Abstract
Influenza viruses impose a constant threat to vertebrates susceptible to this family of viruses. We have developed a new tool to study virus–host interactions that play key roles in viral replication and to help identify novel anti-influenza drug targets. Via the UAS/Gal4 system we ectopically expressed the influenza virus M2 gene in Drosophila melanogaster and generated dose-sensitive phenotypes in the eye and wing. We have confirmed that the M2 proton channel is properly targeted to cell membranes in Drosophila tissues and functions as a proton channel by altering intracellular pH. As part of the efficacy for potential anti-influenza drug screens, we have also demonstrated that the anti-influenza drug amantadine, which targets the M2 proton channel, suppressed the UAS-M2 mutant phenotype when fed to larvae. In a candidate gene screen we identified mutations in components of the vacuolar V1V0 ATPase that modify the UAS-M2 phenotype. Importantly, in this study we demonstrate that Drosophila genetic interactions translate directly to physiological requirements of the influenza A virus for these components in mammalian cells. Overexpressing specific V1 subunits altered the replication capacity of influenza virus in cell culture and suggests that drugs targeting the enzyme complex via these subunits may be useful in anti-influenza drug therapies. Moreover, this study adds credence to the idea of using the M2 “flu fly” to identify new and previously unconsidered cellular genes as potential drug targets and to provide insight into basic mechanisms of influenza virus biology.
INFLUENZA viruses, of the family Orthomyxoviridae, infect vertebrates and are the causative agents of the respiratory disease influenza. Influenza virus can infect a wide variety of animals and can thus spread from population to population. The outcomes of an influenza virus outbreak can be catastrophic, as evidenced by the 1918 Spanish flu, which caused approximately 50 million deaths worldwide (Palese and Shaw 2007; Wright et al. 2007).
Influenza viruses infect and replicate in cells of the upper and lower respiratory tract. Successful entry of the virus into a cell requires two influenza virus proteins: hemagglutinin (HA), which binds to sialic acids on the surface of cells for attachment, and matrix protein 2 (M2), which lowers the pH inside the virion, allowing for viral uncoating (Palese and Shaw 2007). M2 modulates pH due to its proton-transport activity. M2 is a small, type III integral membrane protein that acts as a proton pump (Pinto and Lamb 2006; Palese and Shaw 2007). M2 functions as a tetramer and can be found inserted into a variety of membrane systems within the cell, such as the plasma membrane and Golgi. However for the M2 channel to be active, the N-terminal ectodomain of M2 must be located in a compartment with a pH lower than 7.0 (Pinto and Lamb 2006). In the case of viral uncoating, the influenza virus is imported into the cell within an endocytic vesicle, which contains a lowered pH. M2 becomes active and protons flow via the channel into the virion core, allowing the viral genome and its associated proteins to disassociate (Palese and Shaw 2007).
As M2 and HA are both membrane proteins, they are processed and targeted to the plasma membrane through the secretory pathway. HA is pH sensitive and changes conformation at a lowered pH. This change in HA conformation allows for the fusion of the viral and vesicle membranes (Palese and Shaw 2007). When traveling through the secretory pathway, the ectodomain of M2 is located within the Golgi lumen, which has a lowered pH, and M2 can become active and pump protons out of the Golgi, raising the pH of the Golgi lumen. For strains of influenza virus with acid-sensitive HAs (including H5 and H7), this activity prevents HA from prematurely adopting a lower-pH conformation (Palese and Shaw 2007).
Currently there are four commonly used anti-influenza virus drugs available: amantadine and rimantadine, which target the influenza virus M2 protein, and zanamivir and oseltamivir, which target the influenza viral protein neuraminidase (NA) (Palese and Shaw 2007). Unfortunately, these drugs are not always effective against influenza, since they are viral-strain specific. Resistance to amantadine is of particular concern, as many common influenza virus strains are resistant to amantadine (an example found in Lan et al. 2010). Therefore the need for new anti-influenza drugs is paramount, as is the need to develop new and novel strategies to develop antiviral drugs as a whole. In this study we develop a genetic system that allows for the identification of host genes that modulate influenza virus M2 activity. The identification of host genes involved with M2 activity will not only aid in the understanding of how cellular machinery affects M2’s role in the intracellular propagation of the influenza virus, but also provide new targets for anti-influenza drug therapy.
The Drosophila model system has been previously used to study a variety of human diseases including neurodegenerative diseases such as Alzheimer’s and Parkinson’s diseases, neuromuscular diseases, mitochondrial diseases, cancer, diabetes, and prions, to name a few (Chan and Bonini 2000; Fortini and Bonini 2000; Lasko 2002; Hafen 2004; Vidal and Cagan 2006; Botella et al. 2009; Lu and Vogel 2009; Hirth 2010; Iijima-Ando and Iijima 2010; Lloyd and Taylor 2010; Rea et al. 2010; Rincon-Limas et al. 2010). In addition, a few human viruses including Epstein–Barr virus, cytomegalovirus, and HIV have been studied in transgenic flies (Adamson et al. 2005; Spresser and Carlson 2005; Steinberg et al. 2008). By expressing the influenza A virus M2 protein in Drosophila melanogaster we have created a new research tool to study the influenza virus, which we call the M2 “flu fly.” Expression of the influenza virus M2 gene in specific fly tissues resulted in significant and obvious mutant phenotypes. Characterization of the M2 phenotypes within these Drosophila tissues indicated that the M2 protein has correctly localized in these cells and functions properly as a proton channel as well. We have performed a candidate genetic screen to identify genetic modifiers of M2 function, showing that some, but not all, of the subunits of the cellular V1V0 ATPase proton pump act as modifiers of M2-mediated phenotypes. To show the conservation of these genetic interactions, we further demonstrated that alteration of V1V0 ATPase proton pump activity in mammalian cultured cells also affects influenza viral infection and replication within these cells.
We believe that our M2 flu fly system is valuable because we can examine the effects of overexpression of a viral gene within an intact organism, at any stage of development or in any tissue. Furthermore, the modification of M2 activity by either a genetic modifier or a drug compound can be easily determined by visually examining adult flies. The screen that we have performed for this work has yielded specific and interesting results.
Materials and Methods
Fly culture
Flies were maintained at either room temperature (22°) or 29° in plastic vials on a medium of cornmeal, yeast, molasses, and agar with methyl-4-hydroxybenzoate added as a mold inhibitor.
Cell culture
Madin–Darby canine kidney NBL-2 (MDCK) cells were maintained in Dulbecco’s modified Eagle’s medium H (DMEM) supplemented with 10% fetal calf serum along with antibiotic and antifungal agents. Subconfluent cells were infected with influenza A H1N1 virus (A/PR/8/34; from ATCC) at an MOI of 1. Infection was carried out in DMEM with no serum or additives, along with 1 μg/ml trypsin, 0.125% bovine serum albumin (BSA), and 1% HEPES for 48 hr at 37°. Bafilomycin-treated cells had bafilomycin (Sigma) added to the media 24 hr prior to infection.
