Abstract
Hydrogen sulfide (H2S), an endogenously produced small molecule, protects animals from various stresses. Recent studies demonstrate that animals exposed to H2S are long lived, resistant to hypoxia, and resistant to ischemia–reperfusion injury. We performed a forward genetic screen to gain insights into the molecular mechanisms Caenorhabditis elegans uses to appropriately respond to H2S. At least two distinct pathways appear to be important for this response, including the H2S-oxidation pathway and the hydrogen cyanide (HCN)-assimilation pathway. The H2S-oxidation pathway requires two distinct enzymes important for the oxidation of H2S: the sulfide:quinone reductase sqrd-1 and the dioxygenase ethe-1. The HCN-assimilation pathway requires the cysteine synthase homologs cysl-1 and cysl-2. A low dose of either H2S or HCN can activate hypoxia-inducible factor 1 (HIF-1), which is required for C. elegans to respond to either gas. sqrd-1 and cysl-2 represent the entry points in the H2S-oxidation and HCN-assimilation pathways, respectively, and expression of both of these enzymes is highly induced by HIF-1 in response to both H2S and HCN. In addition to their role in appropriately responding to H2S and HCN, we found that cysl-1 and cysl-2 are both essential mediators of innate immunity against fast paralytic killing by Pseudomonas. Furthermore, in agreement with these data, we showed that growing worms in the presence of H2S is sufficient to confer resistance to Pseudomonas fast paralytic killing. Our results suggest the hypoxia-independent hif-1 response in C. elegans evolved to respond to the naturally occurring small molecules H2S and HCN.
HYDROGEN sulfide (H2S) is an endogenously produced molecule with profound physiological effects (Kabil and Banerjee 2010). While first noted for its potent toxicity, it is now clear that organisms and H2S have long had an intimate relationship (Philippot et al. 2007). Prior to the appearance of oxygen as a component of the earth’s atmosphere, H2S was abundant at the earth’s surface (Reinhard et al. 2009). Sulfur was likely important for redox biochemistry in early life, similar to the role of oxygen today (Philippot et al. 2007). Sulfur-containing redox reactions are still essential in modern organisms in the form of glutathione and thioredoxin (Holmgren et al. 2005) and in iron–sulfur clusters in the electron transport chain and aconitase in the tricarboxylic acid (TCA) cycle (Rouault and Tong 2008). Recent molecular phylogenetic data confirm that H2S-interacting enzymes predated oxygen-binding enzymes (David and Alm 2011). H2S is also essential for the existence of sulfur-containing amino acids because cysteine synthase uses H2S as the sulfur source during de novo cysteine biosynthesis (Feldman-Salit et al. 2009).
In animals, H2S is generated by dissimilating H2S from cysteine (Kabil and Banerjee 2010). The numerous activities ascribed to H2S in animals include its function as a neuromodulator in the brain and as a mediator of vascular tone and smooth muscle of the heart (Li et al. 2011). The ability of H2S to affect mammalian tissue has recently been exploited for therapeutic uses (Mustafa et al. 2009). When rodents are exposed to H2S, they enter a reversible state of decreased metabolic rate and core body temperature (Blackstone et al. 2005). Exogenous H2S has shown benefit in a number of animal-disease models. For example, H2S protects mice from hypoxic death (Blackstone and Roth 2007), helps rats survive severe hemorrhage (Morrison et al. 2008), and diminishes ischemia–reperfusion injury (Elrod et al. 2007).
Our understanding of the mechanism by which H2S affects biology is incomplete. To fill these gaps, we have developed C. elegans as a genetically amenable model system for studying the response of animals to H2S. Previous work has shown that C. elegans becomes longer lived and thermotolerant when grown in H2S (Miller and Roth 2007). We showed that H2S induces the accumulation of the hypoxia-inducible factor 1 (HIF-1) (Budde and Roth 2010). In turn, hif-1 is required for survival in the presence of H2S, and keeping HIF-1 levels constitutively high confers resistance to otherwise lethal H2S concentrations. HIF-1 is a transcription factor and thus the observed protective effects are likely to be mediated by increased expression of genes regulated by HIF-1. To determine the molecular mechanism by which tolerance to H2S is conferred, we performed a forward genetic screen for C. elegans strains sensitive to H2S and report on two hif-1 target genes that function in the first steps of two distinct pathways important for survival in H2S.
Materials and Methods
Strains
C. elegans were grown at room temperature on nematode growth medium plates seeded with live Escherichia coli OP50 food (Brenner 1974). The following mutant strains were obtained from the Caenorhabditis Genetics Center: cysl-1(ok762), cysl-2(ok3516), cysl-4(ok3359), cbs-2(ok666), hif-1 (ia04), sqrd-2 (ok3440), and CB4856. The following strains were provided by Shohei Mitani of the National BioResource Project: sqrd-1(tm3378) and ethe-1(tm4104). Pseudomonas aeruginosa strain PA01 was obtained from Colin Manoil (University of Washington, Seattle, WA).
Atmospheric chambers and viability tests
H2S-containing atmospheric chambers were generated by mixing air with small amounts of defined compressed H2S balanced with N2 (Budde and Roth 2010). The resulting gas mixture is essentially room air containing hydrogen sulfide (RA/H2S). Unless otherwise stated, RA/H2S is room air containing 50 ppm of H2S. All survival experiments were performed in 1000-ml chambers, with a gas flow of 1000 ml/min. HCN-containing atmospheres were created in the same way as the H2S atmospheres except HCN balanced with N2 was used, and they are referred to as RA/HCN (essentially room air containing hydrogen cyanide). Unless otherwise stated, RA/HCN is room air containing 5 ppm of HCN.
Viability tests were performed by picking L4 larval stage worms onto OP50-seeded NGM plates and placing them into the gaseous atmosphere for 24 hr (Budde and Roth 2010). For RA/H2S exposures, all animals observed after 24 hr were either healthy adults or appeared to have perished shortly after initiation of the experiment. For RA/HCN exposures, developmentally arrested L4 animals with very slow movement were occasionally observed. If maintained in RA/HCN, these animals invariably perished after several days as L4. If removed from RA/HCN after 24 hr, some of these animals eventually recovered to become fertile adults. For the data in this article, animals with this developmental arrest were scored as not surviving the RA/HCN exposure. All error values presented are standard deviation (SD).
Video acquisition and automated worm speed analysis
A Quickcam Communicate STX (Logitech) webcam was affixed to the eyepiece of a stereomicroscope. Images were acquired every 12 sec and analyzed with the Multi-Worm Tracker (MWT) (Swierczek et al. 2011). The video was analyzed three times on the MWT and the speed was averaged every 2.5 min.
Genetic screen for H2S-sensitive mutants
Ethyl methanesulfonate (EMS) mutagenesis was performed as previously described (Brenner 1974) and was optimized to maximize the number of unique mutations observed (Shaham 2007). N2 animals at the L4 or young-adult stage were incubated with EMS for 4 hr. The worms were then washed once, aliquoted onto four OP50-seeded NGM plates and allowed to lay eggs overnight. The next day, the adults were washed off the plates and the F1 animals were grown to adults. Each pool of 25,000 F1 adults was bleached to generate synchronized F2 animals. A total of 100,000 F2 L4 animals were screened (25,000 from each pool).
