (GFP) is a reporter gene commonly employed. We report a significant and consistent reduction in the expression of GFP with the culture of mesenchymal stem cells (MSCs) isolated from the bone marrow of GFP+ transgenic rodents. After MSC isolation and immunophenotype characterization, along with co-localization with GFP, MSCs were evaluated for GFP expression through flow cytometry and fluorescent microscopy, revealing that only 50% of the cells expressed GFP. Differentiation of the cells to adipocytes did not alter the GFP expression significantly. Incubation with an anti-GFP antibody increased the fluorescent intensity of the GFP-expressing and some of the GFP nonexpressing cells. Incubation of MSCs with a histone deacetylase inhibitor, trichostatin A, did not significantly alter GFP expression, while incubation with a DNA demethylation reagent, 5-azacytidine, increased GFP expression, suggesting that epigenetic modification by DNA methylation may play a role in GFP expression among MSCs.
Introduction
The ability to reliably label and identify cells, without altering their growth and cell–cell interface characteristics, is central to the investigation of cellular based therapies, including cell migration, survival, and microenvironment interactions. Recently, the use of thymidine analogs such as BrdU have come under scrutiny as cell labels due to the finding that donor transplanted cells could transfer the label to recipient cells [1]. Green fluorescent protein (GFP) is one reporter gene system commonly used for cellular identification because it can be detected with high sensitivity and specificity, combined with its relative ease of insertion, expression, and detection [2,3].
There is considerable variability in GFP expression among different transgenic animal strains, and even inconsistency in GFP expression among various tissues within a given animal [4]. Additionally, the optimal method of GFP detection is dependent on the tissue or cell type containing the gene, along with the method used to detect the presence of GFP (microscopy, flow cytometry, etc.), and may require an anti-GFP secondary antibody to clearly distinguish positive cells from autofluorescence [4].
Bone marrow–derived mesenchymal stem cells (MSCs) have been the focus of significant research, due to certain chemotactic properties, their ability to differentiate to stromal tissue components, and their ability to be expanded rapidly, and have shown diverse promise as agents of cell therapy [5], gene therapy [6], and tissue engineering [7]. Mesenchymal stem cells are emerging as a readily available cell population that may, through currently poorly understood mechanisms, alter pathophysiologic processes such as cardiac failure [8] or traumatic brain injury [9]. Previous publications have specified the criteria for defining human MSCs [10] and our methods for rodent MSC characterization are published elsewhere [11]. Although the details of MSC isolation, characterization, and expansion have been discussed extensively, the consequences of this ex vivo processing on GFP expression after isolation from the bone marrow of GFP+ transgenic rodents is previously unknown. Herein, we report a consistent and significant decrease in the expression of GFP by MSCs after isolation from transgenetically produced GFP+ rats. This expression appears to remain unchanged after differentiation. Further, we show that DNA methylation may play a more prominent role than histone acetylation in silencing the GFP gene.
Materials and Methods
All protocols involving the use of animals were in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee (protocol HSC-AWC-06-038).
Mesenchymal stem cell isolation
We isolated MSCs from 200–250 g male Sprague Dawley (SD) GFP+ transgenic rats (Rat Resource and Research Center, Columbia, MO, USA) using previously reported techniques [11–13]. We used a transgenic strain of rats that contain the enhanced GFP gene under the control of the human ubiquitin-C promoter with a woodchuck hepatitis virus posttranscriptional regulatory element [14]. This transgenic strain was made by injecting the lentivirus vector containing the GFP construct into SD rat embryos [15]. The offspring were mated to wild-type SD rats. One line was selected and male offspring were backcrossed to SD females at least four generations until a single insertion site was demonstrated. The line was continued by mating carrier males to wild-type SD females. Phenotypic expression of each offspring is evaluated through epifluorescence microscopy and the genotype is confirmed through polymerase chain reaction.