Plaque assay
MDCK cells were plated to near confluence and received treatments as indicated. The cells were infected 24 hr post-treatment with influenza A virus as described above. After 24 hr of infection, cells were overlayed with DMEM plus 2% fetal calf serum and 0.4% agarose. Plaques were visualized by staining with Neutral Red and counted 4 days post-infection.
Plasmids and transfection
The V1B plasmid contains the human ATP6V1B1 cDNA in the pCMV6-XL4 vector; the V1A plasmid contains the human ATP6V1A cDNA in the pCMV6-XL5 vector; the V1C plasmid contains the human ATP6V1C1 cDNA in the pCMV6-XL5 vector (all from Origene). The GFP vector contains the GFP cDNA modified to contain a plasma membrane localization sequence. MDCK cells were transfected as follows: 1 μg of each plasmid (for a total of 2 μg) was diluted in 500 μl DMEM with no serum or additives, and 5 μl lipofectamine 2000 (Invitrogen) was diluted in 500 μl DMEM with no serum or additives; both sets were incubated 5 min at RT. The DNA and lipofectamine 2000 dilutions were combined and incubated for 20 min at RT, after which they were added to cells plated in 60 mm plates containing 5 ml DMEM with no serum or additives.
UAS-M2 flies
The influenza A virus M2 cDNA (from the A/Udorn/72 virus) (Zebedee et al. 1985) was cloned into pCaSpeR 3, an upstream activation sequence (UAS) P-element vector, which included the mini-white+ marker gene. This vector was injected into Drosophila melanogaster w1118 embryos as previously described (Spradling and Rubin 1982). These embryos were allowed to develop into adults and mated back to w1118 flies. In the next generation, the flies that had the P-element vector inserted in their genome were identified by their eye color, which ranged from orange to red. Ten UAS-M2 lines were produced, containing UAS-M2 insertions on either the second or third chromosome. When crossed to the Gal4 drivers (see Fly crosses, below) each line yielded identical mutant phenotypes (all temperature and dose sensitive). UAS-M2 flies may be requested by contacting the corresponding author.
Fly crosses
Wild-type flies are w1118. UAS-M2 flies were crossed to an engrailed driver (en-Gal4) for expression in wings, to a Glass-mediated response driver (GMR-Gal4) for expression in eyes, and to the C135-Gal4 driver for expression in fat body and salivary glands. Expression of UAS-M2 via the C135-Gal4 driver resulted in bloated larvae that died as pupae. For the colocalization studies, we used the following stocks: w+; P{UAS-GFP.KDEL}11.1/CyO, endoplasmic reticulum-targeted GFP (Bloomington Stock Number 9898); w+; P{UAS-Grasp65-GFP}2, Golgi-targeted GFP (Bloomington Stock Number 8507); w1118; P{UAS-mitoGFP.AP}3, mitochondria-targeted GFP (Bloomington Stock Number 8443); w1118; P{UASp-Act5C.T:GFP}3, actin-tagged GFP (Bloomington Stock Number 7311); w1118; P{UAS-EGFP-Clc}3, clathrin-tagged GFP (Bloomington Stock Number 7107). Larvae with one copy of GFP-actin, -Golgi, -clathrin, or -ER plus the C135-Gal4 driver plus UAS-M2 were selected on the basis of expressing GFP plus having the bloated larval phenotype, which was a phenotype associated with expression of UAS-M2 in certain cells within the gut and within the fat body. UAS-M2 and GMR-Gal4 were recombined onto a single chromosome for the candidate gene screen. Female GMR-Gal4:UAS-M2/CyO flies were crossed to males of the candidate stocks (see Table 3), and incubated at either room temperature (22°) or 29°. Fly stocks were purchased from the Bloomington Stock Center.
Table 3 . Genetic Modifiers of the M2 eye phenotype.
| Gene | Protein namea | Allelesb | Modification of UAS-M2 phenotype |
|---|---|---|---|
| vha16-1 | ATP6V0C | vha16-1B147 (PBAC insertion) | Slight enhancer in females at 29° |
| vhaAC39-1 | ATP6V0D1 | vhaAC39e04316 (PBAC upstream) | Slight enhancer at 29° |
| vhaM9.7-b | ATP6V0M9.7-2 | Df(3L)Pc-2q (deletes 78C5-79A1) | Slight enhancer at 29° |
| vha13 | ATP6V1G | Df(3R)ED5942 (deletes 91F12-92B3) | Strong enhancer at 29° |
| vha14-1 | ATP6V1F | vha14-1EY12998 (P element insertion) | Slight enhancer in males at 29° |
| Df(2R)Exel7137 (deletes 52A13-C8) | Slight enhancer in males at 29° | ||
| vha26 | ATP6V1E1 | Df(3R)Exel6144 (deletes 83A6-B6) | Strong enhancer at RT and 29° |
| vha36-1 | ATP6V1D | vha36-1K07207 (P-element insertion) | Strong enhancer at 29° |
| vha44c | ATP6V1C | vha44KG00915 (P-element insertion); | Strong enhancer at 29° |
| vha44KG07119 (P-element insertion); | Slight enhancer at 29° | ||
| vha44EY02202 (P-element insertion); | Strong enhancer at RT and 29° | ||
| Df(2R)Exel6063 (deletes 52F6-53C4) | Strong enhancer at 29° | ||
| vha55c | ATP6V1B1 | vha5512 (EMS, hypomorphic); | Strong enhancer at 29° |
| vha5514 (point mutation); | Strong enhancer at 29° | ||
| vha5516 (EMS); | Strong enhancer at RT and 29° | ||
| vha55j2E9 (P-element insertion, amorphic); | Strong enhancer at RT and 29° | ||
| Df(3R)ED5591 (deletes 87B7-C7) | Strong enhancer at 29° | ||
| vha68-1c | ATP6V1A | vha68-1EY02923 (P-element insertion); | Strong enhancer at 29° |
| Df(2L)Exel7055 (deletes 34A2-A7) | Strong enhancer at RT and 29° | ||
| vha68-2c | ATP6V1A | vha68-201510 (P-element insertion); | Slight enhancer at 29° |
| vha68-2EP2364 (P-element insertion); | Slight suppressor at 29° | ||
| Df(2L)Exel7055 (deletes 34A2-A7) | Strong enhancer at 29° | ||
| vhaSFD | ATP6V1H | Df(2L)Exel7066 (deletes 36A1-A12); | Strong enhancer at 29° |
| vhaSFDEY04644 (P-element, may overexpress gene) | Slight suppressor at 29° | ||
| CG2968 | ATP5D | l(1)G0230f04497 (PBAC downstream) | Slight enhancer in males at 29° |
| CG34123 | Ion transport | trpmEY01618 (P-element insertion, hypomorphic) | Slight enhancer in males at 29° |
| vhaAC45 | H+ transport | P{SUP or-P}KG02272 (P-element upstream) | Slight enhancer in males at 29° |
| CG5389 | H+ transport | Df(3L)th102 (deletes 72A2-D10) | Strong enhancer at 29° |
ATP5 genes are part of the mitochondrial F1F0 ATP synthase; ATP6 genes are part of the V1V0 ATPase.
All information from FlyBase. PBAC insertion, piggyBAC element inserted into coding region; P-element insertion, P-element inserted into coding region of gene; EMS, ethyl methanesulfonate.
