Abstract
We revisit the classical problem of nucleated polymerisation and derive a range of exact results describing polymerisation in systems intermediate between the well-known limiting cases of a reaction starting from purely soluble material and for a reaction where no new growth nuclei are formed.
Keywords: protein, aggregation, amyloid
1. Introduction
The classical theory of nucleated polymerisation [1] describes the growth of filamentous structures formed through homogeneous (primary) nucleation [2–7]. This framework was initially developed by Oosawa and coworkers in the 1960s [1,8] to describe the formation of biofilaments, including actin and tubulin. This theory has been generalised to include heterogeneous (secondary) nucleation processes, such as surface-catalysed nucleation, by Eaton and Ferrone [9–11] in the context of their pioneering work elucidating the polymerisation of sickle haemoglobin, and by Wegner [12,13] in order to include fragmentation processes into the growth model for actin filaments; these processes differ from primary nucleation in that they produce new aggregates at a rate that is dependent on the aggregate concentration, whereas the rate of primary nucleation is dependent on only the concentration of monomeric peptide.
For irreversible growth in the absence of pre-formed seed material and secondary nucleation pathways, in 1962 Oosawa presented solutions to the kinetic equations, which were very successful in describing a variety of characteristics of the polymerisation of actin and tubulin [8]. The other limiting case, namely where seed material is added at the beginning of the reaction and where no new growth nuclei are formed during the reaction, is also well-known and results in a rate law given by a single exponential that is characteristic of first order kinetics. In this paper, we present exact results for filament systems proliferating through primary nucleation which encompass all cases between these limiting scenarios, extending the results of Oosawa for a system dominated by primary nucleation to the case where an arbitrary concentration of pre-formed seed material is present. We also discuss a range of general closed form results from the Oosawa theory for the behaviour of a system of biofilaments growing through primary nucleation and elongation. We then compare the behaviour of systems dominated by primary nucleation to results derived recently for systems dominated by secondary nucleation.
2. Results and Discussion
2.1. Derivation of the Rate Laws for the Polymer Number and Mass Concentrations
The theoretical description of the polymerisation of proteins such as actin and tubulin to yield functional biostructures was considered in the 1960s [8]. For a system that evolves through primary nucleation of new filaments, elongation of existing filaments, and depolymerisation from the filament ends, the change in concentration of filaments of size j, denoted f(j; t), is given by the master equation [1,8]:
| (1) |
where k+, koff, kn are rate constants describing the elongation, depolymerisation and nucleation steps and m(t) is the concentration of free monomeric protein in solution. The factor of 2 in Equation (1) originates from the assumption of growth from both ends. The first two terms in Equation (1) describe the change in the population of filaments of size j due to monomer addition, whilst the third and fourth terms relate to monomer dissociation. The final term in Equation (1) describes the creation of a growth nucleus consisting of nc monomers at a rate proportional to the monomer concentration raised to this power. In addition to the direct coalescence of nc monomers, pre-equilibrium and other schemes also result in the homogeneous nucleation term taking this form [14–25].
For the case of irreversible biofilament growth, the polymerisation rate dominates over the depolymerisation rate; from Equation (1), the rate of change of the number of filaments, P(t), and the free monomer concentration, m(t), were shown [1,8] under these conditions to obey:
| (2) |
| (3) |
Combining Equations (2) and (3) yields a differential equation for the free monomer concentration [1]:
| (4) |
Here, we integrate these equations in the general case where the initial state of the system can consist of any proportion of monomeric and fibrillar material; this calculation generalises the results presented by Oosawa to include a finite concentration of seed material present at the start of the reaction. Beginning with Equations (2) and (3), the substitution z(t) := log(m(t)) followed by multiplication through by dz/dt yields:
| (5) |
Integrating both sides results in:
| (6) |
This is a separable equation for dz/dt, for which the solution is given by:
| (7) |
Integration and exponentiation yields the expression for m(t):
| (8) |
The values of the constants A and B are fixed by inserting the appropriate boundary conditions in terms of m(0) and P(0). The mass concentration of polymers, M(t), is then provided through conservation of mass, M(t) = mtot − m(t) with mtot being the total monomer concentration. This results in the exact integrated rate law:
| (9) |
where the effective rate constant λ is given by and β = 2/nc, , ν = arsinh (γ) for .
