Summary
Metabolomics describes the measurement of the full complement of the products of metabolism in a single biological sample and correlating these metabolomic profiles with known physiological or pathological states. The metabolome offers the possibility of finding unique fingerprints responsible for different phenotypes. Analytical techniques such as nuclear magnetic resonance or mass spectrometry measure thousands of compounds within the metabolome simultaneously and appropriate data mining and database tools allow the finding of significant correlations between the measured metabolomes. The first direct outcome of nutritional metabolomics will be the discovery of biomarkers, which can reveal changes in health and disease but also indicate short term and long-term dietary intake. The concerted actions of nutrigenomics and metabolomics will play a crucial role in understanding how specific interactions of single nucleotide polymorphisms (SNP) influence a person’s response to a diet. Finally, systems biology approaches to human nutrition combine transcriptomics, proteomics and metabolomics with the aim of understanding how diets interact within the human being.
Keywords: metabolomics, metabolome, data mining, multivariate statistics, databases, mass spectrometry, biomarkers, nutrigenomics, systems biology
Introduction
Nutrition and clinical scientists are routinely profiling human biofluids for a range of physiological markers, including many macro- and micronutrients as well as a limited number of known metabolites that can be related to nutrition. These markers then provide information on nutritional status and risk factors for health and disease, such as on the bioavailability of nutrients from food sources, on the interrelationships of selected metabolites and their correlation with disease, or on deficiencies that individuals may have of essential nutrients etc. Importantly, these targeted profiling approaches usually only test one specific nutritional hypothesis, measure a specific marker or investigate a specific biochemical pathway, for which results are correlated or compared with predicted outcomes or recommended intake values. Today, however, analytical technologies have emerged that permit the measurement of thousands of metabolites and nutrients simultaneously from a single sample. This opens up numerous possibilities for studying nutritional effects and interactions of metabolites, with important applications such as the determination of disease and pre-disease biomarkers or the identification of the individual metabolism of persons, allowing for personalised nutrition to address specific health problems. This integrative approach of measuring the distribution and concentration of a wide range of metabolites in a body sample (referred to as the metabolome), resulting from the gene and protein expression, and correlating the information with a known physiological or pathological state, is called metabolomics. Nutritional metabolomics focuses on those active metabolites, nutrients (or non-nutrients) among the thousands of components in the metabolome that are associated with the effects that different diets have on us.
Systems biology
Unfortunately, metabolites and genes alone are not sufficient to fully describe the effects of nutrition. Genes are transcribed into RNA and RNA is translated into proteins and a comprehensive picture of nutrition must include all these steps. Different large scale, multi-centre projects are trying to tackle nutritional research questions through systems biology approaches, combining transcriptomics, proteomics and metabolomics with the aim of understanding how diets interact within the human being. These highly complex and difficult to design experiments require significant budgets to cover the required complexity. Moreover, the bioinformatics tools to connect and integrate the different data sets are still under development and data crunching often takes several years to complete. In other fields of science, data is made publicly available, such as gene expression data through the Gene Expression Omnibus (GEO) of the National Center of Biotechnological Information (http://www.ncbi.nlm.nih.gov/geo), enabling the bioinformatics community worldwide to develop and test new algorithms for data conversion (Shaik & Yeasin 2008). Presently, systems biology studies addressing the complexity of nutritional research are very difficult to perform because of the immense budget requirements. Further technological advancements in all omics areas, however, will undoubtedly enable such complex studies in the near future.
Opportunities for human nutrition
Metabolomics offers a drastically different approach to nutritional science, as already seen in other areas of biomedical research, e.g. pharmacology and toxicology. Nutrition research will be significantly impacted by metabolomics, for several reasons. Firstly, metabolism and nutrition are closely linked and thus metabolomics is the logical first choice for application among the various omics technologies (genomics, transciptomics, proteomics, and metabolomics). Another important advantage is that metabolomics – and metabolomics techniques – address and consider the entire metabolic complexity of diets. Classical reductional, hypothesis-driven nutrition research tests one compound at a time, which has lead to key findings such as the importance of vitamins or minerals. But this strategy struggles with food components or diets, which have more subtle, less explicit biological functions. In the classical approach, the complexity of dietary patterns is reduced to one or several metabolites, which are thought to represent the larger group. For example, in fish oil studies, eicosapentaenoic acid (EPA) is often used to describe the entire polyunsaturated fatty acid (PUFA) group, or in the case of the Mediterranean diet, flavonoids are represented as a single compound, quercetin. In these cases, the positive effects for health or for the treatment of a disease become extremely difficult to prove in intervention studies. A metabolomics approach is much more powerful because it has the potential to reveal which components in the diet are responsible for which effect. Finally, metabolomics will have a tremendous impact on nutrition research through the discovery of important new biomarkers. In fact, biomarker research will be the driving force in the rapid progress of nutrition research in the future because nutrition deals with diseases and the health of humans. Whereas the pathology of disease and clinical outcomes can usually be clearly defined, health is much more difficult to describe and improvements in health can rarely be measured. Appropriate biomarkers will enable us to precisely determine health and changes in health in the future. Biomarkers can also reveal dietary intake. Developing methodologies to determine exactly what people are eating has always been essential in nutrition research. Biomarkers of nutritional intake will be extremely important for this area, but it is expected to require a significant amount of focussed metabolomics research.