Significant modifier.
Immunohistochemistry
Tissues from third instar larvae (eye imaginal discs, salivary glands, fat body) were dissected in PBS and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. Tissues were incubated in incubation mix (5% goat serum, 0.3% BSA, 0.1% Triton X, in PBS) for 30 min and incubated in anti-M2 antibody (14C2, Santa Cruz), diluted 1:200 in incubation mix, overnight at room temperature. Tissues were washed six times for 10 min each in wash solution (0.5% BSA in PBS) and incubated in goat anti-mouse CY3 antibody (Jackson Immunoresearch), diluted 1:2000 in incubation mix, for 2 hr at room temperature. After washing six times for 10 min each in Wash solution, tissues were mounted in fluorescent mounting media (DakoCytomation) and were imaged with confocal microscopy.
Immunocytochemistry
Cells were rinsed in 1× PBS, fixed in 4% paraformaldehyde in 1× PBS for 20 min, and incubated in incubation mix for 10 min (5% goat serum, 0.3% BSA, 0.1% Triton X, 1xPBS). Cells were incubated in a 1:200 dilution of anti-HA antibody (C102; Santa Cruz) at 37° for 1 hr, washed four times with 1xPBS, and incubated in goat-anti-mouse-CY3 (1:400 dilution, Jackson Immunoresearch) at 37° for 40 min. Cells were washed four times with 1xPBS, with the last wash containing a 1:2000 dilution of Hoechst stain, and mounted in fluorescent mounting media (DakoCytomation). Images were taken with a confocal microscope and quantified with MicroSuite software.
SNARF-1/BCECF analysis
Fat body tissue from larvae of the appropriate genotype, C135-Gal4 (control) or C135-Gal4/UAS-M2 (experimental), was dissected in S2 cell media and transferred into fresh S2 media containing either 10 μM BCECF-AM (Invitrogen B1150) or 10 μM of SNARF-1 (Invitrogen, C1271) (Han and Burgess 2010). Fat body tissues were incubated for 30 min in either solution and then rinsed three times in fresh S2 cell media. The tissues were imaged immediately with a confocal microscope. For SNARF-1-treated samples, we used an argon laser to excite the samples with a 488-nm wavelength and examined the emissions at 580 and 635 nm. The intensity of emission was measured using the Olympus MicroSuite software package and the ratio of 635 nm/580 nm was calculated. For BCECF-AM-treated samples, we used a blue diode laser to excite the samples with a 405-nm wavelength and the argon laser to excite the samples at 488 nm, and then examined the emission at 535 nm from both excitation wavelengths. The ratio of 535 nmex 488/535 nm ex 440 was calculated. Data were analyzed with the t-test statistical package of Excel.
Amantadine treatment
GMR-Gal4:UAS-M2/CyO second instar larvae were placed on our regular fly food supplemented with a combination of water and amantadine hydrochloride (Sigma) to total 0, 10, 20, 30, or 40 μg/ml amantadine hydrochloride. Amantadine was added daily for 7 days. Flies were kept at 29° for the entire period, up to eclosion.
Results
Creation of M2 transgenic flies
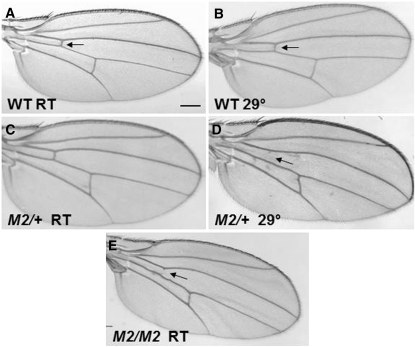
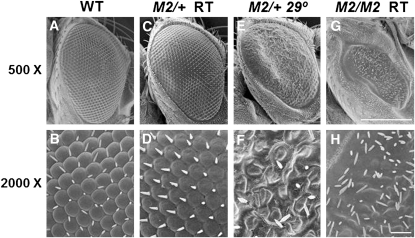
Transgenic flies containing the influenza A virus M2 gene were created as detailed in the Materials and Methods. The expression of M2 was controlled by the UAS element. The UAS element drives transcription of a downstream gene when bound by the protein Gal4. Gal4 expression was controlled by tissue-specific promoter elements. UAS-M2 flies were crossed to both engrailed-Gal4 (en-Gal4) and Glass-mediated response-Gal4 (GMR-Gal4) to express M2 in the wings and eyes, respectively. At room temperature there was little to no alteration of the wings and eyes in en-Gal4/+; UAS-M2/+ and GMR-Gal4/+; UAS-M2/+ flies; however, we found that incubation of these flies at 29° resulted in altered phenotypes (Figures 1 and 2). The nature of this temperature sensitivity remains unclear. As shown in Figure 1D, M2 expression caused wing-vein defects, such as the loss of the anterior cross vein. This phenotype is dose sensitive, as en-Gal4/en-Gal4; UAS-M2/UAS-M2 yielded a phenotype at room temperature (Figure 1E). Figure 2, E and F, shows that at 29° GMR-Gal4/+; UAS-M2/+ flies expressed a rough eye phenotype that was dose sensitive and temperature sensitive. While flies with one copy of UAS-M2 showed little or no mutant phenotype at room temperature (Figure 2, C and D), GMR-Gal4/GMR-Gal4; UAS-M2/UAS-M2 flies did display a striking mutant phenotype at room temperature (Figure 2, G and H). Two copies of UAS-M2 at 29° caused an even more severe eye phenotype, causing a total loss of eye tissue (data not shown). Furthermore, males were found to have more severe eye phenotypes than females (data not shown).
Figure 1 .
M2 expression in wings causes a mutant phenotype. (A) Wild-type wing, room temperature. (B) Wild-type wing, 29°. (C) Wing of en-Gal4/+; UAS-M2/+ reared at room temperature. (D) Wing of en-Gal4/+; UAS-M2/+ reared at 29°. (E) Wing of en-Gal4/en-Gal4; UAS-M2/UAS-M2 reared at room temperature. Arrows refer to anterior cross vein. All images are from female flies. Scale bar, 200 μm.
Figure 2 .
M2 expression in eyes causes a mutant phenotype. (A, C, E, G) at 500× (scale bar, 200 μm), (B, D, F, H) at 2000× (scale bar, 20 μm) magnifications. (A and B) Control eye (GMR-Gal4 only) reared at 29°. (C and D) Eye of GMR-Gal4/+; UAS-M2/+ reared at room temperature. (E and F) Eye of GMR-Gal4/+; UAS-M2/+, reared at 29°. (G and H) Eye of GMR-Gal4/GMR-Gal4; UAS-M2/UAS-M2 reared at room temperature. All images are from female flies.
UAS-M2 expression and M2 proton channel activity in Drosophila tissues
Within influenza virus-infected cells, the M2 protein would be expected to be localized to membrane systems including the plasma membrane and the endoplasmic reticulum/Golgi apparatus. Since the N termini of the M2 proton channels would be present in the lumens of intracellular organelles and vesicles, protons would be transferred out of these compartments and into the cytoplasm, leaving the lumens of the intracellular organelles and vesicles more basic than those of uninfected cells.
