Abstract
In the nematode Caenorhabditis elegans, temperature-sensitive mutants of emb-1 arrest as one-cell embryos in metaphase of meiosis I in a manner that is indistinguishable from embryos that have been depleted of known subunits of the anaphase-promoting complex or cyclosome (APC/C). Here we show that the emb-1 phenotype is enhanced in double mutant combinations with known APC/C subunits and suppressed in double mutant combinations with known APC/C suppressors. In addition to its meiotic function, emb-1 is required for mitotic proliferation of the germline. These studies reveal that emb-1 encodes K10D2.4, a homolog of the small, recently discovered APC/C subunit, APC16.
UNIDIRECTIONAL progression through the cell cycle is driven by a precisely regulated combination of protein phosphorylation and protein degradation events. The anaphase-promoting complex or cyclosome (APC/C) is a key promoter of cell cycle progression. This large, multisubunit E3 ubiquitin ligase drives both the metaphase-to-anaphase transition and M-phase exit by polyubiquitylating its various substrates and thus targeting them for proteasome-mediated degradation (for reviews, Pesin and Orr-Weaver 2008; Simpson-Lavy et al. 2010). The various mitotic and meiotic substrates of the APC/C include both securin, whose destruction releases separase to cleave the cohesin complex between sister chromatids, and cyclin B, whose destruction allows M-phase exit. Depletion or repression of APC/C activity prevents the first step of securin destruction and results in a metaphase arrest.
Like all aspects of the cell cycle, the activity of APC/C is precisely regulated. To be active and facilitate substrate recognition, the APC/C must be bound to one of its WD (tryptophan-aspartate) repeat-containing activators (for review, Pesin and Orr-Weaver 2008). In most mitotic cells, complex biochemical regulatory loops ensure that its two major activators function sequentially; Cdc20/Fizzy drives the metaphase-to-anaphase transition, while the tumor suppressor Cdh1 not only drives mitotic exit but also represses subsequent cell cycle entry and promotes somatic cell differentiation (reviewed in Peters 2006; Thornton and Toczyski 2006; Wasch et al. 2010). Cdc20 alone can mediate the destruction of critical mitotic substrates in yeast (Schwab et al. 1997; Visintin et al. 1997), human cell cultures (Qi and Yu 2007), and embryonic cells that lack a G1 phase (Lorca et al. 1998; Zhou et al. 2002; Li et al. 2007). During meiosis, APC/C activity is also regulated by meiotic-specific activators such as Ama1 in Saccharomyces cerevisiae (Cooper et al. 2000; Diamond et al. 2009) and Fzr1/Mfr1 in Schizosaccharomyces pombe (Asakawa et al. 2001; Blanco et al. 2001). Up until metaphase I, these meiotic-specific activators work in conjunction with Cdc20; subsequently they assume nonredundant roles in various meiotic-specific processes such as sporulation. Since precocious APC/C activity results in aberrant chromosome segregation, APC/C activity is negatively regulated by the spindle assembly checkpoint (SAC). The SAC blocks the metaphase-to-anaphase transition unless all of the kinetochores are both attached to microtubules and under tension from bipolar spindle forces (reviewed in Zich and Hardwick 2010). Single-particle electron microscopy indicates that some SAC components function by binding to the Cdc20-bound APC/C complex and locking the otherwise flexible APC/C in a “closed” state (Herzog et al. 2009).
Our initial understanding of the APC/C as a multisubunit complex arose from a combination of genetic studies in budding yeast and biochemical studies in clam and Xenopus eggs that sought to characterize the enzyme responsible for ubiquitylating cyclin B. In yeast, APC/C mutants were isolated as G2/M cell-cycle arrest mutants, while biochemical studies found that the enzyme activity was associated within a multisubunit 20S particle (Hershko et al. 1994; King et al. 1995; Sudakin et al. 1995). Individual subunits of the APC/C were then isolated either directly or in conjunction with known subunits using epitope tagging and immunopurification technology (for review, Zachariae and Nasmyth 1999; Zachariae et al. 1998). More recently, computationally intensive proteomics in tandem with new methods for identifying in vivo protein interactions (Hubner et al. 2010; Hutchins et al. 2010; Kops et al. 2010; Ohta et al. 2010) and studying gene networks (Green et al. 2011) have identified additional, small molecular weight subunits that had been missed using standard biochemical and genetic approaches (Table 1).
Table 1 . List of APC/C subunits in a variety of species.
| S. cerevisiae | S. pombe | D. melanogaster | C. elegans | Mammals |
|---|---|---|---|---|
| Apc1 | Cut4 | Shattered | MAT-2 | Apc1 |
| Apc2/Rsi1 | Apc2 | Morula | APC-2 | Apc2 |
| Cdc27/Apc3 | Nuc2 | Makos | MAT-1 | Cdc27 |
| Apc4 | Lid1 | Apc4 | EMB-30 | Apc4 |
| Apc5 | Apc5 | Ida | SUCH-1 and GFI-3 | Apc5 |
| Apc6/Cdc16 | Cut9 | Cdc16 | EMB-27 | Apc6 |
| NI | NI | Apc7 | NI | Apc7 |
| Cdc23/Apc8 | Cut23 | Cdc23 | MAT-3 | Apc8 |
| Apc9 | NI | NI | NI | NI |
| Doc1/Apc10 | Apc10 | CG11419 | APC-10 and Y48G1C.12 | Apc10 |
| Apc11 | Apc11 | Lemming | APC-11 | Apc11 |
| Cdc26 | Hcn1 | NI | MAT-4 | Cdc26 |
| Swm1/Apc13 | Apc13 | Apc13 | F59E12.11a | Apc13 |
| NI | Apc14 | NI | NI | NI |
| Mnd2/Apc15 | Apc15 | NI | NI | NI |
| NI | NI | NI | K10D2.4 (EMB-1)b | Apc16/c10orf104c |
| NI | NI | NI | C09H10.7 (APC-17)d | NI |
| 13 | 13 | 11 | 15 | 13 |
NI, not identified to date. Numbers in boldface type are totals.