Analytical technologies and strategies for metabolomics
Metabolomics has emerged as a complementary technology to the other omics disciplines (in particular genomics, transciptomics and proteomics), which are concerned with the measurements of DNA, mRNA, proteins and their interactions. Unlike these disciplines, which apply a single analytical technique or measure, at least in theory, a single chemical class of compounds, metabolomics profiles entire populations of chemically very diverse, low molecular weight metabolites. Moreover, the relative concentration levels of these compounds range from ultra-trace (picomolar or less) to millimolar or even higher levels, thus most likely exceeding the linear dynamic ranges of the applied analytical techniques.
Because of these analytical challenges, the field of metabolomics is currently strongly driven by technological developments. The two most common techniques used are nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). Mass spectrometry is almost always combined with a preliminary chromatographic separation step, either gas chromatography (GC-MS) or liquid chromatography (LC-MS or LC-MS/MS, tandem mass spectrometry). It is outside the scope of this article to discuss technical details of the analytical technologies. The interested reader is referred to several excellent review articles on this subject (e.g. Coen et al. 2008, Schlotterbeck et al. 2006).
It is important to consider that some of the new analytical technologies are so sensitive that they will likely identify previously undetected compounds, thus yielding results that possibly conflict with outcomes of earlier studies. Also, in many cases, the structures of candidate metabolites will be unknown at the discovery stage of the metabolomics study, therefore necessitating extensive structure elucidation work at the subsequent validation stage.
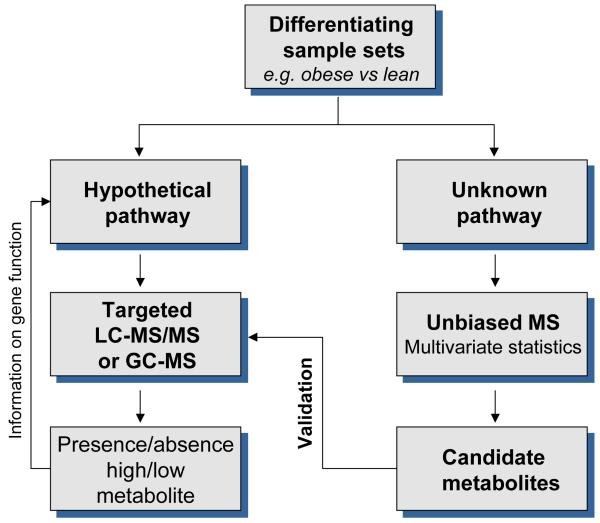
In most metabolomics studies, differential sets of samples are analysed by directly comparing two situations and looking for significant differences between them; for example, obese versus lean or diabetic versus non-diabetic. Metabolomics can be either biased or unbiased (targeted or untargeted), depending on whether a hypothetical pathway is investigated with a target group of known metabolites or whether the pathway is totally unknown and the analysis must cover the entire complement of the metabolome in question. A typical workflow for metabolomics studies is illustrated in Figure 1.
Figure 1.
Workflow of biased and unbiased metabolomics studies
(Abbreviations: LC-MS/MS, liquid chromatography-tandem mass spectrometry; GC-MS, gas chromatography-mass spectrometry).
Equally important to the analytical technologies are the methods used to find the significant correlations between the metabolomes under investigation. State-of-the-art bioinformatics, data mining and database tools are rapidly emerging to manage and interpret the massive data sets from these experiments.