Characterization of the GFP+ MSC population
Cells were collected, and flow cytometric analysis was performed at passage zero (p = 0, or bone marrow) and p = 1, 2, 5, and 9. Flow cytometry analysis was performed using a BD LSR II (BD Biosciences, San Jose, CA, USA). The percentage of cells expressing GFP was identified in the 500–530 nm emission wavelengths (fluorescein isothiocyanate). The standard MSC phenotype and cell-growth patterns were observed. Immunophenotyping was used to confirm the expression of the following six cell surface molecules: CD11b, CD45, CD49e, CD73, CD90 (all from BD Biosciences), and CD29 (BioLegend, San Diego, CA, USA). In addition, four surface markers (CD11b, CD45, CD29, and CD90) were co-localized with GFP expression. Inverted fluorescent microscopy, with automated cell counts, was used to visualize and count the cells while adherent to the plate. Finally, an anti-GFP 1° antibody (Invitrogen, Carlsbad, CA, USA), followed by a CY2 2° antibody (Jackson Immunology, West Grove, PA, USA) and subsequent flow cytometric analysis was used to enhance and assess GFP expression.
GFP expression after differentiation
Mesenchymal stem cells were differentiated to adipocytes using previously reported techniques [11]. Characterization of adipocytes was performed by Oil red O staining (Diagnostic Biosystems, Pleasanton, CA, USA) of the intracellular lipid-rich vacuoles. Differentiated cells were assessed for GFP expression using fluorescent microscopy.
Assessment of histone acetylation and DNA methylation as mechanisms of gene silencing
After isolation of MSCs from GFP+ transgenic rodents and identification of 50% expression of GFP by fluorescent microscopy and flow cytometry, we used a fluorescent activated cell sorter (BD FACSAria) to separate the GFP+/+ cells (cells that were detectably green) from the GFP+/− cells (cells that were not detectably green). We also maintained the wild-type GFP+ cells (GFP+WT; the normal ∼50:50 mixture of MSCs expressing and not expressing GFP). These three cell populations (GFP+WT, GFP+/+, and GFP+/−) were then expanded over several passages.
To assess the role of histone acetylation in GFP expression, the expanded MSCs were then grown in the presence of trichostatin A (TSA), a histone deacetylase inhibitor, at a concentration of 0.01, 0.1, 0.5, and 1.0 μM for 12, 24, or 48 h. These concentrations were selected based on previous work [16,17]. Antibodies were specific for mono-methylated lysine 4 of histone H3 (Abcam, Cambridge, MA, USA) and the mono-methyl (K20) form of Histone H4 (Abcam). Cells were fixed for 15 min on ice in 4% paraformaldehyde in phosphate-buffered saline (PBS) and subsequently refixed in 70% ETOH for 1 h on ice. Cells were permeabilized in 0.1% Triton X-100 + 2% fetal bovine serum in PBS for 15 min at room temperature (RT). Cells were incubated with antibodies (1:100) for 1 h at RT, followed by a phycoerythrin conjugated secondary antibody (Abcam; 1:200) for 1 h at RT. Histone H3 and histone H4 acetylation, as well as GFP expression, were then assessed using flow cytometric techniques described previously [18].
To assess the role of DNA methylation in GFP expression, the expanded MSCs were then grown in the presence of 5-azacytidine (5-AzaC), a DNA demethylation reagent, at a concentration of 1, 5, and 10 μM for 12, 24, or 48 h. These concentrations were selected based on previous work [16,19,20]. GFP expression was then assessed using flow cytometry.