We note that this expression only depends on two combinations of the microscopic rate constants, k0 = 2k+P(0) and λ. The result reveals that λ controls the aggregation resulting from the newly formed aggregates, whereas k0 defines growth from the pre-formed seed structures initially present in solution. In the special case of the aggregation reaction starting with purely soluble proteins, P(0) = 0, m(0) = mtot, these expressions reduce to μ → 1 and ν → 0, and Equation (9) yields the result presented by Oosawa [1] and the single relevant parameter in the rate equations is λ. Interestingly, generalisations of Equation (9) which include secondary pathways, maintain the dependence on λ and k0 but introduce an additional parameter analogous to λ for each active secondary pathway [26–29].
An expression for the evolution of the polymer number concentration, P(t) may be derived using Equation (9). Direct integration of Equation (2) gives the result for P(t):
| (10) |
Equations (9) and (10) give in closed form the time evolution of the mass and number concentrations of biofilaments growing through primary nucleation and filament elongation.
2.2. Characteristic Features of Growth Involving Pre-Formed Seed Material
Insight into the early time behaviour of the polymer mass concentration can be obtained by expanding Equation (9) for early times to yield:
| (11) |
This expression recovers the characteristic ~t2 dependence of the Oosawa theory and has an additional term linear in time relating to the growth of pre-formed aggregates.
In many cases, Equation (9) describes a sigmoidal function with a lag phase. The time of maximal growth rate, tmax, can be found from the inflection point of the sigmoid from the condition d2M/dt2 = 0:
| (12) |
such that a lag phase exists only for:
| (13) |
Using the composition reduces this to the simple condition:
| (14) |
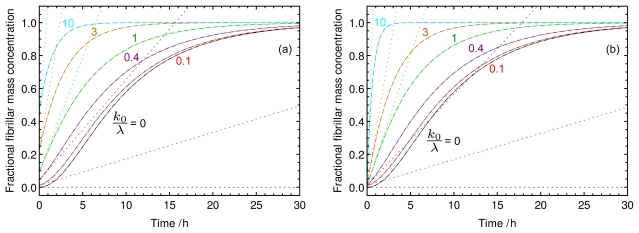
In other words, a point of inflection exists if the growth through elongation from the ends of pre-existing seeds, k0, is less effective that the proliferation through nucleation and elongation of new material λ. This result implies that an increased nucleation rate promotes the existence of an inflection point, whereas an increased elongation rate or an increased level of seeding tends to disfavour its existence. In particular, we also note that in the absence of nucleation, an inflection point cannot exist in the polymer mass concentration as a function of time. This result is illustrated in Figure 1 where kinetic profiles are shown for increasing levels of pre-formed seed material added at the beginning of the reaction; a lag-phase is only observed when k0/λ < 1, and the rate profile transforms from a sigmoidal shape at low pre-seeding levels to a concave form at high levels of seeding. Interestingly, the result Equation (14) is analogous to the criterion applicable for fragmentation dominated growth [26] where a lag phase exists only when the parameter controlling fragmentation-related secondary nucleation is larger than ek0.
Figure 1.
Nucleated polymerisation in the presence of seed material. The thick dashed lines are the exact solution to the rate equations Equation (9); the thin solid lines are calculated from numerical simulations of the master equation Equation (1). The dotted lines are the initial gradients dM/dt|t=0 = M(0) + 2k+m(0)P(0)t; a lag-phase exists when the initial gradient is not the maximal gradient. The numbers accompanying each curve are k0/λ; Equation (14) predicts that a lag-phase only exists when this ratio is less than unity. (a): Nucleated polymerisation in the presence of an increasing quantity of seed material of a fixed average length (5000 monomers per seed) added at the beginning of the reaction. The seed concentrations given as a fraction of the total concentration of monomer present are right to left): 0, 0.01, 0.04, 0.1, 0.2, 0.5; (b): Nucleated polymerisation in the presence of a fixed quantity (1% of total monomer in the system) of seed material of varying average length. The average number of monomer per seed are (right to left): N/A (unseeded), 5000, 1000, 500, 200, 50. The other parameters for both panels are: mtot = 10 μM, nc = 3, knm totnc−1 = 1 · 10−9 s−1, k+ = 1 · 105 M−1 s−1.