Data mining
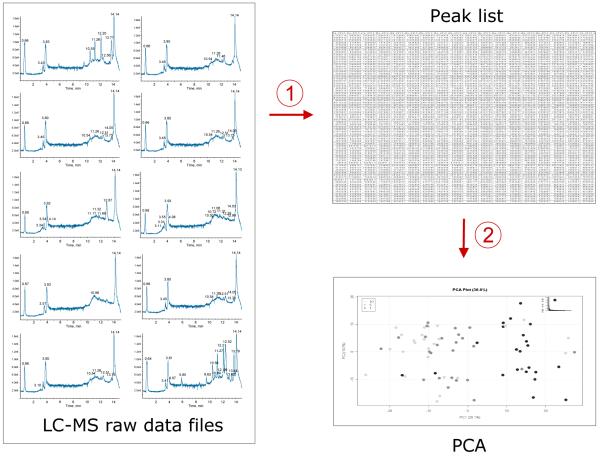
Metabolomics research is relatively fast and inexpensive as compared to other omics disciplines, e.g. transcriptomics and proteomics. Importantly, it facilitates the inclusion of experiments at various time points and the use of a large number of variables and replicates, whereas for micro-arrays and MudPIT (multidimensional protein identification technology) proteomics experiments, samples are usually pooled to reduce time and costs (Usaite et al. 2008). Using larger sample numbers and variables, however, results in vast quantities of data. One of the major challenges in metabolomics therefore is the conversion of this complex raw data into useful information. The first step is data reduction (see Figure 2). Most metabolomics experiments focus on the conversion of LC-MS raw data files to peak lists, where each peak has one or more identifiers (retention time and m/z or tandem mass spectrometry MS2 spectra) and an intensity or level, which should relate to the amount of compound responsible for that peak. Where LC-MS data are continuous and multidimensional, peak lists are usually 2-dimensional and can be easily converted into spreadsheets. Principle component analysis (PCA) and related strategies will usually be applied first for data interrogation. PCA is an unsupervised technique that determines correlation differences between sample sets, which can be caused by either a biological difference or a methodological bias. In most cases, it is a highly informative way for obtaining a first impression of the quality of the acquired data, independent of the actual research question. For example, PCA analysis can differentiate between the metabolic profile of men and women as well as vegetarians and non-vegetarians (Holmes et al., 2008). When the data reveals analytical run dependent effects, this is usually the result of an inadequate data normalisation method. Optimal normalisation processes are extremely difficult to achieve without proper control samples, which are therefore an essential part of any metabolomics experiments.
Figure 2.
Data mining strategy in metabolomics: the first step is data reduction, the second step is data interrogation
(Abbreviations: LC-MS, liquid chromatography-mass spectrometry; PCA, principal component analysis).
In a follow-up step, supervised multivariate statistics can be used, for which partial least square discriminant analysis (PLSDA) is the most commonly applied method. Supervised techniques use prior knowledge about the samples; i.e. which samples are known to belong to a certain group. Nevertheless, for much larger numbers of variables than numbers of samples, there is a severe chance of over-fitting the data (Westerhouse et al. 2008). In addition to the concept of differentiating peaks, it is also possible to use data-mining tools to look for correlating peaks (either correlating in a linear or a non-linear fashion). This results in correlation networks that give additional information on the interconnectivity of peaks and metabolites.
In general, the aim at this stage is to reduce the complete peak list to a much shorter list of ‘interesting’ peaks. The relevance of these interesting peaks has to be assessed, which in most cases is initially conducted based on chemical information. The peak shape and behaviour across the sample will be interrogated to check whether a real metabolite is present or whether an artefact of the method caused the peak. In addition, chemical information will be used to classify or identify the responsible metabolite.
Databases
One of the main driving forces in the rapid technical developments taking place in the post-genomics era are open access databases. A wealth of information can be readily obtained today by aligning protein or DNA sequences with these internet-based databases. Although there is still a reasonable chance that sequences are returned as unknown “open reading frames” (ORF), in many cases the search will show that the protein or gene is either a known gene or similar to a known gene, with a known function, facilitating the biological interpretation. Metabolites, however, are intrinsically different. Where proteins and genes with the same function are similar in structure across organisms; metabolites have the same structure but similar or different functions across organisms. This is not the only drawback encountered in metabolomics, because structural similarity is much more difficult to assess with small molecules as they can not be reduced to a sequence of building blocks, in the way DNA is a string of nucleotides and proteins are a string of amino acids. Even the unambiguous assignment of a chromatographic peak to a metabolite is a laborious process. This is one of the main reasons why many metabolomics groups rely heavily on NMR analysis for metabolic profiling because it yields spectra that are reasonably similar across different institutes and different instrument manufacturers. LC-MS/MS on the other hand does not generally provide these universal and standardised results. Chromatographic retention times are difficult to reproduce and mass spectra of the same metabolite are often quite mass spectrometry vendor-specific. There are, however, a number of open access databases containing mass spectral information, e.g. HMDB (human metabolome database), Massbank and Metlin (Wishart et al. 2007). The increasing speed and resolution of mass spectrometers will further reduce the number of possible candidates for each chromatographic peak and when these results are combined with other spectral information (in particular high resolution MSn data), it is foreseeable that future mass spectrometers will be able to yield enough information with sufficient accuracy for rapid database searching and metabolite classification, overcoming current differences in machine type and brand. Even though automated database searching is not universally available yet for metabolomics applications, different research groups have started to establish large-scale databases on metabolites, e.g. the Kyoto Encyclopaedia on Genes and Genomes (KEGG), which integrates metabolites with proteins and genes. This information is relevant in metabolomics experiments and the ability to search rapidly for the function of a specific gene helps in understanding the relevance of a particular metabolite in the experiment.