Results
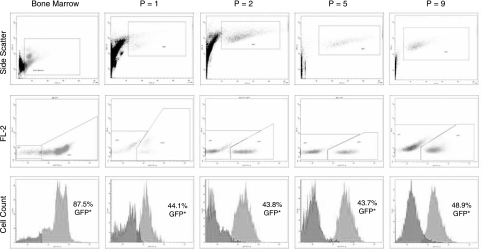
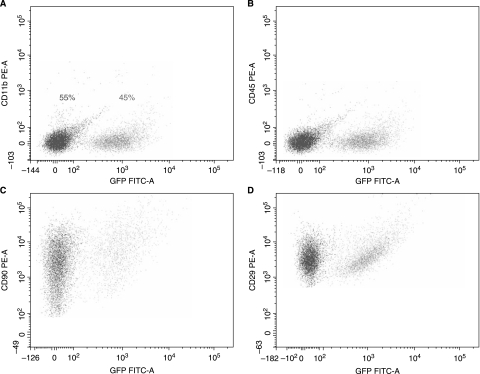
Immediately following isolation of the bone marrow, 87.5% of the total cellular population was found to be GFP+ (Fig. 1). After 7–10 days of culture, the first passage (p = 1) yielded a mixed population of cells typical for an early passage of unsorted cells. The cells were 40% CD11b+, 27% CD45+, 50% CD 29+, 13% CD 49e+, 59% CD 90+, and 2% CD73+. Unexpectedly, 44.1% of the cells were noted to be GFP+ (Fig. 1). At passage 2 (p = 2) the cells were more homogeneous and consistent with MSCs: 6% CD11b+, 4% CD45+, 96% CD 29+, 98% CD 49e+, 95% CD 90+, and 53% CD73+. Once again, ∼50% of the cell population expressed GFP (Fig. 1). As the MSC population was expanded in culture over subsequent passages, the proportion of MSCs expressing GFP remained around 50% (Fig. 1). To ensure that these cells are MSCs, co-localization of GFP+ and CD29, CD90, CD45, and CD11b is shown in Fig. 2.
FIG. 1.
Characterization of the GFP+ MSC population over passage. The top row illustrates the cell scatter distribution (forward scatter vs. side scatter); the middle row illustrates dual-parameter dot plots (GFP vs. FL-2), and the bottom row illustrates histograms (GFP vs. cell counts) of GFP positivity of the MSC population over passages. Freshly isolated bone marrow cells were found to be highly positive for GFP. After in vitro cell culture, however, MSCs were noted to be ∼50% GFP+. This 50% expression was observed to remain the same after subsequent passage (ie., there was not continued decline in expression over time or with multiple cell passage).
FIG. 2.
Co-localization of MSC markers and GFP. Here we correlate the GFP expression with the following MSC markers: CD11b− (A), CD45− (B), CD90+ (C), and CD29+ (D). There are ∼45% GFP+ and ∼55% GFP− cells for each marker.
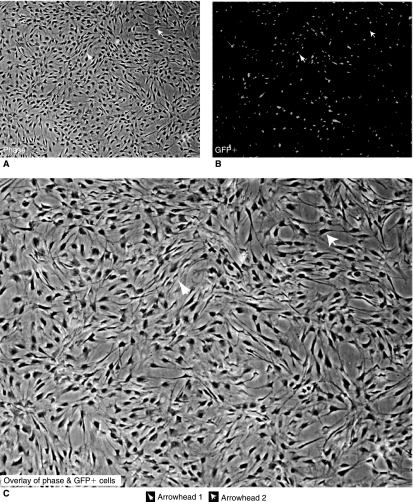
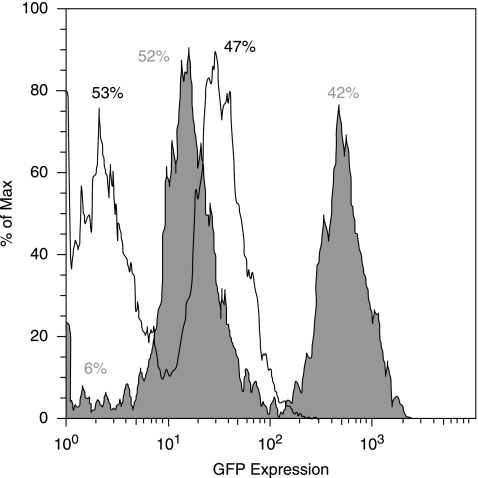
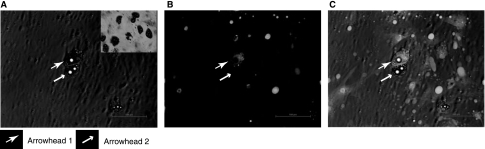
The expression of GFP (Fig. 3). We found 873 ± 42 cells/4× field and only 307 ± 51 GFP+ cells were identified (36 ± 7%). After the addition of a GFP+ antibody, there was a significant increase in fluorescent intensity, among previously positive and some previously negative MSCs, detected by flow cytometry (Fig. 4).
FIG. 3.