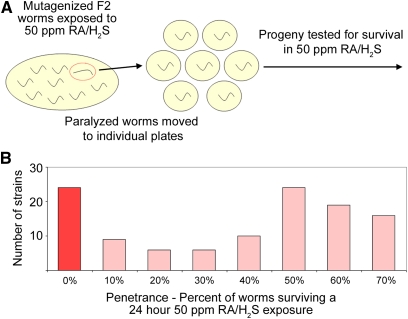
The screen for nematodes that are sensitive to H2S was performed as follows. When the F2 worms entered the L4 larval stage, the plate of worms was exposed to RA/H2S for 4 hr. The worms were moved to room air for 5 min and then all paralyzed worms were picked from the plate within 15 min of removal from RA/H2S. From this screen, 791 F2 animals were singled out to generate 544 viable strains. Of these viable strains, 124 strains exhibited a lethality phenotype during a 24-hr exposure to RA/H2S. Of the 124 sensitive strains, 23 were 100% lethal when exposed to RA/H2S for 24 hr as L4 and were annotated as strongly penetrant (Figure 2).
Figure 2 .
H2S sensitivity screen method and results. (A) Schematic of initial paralysis screen and lethality rescreen. (B) Histogram of the number of strains isolated for each penetrance bin. The strains with zero survivors after sulfide exposure are highlighted in red. These strains were subsequently characterized.
Mapping mutations to chromosomes and complementation tests
Prior to complementation testing, strongly penetrant strains were crossed to the Hawaiian strain CB4856 and SNP enrichment analysis was performed (Davis et al. 2005). Male CB4856 worms were mated into the 23 mutant strains. F1 animals were isolated and synchronous F2 populations were created by egg lays. When F2 animals reached L4 larval stage, the plates were exposed to RA/H2S for 4 hr. Thirty paralyzed animals were picked as phenotypic, and 30 motile animals were picked as aphenotypic. In most cases, ∼25% of F2 worms became paralyzed when exposed to RA/H2S and were annotated as recessive. In four cases, ∼75% of worms became paralyzed and these mutants were annotated as dominant.
After the restriction digests were separated by gel electrophoreses, the gel was visually examined for genomic regions where the DNA from phenotypic and aphenotypic animals did not cut equally well with DraI. Ethidium bromide fluorescence was quantified with Quantity One 1-D analysis software (Bio-Rad) and a fluorescence ratio of cut/uncut DNA was generated for each SNP on the chromosome of interest. The SNPs on two neighboring chromosomes are presented as a background control. Strains that appeared to show similar enrichment patterns were grouped together as suspected complementation groups (Figure 3).
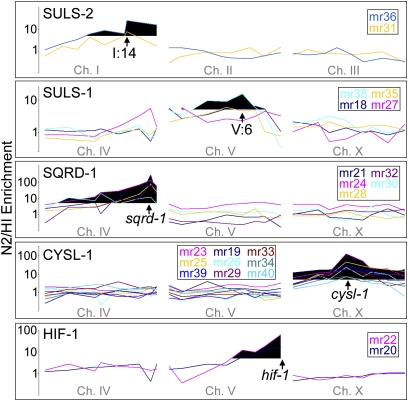
Figure 3 .
Enrichment of H2S-sensitive phenotype after crossing mutant lines with the Hawaiian strain CB4856. SNP mapping data show five apparent complementation groups. For suls-1 and suls-2 the apparent location of mutation is indicated by an arrow. The known locations of sqrd-1, cysl-1, and hif-1 are also indicated by arrows. Black indicates an N2/HI ratio >5.
Complementation tests were carried out by crossing homozygous males of one strain into another strain and testing the F1 progeny for survival in RA/H2S. Sequencing of cysl-1 and sqrd-1 was accomplished by PCR amplification of genomic DNA and then using nested primers for the labeling reaction. After the point mutations were identified, SIFT-BLink (Ng and Henikoff 2001) (http://sift.bii.a-star.edu.sg/www/SIFT_BLink_submit.html) was used to determine whether the mutations are predicted to be tolerated or not tolerated. In parallel, all splice mutations were annotated as not tolerated.
Protein homology
Protein–protein Basic Local and Alignment and Search Tool (BLASTp) was used to locate homologous genes. Potential homologs were reciprocally BLAST searched to ensure specificity. Amino acid alignment was performed with ClustalX2 using the following National Center for Biotechnology Information reference sequence IDs: Arabidopsis thaliana CAS, NP_191703.1; A. thaliana CYS, NP_193224.1; Solanum tuberosum CAS, BAB18760.1; S. tuberosum CYS, AAC25635.1; Zea mays CAS, ADG60236.1; Z. mays CYS, NP_001105469.1; Mus musculus CBS, NP_659104.1; Homo sapiens CBS, NP_000062.1; Ciona intestinalis CBS, XP_002120247.1; and Drosophila melanogaster CBS, NP_608424.1.
Psuedomonas aeruginosa dependent paralysis
Pseudomonas experiments were performed with strain PA01 as previously described (Shao et al. 2010). Briefly, a full lawn of P. aeruginosa was grown on brain heart infusion agar (37 g Difco BHI, 20 g agar per liter) on 3.5-cm plates for 24 hr at 37°. The plates were cooled to room temperature and 10 L4 animals were placed onto the bacterial lawn and the plate was sealed with parafilm. The number of animals moving and not moving after 6 hr was recorded.
mRNA detection by QRT–PCR
Quantitative reverse transcriptase PCR was performed on a Bio-Rad C1000 Thermal Cycler with a CFX96 real-time system, with iQ SYBR green supermix (Bio-Rad) as previously described (Budde and Roth 2010). Experimental mRNA was compared to the average mRNA level of the housekeeping genes tba-1 and tbb-2. The primer sequences are listed in Supporting Information, Table S1.
Western blotting
Antibody was generated and affinity purified using the amino acids 419–431 of SQRD-1 (wormbase.org WP:CE17629), amino acids AMETTPFDQSKPTY, by ProSci Incorporated (San Diego, CA). Western blotting was performed as previously described (Budde and Roth 2010) with the following changes: primary antibody was diluted 1:500. Goat antirabbit IgG (H+L)-horseradish peroxidase (catalog no. 111-035-003; Jackson ImmunoReseach Laboratories, West Grove, PA) was used as the secondary antibody.
Fosmid recombineering
GFP was inserted into fosmid WRM066CH11 as previously described (Dolphin and Hope 2006). The oligonucleotides used to insert the RT cassette were:
RT cassette forward -gtttgattaaaggatactggaatggaccagctacactcagaaattgtacaTCGCTGTCGAGATATGACGGTG
RT cassette reverse -acaaataaaaacgacaaagtgggaaatatctatttagactttaccaatcgGATGATAAGCTGTCAAACATGAG
GFP forward -acaaataaaaacgacaaagtgggaaatatctatttagactttaccaatcgTTTGTATAGTTCATCCATGCCATGTG
GFP reverse -gtttgattaaaggatactggaatggaccagctacactcagaaattgtacaATGAGTAAAGGAGAAGAACTTTTC
Uppercase letters indicate primers for the PCR reaction.