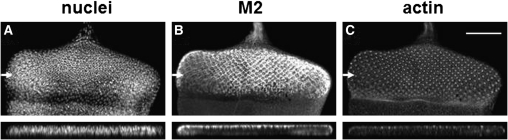
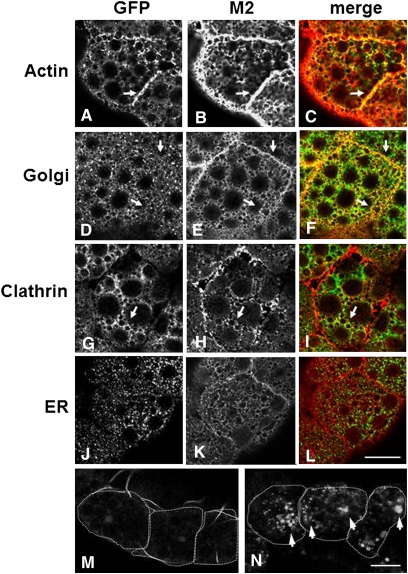
To examine the M2 protein localization within cells of our Drosophila system, we immunostained larval tissues expressing the M2 protein, in different tissues and under different Gal4 drivers, with an anti-M2 antibody. In third instar eye imaginal discs, UAS-M2 was expressed under the GMR-Gal4 driver in cells posterior to the morphogenetic furrow (Figure 3B). The localization of M2 was found to be apical in relation to nuclei, as was actin (Figure 3). We expressed UAS-M2 in the epithelial cells of the larval fat body using the C125-Gal4 driver; in fat body, the M2 protein was targeted to the plasma membrane (Figure 4B). In addition, M2 was found localized to intracellular membrane systems (Figure 4, B, E, H, K). To examine the intracellular localization of the M2 protein in relation to specific intracellular compartments in the fat body, we compared M2 localization to the localizations of GFP-tagged actin, Golgi, clathrin, and endoplasmic reticulum (ER) (Figure 4, A, D, G, J). M2 clearly colocalized with actin, especially at the plasma membrane (Figure 4C). M2 also colocalized with the Golgi, especially at the cell periphery (Figure 4F). Similarly, M2 shared some colocalization with clathrin–GFP (Figure 4I). Clathrin is a vesicle protein coat that is found on vesicles budding from the trans-Golgi and from the plasma membrane. M2 did not appear to colocalize with the ER to a great extent (Figure 4L). M2 localization was also examined in Drosophila salivary glands (supporting information, Figure S1). Interestingly, the localization of M2 in salivary glands was found to be basal instead of apical. This basal membrane localization appeared to colocalize with basal membrane actin (Figure S1). Overall, these experiments demonstrated that the M2 protein was targeted to the plasma membrane and to the membranes of intracellular organelles or vesicles, as would be found in mammalian cells.
Figure 3 .
M2 localization in the Drosophila eye is apical. Eye imaginal disc from a GMR-Gal4/+; UAS-M2/+ third instar larva stained with Hoescht stain for DNA (A), anti-M2 antibody (B), and phalloidin for actin (C). Below each image is a cross-section of each disc (taken at plane noted by arrows). Note that M2 localization is mostly apical. Scale bar, 200 μm.
Figure 4 .
M2 is localized to the plasma membrane and intracellular compartment membranes in Drosophila fat body. Fat body from a C135-Gal4/+; UAS-M2/+ third instar larvae that coexpressed either actin-GFP (A–C), Golgi-GFP (D–F), Clathrin-GFP (G–I), or ER-GFP (J–L). GFP fluorescence is shown in A, D, G, and J. M2 staining is shown in B, E, H, and K. The merge of both GFP and M2 is shown in C, F, I, and L. Arrows refer to areas of colocalization. Wild type (C135-Gal4 alone) (M) and C135-Gal4/+; UAS-M2/+ (N) fat body were incubated in BCECF and imaged as detailed in Materials and Methods. Arrows in N refer to compartments with increased fluorescence and thus higher pH. Individual cells are outlined with dashed lines. Scale bars, 200 μm.
To determine whether the expressed M2 protein functioned as a proper proton channel within Drosophila tissue, we examined the relative pH of intracellular organelles and vesicles, comparing M2-expressing and control tissues. Since the M2 proton channel transports protons from a region of higher proton concentration to a region of lower proton concentration, thus raising the pH in one compartment and lowering it in another, we predicted that the pH in acidic compartments in cells (for example the Golgi and vesicles) expressing M2 would become higher (more basic) than in the wild-type control, as the M2 channels would transport protons from inside the compartments out into the cytosol.
We determined the relative changes in intracellular pH using two vital dyes, SNARF-1 and BCECF, whose fluorescent properties change in response to changes in intracellular pH (Han and Burgess 2010). SNARF-1 is a fluorescent dye that excites with a light wavelength of 488 nm but whose emission changes according to the pH. In more acidic solutions, SNARF-1 fluoresces with an emission peak at around 580 nm, while in more basic solutions the emission peak rises to 640 nm. By determining and comparing the ratios of 640 nm/580 nm emissions between intracellular compartments of wild-type (the C135-Gal4 driver alone) larval fat body and intracellular compartments within fat body tissue expressing M2, we were able to determine changes in relative pH; the higher the ratio number the more basic the pH within the compartments. We observed a significant increase in the 640 nm/580 nm SNARF-1 emission ratios of intracellular compartments in M2-expressing cells, which indicated an increase in the pH within these compartments (Table 1).
Table 1 . M2 increased the pH within intracellular compartments of fat body tissues.
| Genotype | n | Average ratio: 640 nm/580 nm emissiona |
|---|---|---|
| SNARF-1 analysis | ||
| C135 Gal4 | 15 | 0.43 ± 0.11 |
| C135 Gal4; UAS M2 | 15 | 0.70 ± 0.16* |
| Genotype | n | Average ratio: 640 nm/580 nm emissiona |
| Genotype | n | Average ratio of emissions at 535 nm with dual 490 nm/440 nm excitationa |
| BCECF analysis | ||
| C135 Gal4 | 44 | 1.29 ± 0.85 |
| C135 Gal4; UAS M2 | 37 | 2.28 ± 1.83* |
Significant to a P < 0.05 level using the student t-test.
The higher the ratio, the higher the pH within the vesicles examined.
As a complement to these experiments we utilized another vital dye, BCECF, which functions in a slightly different manner. Unlike SNARF-1, BCECF has two excitation wavelengths that are pH dependent, but emits at a single wavelength of 535 nm. Under acidic conditions, the major excitation wavelength is 440 nm while under more basic conditions the excitation wavelength shifts to 490 nm. By comparing the ratios of 535 nm emission from both excitation wavelengths, we examined the changes in pH within intracellular compartments, in wild-type (the C135-Gal4 driver alone) fat body tissue and M2-expressing fat body tissues. Akin to the SNARF-1 results, we observed a significant increase in the 490 nm/440 nm excitation ratios in the compartments within cells expressing M2, which demonstrated that the pH of intracellular compartments had increased relative to the control (Table 1). Figure 4, M and N, shows the difference in BCECF fluorescence intensity between control and M2-expressing cells. When the cells were exposed to an excitation wavelength of 490 nm the control compartments did not fluoresce (as they were still acidic) (Figure 4M) but the M2-expressing compartments did fluoresce (as they had become more basic) (Figure 4N).