Fluorescent microscopy showing expression of GFP among MSCs. All MSCs shown under 4× light microscopy (A), GFP+ cells only (B), and an overlay of the two populations (C). An example of a GFP+ MSC (arrowhead 1) and a GFP− MSC (arrowhead 2) is identified in each image. These images show that ∼45% of the total MSC population is expressing GFP.
FIG. 4.
GFP expression after incubation with an anti-GFP antibody. Mesenchymal stem cells (MSCs) were incubated with an anti-GFP 1° antibody, followed by a CY2 2° antibody. Flow cytometric analysis revealed a significant increase in GFP expression (green area) compared to control GFP+ MSCs (transparent area). Even with the addition of the anti-GFP antibody, 15–20% of the MSC population expressed fluorescence levels equal or below that of GFP− MSCs.
After differentiation to adipocytes, there was no appreciable change in GFP expression. Approximately 50% of the differentiated cells continued to express GFP (Fig. 5).
FIG. 5.
Differentiated mesenchymal stem cells (MSCs) continue to express GFP. MSCs were differentiated to adipocytes and stained for intracellular lipid-rich vacuoles by Oil Red O (inset). Phase microscopy (A), fluorescent microscopy in the 500–530 nm emission wavelengths (B), and an overlay of both (C) is used to show two MSCs differentiated to adipocytes: arrowhead 1 shows a cell that maintained GFP expression and arrowhead 2 shows a cell that maintained GFP nonexpression.
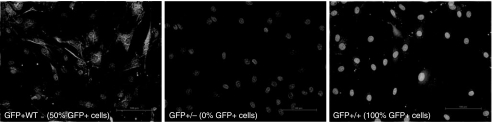
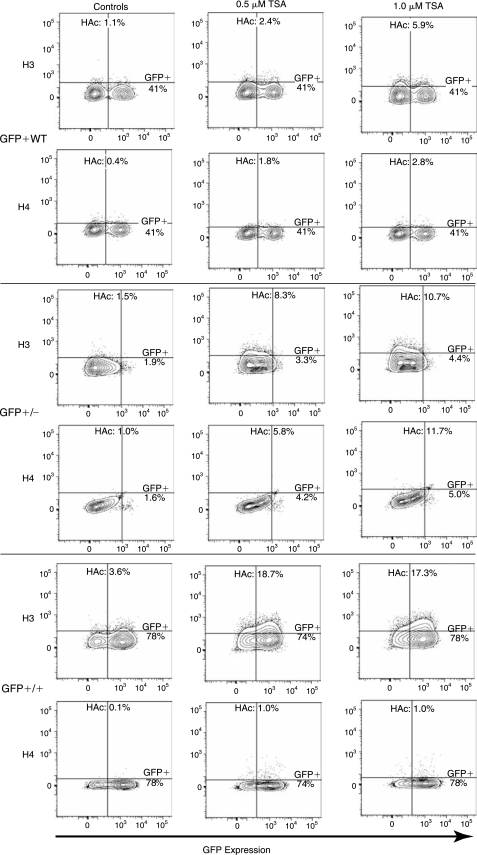
After sorting and subsequent expansion of all three cell populations (GFP+WT, GFP+/+, and GFP+/−; Fig. 6) in the presence of the higher concentrations of TSA (0.5, and 1.0 μM), both H3 and H4 acetylation were increased after 48 h in culture (Fig. 7). The increase of H3 acetylation was more pronounced than the increases in the H4 acetylation (9.2 ± 2.5% vs. 4.7±3.0%). Among the three cell populations the GFP+/+ population showed the greatest increase in H3 acetylation (13.7%), the GFP+/− population increased 9.1%, and the GFP+WT population increased 4.8%. Despite these increases in histone acetylation, H3 greater than H4, no increase in GFP expression was identified among the GFP+WT and GFP+/+ populations, and only a 2.5–3.4% increase was seen among the GFP+/− population. Among the GFP+WT population, no disparity in GFP expression between the GFP-expressing and GFP-nonexpressing populations was detected after incubation with TSA. No increases in the mean fluorescent intensities (MFIs) of any cell population were identified.
FIG. 6.