Generation of transgenic animals
Wild-type C. elegans were injected with 50 ng/μl fosmid together with 100 ng/μl pRF4, which confers a dominant roller [rol-6(su1006)] phenotype (Mello et al. 1991). Rolling F1 animals were picked onto single plates to generate transgenic lines. Animals were exposed to 50 ppm RA/H2S prior to confocal fluorescent microscopy, as previously described (Budde and Roth 2010).
RNAi experiments
RNAi was performed as previously described (Frazier and Roth 2009) using WBRNAi00013759 to target egl-9. L4 animals were moved onto the RNAi food and the progeny were moved onto NGM-OP50 or BHI-PA01 plates and immediately assayed.
Results
We showed previously that hypoxia-inducible factor, hif-1, is required for growth and survival in H2S (Budde and Roth 2010). HIF-1 protein concentration and activity are increased when C. elegans are moved into RA/H2S, and egl-9 mutant animals, which have constitutively high HIF-1 expression and activity, are resistant to H2S.
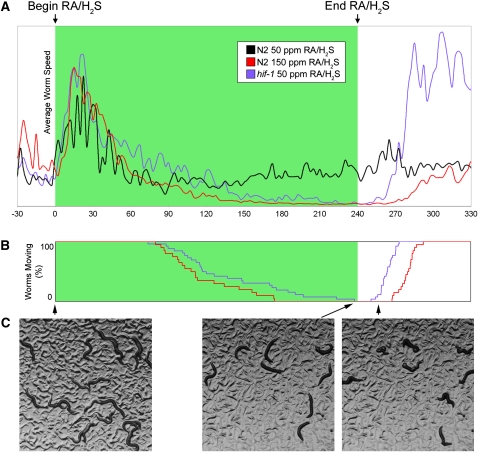
During our investigations with hif-1 mutants, we noticed that although hif-1 mutant animals are unable to survive prolonged exposure to RA/H2S (24 hr), a brief exposure (4 hr) causes a reversible paralysis (Figure 1A). Roughly 2 hr after entering an environment containing RA/H2S, hif-1 mutant L4 animals enter a state of suspended animation, characterized by a complete cessation of movement and feeding (Figure 1B), and the animals lose their characteristic sinusoidal shape (Figure 1C). If removed to fresh air during this suspended state, the animals resume moving after ∼20 min and proceed to become fertile adults. This phenotype is also present in wild-type animals exposed to 150 ppm RA/H2S (Figure 1, A and B; File S1). We used this phenotype as a basis for screening mutant C. elegans that are sensitive to H2S, in an attempt to find genes that confer resistance to H2S (Figure 2A).
Figure 1 .
Wild-type nematodes exposed to 50 ppm RA/H2S exhibit a brief increase in movement initially and then continue to move (A, black line). When either wild-type or hif-1 mutant animals are exposed to 150 ppm RA/H2S or 50 ppm RA/H2S, respectively, they stop moving and when the RA/H2S is removed, they resume movement (blue and red lines, A and B). Examples of Hif-1 moving, stopped, and reanimated worms are shown in C.
About 200,000 mutant haploid genomes were screened for H2S sensitivity. Through screening, we obtained 124 lines that were sensitive to H2S (Figure 2B). Of these lines, 23 were completely penetrant, where penetrance is defined as the percentage of worms unable to survive a 24-hr RA/H2S exposure. These mutant strains were chosen for further characterization.
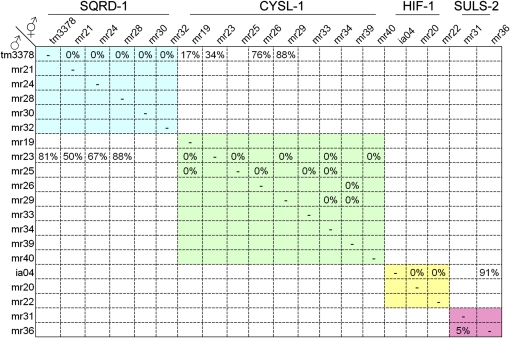
The 23 strongly penetrant mutant lines were crossed with the Hawaiian CB4856 strain for mapping the sensitivity trait to chromosomal regions using enrichment of SNPs. The SNP analysis revealed that 22 mutants map to only five chromosomal regions (Figure 3, Table 1); only mr37 showed no Mendelian inheritance of the trait. These mutants were then crossed to confirm that each chromosomal region contained a single complementation group (Figure 4). One group on chromosome V (mr18, mr27, mr35, and mr38), exhibits dominant genetics and thus could not be further tested for complementation. We named this gene suls-1 (sulfide sensitive) and suls-1 (mr35) was mapped to the 4-cM region between SNP pkP5097 and pkP5068. The two alleles on chromosome I were named suls-2. The genes mutated in suls-1 and suls-2 strains remain to be identified. Two alleles on chromosome V (mr20 and mr22), mapped near the hif-1 locus and failed to complement the hif-1(ia04) mutation, which confirms that we successfully screened for our phenotype of interest.
Table 1 . Alleles unable to survive in RA/H2S.
| Alive/total (% surviving) | ||||||
|---|---|---|---|---|---|---|
| Gene name | Chromosome | Allele | 32 ppm RA/H2S | 15 ppm RA/H2S | 7.5 ppm RA/H2S | Dominance |
| N2 | — | — | 12/12 (100) | 12/12 (100) | 13/13 (100) | — |
| cysl-1 | X | ok762 | 0/20 (0) | 0/18 (0) | 0/11 (0) | — |
| cysl-1 | X | mr19 | 0/13 (0) | 0/12 (0) | 0/10 (0) | Recessive |
| cysl-1 | X | mr23 | 0/16 (0) | 0/10 (0) | 0/10 (0) | Recessive |
| cysl-1 | X | mr25 | 0/13 (0) | 0/10 (0) | 0/6 (0) | Recessive |
| cysl-1 | X | mr26 | 0/12 (0) | 0/10 (0) | 0/11 (0) | Recessive |
| cysl-1 | X | mr33 | 0/15 (0) | 5/9 (56) | 10/10 (100) | Recessive |
| cysl-1 | X | mr34 | 0/10 (0) | 0/11 (0) | 1/10 (10) | Recessive |
| cysl-1 | X | mr29 | 0/11 (0) | 0/10 (0) | 1/13 (8) | Recessive |
| cysl-1 | X | mr39 | 0/10 (0) | 0/10 (0) | 13/13 (100) | Recessive |
| cysl-1 | X | mr40 | 0/10 (0) | 0/10 (0) | 1/11 (9) | Recessive |
| sqrd-1 | IV | tm3378 | 5/10 (50) | 10/10 (100) | 10/10 (100) | — |
| sqrd-1 | IV | mr24 | 10/14 (71) | 11/11 (100) | — | Recessive |
| sqrd-1 | IV | mr30 | 2/13 (15) | 9/9 (100) | — | Recessive |
| sqrd-1 | IV | mr32 | 12/13 (92) | 10/10 (100) | — | Recessive |
| sqrd-1 | IV | mr21 | 6/15 (40) | 10/10 (100) | — | Recessive |
| sqrd-1 | IV | mr28 | 5/9 (56) | 10/10 (100) | 11/11 (100) | Recessive |
| hif-1 | V | ia04 | 0/15 (0) | 0/11 (0) | 10/10 (100) | — |
| hif-1 | V | mr20 | 0/14 (0) | 0/10 (0) | 10/13 (77) | Recessive |
| hif-1 | V | mr22 | 0/15 (0) | 1/11 (9) | 8/10 (80) | Recessive |
| suls-1 | V | mr18 | 6/12 (50) | 11/12 (92) | 10/10 (100) | Dominant |
| suls-1 | V | mr27 | 2/9 (22) | 10/10 (100) | — | Dominant |
| suls-1 | V | mr35 | 0/5 (0) | 0/10 (0) | 0/10 (0) | Dominant |
| suls-1 | V | mr38 | 0/9 (0) | 8/8 (100) | — | Dominant |
| suls-2 | I | mr31 | 7/8 (88) | 8/8 (100) | — | Recessive |
| suls-2 | I | mr36 | 8/9 (89) | 11/11 (100) | — | Recessive |
| — | — | mr37 | 11/12 (92) | 11/11 (100) | — | — |
Figure 4 .