(W. Zachariae, personal communication; Green et al. 2011).
(This study; Green et al. 2011; Kops et al. 2010).
Although the exact number of APC/C subunits appears to vary among different species (Table 1), the APC/C of both vertebrates and budding yeast have ≥13 subunits (Zachariae et al. 1996, 1998; Yoon et al. 2002). However, recent structural (Dube et al. 2005; Passmore et al. 2005; Ohi et al. 2007; Herzog et al. 2009) and evolutionary (Seidl and Schultz 2009) studies suggest that 9 of these function as core subunits within three subcomplexes: the catalytic arm (APC2, APC11, and APC10/Doc1), the structural part (APC1, APC4, and APC5), and the activator and substrate binding tetratricopeptide repeat (TPR) arm (APC8, APC6, and APC3).
The Caenorhabditis elegans genome contains 15 identifiable APC/C orthologs and has the unusual distinction of having two Apc5-like subunits and two Apc10-like subunits (Table 1). Five of these subunits (emb-27, emb-30, mat-1, mat-2, and mat-3) are represented by temperature-sensitive (ts) mutants whose embryos hatch and develop at 15°, but arrest as meiotic one-cell embryos at the nonpermissive temperature of 25° (Furuta et al. 2000; Golden et al. 2000; Davis et al. 2002; Shakes et al. 2003). Like wild-type embryos, these mutant APC/C embryos are fertilized just after the breakdown of the oocyte’s nuclear envelope. Unlike wild-type embryos, they arrest with their oocyte chromosomes locked in metaphase of the first meiotic division. In addition, spindle positioning fails and subsequent events in both the meiotic cell cycle and the oocyte-to-embryo transition fail to occur (Golden et al. 2000; Davis et al. 2002; Yang et al. 2003; Stitzel et al. 2007). Identical meiotic arrest phenotypes have been generated through RNAi mediated depletion of any of the following: (a) any 1 of 11 different APC/C subunits (Davis et al. 2002; Dong et al. 2007; Kops et al. 2010; Green et al. 2011), (b) the CDC20 ortholog, fzy-1, or (c) any one of several proteasome subunits or the two ubiquitin genes, ubq-1 and ubq-2 (Sonnichsen et al. 2005). To date, the less severe RNAi depletion phenotypes of the Apc10-like subunits remain poorly characterized, and those of the two Apc5-like subunits (SUCH-1 and GFI-3) suggest that they function redundantly during meiosis (Stein et al. 2010).
Yet despite the many new insights that these various biochemical, genetic, and structural studies are revealing in regard to the mechanistic details of the APC/C, both the composition and regulation of the APC/C remain incompletely understood, particularly as APC/C studies are extended into additional species and cell types. Here we present genetic evidence that emb-1 functions as an essential component of the APC/C. When emb-1 mutants were originally isolated in a genetic screen for ts, maternal effect lethal mutants, the emb-1 phenotype was described as having a one-cell arrest phenotype (“fertilized eggs . . . do not divide”) (Schierenberg et al. 1980), a phenotype that we more specifically characterized as a metaphase I arrest (Golden et al. 2000). In this study, we show that emb-1 behaves both genetically and phenotypically like other APC/C mutants, both alone and in combination with other APC/C mutants. It is also weakly suppressed by mutations that are known to suppress certain APC/C alleles. Our studies reveal that emb-1 encodes K10D2.4, a gene that was recently identified by others as exhibiting an APC/C-like phenotype in a large scale RNAi survey of C. elegans genes with sterility defects (Green et al. 2011). The human ortholog of this gene was also found to co-immunoprecipate in association with SAC–APC/C complexes (Kops et al. 2010). Thus, our studies provide the complementary genetic evidence that K10D2.4 is functioning as a subunit of the APC/C and reveal emb-1(hc62ts) as the first temperature-sensitive mutant in this small, 81-amino-acid subunit of the APC/C.