Fluorescent-activated cell-sorting isolation of three cell populations. A fluorescent-activated cell sorter was used to separate the GFP+/+ cells (cells that were detectably green) and GFP+/− cells (cells that were not detectably green) from the original “wild-type” GFP+ cells (GFP+WT), the normal ∼50:50 mixture of MSCs expressing and not expressing GFP.
FIG. 7.
Flow cytometric assessment of the role of histone acetylation in GFP expression. Mesenchymal stem cells were sorted, expanded, and grown in the presence of trichostatin A (TSA), a histone deacetylase inhibitor, at a concentration of 0.01, 0.1, 0.5, and 1.0 μM for 12, 24, or 48 h. Only the higher concentrations at the later time points lead to increases in histone acetylation and are shown here. Although increases in histone acetylation, H3 > H4, are observed, there are minimal increases in GFP expression. H3: histone H3; H4: histone H4; HAc: histone acetylation.
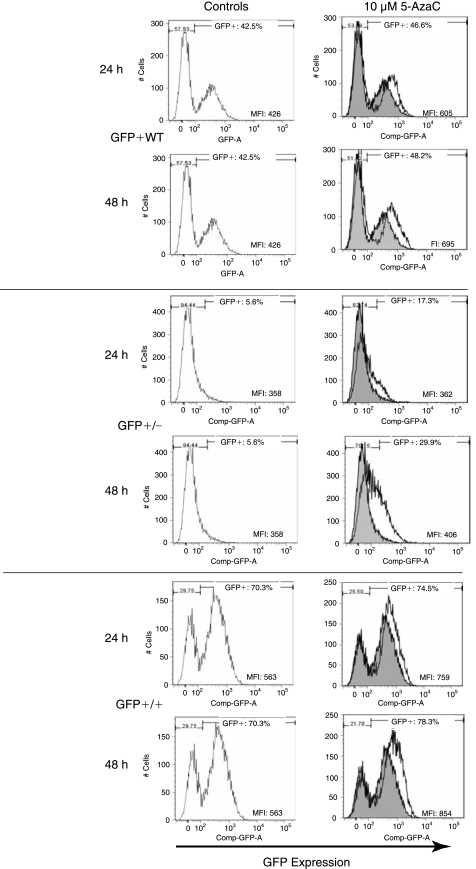
After culture of all three cell populations in the presence of the high concentration of 5-AzaC (10 μM), increases in GFP expression, or reactivation from silencing, were identified at 24 and 48 h. The GFP+/− population showed the greatest increase, 24.3%, at 48 h. The GFP+/+ population increased 8.1% and the GFP+WT population increased 5.7%. Increases in MFI at 48 h were also noted among all three cell populations compared to controls. The MFI of the GFP+/+ population increased from 563 to 864, the GFP+/− population increased from 358 to 406, and the GFP+WT population increased from 426 to 695 (Fig. 8). Among the GFP+WT population, no disparity in GFP expression between the GFP-expressing and GFP-nonexpressing populations was detected after incubation with 5-AzaC.
FIG. 8.
Flow cytometric assessment of the role of DNA methylation in GFP expression. Mesenchymal stem cells were sorted, expanded, and grown in the presence of 5-azacytidine, a DNA demethylation reagent, at a concentration of 1, 5, and 10 μM for 12, 24, or 48 h. The highest concentration at the later time points lead to the largest increases in GFP expression and are shown here. Significant increases in GFP expression (GFP+) and mean fluorescent intensity (MFI) are seen with all three cell populations. Greater increases in GFP expression were identified with longer 5-AzaC incubation.
Discussion
Our results show that while 80–90% of the bone marrow cell population of a GFP+ transgenic rodent may be GFP+, only ∼50% of the expanded MSC population is GFP+, irrespective of passage number. In the presence of a GFP-specific antibody, the intensity of the GFP expressing and a subset of the nonexpressing cells is increased. After differentiation to adipocytes, no major alteration of GFP expression was noted. An analysis of sorted GFP-expressing, GFP-nonexpressing, and GFP wild-type populations revealed that epigenetic modification through DNA methylation is more likely than histone acetylation to play a role in the observed genetic downregulation.