Amantadine rescues the M2 phenotype
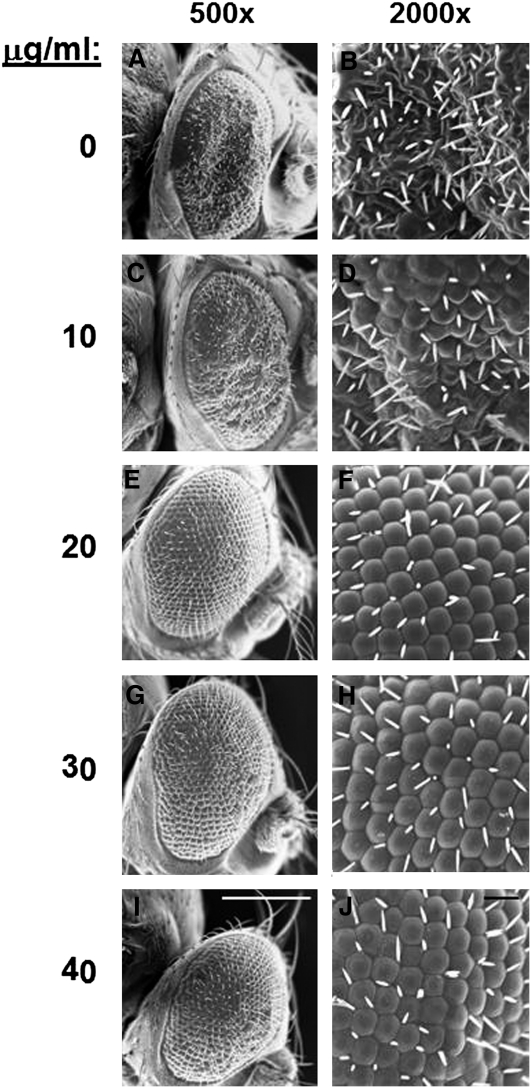
Amantadine is an anti-influenza A virus drug that targets the M2 ion channel. Amantadine plugs the M2 channel and thus prevents proton flow (Wang et al. 1993; Duff et al. 1994). To examine whether amantadine would affect M2 activity in our Drosophila model system, we added amantadine hydrochloride to fly food and fed this to GMR-Gal4:UAS-M2/+ larvae (from second instar through pupation) kept at 29°. Upon eclosion, adults were scored for a rough eye phenotype. Flies with any form of the rough eye phenotype were scored as “rough.” We found that amantadine was able to revert the UAS-M2 rough eye phenotype, both quantitatively and qualitatively, toward a wild-type appearance (Table 2 and Figure 5). This effect was dose dependent and exhibited a phenotypic threshold effect: while the lower dose of 10 μg/ml yielded minimal changes to the phenotype, 20, 30, and 40 μg/ml amantadine restored the phenotype to near wild type (Table 2 and Figure 5). The number of flies with a rough eye phenotype decreased as the amantadine concentration increased, so that with the 40 μg/ml concentration only 9% of the flies had the rough eye phenotype shown in Figure 5; the other 91% of flies resulting from this treatment were wild type in appearance. By demonstrating that an anti-M2 drug works in our fly system, we further validate the study of the influenza virus using our Drosophila model system.
Table 2 . Amantadine reversed the M2 eye phenotype.
| Amantadine (μg/ml) | No. rough/total | % rough |
|---|---|---|
| 0 | 26/26 | 100 |
| 10 | 22/31 | 71* |
| 20 | 13/32 | 41* |
| 30 | 10/28 | 35* |
| 40 | 3/32 | 9* |
Significant to a P < 0.05 level using the student t-test when compared to the control (0 μg/ml).
Figure 5 .
Amantadine reverses the M2 eye phenotype. Second instar larvae of GMR-Gal4/+; UAS-M2/+ were placed on fly food containing varying amounts of amantadine hydrochloride, as shown. The resulting adults were assessed for a rough eye phenotype. The rough eye phenotype for each amantadine concentration is presented. (A, C, E, G, I) Scale bars, 200 μm; (B, D, F, H, J) scale bars, 20 μm.
Candidate screen for modifiers of M2 phenotype
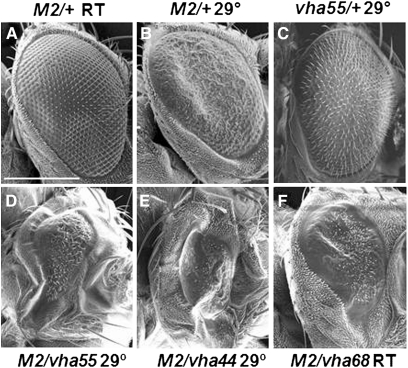
Since M2 transfers protons and alters the pH across various membrane systems within a cell, the function (or dysfunction) of endogenous proton pumps in the cell may affect the efficiency in which M2 alters pH. Therefore we performed a candidate gene screen to identify genetic modifiers of the UAS-M2 phenotype. We initially chose to screen mutations in ion transport/pH homeostasis-related genes as they would most likely be able to affect M2 activity. Even though it is known that loss of the V1V0 ATPase pump affects influenza viral infection, we were interested in dissecting this further, by looking at the individual protein subunits and how knocking down each subunit by 50% would affect M2 activity. To this end, we crossed GMR-Gal4:UAS-M2/+ females to males that harbored deficiencies and mutations in genes that are predicted to encode proteins involved in ion transport and/or pH homeostasis in the cell. In addition to the deletions, which would be null alleles, most of the alleles tested contained P-element insertions within the coding regions (http://Flybase.org; Table 3). Where documented, many alleles cause a reduction or loss of V1V0 ATPase pump activity and are recessive lethal (Davies et al. 1996; Dow 1999; Dow et al. 1997; Allan et al. 2005) (http://FlyBase.org). Crosses were maintained at both 22° and 29°. We then scored the progeny for alterations of the UAS-M2 phenotype. Modifications included slight enhancers, where the eye phenotype was more rough, had areas of black pigmentation, and/or were slightly smaller than UAS-M2/+ eyes. Strong enhancers had very small, narrow eyes that were smooth with no/few ommatidia. Slight suppressors included eyes that had more wild-type-appearing ommatidial structure (less rough). Alleles that modified the UAS-M2 phenotype are listed in Table 3; all alleles tested are listed in Table S1. We found that mutations in the genes whose proteins encode subunits of the vacuolar V1V0 ATPase were consistent modifiers of the UAS-M2 eye phenotype. Notably, all V1 subunits had at least one mutant allele that modified the UAS-M2 phenotype, while 3 of the 11 V0 subunit mutants modified the UAS-M2 phenotype. The strongest modifiers were vha44, vha55, vha68-1, and vha68-2 (Table 3 and Figure 6). These were concluded to be the best modifiers of the UAS-M2 phenotype because most or all alleles tested for each gene strongly enhanced the UAS-M2 phenotype and did so at room temperature for at least one allele. These four genes encode for the V1C, V1B, and V1A subunits, respectively, of the vacuolar V1V0 ATPase.