Materials and Methods
Genetics
Strains:
The wild type was the Bristol strain N2. All strains were cultured using standard techniques (Brenner 1974). Temperature-sensitive strains were maintained at 15°; all other strains were maintained at 20°. Other strains used are listed as follows: MJ57: emb-1(hc57ts)III, MJ62: emb-1(hc62ts)III, MT2330: dpy-17(e164) lon-1(e1820)III, CB270: unc-42(e270)V, CB933: unc-17(e245)IV, CB450: unc-13(e450)I, AG164: mdf-1(av19) unc-42(e270)V, AG166: mdf-2(av16) unc-17(e245)IV, AG165: san-1(av31) unc-13(e450)I, BC4697: sDf121(s2098) unc-32(e189)III; sDp3 (III;f), AG178: emb-27(ax81ts) unc-4(e120)II, AG179: rol-6 (e187) mat-2 (or170ts)II, AG180: dpy-10 (e128) mat-2 (ax102ts)II, AG181: mat-3 (or180ts) dpy-1 (e1)III, AG205: mat-1(ax144ts) dpy-5(e61)I, AG206: mat-1(ax227ts) dpy-5(e61)I, AG207: mat-1(ax212ts)I; him-8(e1489)IV, AG182: fzy-1(h1983) dpy-10(e128)II, DS96: emb-1(hc62ts) unc-32(e189)III, DS129: emb-1(hc62ts) lon-1(e185)III, AG168: fzy-1(av15gf) unc-4(e120)II, CB120: unc-4(e120)II, DS77: mat-1(ax161ts)I; him-8(e1489)IV, DG627: emb-30(tn377ts)III, DS154: emb-1(hc62ts); ojIs1[pJH4.66: unc-119(+) + pie-1::gfp::tbb-2], AG200: cid-1(tm936)III, AG201: cid-1(tm1021)III, AG202: ok2757/dpy-17(e164) lon-1(e1820)III, AG203: ok2757/hT2 (I;III); him-8(e1489)IV, AG204: ok2759/dpy-17(e164) lon-1(e1820)III, AG218: apc-11(gk37)/unc-93(e1500) dpy-17(e164).
Complementation test with sDf121:
emb-1(hc62ts); him-8(e1489) males were crossed into Unc sDf121(s2098) unc-32(e189); sDp3 hermaphrodites and non-Unc progeny were shifted to either 15°, 20°, or 24°. The free duplication (sDp3) chromosome balances the deficiency (sDf121) but not unc-32. Non-Unc cross progeny were either emb-1(hc62ts)/sDf121unc-32 with or without sDp3. Hemizygotes that produced viable progeny at 24° also segregated Unc animals, thus confirming the presence of sDp3.
Three-factor mapping of emb-1:
Three-factor mapping was carried out with emb-1(hc62ts) unc-32(e189)/lon-1(e185) animals. Of 30 Unc non-Emb recombinant animals isolated, 26 were marked with lon-1, suggesting that lon-1 is close to, but to the right of, emb-1. Three-factor mapping was carried out with emb-1(hc62ts)/dpy-17(e164) lon-1(e1820). Since the dpy-17 mutation is epistatic to lon-1 (dpy-17lon-1 animals are Dpy), only Lon non-Dpy recombinants can be picked. Lon non-Dpy recombinant animals were isolated and animals homozygous for the lon-1 chromosome were assayed for the presence of emb-1 by shifting L4 animals to 24°. Of 61 Lon non-Dpy recombinants isolated, 50 contained emb-1, putting emb-1 at a genetic map position of approximately −2.06 on linkage group III (based on WormBase Release WS207; www.wormbase.org). Together, these mapping results suggest that emb-1 is between dpy-17 and lon-1, two genes that have been cloned and are 0.5 map units apart.
Genetic enhancement by mat and fzy mutants (all performed at 15°):
mat-1(ax212ts); him-5(e1490) males were crossed into emb-1(hc62ts) unc-32 (e189) hermaphrodites and non-Unc F1 were picked. Sixteen Unc F2 L4s were isolated and scored for fertility and viability of their progeny. mat-1(ax144ts or ax227ts) dpy-5(e61) animals were crossed with him-8(e1489) males. F1 cross males were then crossed into emb-1(hc62ts) unc-32(e189) animals and non-Unc cross progeny were picked. From them, DpyUnc progeny were isolated (as L4s) and scored for fertility and viability of their progeny.
emb-27(ax81ts) unc-4(e120)/+ + males were crossed with emb-1(hc62ts) lon-1(e1820) hermaphrodites. rol-6(e187) mat-2(or170ts)/++ males were crossed with emb-1(hc62ts) unc-32 (e189). emb-1(hc62ts) lon-1(e1820)/+ + males were crossed with mat-2(ax102ts) dpy-10(e128) or fzy-1(h1983) dpy-10(e128) hermaphrodites. Cross progeny from the above crosses were picked as non-Lon, non-Unc, or non-Dpy, and their doubly marked progeny were picked to 15° to score for fertility and embryonic viability.
emb-1(hc62ts) unc-32 (e189) hermaphrodites were crossed with emb-1(hc62ts); him-8(e1489) males and progeny males (emb-1unc-32/emb-1 +) were crossed into dpy-10(e128) mat-2(ax102ts), rol-6(e187) mat-2(or170ts), and fzy-1(h1983) dpy-10(e128) hermaphrodites. Cross progeny were picked and from those that segregated DpyUncs or RolUncs; their doubly marked progeny were isolated and scored at 15°. A similar approach was taken starting with emb-1(hc62ts) lon-1(e1820) hermaphrodites crossed with emb-1(hc62ts); him-8(e1489) males. The subsequent males were mated into emb-27(ax81ts) unc-4(e120) hermaphrodites.