Complementation data showing that each genetic locus consists of only one complementation group (blue, green, yellow, and pink shades). suls-1 is not shown because it is dominant. sqrd-1, cysl-1 double heterozygotes are slightly sensitive to RA/H2S.
The 23 strongest mutant lines were next tested for survival in room air containing lower H2S concentrations in an effort to phenotypically discriminate between the complementation groups. Table 1 shows that complementation groups have differences in H2S sensitivity, with the group mapping to the X chromosome being the most sensitive and suls-2 being the least sensitive. In contrast to the other complementation groups, suls-1 alleles vary in sensitivity to H2S.
A cysteine synthase homolog, cysl-1, is required for survival in H2S
mr23 was used as a representative allele of the group of alleles on the X chromosome (mr19, mr23, mr25, mr26, mr29, mr33, mr34, mr39, and mr40) and was mapped to a 0.2-cM region between SNPs pas23398 and snp_F41E7[3]. This region contains 70 predicted genes. Of the deletion mutants available for that region, ok762 phenocopies and fails to complement these H2S-sensitive mutations. The ok762 allele deletes five of the six exons of the gene C17G1.7. Sequencing a portion of C17G1.7 of our isolated strains revealed adenine-to-guanine point mutations, which are characteristic of EMS mutagenesis (Figure S1). All identified mutations were either missense or splice mutations.
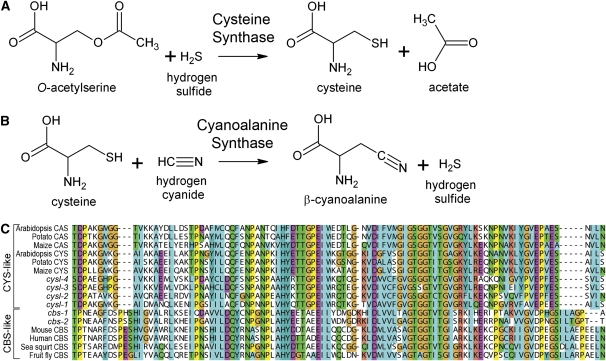
C17G1.7 is a previously uncharacterized gene encoding a member of the cystathionine β-synthase (CBS)-like subgroup, which is made up of enzymes important in cysteine production, CBS and cysteine synthase (CYS) (OAS-TL and CysK) (Feldman-Salit et al. 2009). Animals generally have CBS homologs, and bacteria and plants tend to have CYS homologs (Hell et al. 2008). CBS is critical for generating cysteine in animals, which make cysteine from dietary methionine (Finkelstein et al. 1988). CBS is also one of the only H2S-generating enzymes in mammals (Kabil and Banerjee 2010). In plants and bacteria, which are cysteine prototrophs, CYS combines H2S with O-acetylserine (OAS) to generate cysteine (Figure 5A) (Wirtz et al. 2010). Humans and mice each have one CBS-like family member. In contrast, C. elegans has six: C17G1.7, K10H10.2, R08E5.2, F59A7.9, ZC373.1, and F54A3.4 (Table 2).
Figure 5 .
(A) Cysteine synthase (CYS) catalyzes the formation of cysteine and acetate from OAS and H2S. (B) Cyanoalanine synthase (CAS) catalyzes the formation of β-cyanoalanine and H2S from cysteine and HCN. (C) Amino acid alignment of a selected region of CYS and CBS homologs. C. elegans Cysl genes are homologous to CYS, whereas C. elegans Cbs genes are homologous to cystathionine β-synthase (CBS).
Table 2 . Genes named in this article.
| Gene name | Gene ID | Allele tested | 50 ppm RA/H2S survival (%) | 5 ppm RA/HCN survival (%) |
|---|---|---|---|---|
| cysl-1 | C17G1.7 | ok762 | 0 ± 0* | 0 ± 0* |
| cysl-2 | K10H10.2 | ok3516 | 100 ± 0 | 0 ± 0* |
| cysl-3 | R08E5.2 | — | — | — |
| cysl-4 | F59A7.9 | ok3359 | 92 ± 11 | 100 ± 0 |
| cbs-1 | ZC373.1 | — | — | — |
| cbs-2 | F54A3.4 | ok666 | 100 ± 0 | 100 ± 0 |
| sqrd-1 | F02H6.5 | tm3378 | 0 ± 0* | 97 ± 5 |
| ethe-1 | C33A12.7 | tm4101 | 0 ± 0* | 100 ± 0 |
*P < 0.05 as compared to N2 wild-type animals.
C17G1.7 (cysl-1), K10H10.2 (cysl-2), R08E5.2 (cysl-3), and F59A7.9 (cysl-4) are more similar to CYS from plants and bacteria, whereas ZC373.1 (cbs-1) and F54A3.4 (cbs-2) are more similar to CBS from animals (Figure 5C). A BLAST search reveals that the genus Caenorhabditis contains the only known animal CYS homologs. We have renamed these genes cysl-1 (cysteine synthase-like), cysl-2, cysl-3, cysl-4, cbs-1, and cbs-2 to reflect this homology. Depletion of H2S levels, as it is used as a substrate by CYS, may explain why cysl-1 mutant animals are sensitive to H2S. Nematodes appear to have acquired the ability to assimilate H2S, and cysl-1 is likely using this reaction to drive down internal H2S levels.
cysl-2 mediates immunity to fast paralytic killing by Pseudomonas
The existence of three additional Cysl homologs in C. elegans raises the possibility that these other Cysl enzymes are also important in H2S detoxification. This possibility is especially intriguing, because we have shown a hif-1–dependent increase in cysl-2 transcript levels during RA/H2S exposure (Budde and Roth 2010). We obtained the available knockout strains of cysl-2, cysl-4, and cbs-2 and tested them for RA/H2S sensitivity, but none showed a RA/H2S sensitivity phenotype (Table 2).