The maximal growth rate, rmax, is given by:
| (15) |
which occurs at a polymer mass concentration Mmax given from Equation:
| (16) |
The lag time, τlag := tmax − M(tmax)/rmax, is then given by:
| (17) |
Interestingly, from Equation (17), we note that a point of inflection can never exist for P(t) for simple nucleated polymerisation. By contrast, when secondary pathways are active, an inflection point can frequently be present [27].
2.3. Comparison between Nucleated Polymerisation in the Presence and Absence of Secondary Pathways
Many systems that evolve through nucleated polymerisation display characteristic scaling behaviour, including power-law relationships between phenomenological parameters, such as the lag-time and maximal growth rate, and the initial concentration of monomeric peptide [26–31]. This behaviour can be seen to be a consequence of the fact that under many conditions, the rate equations are dominated by a single parameter that corresponds to the dominant form of nucleation: λ for classical nucleated polymerisation through primary nucleation, Equation (9), or an analogous parameter [21,26,29] κ− or κ2, respectively, for polymerisation in the presence of filament fragmentation of monomer-dependent secondary pathways [11,21,26,29,32–34], Table 1. These parameters have the general form where kN = kn; k−; k2 corresponds to the dominant nucleation process, kn for primary nucleation, k− for filament fragmentation, k2 for monomer-dependent secondary nucleation, and n is related to the monomer dependence of this process: n = nc − 1, where nc is the critical nucleus size for primary nucleation, n = 0 for fragmentation driven growth and n = n2, the secondary nucleus size in cases where monomer-dependent secondary nucleation is dominant. The dominance of a single combination of the rate constants implies that many of the macroscopic system observables will be correlated since they are dependent on the same parameter. A striking examples of this behaviour is provided by the very general correlation between the lag-time and the maximal growth rate [29,30,35,36], which is manifested in the present case in Equations (15) and (17) as rmax ~ λ and τlag ~ λ−1.
Table 1.
Comparison of biofilament growth dominated by primary and secondary nucleation pathways. Primary nucleation processes create new aggregates at a rate that depends only on the concentration of monomeric peptide, whereas fragmentation creates new aggregates at a rate that depends only on the concentration of existing aggregates; monomer-dependent secondary nucleation creates new aggregates at a rate that depends on both the concentration of monomeric peptide and the concentration of existing aggregates. The dependencies of the latter two (secondary) nucleation processes on the existing aggregate concentration results in positive feedback: as the reaction proceeds, and proliferation through these mechanisms increases the concentration of aggregates, the rate at which these processes occur further is increased.
| Primary nucleation | Fragmentation | Monomer-dependent secondary nucleation | |
|---|---|---|---|
| Kinetic parameters | λ, k+ | λ, κ−, k+ | λ, κ2, k+ |
| Early time growth | Polynomial | Exponential | Exponential |
| Scaling behaviour (lag time, max growth rate) | Yes with λ | Yes with κ− | Yes with κ2 |
| Positive feedback | No | Yes | Yes |
Interestingly the rate equations describe sigmoidal curves both in the presence and in the absence of secondary nucleation processes. For more complex primary nucleation pathways [19,21] the polynomial form for the early time solution is maintained, but higher-order terms are obtained. This observation implies that in the absence of secondary processes the lag-phase is, in general, less marked since the early time rise is a slower polynomial relationship rather than the exponential onset characteristic of secondary pathways [21]. However, the difference between a high-order polynomial and an exponential may not be apparent in experimental data in the presence of noise, and therefore a global analysis of the system under different conditions is required in order to obtain robust mechanistic information. This might involve, for example [29], experimental measurements at varying initial concentrations of the monomeric peptide; a global analysis of the measured reaction profiles in terms of integrated rate laws and the related scaling behaviours can then be used to establish information about the microscopic mechanisms of proliferation [29].