To make the mat-3(or180) emb-1(hc62ts) double mutant, emb-1(hc62ts) unc-32(e189)/emb-1(hc62ts) +; him-8(e1489)/+ males were mated into mat-3(or180ts) dpy-1(e1) hermaphrodites. Non-Dpy progeny [mat-3dpy-1/emb-1unc-32 (or ++)] were picked to separate plates. From those plates segregating Uncs, 10 Uncs were picked to a new plate and screened for the presence of DpyUncs that would be a result of recombination. DpyUnc animals were maintained at 15° and scored for phenotypes.
Genetic suppression by mdf and fzy mutants:
To test for suppression effects, emb-1(hc62ts) lon-1(e1820)/emb-1(hc62ts) +; him-8(e1489)/+ males were crossed with any one of the following hermaphrodites: mdf-1(av19) unc-42(e270), mdf-2(av16) unc-17(e245), mdf-3(av31) unc-13(e450), fzy-1(av15gf) unc-4(e120), unc-42(e270), unc-17(e245), unc-13(e450), or unc-4(e120). Doubly marked L4 larvae from the F2 generation were shifted to 24° and their embryos were scored for hatching.
Deletion alleles:
The deletion alleles ok2757 and ok2759 were obtained from the C. elegans Reverse Genetics Core Facility in Vancouver via the Caenorhabditis Genetics Center as balanced heterozygous lines, VC2189: ok2757/hT2 [bli-4(e937) let-?(q782) qIs48 (an insertion of ccEx9747 which carries myo-2::gfp + pes-10::gfp + gut enhancer::gfp)] (I; III) and VC2216: ok2759/hT2 [bli-4(e937) let-?(q782) qIs48]. These animals were crossed with N2 males and non-GFP (ok2757/+ or ok2759/+) males were crossed with dpy-17lon-1 hermaphrodites. The males from this cross (Δ/dpy-17lon-1 or +/dpy-17lon-1) were singly mated into dpy-17lon-1 hermaphrodites. After 2 days of mating, the males were subjected to single-animal PCR to determine which carried the deletion. From plates in which deletion males had been confirmed by PCR, 6–10 non-Dpy males were picked and crossed with 3–4 dpy-17lon-1 hermaphrodites. From such crosses, non-Dpy males were picked again and mated with dpy-17lon-1 hermaphrodites until 10 outcrosses were completed. The outcrossed strains were maintained as Δ/dpy-17lon-1 since ok2757 and ok2759 homozygotes are not viable. Sequencing the breakpoints of the ok2757 allele revealed that this deletion removes both exons of emb-1 and the first exon of the downstream cid-1 gene. Sequencing the breakpoints of the ok2759 allele revealed that this deletion removes both exons of emb-1 and most of the 3′-UTR of the upstream K10D2.5 gene. Both deletion alleles should thus be considered small deficiency chromosomes.
Two cid-1 deletion alleles were obtained from the National BioResource Project (NBRP) at the Tokyo Women’s Medical University School of Medicine in Japan, tm936 and tm1021. Both strains were homozygous and viable. Both alleles were outcrossed 10 times by the following protocol. dpy-17lon-1/++ males were crossed with each deletion allele homozygote. Cross males (∆/+ or ∆/dpy-17lon-1) were singly mated to dpy-17lon-1 hermaphrodites. From plates segregating F1 Dpy males, 6–10 non-Dpy males (∆/dpy-17lon-1) were mated to 3–4 dpy-17lon-1 hermaphrodites until 10 outcrosses were completed. Outcrossed animals were maintained as homozygotes and confirmed by PCR analysis.
emb-1(hc62ts) and ok2757 complementation tests: emb-1(hc62ts) lon-1(e1820) hermaphrodites were mated with emb-1(hc62ts); him-8(e1489) males. Cross males (emb-1lon-1/emb-1 +; him-8/+) were mated with ok2757/hT2, and non-GFP L4 cross progeny (emb-1lon-1/ok2757 or emb-1/ok2757) were picked to different temperatures. At 15° and 20°, the majority of embryos hatched. At 24°, all the embryos arrested as one-cell embryos. The same was done with the ok2759 deletion allele.
In an alternative approach, emb-1(hc62ts); him-8(e1489) males were crossed with ok2757/hT2 hermaphrodites, and non-GFP larvae were picked to 20° and 24°. At 20°, the majority of embryos hatched (85% hatched; 1046/1226). At 24°, all the embryos arrested as one-cell embryos (n > 400). The same was done with ok2759/hT2. At 20°, the majority of embryos hatched (81% hatched; 569/702). At 24°, all the embryos arrested as one-cell embryos (n > 800).
Molecular biology
Sequencing of yk1426h04 cDNA clone:
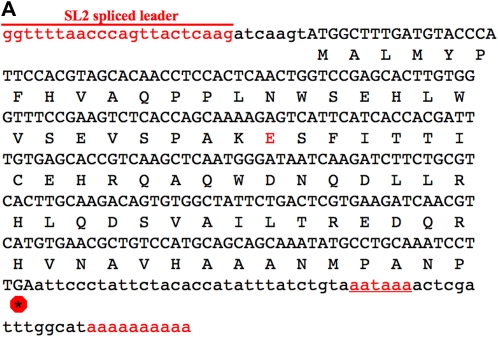
The single cDNA clone for K10D2.4 listed in WormBase was obtained from Yuji Kohara (National Institute of Genetics, Mishima, Japan). C. elegans mRNAs are often trans-spliced to a short leader sequence called SL1 or SL2. Single genes and the first gene of an operon are often SL1 trans-spliced, while downstream genes in an operon are SL2 trans-spliced (Blumenthal 1995). The yk1426h04 cDNA was sequenced and found to be SL2 trans-spliced 7 bp before the initiation codon. A poly(A) tail is located 50 bp downstream of the stop codon (Figure 2A).