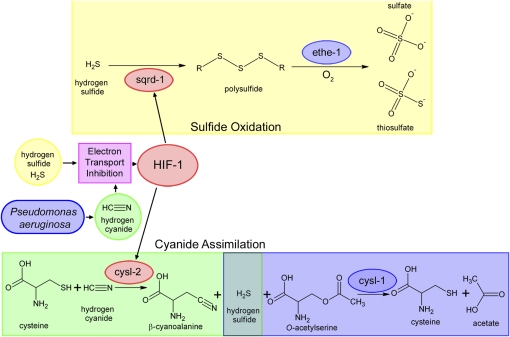
In plants, CYS and cyanoalanine synthase (CAS) are closely related and have overlapping catalytic activities (Figure 5, A and B). The CAS reaction combines cysteine and HCN to produce β-cyanoalanine and H2S, lowering cellular HCN levels (Figure 5B). Furthermore, cysl-2 mRNA was previously shown to be upregulated by HIF-1 (Shen et al. 2005; Shao et al. 2010; Budde and Roth 2010), and high HIF-1 activity has been shown to create resistance to HCN-mediated fast paralytic killing by P. aeruginosa (Darby et al. 1999; Gallagher and Manoil 2001; Shao et al. 2010). Therefore, cysl-2 may be induced not to effect H2S levels, but instead may be the mediator of resistance to HCN-dependent fast paralytic killing. To test this hypothesis, we exposed cysl-1, cysl-2, and cysl-4 mutant animals to RA/HCN and found that both cysl-1 and cysl-2 mutant animals are sensitive to RA/HCN (Table 2). The specificity of cysl-2 mutant sensitivity to cyanide suggests that this enzyme is directly acting to diminish HCN levels rather than to affect H2S levels. The sensitivity of cysl-1 mutants to both RA/H2S and RA/HCN suggest that cysl-1 acts downstream of cysl-2, and cysl-1 mutants are being overwhelmed by the H2S produced by cysl-2 during HCN detoxification (Figure 8).
Figure 8 .
Proposed mechanism. Both H2S and HCN can inhibit electron transport. Electron transport inhibition causes induction of HIF-1 activity resulting in high expression of SQRD-1 and CYSL-2. SQRD-1 catalyzes the first step in the H2S-oxidation pathway, ultimately resulting in production of sulfate and thiosulfate. CYSL-2 catalyzes the first step in the HCN assimilation pathway, producing H2S. The resultant H2S is detoxified by CYSL-1.
Next, we tested whether these enzymes are responsible for HIF-1–mediated innate immunity to P. aeruginosa. We showed that, similar to previous studies (Gallagher and Manoil 2001; Shao et al. 2010), egl-9–deficient animals have increased innate immunity to P. aeruginosa (Table 3). Next, we demonstrated that cysl-1 and cysl-2 are both required for the observed innate immunity to P. aeruginosa, demonstrating a mechanism of innate immunity to Pseudomonas-dependent fast paralytic killing. Since we previously showed that animals grown in RA/H2S have increased cysl-2 mRNA expression, we assayed whether growth in H2S confers resistance to P. aeruginosa. Indeed, our results show that growth in H2S increases the innate immunity of C. elegans to P. aeruginosa (Table 3).
Table 3 . Innate immunity to P. aeroginosa requires hif-1, cysl-1, and cysl-2.
| Strain | Pretreatment | Allele | Survival on P. aeruginosa (%) |
|---|---|---|---|
| N2 | Control RNAi | — | 0 ± 0 |
| N2 | Egl-9 RNAi | — | 89 ± 19 |
| hif-1 | Egl-9 RNAi | ia04 | 10 ± 16* |
| egl-9 | Egl-9 RNAi | sa307 | 100 ± 0 |
| cysl-1 | Egl-9 RNAi | ok762 | 4 ± 7* |
| cysl-2 | Egl-9 RNAi | ok3516 | 0 ± 0* |
| sqrd-1 | Egl-9 RNAi | tm3378 | 100 ± 0 |
| ethe-1 | Egl-9 RNAi | tm4101 | 100 ± 0 |
| N2 | H2S grown | — | 83 ± 29† |
*P < 0.05 as compared to egl-9 knockdown in N2 animals. †P < 0.05 as compared to untreated control RNAi N2 animals.
sqrd-1 is required for survival in H2S
The group of alleles on chromosome IV, mr21, mr24, mr28, mr30, and mr32 were mapped near the SNP pkP4095 located at the genetic location IV:12. Analysis of the region revealed a gene, F02H6.5, encoding an apparent homolog of the protein sulfide:quinone reductase dehydrogenase (SQRD), which we have named sqrd-1 to reflect this homology. SQRD was initially described in prokaryotes as oxidizing sulfide and transferring the electrons to ubiquinone for generation of ATP during oxidative phosphorylation (Vande Weghe and Ow 1999). The available deletion allele of sqrd-1 (tm3378) phenocopies and fails to complement these mutants. Sequence analysis revealed A-to-G transitions in our isolated mutants (Figure S1B).
The C. elegans gene sqrd-1 encodes a member of the group II SQRD enzymes, which also includes human, mouse, and Drosophila homologs. The C. elegans genome is predicted to encode another SQRD homolog, Y9C9A.16 (sqrd-2), which was initially classified as a pseudogene. We obtained the deletion allele of sqrd-2 and found that it is not sensitive to RA/H2S.
Transcriptional regulation of identified genes
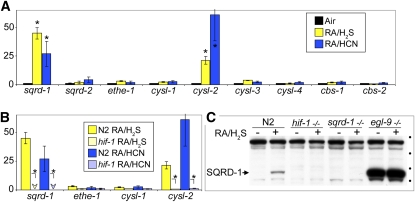
We undertook this screen to determine the mechanism by which HIF-1 confers resistance to H2S. Therefore, we were interested to learn whether either cysl-1 or sqrd-1 is transcriptionally regulated by HIF-1 in response to RA/H2S exposure. Figure 6A shows that sqrd-1 and cysl-2 mRNA levels increase during RA/H2S and RA/HCN exposure, but cysl-1 mRNA levels remain unchanged. Furthermore, hif-1(ia04) mutant animals fail to increase sqrd-1 and cysl-2 mRNA levels during RA/H2S exposure (Figure 6B), suggesting that upregulation of sqrd-1 by HIF-1 is a mechanism for surviving in the presence of H2S.
Figure 6 .
(A) Both sqrd-1and cysl-2 mRNA levels increase relative to untreated animals as measured by quantitative reverse transciptase PCR. Y axis is relative mRNA concentration compared to room air-treated control animals. *P < 0.05 as compared to animals grown in room air. (B) hif-1 is required for sqrd-1 and cysl-2 mRNA induction when exposed to RA/H2S or RA/HCN. Arrowheads indicate that mRNA levels were observed at very low levels. *P < 0.05 when comparing wild-type and hif-1 mutant animals. (C) Western blot shows SQRD-1 protein induction in RA/H2S. This induction requires hif-1. No immunoreactivity is observed in sqrd-1 mutant animals. SQRD-1 protein is abundant in egl-9 mutant animals. Circles indicate protein size markers of 116, 82, 62, and 49 kDa from top to bottom. The calculated molecular weight of 53 kDa corresponds well with the observed relative migration.