Nutritional metabolomics, however, is even more complex. Humans are heterotrophic organisms; that is, they feed on other organisms, each with its own metabolome. The number of different metabolomes that we consume daily in our diet is significant, making nutritional metabolomics drastically more complex than other metabolomics disciplines. Consider a simple example: the amount of calories or the protein, fat and carbohydrate content of a bowl of cereal can be easily found in the literature. The metabolic profile of the same bowl of cereal, however, is extremely complex and will probably consists of several thousands (if not ten thousands) of different metabolites, most of which are not known or not available in any open-access database.
Validation
The next step in a metabolomics experiment, after the raw data is reduced to a short-list of significantly differentiating metabolites, which are either fully identified or at least classified metabolites, is the interpretation and correlation with phenotype. In most cases, however, where unbiased or untargeted methods are used, it is highly desirable to further validate the results. It is very difficult to validate a metabolic profiling method, as differences found between sample sets may simply be the result of artefacts of the methodology rather than genuine results. It is therefore essential to confirm the results with a targeted analytical method. At this stage, the differentiating peaks are assumed to be originating from known or classified metabolites, which can be easily confirmed by re-analysing the samples specifically for those compounds, using authentic standards and/or previously published and validated methods. This essential step in metabolomics is very often omitted, but is clearly required to avoid speculation (Villas-Boas et al. 2007).
Genetics, genomics and metabolomics
Pharmacogenomics and nutrigenomics are removing the idealised concept that all humans react the same way in an experiment. Metabolomics shows that metabolic phenotypes can be specific to persons (Assfalg et al. 2008). Differences in genes lead to differences in metabolism and differences in efficacy of compounds. These differences can have dramatic effects in pharmacology but the same applies to nutrition. International collaborations like the “haplotype map of the human genome” (HAPMAP) are mapping the single nucleotide polymorphisms (SNP) in the human genome and studies on the effect of SNPs on human metabolism are rapidly advancing (Marvelle et al. 2008, Méplan et al. 2008). At the same time, technological developments make it possible to rapidly and inexpensively map all SNPs in a person and it is expected that within a few years this SNP mapping will be an essential step in every nutritional study involving humans. Although the information is already available to predict the effect of one SNP (Yang et al. 2007), the ability to forecast the effects of interacting SNPs is still years from being substantiated, especially in relation to metabolism. Metabolomics will play a crucial role in understanding this interaction and it will help to elucidate how specific interacting SNPs influence a person’s response to a diet.
The development of genetic markers and chips has also accelerated the research in human genetics. Large studies involving ten thousands or more subjects enable scientists to determine which regions of the genome are responsible for a disease or the predisposition to a disease (e.g. obesity or metabolic syndrome). The interpretation of these results are always obscured by what are called epistatic effects, which describe the specific interactions of regions in the genome that hinder the ability to link a specific trait to a specific region (Boone et al. 2007). Geneticists, however, are dependent on phenotypic details (traits) to be able to map them to the genome. Currently this is mainly done with traits that are clinically relevant and measurable, such as weight, BMI or glucose levels in urine. Combining nutritional and metabolomics data to phenotype humans will allow geneticists to reveal how genes interact with food in much more detail, and how this information can be used for health changes. Such projects demand a concerted action of different research disciplines and are strongly dependent on a large enough number of subjects and sufficient experimental detail to make significant progress in the future. Nutritional research and nutrition in general would benefit tremendously from such projects, however, as it opens up the possibility for personalised nutrition given by nutrition clinicians according to people’s specific genetics. In the same context, it is worthwhile mentioning that the food industry will recognise soon that specific genetic profiles will open up new markets.
Outlook to the future
Metabolomics is rapidly changing nutrition science, the same way it changed other areas of biomedical research over the past several years. Currently, the field of nutritional metabolomics is driven by tech-savvy bioanalytical chemists, interested in method and technology development, by bioinformaticians developing novel data mining and database tools, and by nutrition biochemists wanting to learn more about the influence of different types of diets on a molecular level. This will most likely be the case for a few more years until the technology for metabolomics is mature, user-friendly and powerful enough.