Figure 2 .
cDNA and protein sequences. (A) The emb-1 cDNA sequence from clone yk1426h04 (gift from Y. Kohara). The cDNA contains a SL2 trans-spliced leader sequence at its 5′ end and a poly(A) tail at its 3′ end, which is preceded by a polyadenylation signal (underlined), all indicated in red. Asterisk in a red octagon indicates the stop codon. Residue that is altered in the hc62ts allele is also in red (codon 30; glutamic acid). In hc62ts animals, this residue is a lysine. (B) EMB-1 is a conserved protein among Caenorhabditis species. Amino acid sequence alignment of the EMB-1 protein are from C. elegans, C. briggsae, C. remanei, and C. brenneri (www.wormbase.org). Those residues in red are identical in at least three of the four species. Indicated above the sequence is the amino acid change in residue 30. (C) Genomic region of emb-1. Shown is a schematic from www.wormbase.org of ∼5 kb of the genomic region that contains the emb-1 gene (K10D2.4). Shown in pink are the three genes that make up the operon, with emb-1 located in the middle. The deletion alleles discussed in this report (ok2759 and ok2757) are shown below as red and blue bars. The genomic fragment used for rescue is shown as a green rectangle below the operon.
Sequencing of hc58ts and hc62ts alleles:
A 0.9-kb fragment of genomic DNA was PCR amplified from wild-type, emb-1(hc58ts), and emb-1(hc62ts) lysates with primers “K10 F1”: 5′-GGCACCGATTTGTGCTAGG and “K10 R1”: 5′-ACCACTATTCTGGCTATTTCC. The PCR product was sequenced with primers “K10 F2”: 5′-ATATCCATTCGTCGGACACG and “K10 R2”: 5′-TTCCAGACCTTTGCTCATCG by SeqWright (Houston, TX). All oligonucleotides in this study were synthesized by Integrated DNA Technologies (Coralville, IA).
emb-1 rescuing construct:
An ∼1.98-kb genomic fragment (corresponding to ∼750 bp of 5′ regulatory sequences, K10D2.5, K10D2.4, and ∼230 bp beyond the stop codon of K10D2.4) was PCR amplified from wild-type animals and recombined into Gateway vector pDONR P4P3 (Invitrogen, Carlsbad, CA). Primers “EMB-1 F3” contained an attB4 site at the 5′ end (5′-GGGGACAACTTTGTATAGAAAAGTTGCTCCTGAAAGTAGCAAAATATACACG) and “emb R2” included an attB3 sites on its 5′ end (5′-GGGGACAACTTTGTATAATAAAGTTGCTCACAACTCTTTTTAATAATTACAG).
This entry clone was then recombined into pCR319, a destination vector containing a C. elegans unc-119 genomic fragment capable of rescuing unc-119 mutants. The final expression clone, pAG-109, was used for microparticle bombardment into unc-119(ed3) animals. This construct essentially carries the 5′ regulatory sequences and the first two genes (of three) of the CEOP3264 operon (WormBase).
Microparticle bombardment:
Transgenic lines using pAG-109 were generated by the C. elegans ‘Worm’ Core Facility at The University of Utah, directed by Dr. Colin Thacker. Microparticle bombardment was performed with unc-119(ed3) animals using the published protocol developed by Praitis et al. (2001). Three integrated lines were identified by this method.
PCR monitoring of deletion alleles and hc62ts:
In double mutants in which emb-1 was unmarked, the presence of hc62ts was confirmed via PCR and restriction enzyme digestion. PCR was carried out with primers “EMB-1 F6”: 5′-GACTTTGATTCAAAAAACCACG and “EMB-1 R4”: 5′-CGAGTCAGAATAGCCACACT to amplify an ∼290-bp fragment. The hc62ts allele contains a mutation that alters a HinfI restriction site; PCR-amplified hc62ts sequences are not cut, while sequences from wild-type animals yield two bands.
The ok2757 deletion allele was monitored by PCR using primers “K10 F4”: 5′- AAATCTCAGCGGGAGTTTGA and “K10 R3”: 5′-CATCAATGGTTGTACAGCGG.
The ok2759 deletion allele was monitored by PCR using primers “EMB-1 F7”: 5′-CTGCAGTGGAGCGTACTTGC and “EMB-1 R2”: 5′-GGGGACAACTTTGTATAATAAAGTTG.
The cid-1(tm936) allele was monitored with primers “cid-1 F2”: 5′-CCTTGGTTGCCGCTGTACAA and “CID-1 R2”: 5′-CTCACATCTCGACTCATTGG and the cid-1(tm1021) allele with primers “CID-1 F1”: 5′-TCTGCGTCACTTGCAAGACA and “CID-1 R1”: 5′-TCCGGAAGTGTGACGTCATA.
Immunostaining:
Phosphohistone H3 staining was performed as previously described (Golden et al. 2000). Images were obtained on an Olympus BX-60 microscope equipped with a Cooke Sensicam.