Figure 6 .
Specific V1V0 ATPase subunit mutants modify the M2 phenotype. (A) GMR-Gal4/+; UAS-M2/+ eye, reared at room temperature. (B) GMR-Gal4/+; UAS-M2/+ eye, reared at 29°. (C) GMR-Gal4/+; vha5516/+, reared at 29°. This control shows the slightly rough phenotype representative of vha55, vha44, and vha68 mutants when Gal4 is also expressed. (D) GMR-Gal4/+; UAS-M2/vha5516 eye, reared at 29°. (E) GMR-Gal4/vha44EY02202; UAS-M2/+ eye, reared at 29°. (F) GMR-Gal4/Df(2L)Exel7055; UAS-M2/+ eye, reared at room temperature. The Df(2L)Exel7055 deficiency removes both vha68-1 and vha68-2. Scale bar, 200 μm.
Alteration of V1V0 ATPase activity alters influenza virus replication in cell culture
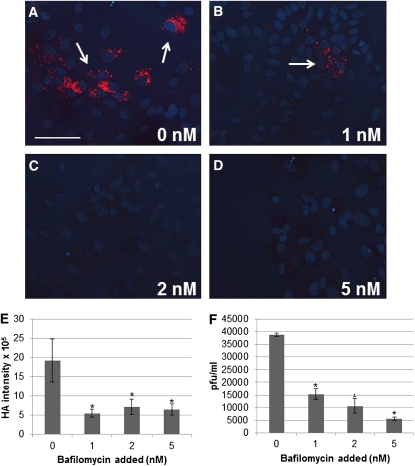
To explore how our genetic screen results may be translated to influenza virus replication in mammalian cells, we performed two sets of experiments. First we examined the effects of loss of the V1V0 ATPase pump upon influenza viral infection. We treated MDCK cells with 0, 1, 2, or 5 nM bafilomycin 24 hr prior to infecting the cells with H1N1 influenza virus. Bafilomycin specifically targets the V1V0 ATPase and inhibits its activity. At 48 hr postinfection we immunostained cells with anti-HA antibody and found that the number of cells becoming infected with influenza virus significantly decreased in the treated cells in comparison to the untreated control cells (Figure 7, A–E). This was confirmed with plaque assays (Figure 7F) and had been previously observed in Perez and Carrasco (1994), Guinea and Carrasco (1995), and Ochiai et al. (1995).
Figure 7 .
Bafilomycin decreases influenza viral infection of MDCK cells. (A–D) Cells were treated with 0, 1, 2, or 5 nM bafilomycin (as indicated) 24 hr prior to infection then immunostained with anti-HA antibody (red) and treated with Hoechst stain for DNA (blue). Arrows refer to infected cells. Magnification 200×; scale bar, 50 μm. (E) Graph showing quantification of the HA staining for cells that had become infected. (F) Graph showing the plaque-forming unit per milliliter determined via plaque assays. Cells were treated and infected as above and plaques counted four days postinfection. Plaque assays were each performed three times. (*) P value ≤ 0.05 when compared to 0 nM.
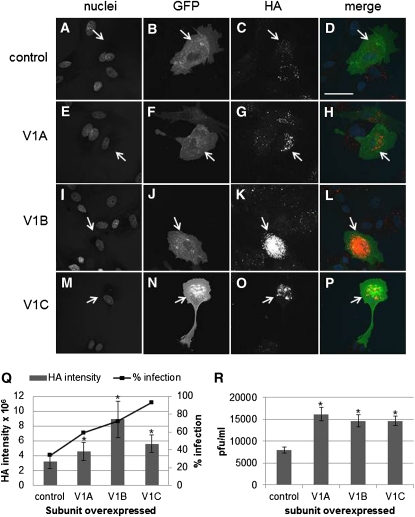
Using a second approach, we overexpressed the specific subunits of the V1V0 ATPase that we identified as significant in our genetic screen within cells and examined subsequent viral infection. MDCK cells were transfected with expression vectors for human V1A, V1B, or V1C, along with a vector expressing membrane-localized GFP to track transfected cells. Cells were infected with H1N1 virus 24 hr later, and after 48 hr underwent immunocytochemistry with an anti-HA antibody to detect viral infection and replication levels. We found that overexpression of any of the subunits produced an increase in the number of infected cells, as well as an increase in replication within cells, as judged by HA levels within cells (Figure 8). Figure 8, A–P shows representative cell staining for each of the overexpressed subunits. The number of cells that became infected with influenza virus subsequent to overexpression of any of the subunits increased significantly, along with the level of HA protein within these cells (Figure 8Q). These results were also corroborated by plaque assays, which showed that overexpression of any of the three subunits lead to greater influenza viral infection (Figure 8R).
Figure 8 .
Overexpression of V1A, V1B, or V1C subunits increases influenza viral infection of MDCK cells. Cells were transfected with plasmids to express GFP alone (A–D), GFP plus V1A (E–H), GFP plus V1B (I–L), or GFP plus V1C (M–P) prior to infection and then immunostained with anti-HA antibody (C, G, K, O; red in merge) and treated with Hoechst stain for DNA (A, E, I, M; blue in merge). GFP is noted in B, F, J, N (green in merge). Magnification 400×; scale bar, 50 μm. (Q) Graph showing the quantification of HA staining. Line graph shows the percentage of transfected cells that had become infected; columns show the intensity of HA staining within transfected cells that had become infected. (R) Graph showing the plaque-forming unit per milliliter determined via plaque assays. Cells were transfected with the subunit plasmids as above, infected, and plaques counted 4 days postinfection. Plaques assays were performed three times. (*) P value ≤ 0.05 when compared to control.
Discussion
The influenza virus has been the root cause for a number of deadly pandemics throughout history. We have developed a Drosophila model to study the genetics of influenza viral/host interactions. Using the UAS/Gal4 system we expressed the influenza A virus M2 gene in a variety of Drosophila tissues and found that M2 maintained both its proper subcellular localization to cellular membranes and function, as a pH modulator. We established that the localization of M2 within the eye tissue was apical (see Figure 3), which mirrors the localization of M2 within human cells of the respiratory tract (Hughey et al. 1992). Furthermore, in all fly tissues tested, M2 also colocalized with actin at the plasma membrane. Similar observations have been noted in mammalian cells where influenza previral particles (i.e., genomes and envelope proteins) congregate at the plasma membrane, aided by actin microfilaments, for viral particle assembly and budding (Simpson-Holley et al. 2002; Nayak et al. 2009).
Expression of M2 within specific Drosophila tissues via the bipartite UAS/Gal4 system resulted in specific mutant phenotypes in adult flies. M2 expression in wings via the engrailed-Gal4 (en) driver led to a wing phenotype dominated by the loss of the anterior cross veins; M2 expression in eyes via the Glass-mediated-response-Gal4 (GMR) driver caused the eyes to be smaller and rough. These phenotypes were both temperature sensitive and dose dependent and served as powerful tools for the investigation of the interactions between M2 and host cellular genes.