3. Conclusions
In this paper, we have provided results for the time course of nucleated polymerisation for systems that are initially in a mixed state and contain both monomeric and fibrillar material. These results generalise the classical Oosawa theory that describes the formation of biofilaments to cases where an arbitrary amount of pre-formed seed material is present in the system. Furthermore, these results represent a reference to which polymerisation driven by secondary pathways can be compared.
Acknowledgements
We are grateful to the Schiff Foundation (SIAC), and to the Wellcome (MV, CMD, TPJK) and Leverhulme Trusts (CMD) for financial support.
References
- 1.Oosawa F, Asakura S. Thermodynamics of the Polymerization of Protein. Academic Press; Waltham, MA, USA: 1975. [Google Scholar]
- 2.Gibbs JW. On the equilibrium of heterogeneous substances. Trans Conn Acad Arts Sci. 1878;3:108–343. [Google Scholar]
- 3.Volmer M, Weber A. Keimbildung in übersättigten Gebilden. Z Phys Chem. 1926;119:277. [Google Scholar]
- 4.Kaischew R, Stranski IN. The theory of the linear rate of crystallisation. Z Phys Chem. 1934;A170:295. [Google Scholar]
- 5.Stranski IN, Kaischew R. Crystal growth and crystal nucleation. Z Phys. 1935;36:393. [Google Scholar]
- 6.Becker R, Döring W. Kinetische Behandlung der Keimbildung in übersättigten Dämpfen. Ann Phys. 1935;24:719–752. [Google Scholar]
- 7.Avrami M. Kinetics of phase change. I General theory. J Chem Phys. 1939;7:1103–1112. [Google Scholar]
- 8.Oosawa F, Kasai M. A theory of linear and helical aggregations of macromolecules. J Mol Biol. 1962;4:10–21. doi: 10.1016/s0022-2836(62)80112-0. [DOI] [PubMed] [Google Scholar]
- 9.Ferrone FA, Hofrichter J, Sunshine HR, Eaton WA. Kinetic studies on photolysis-induced gelation of sickle cell hemoglobin suggest a new mechanism. Biophys J. 1980;32:361–380. doi: 10.1016/S0006-3495(80)84962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop MF, Ferrone FA. Kinetics of nucleation-controlled polymerization. A perturbation treatment for use with a secondary pathway. Biophys J. 1984;46:631–644. doi: 10.1016/S0006-3495(84)84062-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrone FA, Hofrichter J, Eaton WA. Kinetics of sickle hemoglobin polymerization. II. A double nucleation mechanism. J Mol Biol. 1985;183:611–631. doi: 10.1016/0022-2836(85)90175-5. [DOI] [PubMed] [Google Scholar]
- 12.Wegner A, Savko P. Fragmentation of actin filaments. Biochemistry. 1982;21:1909–1913. doi: 10.1021/bi00537a032. [DOI] [PubMed] [Google Scholar]
- 13.Wegner A. Spontaneous fragmentation of actin filaments in physiological conditions. Nature. 1982;296:266–267. doi: 10.1038/296266a0. [DOI] [PubMed] [Google Scholar]
- 14.Hofrichter J, Ross PD, Eaton WA. Kinetics and mechanism of deoxyhemoglobin S gelation: A new approach to understanding sickle cell disease. Proc Natl Acad Sci USA. 1974;71:4864–4868. doi: 10.1073/pnas.71.12.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firestone MP, De Levie R, Rangarajan SK. On one-dimensional nucleation and growth of ”living” polymers. I. Homogeneous nucleation. J Theor Biol. 1983;104:535–552. doi: 10.1016/0022-5193(83)90244-8. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein RF, Stryer L. Cooperative polymerization reactions. Analytical approximations, numerical examples, and experimental strategy. Biophys J. 1986;50:583–599. doi: 10.1016/S0006-3495(86)83498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ataka M, Ogawa T. Nucleation and growth of oxide precipitates in CZ-Si wafers. J Mater Res. 1993;11:2889–2992. [Google Scholar]