RNAi:
Bacterial feeding clones were from the Ahringer feeding library (Kamath and Ahringer 2003) (Geneservice, Cambridge, UK). L4 larvae were fed bacteria containing the RNAi construct for 24–28 hr (Timmons et al. 2001) and then moved to a new RNAi plate for another 24 hr. Hermaphrodites were then removed. The second RNAi plate was scored ≥24 hr later for embryonic lethality or hatching. For the K10D2.4 clone, dsRNA was synthesized from the Ahringer library clone using an Ambion MEGAscript T7 kit (Austin, TX), precipitated, resuspended in water, microinjected into L4 animals by standard methods (Fire et al. 1998), and scored for embryonic lethality as described above.
Whole animal DAPI staining:
Whole animals were mounted on slides and fixed in Carnoy’s II fixative (6:3:1 ethanol/acetic acid/chloroform) for 16 hr at 22°, rehydrated through a series of ethanol/PBS rinses, and then stained with 1 μg/ml 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI) in PBS. Germline and embryonic nuclei were then examined by UV epifluorescence.
Results
Temperature-sensitive alleles of emb-1 result in arrest in metaphase I
The two ts alleles of emb-1 were originally described as recessive, maternal-effect lethal mutants whose embryos arrested at the one-cell stage of embryogenesis when raised at the restrictive temperature of 25° (Schierenberg et al. 1980). Later studies revealed that the embryos specifically arrest at metaphase of meiosis I with a phenotype similar to that of mutants in known APC/C subunits (Golden et al. 2000). emb-1 animals expressing a maternally expressed tubulin::GFP transgene accumulate meiotic one-cell embryos with barrel-shaped meiotic spindles (Figure 1). Temperature-shift experiments suggest that EMB-1 is synthesized prior to the one-cell stage; embryos produced by permissively grown mothers develop normally and hatch when up-shifted to the nonpermissive temperature at the one-cell stage (Miwa et al. 1980). The mutant defects are maternal specific as emb-1(hc62ts) males are fertile.
Figure 1 .
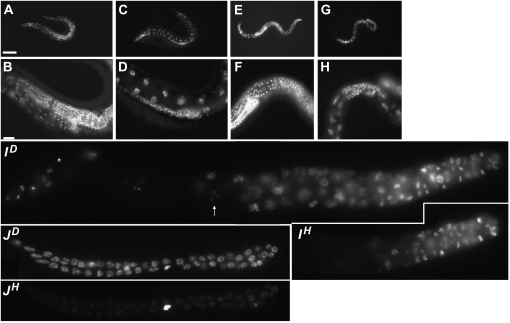
emb-1 mutants arrest at metaphase of meiosis I. (A) emb-1(hc62ts); tubulin::gfp animal at 16°. Bar, ∼20 μm (A and B). (B) emb-1(hc62ts); tubulin::gfp animal at 24°. (C) Embryo from mother injected with K10D2.4 dsRNA and stained with anti–α-tubulin antibodies. Bar, ∼10 μm (C and D). (D) Same embryo stained with TOTO-3 to visualize the oocyte and sperm chromosomes. Oocyte chromosomes in metaphase I are marked with a white arrow.
The hc62ts allele fails to complement a small deficiency chromosome sDf121. emb-1(hc62ts)/sDf121 hemizygotes up-shifted to 24° at the fourth (L4) larval stage (n = 16) produced all one-cell progeny. Hemizygotes up-shifted to 24° at the L1 larval stage exhibited additional male tail defects similar to those previously described in APC/C mutants (Shakes et al. 2003). At 15° and 20°, hemizygous hermaphrodites produced viable embryos. Although all of these phenotypic studies were carried out with the hc62ts allele, sequencing results shown below reveal that the hc62ts and hc57ts alleles bear the same molecular lesion and thus are unlikely to represent independently derived mutations.
The phenotype of emb-1 is enhanced by mutations in APC/C subunit genes
Given the phenotypic similarities between emb-1 and known APC/C subunit mutants, we tested whether the emb-1 mutant would exhibit genetic interactions similar to those observed for the APC/C subunit mutants. Though all the existing ts APC/C subunit mutants are viable and healthy at 15°, we have been unable to construct a double mutant of two APC/C subunits that could be maintained at the permissive temperature of 15° (Shakes et al. 2003; Stein et al. 2007; our unpublished observations). Typically such APC/C double mutants are either sterile or if fertile, produce broods of multicellular embryos that fail to hatch. The phenotype of emb-1 in combination with the reduction-of-function APC/C mutants emb-27, mat-2, mat-3, or the CDC-20 ortholog, fzy-1, recapitulated what we have seen with APC/C double mutants (Table 2); double mutant lines could not be maintained at the permissive temperature. These enhancement effects suggest that emb-1 encodes either a regulator or novel subunit of the APC/C.