The localization and function of M2 in Drosophila cells remains unchanged from its activity and localization in mammalian cells. In addition, we demonstrated that the antiviral drug amantadine suppressed the UAS-M2 mutant phenotype. The results that we have obtained with amantadine suggest that our M2 flu fly system will also be amenable to performing small molecule drug screens to find inhibitors of M2 activity.
We utilized the dose- and temperature-dependent UAS-M2 eye phenotype to screen for second-site modifiers of the UAS-M2 phenotype. From collection of 57 candidate genes that encoded components of the V1V0 ATPase or proteins predicted to be involved in the regulation intracellular pH homeostasis, we demonstrated that mutations in the genes encoding for the V1V0 ATPase subunits, especially vha68, vha55, and vha44, yielded strong modifications of the UAS-M2 eye phenotype (see Table 3). It is interesting to note that not all V1V0 ATPase subunits tested led to modifications of the UAS-M2 phenotype, suggesting that the strong UAS-M2 modifiers (vha68, vha55, and vha44) play key roles in the regulation and/or assembly of the V1V0 ATPase enzyme complex, in regard to M2 activity, and therefore may serve as especially interesting and specific targets for antiviral drug design.
The V1V0 ATPase is a proton pump that uses ATP to drive transport of protons against the concentration gradient. This pump regulates pH in a variety of intracellular compartments (Jefferies et al. 2008). For example, in late endosomes, the V1V0 ATPase increases the pH within this compartment to facilitate the hydrolysis of endocytosed molecules (Van Dyke 1996). vha68, vha55, and vha44 encode the Drosophila homologs for the mammalian V1A, V1B, and V1C, subunits, respectively. The subunits V1A and V1B are involved in ATP binding and hydrolysis. When ATP is hydrolyzed, conformational changes in V1A drive rotation of the central stalk. This activity drives proton transport. The V1C subunit is located between the V1 and V0 domains and can dissociate from the rest of the pump. Since disassembly of the pump regulates pump activity, C appears to play a regulatory role (Jefferies et al. 2008). Reassembly of the pump requires the joining of the V1 domain, the V0 domain, and subunit V1C. This assembly has been shown to be regulated by glucose concentrations and by the Ras/cAMP/PKA pathway (Bond and Forgac 2008; Jefferies et al. 2008).
Cellular regulation of pH plays at least two critical roles in influenza viral infection. When the influenza virus is imported into the cell within an endocytic vesicle, V1V0 ATPase pumps are situated in the vesicle membrane such that they pump protons inside the vesicle (Van Dyke 1996). This increased pH allows for M2 activation, and M2 transports protons into the interior of the virion (Palese and Shaw 2007). Inhibition of the V1V0 ATPase pump in the late endosome, preventing acidification, would likely inhibit M2 activation and thus inhibit viral uncoating. In fact this has been shown to be true (Perez and Carrasco 1994; Guinea and Carrasco 1995; Ochiai et al. 1995). On the other hand, overexpression of the V1V0 ATPase pump at this stage of influenza viral infection might alter the V1V0 ATPase assembly and perhaps increase the ability of M2 to acidify the interior of the virion to cause increased viral uncoating and replication. Our findings support these hypotheses (Figures 7 and 8). Other work lends supports to our data. In an RNAi screen in an influenza virus-infectable Drosophila S2 cell line, two subunits of the V1V0 ATPase (Drosophila vhaAC39 and vha16, which are homologous to V0D1 and V0C, respectively) were shown to be essential for influenza viral replication (Hao et al. 2008). Curiously no other V1V0 ATPase subunits were identified in this screen, suggesting that either the genes were essential for cell viability (and thus their knockdown resulted in cell death) or that the RNAi system failed to knock down expression low enough to produce an influenza virus replication defect. Given the nonepithelial nature of S2 cells, it is possible that other mechanisms, such as the organization of the cytoskeleton or apical/basal polarity, differentiate requirements for influenza viral replication in a nonepithelial vs. epithelial cell types. More recently, an RNAi screen in human lung epithelial cells identified several human genes necessary for influenza viral replication (Konig et al. 2010). In relation to our work, the V1V0 ATPase subunits V1A, V1B, V0B, and V0C were identified as essential (Konig et al. 2010). It is interesting to note that two of the four subunits that Konig et al. found to be important correspond to two of the three subunits that we determined to be important for M2 activity and influenza virus replication.
In addition to the obvious utility of identifying novel genetic modifiers of M2 proton channel activity for the study of viral and host cell interactions, we believe that our M2 flu fly system can play an important role in the identification of two types of new anti-influenza virus compounds: (1) new inhibitors of the M2 proton pump, and (2) compounds that modulate endogenous V1V0 ATPase activity (or other M2-activity essential mechanisms). As we have shown with amantadine, compounds can be added to fly food, fed to M2-expressing larvae (perhaps using a lethal expression system), and the compounds that had been fed to flies that survived then tested for their abilities to inhibit M2 activity and viral replication in mammalian cell culture.
The modifiers that we have identified (Table 3) are excellent drug targets themselves. Although this idea may seem counterintuitive as most of the V1V0 ATPase subunits are essential genes with recessive lethality, dose-sensitive modifiers are excellent drug targets a priori. Such modifiers are genetically dominant. Thus UAS-M2 modifiers express significant phenotypic effects on the activity of an essential viral gene and therefore affect the viral replication program indirectly and most importantly, not by complete loss of function but through a partial loss of function. Therefore drugs targeting proteins encoded by dose-sensitive modifiers would need only to temporarily/partially effect enzyme function to potentially exert a profound effect on viral infection/replication. In the case of influenza virus, a compound that bound to the V1V0 ATPase subunit V1C and decreased the ability of the V1C subunits to assemble a complete and functional V1V0 ATPase complex may result in a significant reduction of viral infection/replication and therefore slow the progress of the overall viral infection.
In summary, we have established a transgenic model system to study influenza virus M2 protein activity. We have shown that the Drosophila system is an excellent heterologous system with which to study a viral protein that alters intracellular physiology. We have shown that the M2 protein functions and localizes properly within Drosophila cells and we have used this system to identify important host genes that affect the ability of M2 to alter intracellular pH.
The M2 flu fly has proven to be an efficient means of identifying cellular genes that affect influenza viral replication. This model system can also be used to develop and test novel drugs for antiviral therapy. Our results suggest that the Drosophila flu fly system would also benefit by expansion to include other influenza virus genes, with the goal of identifying additional cellular proteins that might become antiviral drug targets.
Acknowledgments
We thank Robert A. Lamb for the M2 cDNA. This work was funded in part by a University of North Carolina—Greensboro Faculty Grant to A.L.A. We acknowledge the Bloomington Stock Center for our fly stocks and the website FlyBase for information on Drosophila genes and stocks.