To determine whether or not SQRD-1 protein levels increase during RA/H2S exposure, we performed a Western blot to determine relative protein concentrations. Figure 6C shows that SQRD-1 protein levels increase during RA/H2S exposure in the parental N2 Bristol strain. No SQRD-1 protein accumulation is observed in the sqrd-1(tm3378) or in the hif-1(ia04) mutant animals. egl-9(sa307) mutant animals have constitutively high levels of SQRD-1 protein, which may explain their ability to survive in 100 times higher RA/H2S concentration than hif-1(ia04) mutant animals.
SQRD-1::GFP protein is expressed during H2S exposure
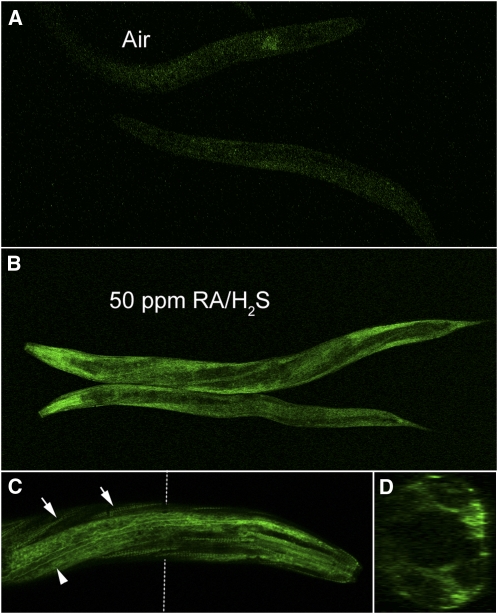
Next, we analyzed the expression pattern of SQRD-1 in C. elegans. We created a SQRD-1::GFP fusion construct because the antibody we created was unable to stain worms by immunofluorescence. The presence of a predicted SL2 alternative splice site upstream of sqrd-1 indicated that a standard promoter fusion might not accurately recapitulate the endogenous expression pattern. Therefore, we used the recombineering approach for GFP fusion construction, which includes a 35-kb region surrounding the 4.5-kb SQRD-1 gene. RA/H2S exposure causes a fluorescence increase in recombinant animals that is most apparent in the anterior of the worm (Figure 7). Expression appears limited to the hypodermis, the body wall muscle, and the anterior bulb of the pharynx (Figure 7C). A cross section through the worm confirms that expression is limited to the most exposed parts of the worm (Figure 7D). It is possible that this SQRD-1 expression pattern reflects a function of SQRD-1 to prevent H2S from reaching the interior tissue.
Figure 7 .
SQRD-1::GFP expression is induced by exposure to RA/H2S. (A) SQRD-1::GFP is not expressed in room air. (B) RA/H2S at 50 ppm induces expression of SQRD-1::GFP. (C) Confocal microscopy at ×40 magnification shows that the expression is brightest in the muscle dense bodies (tailed arrows) and the hypodermis (tail-less arrow). (D) Cross-section (Z-stack) shows expression is most intense in the hypodermis.
We conclude from these experiments that HIF-1 acts to initiate the response of C. elegans to H2S by upregulating the SQRD-1 protein levels. SQRD-1 is an oxidoreductase, oxidizing H2S to polysulfide and reducing ubiquinone (Theissen et al. 2003). The oxidation of sulfide generates polysulfide (Marcia et al. 2009), which can then be further oxidized via the H2S oxidation pathway to yield sulfate and thiosulfate (Kabil and Banerjee 2010). The decrease in H2S caused by SQRD-1 is likely to reduce internal H2S levels to a tolerated range.
A second enzyme required for the H2S oxidation pathway is ETHE1 (Tiranti et al. 2009). ETHE1 is a sulfur dioxygenase required in mammals for oxidation of sulfur to sulfate (Kabil and Banerjee 2010). C. elegans has a single homolog of Ethe1, C33A12.7, which we named ethe-1. We obtained the available ethe-1 (tm4101) deletion allele and tested for H2S sensitivity (Table 2). Similar to sqrd-1 (tm3378), ethe-1 (tm4101) is sensitive to RA/H2S, which suggests that sqrd-1 confers resistance to H2S through the sulfur–oxidation pathway.
Epistasis with egl-9
Egl-9–deficient animals have been shown to be resistant to both H2S and HCN and our data suggest that resistance is mediated by HIF-1 upregulating sqrd-1 and cysl-2, respectively. To test this hypothesis, we used RNAi to knockdown egl-9 in mutant animals, which we have described as H2S and HCN sensitive and tested them for resistance to H2S and HCN. Table 4 shows that egl-9 knockdown does not mediate resistance to HCN in cysl-1 and cysl-2 mutant backgrounds, and therefore cysl-1 and cysl-2 are epistatic to egl-9. Sqrd-1 and ethe-1 mutant animals with knocked down egl-9 are partially resistant to H2S.
Table 4 . Epistasis relationships with egl-9.
| 15 ppm RA/HCN | 250 ppm RA/H2S | |||
|---|---|---|---|---|
| Empty vector (%) | egl-9 RNAi (%) | Empty vector (%) | egl-9 RNAi (%) | |
| N2 | 0 ± 0 | 92 ± 10 | 0 ± 0 | 98 ± 4 |
| hif-1 (ia04) | 0 ± 0 | 0 ± 0* | 0 ± 0 | 0 ± 0* |
| egl-9 (sa307) | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 |
| sqrd-1 (tm3378) | 0 ± 0 | 87 ± 16 | 0 ± 0 | 47 ± 15* |
| ethe-1 (tm4101) | 0 ± 0 | 89 ± 14 | 0 ± 0 | 73 ± 10* |
| cysl-1 (ok762) | 0 ± 0 | 27 ± 35* | 0 ± 0 | 68 ± 30 |
| cysl-2 (ok3516) | 0 ± 0 | 0 ± 0* | 0 ± 0 | 94 ± 6 |
*P < 0.05 as compared to egl-9 RNAi knockdown in wild-type animals. Data are presented as percentage of survival.
Discussion
C. elegans likely encounters H2S in the soil as a result of breakdown of organic sulfur (Morra and Dick 1991) and HCN produced by both bacteria (Gallagher and Manoil 2001) and plants (García et al. 2010). This study has examined the mechanisms that C. elegans uses to respond to high levels of H2S and HCN. RA/H2S and RA/HCN both cause an increase in HIF-1 activity, which increases the expression of sqrd-1 and cysl-2 to catalyze the first steps of the metabolism of H2S and HCN, respectively. sqrd-1 acts to oxidize H2S, ultimately preventing internal H2S levels from exceeding the maximum tolerated concentration. cysl-2 acts in parallel to keep HCN levels low; however, equimolar amounts of H2S are generated as a consequence of cysl-2 activity. cysl-1 is required to reduce H2S levels that are produced by cysl-2 during RA/HCN exposure (Figure 8).