Table 2 . mat mutants are enhanced by emb-1 at the permissive temperature.
| Genotype | N | Phenotype @ 15° |
|---|---|---|
| emb-27(ax81ts) unc-4; emb-1(hc62ts) lon-1 | 9 | Pvl Ste |
| rol-6 mat-2(or170ts); emb-1(hc62ts) unc-32 | 21 | One-cell progeny |
| dpy-10 mat-2(ax102ts); emb-1(hc62ts) unc-32 | 25 | Unh MC (22) or Ste (3) |
| mat-3(or180ts) dpy-1 emb-1(hc62ts) unc-32 | 10 | Ste |
| fzy-1(h1983) dpy-10; emb-1(hc62ts) unc-32 | 36 | Unh MC (34) or Ste (2) |
Double mutants of emb-1 with mat and fzy-1 reduction-of-function alleles. Mothers were identified on the basis of their linked morphological markers. The generation of these double mutants was carried out at 15°. The phenotype indicated is for the mother (Ste, Pvl) or the progeny (MC unh) of the doubly mutant mother. Ste, sterile; Pvl, protruding vulva; unh MC, unhatched multicellular embryos. All single mutant control animals were not Ste, Pvl, nor did they produce a significant number of unhatched embryos at 15°. N = number of adult mothers examined.
The emb-1 mutant is suppressed by mutations in SAC genes and fzy-1
In a reciprocal set of experiments, we tested whether the emb-1 phenotype could be repressed in combination with known suppressors of APC/C mutants. Originally isolated in a screen for genetic suppressors of mat-3(or180ts) at 24° (Stein et al. 2007), these suppressors include gain-of-function alleles of the CDC20 ortholog, fzy-1, and loss-of-function mutations in mdf-1, -2, and -3, orthologs of the MAD genes that function in the SAC (Stein et al. 2007). Presumably, these suppressor mutations allow a compromised APC/C to function more effectively at nonpermissive temperatures. At 24°, double mutant combinations of emb-1(hc62ts) with these suppressor mutations exhibited a partial suppression of the emb-1 one-cell arrest phenotype (Table 3). However, while suppression of several APC/C subunit mutants in combination with mdf-1, -2, -3, or fzy-1 yielded significant numbers of hatching embryos and lines that could be maintained at restrictive temperatures (Stein et al. 2007), suppression of emb-1(hc62ts) barely restored embryonic viability. For emb-1 animals, suppression was scored as the production of embryos that progressed beyond the meiotic one-cell stage to arrest as unhatched multicellular (Unh MC) embryos. The few embryos that did hatch died as larvae or developed into sterile adults. None of the emb-1(hc62ts) suppressed lines could be maintained at 24°.
Table 3 . The emb-1 mutant is suppressed by SAC and fzy-1 mutations.
| Genotype | N | Phenotype @ 24° |
|---|---|---|
| unc-42; emb-1(hc62ts) lon-1 | 400 | One-cell embryos; 0% hatch |
| mdf-1(av19) unc-42; emb-1(hc62ts) lon-1 | 1012 | Unh MC embryos; 0% hatch |
| mdf-1(av19) unc-42; emb-1(hc62ts) | 447 | Unh MC embryos; 3% hatch |
| unc-17; emb-1(hc62ts) lon-1 | 589 | One-cell embryos; 0% hatch |
| mdf-2(av16) unc-17; emb-1(hc62ts) lon-1 | 405 | Unh MC embryos; 2% hatch |
| mdf-2(av16) unc-17; emb-1(hc62ts) | 387 | Unh MC embryos; 1 hatched |
| unc-13; emb-1(hc62ts) lon-1 | 200 | One-cell embryos; 0% hatch |
| mdf-3(av31) unc-13; emb-1(hc62ts) lon-1 | 1567 | Unh MC embryos; 1 hatched |
| mdf-3(av31) unc-13; emb-1(hc62ts) | 236 | Unh MC embryos; 2% hatch |
| unc-4; emb-1(hc62ts) lon-1 | 712 | One-cell embryos; 0% hatch |
| fzy-1(av15gf) unc-4; emb-1(hc62ts) lon-1 | 1395 | Unh MC embryos; 1% hatch |
| fzy-1(av15gf) unc-4; emb-1(hc62ts) | 504 | Unh MC embryos; 1% hatch |
Double mutants of emb-1 with previously identified SAC mutants or a gain-of-function fzy-1 allele. Mothers were identified on the basis of their linked morphological markers. The generation of these double mutants was carried out at 15°. L4 larvae of the appropriate genotype were shifted to 24° to assay for suppression of the emb-1 one-cell phenotype. The phenotype indicated is for the progeny of the doubly mutant mother. N = number of embryos counted. Unh MC, unhatched multicellular embryos, which indicates suppression at the cellular level.
The emb-1 gene encodes the small APC/C subunit K10D2.4/APC16
To molecularly identify the emb-1 gene product, emb-1 was genetically mapped to a small, ∼370-kb interval on chromosome III between dpy-17 and lon-1. When Lon non-Dpy recombinants were subsequently picked from emb-1(hc62ts)/dpy-17(e164) lon-1(e1820) animals, 50 of 61 Lon non-Dpy animals contained emb-1 based on the phenotypes of progeny shifted to 24°. This mapping narrowed the number of potential emb-1 candidate genes from the ∼80 genes between dpy-17 and lon-1 to ∼20 genes. Of these 20 genes, RNAi data from WormBase and our own feeding RNAi studies revealed a subset of candidate genes whose depletion results in embryonic lethality, but none that led to a one-cell arrest phenotype. However injection of dsRNA from K10D2.4 resulted in a penetrant, meiotic one-cell arrest phenotype. Sequencing of the K10D2.4 locus from emb-1(hc57ts) and emb-1(hc62ts) animals revealed that both alleles contained the same missense mutation in codon 30, resulting in a glutamic acid-to-lysine change (Figure 2B). This locus consists of two exons and encodes an 81-amino-acid protein with no known motifs or domains but was independently discovered as an APC/C subunit in biochemical copurification experiments (Green et al. 2011). Given the size of the C. elegans genome, it is likely that these two alleles are not from two unique and separate hits in the emb-1 gene, but rather represent sisters from the original mutagenesis screen (Miwa et al. 1980).