Although GFP transgene expression is a popular cell-labeling mechanism, it has been shown to have limitations [21]. Swenson and coauthors recently evaluated the expression of GFP in transgenic rodents and found considerable variability of expression both within and between transgenic rodents [4]. Only ∼79.4% of the blood cells isolated from three rodents expressed GFP. Although they isolated cells from various organs, identifying variation in GFP expression within an animal, they did not isolate the bone marrow or, more specifically, MSCs. The gene-silencing phenomenon has been shown to be prominent among stem cell populations [22,23], particularly embryonic and cancer lines. Given rapid growth, expansion, and differentiation, these cells may have more active cellular mechanisms, potentially through epigenetic modification, of repressing foreign DNA [16]. Differentiation of stem cells may lead to changes in the chromatin structure surrounding the lentivirus [19] and, subsequently, contribute to loss of GFP expression [24]. Although we clearly show decreased expression to 40–60% among expanded MSCs, differentiation to adipocytes did not significantly alter, at least acutely, GFP expression.
Given these limitations with isolation of MSCs from the bone marrow of GFP+ transgenic rodents and previously mentioned concerns surrounding BrdU labeling, there are several options, currently available, for labeling and tracking cells in vivo. Thiol-Reactive (CellTracker™; Invitrogen) or Amine-Reactive (CellTrace™; Invitrogen) tracers freely enter live cells and can be identified through several cell divisions. Carbocyanine membrane dyes (DiI) diffuse within the plasma membrane with low cell toxicity. Nanocrystals (Qdot or Qtracker) are taken up into the cytoplasm of live cells and are highly fluorescent over multiple cell generations, spanning several weeks. Alternative vectors (such as DsRed) can be transfected, although epigenetic silencing may apply to these as well. Finally, fluorescent-activated or magnetic-activated cell sorting can be used to isolate the positive cells within a mixed population. Despite the many options, each is fraught with limitations depending on application, timing, and tissue processing, among numerous factors.
Although detection of GFP+ cells through flow cytometry clearly identified a positive and negative population, discrimination of these cell populations through microscopy, particularly in vivo (i.e., in cell therapy studies), is more challenging. The use of anti-GFP antibodies allows significantly clearer discrimination of authentic signal from autofluorescence [4]. We found that the antibody not only increased the intensity of expression among the MSCs that already expressed GFP, but it also allowed some (but not all) of the previously nonexpressing cells to be visualized. Previous work has shown that only 15 amino acids, of the 238 total that compose the GFP protein, can be deleted from the GFP protein without disrupting it's fluorescence and that removal of even a few amino acids can reduce or significantly alter fluorescent activity [25]. Such a high minimal functional domain provides a reasonable explanation as to why transgenic rodents do not ubiquitously express the protein. Additionally, it may explain why addition of an anti-GFP antibody may bind to an epitope of a protein that does not have the minimal domain necessary for fluorescence, rendering a previously invisible cell visible.
The dynamic interplay of multiple epigenetic modifications that lead to transgene expression, or lack thereof, has been the source of significant investigation [19]. Epigenetic modifiers, such as histone acetyltransferase, histone deacetylase, chromatin-remodeling complex, and DNA methyltransferase, may spread from nearby areas or be directly recruited to the site of the GFP gene insertion [16]. Such modifications can lead to variegation, where the switch flickers between on and off, producing subsets of cells that either express GFP or are silent [19]. It is widely, although not universally, accepted that DNA methylation is the first step in a gene-silencing mechanism that ultimately controls chromatin structure [26]. DNA methylation and histone modification are commonly implicated in gene silencing [27,28]. Regions of methylated DNA are characterized by condensed chromatin prohibitive to transcriptional initiation and chromatin associated with hypoacetylated histones is also characterized by a condensed conformation, resistant to digestion by DNAse I [29]. Therefore, if epigenetic modifications, such as DNA methylation and histone acetylation, are responsible for decreases in GFP expression, demethylating agents (such as 5-AzaC) or deacetylase inhibitors (such as TSA) should increase the expression of GFP. Many studies have now indicated both independent and combinatorial roles for DNA methylation and histone modification in gene silencing [16,19,26]. Among MSCs, we found that DNA methylation is likely to play a larger role than histone modification, as cells in the presence of 5-AzaC show significant increases in GFP expression, while those expanded in the presence of TSA showed hyperacetylation of histones without significant increases in GFP expression.