Here, we provide direct evidence that H2S acts through hif-1 to increase both SQRD-1 mRNA and protein expression. This initiates the sulfide–oxidation pathway, which lowers internal H2S levels (Figure 8). SQRD-1 expression appears to be localized to the cells most readily exposed to exogenous H2S in the worm. In mammals, colon epithelial cells that express high levels of SQRDL (Pontén et al. 2008) are likely to do so because a prominent enteric bacteria, Desulfovibrio piger, produces high levels of H2S in the lower intestine (Loubinoux et al. 2002). The requirement for ethe-1 during RA/H2S exposure confirms that the sulfide–oxidation pathway is the essential function of sqrd-1. The sulfide–oxidation pathway requires oxygen, both to maintain a functional electron transport chain (to generate oxidized ubiquinone) and to directly oxidize sulfur via ethe-1. Exposure of mammals to exogenous H2S results in many alterations in physiology, including the induction of a suspended-animation–like state and improved outcome in ischemia reperfusion injury. It will be interesting to learn whether SQRDL expression is induced upon exposure to H2S in mammals.
The question remains as to why SQRD-1 is inducible at all. Surrounded by decaying matter, C. elegans must encounter areas of high H2S in its natural environment. Having constitutively high SQRD-1 levels, as in egl-9 mutant animals, allows survival when wild-type animals with lower HIF-1 activity would perish. However, low levels of H2S are important for biological function, so perhaps nematodes are using SQRD-1 to deliberately titrate the cellular concentration of H2S by controlling its rate of destruction. In mammals, the H2S-generating enzymes CBS, CSE, and 3MP control sulfide levels (Kabil and Banerjee 2010). However, our study suggests that mammals may also modulate cellular H2S levels by regulating SQRDL-dependent destruction.
In addition to furthering our understanding of the sulfide oxidation pathway, this study also provides insight into two new metabolic activities previously unknown in animals, cysteine synthase and cyanoalanine synthase. cysl-1 mutant animals are sensitive to both RA/H2S and RA/HCN, while cysl-2 mutant animals are only sensitive to RA/HCN. This suggests that cysl-1 is a cysteine synthase acting downstream of cysl-2, a cyanoalanine synthase (Figure 8). We hypothesize that cysl-1 mutant animals could be succumbing to H2S produced by CYSL-2 as a result of cyanide assimilation. In this way each molecule of HCN assimilated by CYSL-2 would produce one molecule of H2S. In this reaction mechanism CYSL-1 might regenerate cysteine used in cyanide assimilation. Alternatively, it is possible that CYSL-1 is catalyzing the assimilation of both H2S and HCN, a hypothesis supported by in vitro plant studies (Hell et al. 2002). The function of the two remaining cysteine synthase homologs, cysl-3 and cysl-4, remains to be elucidated, but they may also have important CYS/CAS activity during development or in specific tissues.
To our knowledge, cysl-1 is the first reported functional CYS homolog in animals. Indeed, while no orthologs appear to be present in animals outside of the genus Caenorhabditis, in plants and bacteria CYS uses OAS as the H2S-accepting substrate (Feldman-Salit et al. 2009). Plants and bacteria generate OAS by acetylating serine with the enzyme serine acetyltransferase (SAT) (Wirtz et al. 2010), which has no homolog in C. elegans. How might C. elegans be obtaining OAS? As with many other amino acids, C. elegans is likely obtaining OAS from its bacterial food source. Alternatively, C. elegans might have evolved a mechanism of generating OAS using alternative enzymes. CYS activity is essential for cysteine biosynthesis in prototrophs (Feldman-Salit et al. 2009). Because the critical aspect of CYS appears to be the elimination of H2S instead of the generation of cysteine, perhaps C. elegans is freed from using OAS as the H2S acceptor and is using an alternative H2S acceptor that does not generate cysteine. In mammals the closest mammalian homolog to CYS is CBS. Consistent with the idea that mammals have CYS activity, there is evidence that radiolabeled H2S can be incorporated into cysteine in germ-free rats (Huovinen and Gustafsson 1967).
The protective effect of cysl-2 to HCN explains the previous observation that egl-9 mutant animals are resistant to the pathogenic Pseudomonas fast paralytic killing. P. aeruginosa is a clinically relevant pathogen and can produce toxic cyanide levels in the sputum of cystic fibrosis patients (Anderson et al. 2010). P. fluorescens is a cyanide-producing soil bacteria that has also been shown to exhibit cyanide-dependent fast paralytic killing effect on C. elegans (Romanowski et al. 2011). HIF-1 activates resistance to hypoxia, and this has been proposed to also confer resistance to the cytochrome c oxidase inhibiting effects of cyanide (Gallagher and Manoil 2001; Shao et al. 2010; Romanowski et al. 2011). While this certainly may play some role, we provide evidence here that the resistance of egl-9 mutant animals to P. aeruginosa requires cysl-2, likely by directly detoxifying cyanide to β-cyanoalanine (Figure 8). This reaction produces H2S, which can be further detoxified by cysl-1 and sqrd-1.
While we have demonstrated critical components of the RA/H2S-induced hif-1 response, the mechanism of hif-1 induction remains unclear. The overlapping gene expression patterns between both H2S and HCN suggests that C. elegans has a shared response to these two molecules. In fact, our studies suggest that C. elegans is unable to discriminate between H2S and HCN, leading them to activate the degradation pathways of both molecules when either is presented. Thus, C. elegans treated with RA/H2S or RA/HCN increases both H2S-detoxification enzymes and cyanide-detoxification enzymes (Figure 6A). A recent study showed that inhibition of electron transport through reduction of expression of electron transport proteins is sufficient to induce HIF-1 activity (Lee et al. 2010). H2S and HCN are natural effectors of electron transport and cause increased HIF-1 activity; therefore, H2S and HCN might be natural regulators of electron transport.
Acknowledgments
We thank members of the Roth lab for helpful discussions, Jim Priess and Sue Biggins for allowing gratuitous reagent use, Colin Manoil and Larry Gallagher for Pseudomonas aeruginosa strains, and Shohei Mitani at Tokyo Women's Medical University School of Medicine for providing knockout strains.