- 18.Bessho Y, Ataka M, Asai M, Katsura T. Analysis of the crystallization kintics of lysozyme using a model with polynuclear growth mechanism. Biophys J. 1994;66:310–313. doi: 10.1016/s0006-3495(94)80779-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flyvbjerg H, Jobs E, Leibler S. Kinetics of self-assembling microtubules: An “inverse problem” in biochemistry. Proc Natl Acad Sci USA. 1996;93:5975–5979. doi: 10.1073/pnas.93.12.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomakin A, Teplow DB, Kirschner DA, Benedek GB. Kinetic theory of fibrillogenesis of amyloid beta-protein. Proc Natl Acad Sci USA. 1997;94:7942–7947. doi: 10.1073/pnas.94.15.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrone F. Analysis of protein aggregation kinetics. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- 22.Murphy RM, Pallitto MM. Probing the kinetics of beta-amyloid self-association. J Struct Biol. 2000;130:109–122. doi: 10.1006/jsbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- 23.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Ferrone FA, Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci USA. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews JM, Roberts CJ. A Lumry-Eyring nucleated polymerization model of protein aggregation kinetics: 1. Aggregation with pre-equilibrated unfolding. J Phys Chem B. 2007;111:7897–7913. doi: 10.1021/jp070212j. [DOI] [PubMed] [Google Scholar]
- 26.Cohen SIA, Vendruscolo M, Welland ME, Dobson CM, Terentjev EM, Knowles TPJ. Nucleated polymerization with secondary pathways. I. Time evolution of the principal moments. J Chem Phys. 2011;135:065105:1–065105:16. doi: 10.1063/1.3608916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. Nucleated polymerization with secondary pathways. II. Determination of self-consistent solutions to growth processes described by non-linear master equations. J Chem Phys. 2011;135:065106:1–065106:18. doi: 10.1063/1.3608917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen SIA, Vendruscolo M, Dobson CM, Knowles TPJ. Nucleated polymerization with secondary pathways. III. Equilibrium behavior and oligomer populations. J Chem Phys. 2011;135:065107:1–065107:10. doi: 10.1063/1.3608918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knowles TPJ, Waudby CA, Devlin GL, Cohen SIA, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 30.Fändrich M. Absolute correlation between lag time and growth rate in the spontaneous formation of several amyloid-like aggregates and fibrils. J Mol Biol. 2007;365:1266–1270. doi: 10.1016/j.jmb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Hellstrand E, Boland B, Walsh DM, Linse S. Amyloid β-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chem Neurosci. 2010;1:13–18. doi: 10.1021/cn900015v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wegner A. Kinetic analysis of actin assembly suggests that tropomyosin inhibits spontaneous fragmentation of actin filaments. J Mol Biol. 1982;161:217–227. doi: 10.1016/0022-2836(82)90149-8. [DOI] [PubMed] [Google Scholar]
- 33.Andersen CB, Yagi H, Manno M, Martorana V, Ban T, Christiansen G, Otzen DE, Goto Y, Rischel C. Branching in amyloid fibril growth. Biophys J. 2009;96:1529–1536. doi: 10.1016/j.bpj.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meinhardt J, Tartaglia GG, Pawar A, Christopeit T, Hortschansky P, Schroeckh V, Dobson CM, Vendruscolo M, Fändrich M. Similarities in the thermodynamics and kinetics of aggregation of disease-related Abeta (1–40) peptides. Protein Sci. 2007;16:1214–1222. doi: 10.1110/ps.062734207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auer S, Kashchiev D. Insight into the correlation between lag time and aggregation rate in the kinetics of protein aggregation. Proteins. 2010;78:2412–2416. doi: 10.1002/prot.22762. [DOI] [PubMed] [Google Scholar]