Regardless of the ultimate mechanism, GFP+ transgenic animal–derived bone marrow progenitor cells may not ubiquitously express GFP with cell expansion. A thorough analysis of the particular progenitor cell population isolated must be performed prior to initiation of cell therapy experimentation. Growing the cells in the presence of a demethylating agent or isolation of the GFP-expressing cell fraction, through fluorescent-activated cell sorting or magnetic-activated cell sorting, are alternative ways to help secure a largely GFP+ cell population.
Acknowledgments
This work was supported by grants NIH T32 GM008792-06 (M.T.H.); M01 RR02558; R21 HD 042659-01A1; Texas Higher Education Coordinating Board; Children's Memorial Hermann Foundation.
References
- 1.Burns TC. Ortiz-Gonzalez XR. Gutierrez-Perez M. Keene CD. Sharda R. Demorest ZL. Jiang Y. Nelson-Holte M. Soriano M. Nakagawa Y. Luquin MR. Garcia-Verdugo JM. Prosper F. Low WC. Verfaillie CM. Thymidine analogs are transferred from prelabeled donor to host cells in the central nervous system after transplantation: a word of caution. Stem Cells. 2006;24:1121–1127. doi: 10.1634/stemcells.2005-0463. [DOI] [PubMed] [Google Scholar]
- 2.Okabe M. Ikawa M. Kominami K. Nakanishi T. Nishimune Y. “Green mice”. as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 3.Schmid JA. Neumeier H. Evolutions in science triggered by green fluorescent protein (GFP) Chembiochem. 2005;6:1149–1156. doi: 10.1002/cbic.200500029. [DOI] [PubMed] [Google Scholar]
- 4.Swenson ES. Price JG. Brazelton T. Krause DS. Limitations of green fluorescent protein as a cell lineage marker. Stem Cells. 2007;25:2593–2600. doi: 10.1634/stemcells.2007-0241. [DOI] [PubMed] [Google Scholar]
- 5.Le Blanc K. Pittenger M. Mesenchymal stem cells: progress toward promise. Cytotherapy. 2005;7:36–45. doi: 10.1080/14653240510018118. [DOI] [PubMed] [Google Scholar]
- 6.Hamada H. Kobune M. Nakamura K. Kawano Y. Kato K. Honmou O. Houkin K. Matsunaga T. Niitsu Y. Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Sci. 2005;96:149–156. doi: 10.1111/j.1349-7006.2005.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leo AJ. Grande DA. Mesenchymal stem cells in tissue engineering. Cells Tissues Organs. 2006;183:112–122. doi: 10.1159/000095985. [DOI] [PubMed] [Google Scholar]
- 8.Barbash IM. Chouraqui P. Baron J. Feinberg MS. Etzion S. Tessone A. Miller L. Guetta E. Zipori D. Kedes LH. Kloner RA. Leor J. Systemic delivery of bone marrow-derived mesenchymal stem cells to the infarcted myocardium: feasibility, cell migration, and body distribution. Circulation. 2003;108:863–868. doi: 10.1161/01.CIR.0000084828.50310.6A. [DOI] [PubMed] [Google Scholar]
- 9.Mahmood A. Lu D. Qu C. Goussev A. Chopp M. Long-term recovery after bone marrow stromal cell treatment of traumatic brain injury in rats. J Neurosurg. 2006;104:272–277. doi: 10.3171/jns.2006.104.2.272. [DOI] [PubMed] [Google Scholar]
- 10.Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F. Krause D. Deans R. Keating A. Prockop D. Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 11.Harting MT. Jimenez F. Pati S. Baumgartner J. Cox CS. Immunophenotype characterization of rat bone-marrow derived mesenchymal stem cells. Cytotherapy. 2008;10(3):243–253. doi: 10.1080/14653240801950000. [DOI] [PubMed] [Google Scholar]