Literature Cited
- Adamson A. L., Wright N., LaJeunesse D. R., 2005. Modeling early Epstein–Barr virus infection in Drosophila melanogaster: the BZLF1 protein. Genetics 171: 1125–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan A. K., Du J., Davies S. A., Dow J. A., 2005. Genome-wide survey of V-ATPase genes in Drosophila reveals a conserved renal phenotype for lethal alleles. Physiol. Genomics 22: 128–138 [DOI] [PubMed] [Google Scholar]
- Bond S., Forgac M., 2008. The Ras/cAMP/protein kinase A pathway regulates glucose-dependent assembly of the vacuolar (H+)-ATPase in yeast. J. Biol. Chem. 283: 36513–36521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella J. A., Bayersdorfer F., Gmeiner F., Schneuwly S., 2009. Modelling Parkinson's disease in Drosophila. Neuromol. Med. 11: 268–280 [DOI] [PubMed] [Google Scholar]
- Chan H., Bonini N., 2000. Drosophila models of human neurodegenerative disease. Cell Death Differ. 7: 1075–1080 [DOI] [PubMed] [Google Scholar]
- Davies S. A., Goodwin S. F., Kelly D. C., Wang Z., Sozen M. A., et al. , 1996. Analysis and inactivation of vha55, the gene encoding the vacuolar ATPase B-subunit in Drosophila melanogaster reveals a larval lethal phenotype. J. Biol. Chem. 271: 30677–30684 [DOI] [PubMed] [Google Scholar]
- Dow J. A., 1999. The multifunctional Drosophila melanogaster V-ATPase is encoded by a multigene family. J. Bioenerg. Biomembr. 31: 75–83 [DOI] [PubMed] [Google Scholar]
- Dow J. A., Davies S. A., Guo Y., Graham S., Finbow M. E., et al. , 1997. Molecular genetic analysis of V-ATPase function in Drosophila melanogaster. J. Exp. Biol. 200: 237–245 [DOI] [PubMed] [Google Scholar]
- Duff K. C., Gilchrist P. J., Saxena A. M., Bradshaw J. P., 1994. Neutron diffraction reveals the site of amantadine blockade in the influenza A M2 ion channel. Virology 202: 287–293 [DOI] [PubMed] [Google Scholar]
- Fortini M. E., Bonini N. M., 2000. Modeling human neurodegenerative diseases in Drosophila: on a wing and a prayer. Trends Genet. 16: 161–167 [DOI] [PubMed] [Google Scholar]
- Guinea R., Carrasco L., 1995. Requirement for vacuolar proton-ATPase activity during entry of influenza virus into cells. J. Virol. 69: 2306–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafen E., 2004. Cancer, type 2 diabetes, and ageing: news from flies and worms. Swiss Med. Wkly. 134: 711–719 [DOI] [PubMed] [Google Scholar]
- Han J., Burgess K., 2010. Fluorescent indicators for intracellular pH. Chem. Rev. 110: 2709–2728 [DOI] [PubMed] [Google Scholar]
- Hao L., Sakurai A., Watanabe T., Sorensen E., Nidom C. A., et al. , 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454: 890–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F., 2010. Drosophila melanogaster in the study of human neurodegeneration. CNS Neurol. Disord. Drug Targets 9: 504–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey P. G., Compans R. W., Zebedee S. L., Lamb R. A., 1992. Expression of the influenza A virus M2 protein is restricted to apical surfaces of polarized epithelial cells. J. Virol. 66: 5542–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima-Ando K., Iijima K., 2010. Transgenic Drosophila models of Alzheimer's disease and tauopathies. Brain Struct. Funct. 214: 245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies K. C., Cipriano D. J., Forgac M., 2008. Function, structure, and regulation of the vacuolar (H+)-ATPases. Arch. Biochem. Biophys. 476: 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R., Stertz S., Zhou Y., Inoue A., Hoffmann H. H., et al. , 2010. Human host factors required for influenza virus replication. Nature 463: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y., Zhang Y., Dong L., Wang D., Huang W., et al. , 2010. A comprehensive surveillance of adamantane resistance among human influenza A virus isolated from mainland China between 1956 and 2009. Antivir. Ther. 15: 853–859 [DOI] [PubMed] [Google Scholar]
- Lasko P., 2002. Diabetic flies?: using Drosophila melanogaster to understand the causes of monogenic and genetically complex diseases. Clin. Genet. 62: 358–367 [DOI] [PubMed] [Google Scholar]
- Lloyd T. E., Taylor J. P., 2010. Flightless flies: Drosophila models of neuromuscular disease. Ann. N. Y. Acad. Sci. 1184: e1–e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Vogel H., 2009. Drosophila models of neurodegenerative diseases. Annu. Rev. Pathol. 4: 315–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak D., Balogun R., Yamada H., Zhou Z., Barman S., 2009. Influenza virus morphogenesis and budding. Virus Res. 143: 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H., Sakai S., Hirabayashi T., Shimizu Y., Terasawa K., 1995. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antiviral Res. 27: 425–430 [DOI] [PubMed] [Google Scholar]
- Palese P., Shaw M. L., 2007. Orthomyxoviridae: the viruses and their replication, pp. 1647–1689 Fields Virology, edited by Knipe D. M., Howley P. M. Lippincott, Williams, & Wilkins, Philadelphia, PA [Google Scholar]
- Perez L., Carrasco L., 1994. Involvement of the vacuolar H(+)-ATPase in animal virus entry. J. Gen. Virol. 75: 2595–2606 [DOI] [PubMed] [Google Scholar]
- Pinto L. H., Lamb R. A., 2006. The M2 proton channels of Influenza A and B viruses. J. Biol. Chem. 281: 8997–9000 [DOI] [PubMed] [Google Scholar]
- Rea S., Graham B., Nakamaru-Ogiso E., Kar A., Falk M., 2010. Bacteria, yeast, worms,and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev. Disabil. Res. Rev. 16: 200–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon-Limas D., Casas-Tinto S., Fernandez-Funez P., 2010. Exploring prion protein biology in flies: genetics and beyond. Prion 4: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Holley M., Ellis D., Fisher D., Elton D., McCauley J., et al. , 2002. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 301: 212–225 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M., 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347 [DOI] [PubMed] [Google Scholar]
- Spresser C. R., Carlson K. A., 2005. Drosophila melanogaster as a complementary system for studying HIV-1-related genes and proteins. J. Neurosci. Res. 80: 451–455 [DOI] [PubMed] [Google Scholar]
- Steinberg R., Shemer-Avni Y., Adler N., Neuman-Silberberg S., 2008. Human cytomegalovirus immediate-early-gene expression disrupts embryogenesis in transgenic Drosophila. Transgenic Res. 17: 105–119 [DOI] [PubMed] [Google Scholar]
- Van Dyke R. W., 1996. Acidification of lysosomes and endosomes. Subcell. Biochem. 27: 331–360 [DOI] [PubMed] [Google Scholar]
- Vidal M., Cagan R. L., 2006. Drosophila models for cancer research. Curr. Opin. Genet. Dev. 16: 10–16 [DOI] [PubMed] [Google Scholar]
- Wang C., Takeuchi K., Pinto L. H., Lamb R. A., 1993. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 67: 5585–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P. F., Neumann G., Kawaoka Y., 2007. Orthomyxoviruses, pp. 1691–1740 Fields Virology, edited by Knipe D. M., Howley P. M. Lippincott, Williams, & Wilkins, Philadelphia, PA [Google Scholar]
- Zebedee S. L., Richardson C. D., Lamb R. A., 1985. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J. Virol. 56: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]