Literature Cited
- Anderson R. D., Roddam L. F., Bettiol S., Sanderson K., Reid D. W., 2010. Biosignificance of bacterial cyanogenesis in the CF lung. J. Cyst. Fibros. 9: 158–164 [DOI] [PubMed] [Google Scholar]
- Blackstone E., Roth M. B., 2007. Suspended animation-like state protects mice from lethal hypoxia. Shock 27: 370–372 [DOI] [PubMed] [Google Scholar]
- Blackstone E., Morrison M., Roth M. B., 2005. H2S induces a suspended animation-like state in mice. Science 308: 518. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde M. W., Roth M. B., 2010. Hydrogen sulfide increases hypoxia-inducible factor-1 activity independently of von Hippel-Lindau tumor suppressor-1 in C. elegans. Mol. Biol. Cell 21: 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C., Cosma C. L., Thomas J. H., Manoil C., 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96: 15202–15207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A., Alm E. J., 2011. Rapid evolutionary innovation during an Archaean genetic expansion. Nature 469: 93–96 [DOI] [PubMed] [Google Scholar]
- Davis M. W., Hammarlund M., Harrach T., Hullett P., Olsen S., et al. , 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolphin C. T., Hope I. A., 2006. Caenorhabditis elegans reporter fusion genes generated by seamless modification of large genomic DNA clones. Nucleic Acids Res. 34: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod J. W., Calvert J. W., Morrison J., Doeller J. E., Kraus D. W., et al. , 2007. Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury by preservation of mitochondrial function. Proc. Natl. Acad. Sci. USA 104: 15560–15565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman-Salit A., Wirtz M., Hell R., Wade R. C., 2009. A mechanistic model of the cysteine synthase complex. J. Mol. Biol. 386: 37–59 [DOI] [PubMed] [Google Scholar]
- Finkelstein J. D., Martin J. J., Harris B. J., 1988. Methionine metabolism in mammals. The methionine-sparing effect of cystine. J. Biol. Chem. 263: 11750–11754 [PubMed] [Google Scholar]
- Frazier H. N., 3rd, Roth M. B., 2009. Adaptive sugar provisioning controls survival of C. elegans embryos in adverse environments. Curr. Biol. 19: 859–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., Manoil C., 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183: 6207–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García I., Castellano J. M., Vioque B., Solano R., Gotor C., et al. , 2010. Mitochondrial beta-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana. Plant Cell 22: 3268–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R., Jost R., Berkowitz O., Wirtz M., 2002. Molecular and biochemical analysis of the enzymes of cysteine biosynthesis in the plant Arabidopsis thaliana. Amino Acids 22: 245–257 [DOI] [PubMed] [Google Scholar]
- Hell R., Dahl C., Knaff D. B., Leustek T., 2008. Sulfur Metabolism in Phototrophic Organisms. Springer-Verlag, New York [Google Scholar]
- Holmgren A., Johansson C., Berndt C., Lönn M. E., Hudemann C., et al. , 2005. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem. Soc. Trans. 33: 1375–1377 [DOI] [PubMed] [Google Scholar]
- Huovinen J. A., Gustafsson B. E., 1967. Inorganic sulphate, sulphite and sulphide as sulphur donors in the biosynthesis of sulphur amino acids in germ-free and conventional rats. Biochim. Biophys. Acta 136: 441–447 [DOI] [PubMed] [Google Scholar]
- Kabil O., Banerjee R., 2010. Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 285: 21903–21907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-J., Hwang A. B., Kenyon C., 2010. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20: 2131–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Rose P., Moore P. K., 2011. Hydrogen sulfide and cell signaling. Annu. Rev. Pharmacol. Toxicol. 51: 169–187 [DOI] [PubMed] [Google Scholar]
- Loubinoux J., Valente F. M. A., Pereira I. A. C., Costa A., Grimont P. A. D., et al. , 2002. Reclassification of the only species of the genus Desulfomonas, Desulfomonas pigra, as Desulfovibrio piger comb. nov. Int. J. Syst. Evol. Microbiol. 52: 1305–1308 [DOI] [PubMed] [Google Scholar]
- Marcia M., Ermler U., Peng G., Michel H., 2009. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc. Natl. Acad. Sci. USA 106: 9625–9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L., Roth M. B., 2007. Hydrogen sulfide increases thermotolerance and lifespan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104: 20618–20622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra M. J., Dick W. A., 1991. Mechanisms of H2S production from cysteine and cystine by microorganisms isolated from soil by selective enrichment. Appl. Environ. Microbiol. 57: 1413–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M. L., Blackwood J. E., Lockett S. L., Iwata A., Winn R. K., et al. , 2008. Surviving blood loss using hydrogen sulfide. J. Trauma 65: 183–188 [DOI] [PubMed] [Google Scholar]
- Mustafa A. K., Gadalla M. M., Snyder S. H., 2009. Signaling by gasotransmitters. Sci. Signal. 2: re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S., 2001. Predicting deleterious amino acid substitutions. Genome Res. 11: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot P., Van Zuilen M., Lepot K., Thomazo C., Farquhar J., et al. , 2007. Early Archaean microorganisms preferred elemental sulfur, not sulfate. Science 317: 1534–1537 [DOI] [PubMed] [Google Scholar]
- Pontén F., Jirström K., Uhlen M., 2008. The Human Protein Atlas–a tool for pathology. J. Pathol. 216: 387–393 [DOI] [PubMed] [Google Scholar]
- Reinhard C. T., Raiswell R., Scott C., Anbar A. D., Lyons T. W., 2009. A late Archean sulfidic sea stimulated by early oxidative weathering of the continents. Science 326: 713–716 [DOI] [PubMed] [Google Scholar]
- Romanowski A., Migliori M. L., Valverde C., Golombek D. A., 2011. Circadian variation in Pseudomonas fluorescens (CHA0)-mediated paralysis of Caenorhabditis elegans. Microb. Pathog. 50: 23–30 [DOI] [PubMed] [Google Scholar]
- Rouault T. A., Tong W. H., 2008. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 24: 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S., 2007. Counting mutagenized genomes and optimizing genetic screens in Caenorhabditis elegans. PLoS ONE 2: e1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Zhang Y., Ye Q., Saldanha J. N., Powell-Coffman J. A., 2010. C. elegans SWAN-1 Binds to EGL-9 and regulates HIF-1-mediated resistance to the bacterial pathogen Pseudomonas aeruginosa PAO1. PLoS Pathog. 6.:pii: e1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Nettleton D., Jiang M., Kim S. K., Powell-Coffman J. A., 2005. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J. Biol. Chem. 280: 20580–20588 [DOI] [PubMed] [Google Scholar]
- Swierczek N. A., Giles A. C., Rankin C. H., Kerr R. A., 2011. High-throughput behavioral analysis in C. elegans. Nat. Methods. 8: 592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen U., Hoffmeister M., Grieshaber M., Martin W., 2003. Single eubacterial origin of eukaryotic sulfide: quinone oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Mol. Biol. Evol. 20: 1564–1574 [DOI] [PubMed] [Google Scholar]
- Tiranti V., Viscomi C., Hildebrandt T., Di Meo I., Mineri R., et al. , 2009. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat. Med. 15: 200–205 [DOI] [PubMed] [Google Scholar]
- Vande Weghe J. G., Ow D. W., 1999. A fission yeast gene for mitochondrial sulfide oxidation. J. Biol. Chem. 274: 13250–13257 [DOI] [PubMed] [Google Scholar]
- Wirtz M., Birke H., Heeg C., Müller C., Hosp F., et al. , 2010. Structure and function of the hetero-oligomeric cysteine synthase complex in plants. J. Biol. Chem. 285: 32810–32817 [DOI] [PMC free article] [PubMed] [Google Scholar]