Nutrition places unique demands on scientists developing metabolic fingerprinting techniques for metabolomes, which currently makes nutritional metabolomics one of the most exciting fields of bioanalytical research. Firstly, nutritional metabololomic profiles are time-dependent as metabolism is a dynamic process, thus requiring multiple time-points in dietary interventions to measure the metabolic flux. In addition, the human metabolomes cannot be isolated from ‘interfering’ foreign metabolomes such as those from our gut microflora and those from all the organisms we consume with our diet. These complicating factors necessitate the development of state-of-the-art high-throughput and high-resolution analytical methodologies and equally sophisticated bioinformatics tools to interrogate the massive amount of resulting raw analytical data.
The first promising field of application in nutritional metabolomics is the discovery and application of biomarkers, as fingerprints of metabolites in the metabolome can be correlated with changes in physiology or with the early detection of a pathological state. Importantly, future metabolomics studies will undoubtedly reveal pre-disease biomarkers based on the detection of very subtle changes in the metabolism of healthy individuals, which are early signs of disease. Moreover, metabolomics may eventually reveal a true biomarker for a healthy organism rather than the current biomarkers for diseases. Nutritional treatment can then be used to maintain an optimal metabolism specific for every individual (“personalised nutrition”). Systems biology, nutrigenomics and metabolomics are expected to open the door for personalised nutrition, where the genetic variations of individuals and its influence on metabolism will allow us to use person-specific diets to maintain health and prevent disease.
References
- Assfalg M, Bertini I, Colangiuli D, et al. Evidence of different metabolic phenotypes in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1420–24. doi: 10.1073/pnas.0705685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nature Reviews Genetics. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Coen M, Holmes E, Lindon JC, Nicholson JK. NMR-based metabolic profiling and metabonomic approaches to problems in molecular toxicology. Chemical Research in Toxicology. 2008;21:9–27. doi: 10.1021/tx700335d. [DOI] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Stamler J, Bictash M, Yap IKS, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvelle AF, Lange LA, Qin L, et al. Comparison of ENCODE region SNPs between Cebu Filipino and Asian HapMap samples. Journal of Human Genetics. 2007;52:729–37. doi: 10.1007/s10038-007-0175-9. [DOI] [PubMed] [Google Scholar]
- Méplan C, Crosley LK, Nicol F, et al. Functional effects of a common single-nucleotide polymorphism (GPX4c718t) in the glutathione peroxidase 4 gene: Interaction with sex. American Journal of Clinical Nutrition. 2008;87:1019–27. doi: 10.1093/ajcn/87.4.1019. [DOI] [PubMed] [Google Scholar]
- Ruxton CHS, Reed SC, Simpson MJA, et al. The health benefits of omega-3 polyunsaturated fatty acids: A review of the evidence. Journal of Human Nutrition and Dietetics. 2004;17:449–59. doi: 10.1111/j.1365-277X.2004.00552.x. [DOI] [PubMed] [Google Scholar]
- Schlotterbeck G, Ross A, Dieterle F, et al. Metabolic profiling technologies for biomarker discovery in biomedicine and drug development. Pharmacogenomics. 2006;7:1055–75. doi: 10.2217/14622416.7.7.1055. [DOI] [PubMed] [Google Scholar]
- Shaik JS, Yeasin M. A unified framework for finding differentially expressed genes from microarray experiments. BMC Bioinformatics. 2007;8:347. doi: 10.1186/1471-2105-8-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usaite R, Wohlschlegel J, Venable JD, et al. Characterization of global yeast quantitative proteome data generated from the wild-type and glucose repression Saccharomyces cerevisiae strains: The comparison of two quantitative methods. Journal of Proteome Research. 2008;7:266–75. doi: 10.1021/pr700580m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villas-Boas SG, Koulman A, Lane GA. Analytical methods from the perspective of method standardization. Topics in Current Genetics. 2007;18:11–52. [Google Scholar]
- Wishart DS, Tzur D, Knox C, et al. HMDB: The human metabolome database. Nucleic Acids Research. 2007;35:D521–26. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-S, Yang Y-C, Chen C-L, et al. Adiponectin SNP276 is associated with obesity, the metabolic syndrome, and diabetes in the elderly. American Journal of Clinical Nutrition. 2007;86:509–13. doi: 10.1093/ajcn/86.2.509. [DOI] [PubMed] [Google Scholar]