- 12.Breyer A. Estharabadi N. Oki M. Ulloa F. Nelson-Holte M. Lien L. Jiang Y. Multipotent adult progenitor cell isolation and culture procedures. Exp Hematol. 2006;34:1596–1601. doi: 10.1016/j.exphem.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Schrepfer S. Deuse T. Lange C. Katzenberg R. Reichenspurner H. Robbins RC. Pelletier MP. Simplified protocol to isolate, purify, and culture expand mesenchymal stem cells. Stem Cells Dev. 2007;16:105–107. doi: 10.1089/scd.2006.0041. [DOI] [PubMed] [Google Scholar]
- 14.Rat Resource and Research Center, Columbia, MO. http://www.nrrrc.missouri.edu/ http://www.nrrrc.missouri.edu/
- 15.Lois C. Hong EJ. Pease S. Brown EJ. Baltimore D. Germline transmission and tissue-specific expression of trans-genes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 16.Katz RA. Jack-Scott E. Narezkina A. Palagin I. Boimel P. Kulkosky J. Nicolas E. Greger JG. Skalka AM. High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. J Virol. 2007;81:2592–2604. doi: 10.1128/JVI.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snykers S. Vanhaecke T. De Becker A. Papeleu P. Vinken M. Van Riet I. Rogiers V. Chromatin remodeling agent trichostatin A: a key-factor in the hepatic differentiation of human mesenchymal stem cells derived of adult bone marrow. BMC Dev Biol. 2007;7:24. doi: 10.1186/1471-213X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronzoni S. Faretta M. Ballarini M. Pelicci P. Minucci S. New method to detect histone acetylation levels by flow cytometry. Cytometry A. 2005;66:52–61. doi: 10.1002/cyto.a.20151. [DOI] [PubMed] [Google Scholar]
- 19.Yao S. Sukonnik T. Kean T. Bharadwaj RR. Pasceri P. Ellis J. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Ye NS. Chen J. Luo GA. Zhang RL. Zhao YF. Wang YM. Proteomic profiling of rat bone marrow mesenchymal stem cells induced by 5-azacytidine. Stem Cells Dev. 2006;15:665–676. doi: 10.1089/scd.2006.15.665. [DOI] [PubMed] [Google Scholar]
- 21.Brazelton TR. Blau HM. Optimizing techniques for tracking transplanted stem cells in vivo. Stem Cells. 2005;23:1251–1265. doi: 10.1634/stemcells.2005-0149. [DOI] [PubMed] [Google Scholar]
- 22.Cherry SR. Biniszkiewicz D. van Parijs L. Baltimore D. Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J. Yang Q. Chang LJ. Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J Virol. 2005;79:13497–13508. doi: 10.1128/JVI.79.21.13497-13508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Overstraeten-Schlogel N. Delgaudine M. Beguin Y. Gothot A. Limitations of the use of GFP transgenic mice in bone marrow transplantation studies. Leuk Lymphoma. 2006;47:1392–1393. doi: 10.1080/10428190500513512. [DOI] [PubMed] [Google Scholar]
- 25.Li X. Zhang G. Ngo N. Zhao X. Kain SR. Huang CC. Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence. J Biol Chem. 1997;272:28545–28549. doi: 10.1074/jbc.272.45.28545. [DOI] [PubMed] [Google Scholar]
- 26.Pannell D. Osborne CS. Yao S. Sukonnik T. Pasceri P. Karaiskakis A. Okano M. Li E. Lipshitz HD. Ellis J. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 2000;19:5884–5894. doi: 10.1093/emboj/19.21.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pannell D. Ellis J. Silencing of gene expression: implications for design of retrovirus vectors. Rev Med Virol. 2001;11:205–217. doi: 10.1002/rmv.316. [DOI] [PubMed] [Google Scholar]
- 28.Ellis J. Yao S. Retrovirus silencing and vector design: relevance to normal and cancer stem cells? Curr Gene Ther. 2005;5:367–373. doi: 10.2174/1566523054546233. [DOI] [PubMed] [Google Scholar]
- 29.Swindle CS. Klug CA. Mechanisms that regulate silencing of gene expression from retroviral vectors. J Hematother Stem Cell Res. 2002;11:449–456. doi: 10.1089/15258160260090915. [DOI] [PubMed] [Google Scholar]