The emb-1 locus is the second gene in a three-gene operon (Figure 2C). emb-1 is represented by a single full-length cDNA clone (yk1426h04, a kind gift from Y. Kohara); consistent with its position in an operon, the cDNA has a SL2 leader sequence trans-spliced 7 bp upstream of the methionine codon. On the basis of the cDNA sequence, the mRNA has a short 3′-UTR that includes a polyadenylation signal sequence (AAUAAA) just 14 bp upstream of the poly(A) tail (Figure 2A).
The EMB-1 protein is molecularly conserved among the four Caenorhabditis species sequenced to date (C. elegans, C. briggsae, C. remanei, and C. brenneri). The four predicted proteins are 81–84 amino acids in size. The four proteins are 77% identical (94% similar) in their first 66 residues and more divergent in their carboxyl-terminal tails (Figure 2B). Although basic sequence alignments fail to reveal orthologs in other organisms, K10D2.4 has been proposed to be orthologous to the human C10orf104/APC16 gene (Kops et al. 2010).
Expression of K10D2.4 rescues the emb-1 mutant
In a complementary approach, we also tested whether emb-1(hc62ts) animals could be transgenically rescued. Given that K10D2.4 was part of an operon, we created a rescuing transgene (pAG-109) that contained ∼2 kb of sequence containing K10D2.4 and ∼240 bp of its 3′-UTR as well as the upstream gene K10D2.5 and ∼750 bp of sequence upstream of K10D2.5 (Figure 2C). This transgene lacked the third gene of the operon (cid-1). Homozygous unc-119(ed3) hermaphrodites were transformed by microparticle bombardment with pAG-109, which also contained a wild-type copy of the unc-119 gene as a selectable marker. Non-Unc progeny were isolated and three homozygous integrated lines were generated. The integrated transgene from line UZ839 was crossed into emb-1(hc62) lon-1 animals, and F2 Lon animals were isolated and scored for rescue at 24°. Since there are no visible markers on the transgene, it was necessary to pick many Lon animals, since only one-quarter was predicted to be homozygous for the transgene. Many Lon animals segregated multicellular embryos at 24°, and four animals segregated live embryos from which we were able to establish lines that can be maintained at 24°. PCR analysis of these rescued emb-1(hc62ts) lines demonstrated that the lines were homozygous for the emb-1(hc62ts) allele and contained the transgene (data not shown). These data provide independent confirmation that K10D2.4 is the gene mutated in emb-1 animals.
The zygotic phenotype of emb-1 is sterility
To determine the null phenotype of emb-1, we analyzed the phenotype of two deletion alleles (Figure 2C) generated by the C. elegans Gene Knockout Consortium (Oklahoma Medical Research Center) and the C. elegans Reverse Genetics Core Facility (Vancouver). These alleles, ok2757 and ok2759, cannot be maintained as homozygotes and both fail to complement emb-1(hc62ts). emb-1(hc62ts)/ok2757 and emb-1(hc62ts)/ok2759 L4 animals produce viable embryos at 20° and one-cell arrested embryos at 24°. Analysis of the homozygous deletion phenotypes is complicated as both deletion alleles lack not only emb-1 sequences but also part of one of its flanking genes within the three-gene operon. The ok2757 deletion lacks both exons of K10D2.4 (emb-1), its 3′-UTR, and exon 1 of the downstream K10D2.3 (cid-1) gene. ok2757 deletion homozygotes hatch as embryos, presumably due to maternal rescue, and subsequently arrest as active L2 larvae. As cid-1 was recently shown to play a role in chromosome segregation (van Wolfswinkel et al. 2009), we suspect that the L2 arrest phenotype results from a synthetic interaction between emb-1 and cid-1 (cid-1 deletion alleles alone do not display a L2 arrest phenotype (our unpublished observations). Conversely, the ok2759 deletion lacks both exons of emb-1, its 3′-UTR, and part of the 3′-UTR of the upstream K10D2.5 gene. ok2759 deletion homozygotes hatch and develop into sterile (Ste) adults with a protruding vulva (Pvl) phenotype. This Pvl/Ste phenotype is very similar to the null phenotype of some APC/C subunit genes (Davis et al. 2002).
Since this less severe phenotype of ok2759 deletion homozygotes is more likely to reflect the null phenotype of emb-1, we extended our analysis of its Pvl/Ste phenotype. When ok2759 homozygotes were isolated from balanced heterozygotes (over dpy-17lon-1), all embryos hatched. The resulting larvae were picked to separate plates and development was followed. All grew to adults and were either balanced heterozygotes, Dpy (dpy-17 is epistatic to lon-1) or sterile. This observation suggests that homozygous ok2759 embryos from heterozygous mothers are maternally rescued but fail to develop a productive germline once maternal product is depleted. Similar observations were made when ok2759 homozygous progeny were picked from ok2759/ hT2 [bli-4(e937) let-?(q782) qIs48] mothers; they reached adulthood but lacked a functional germline. Sterile ok2759 hermaphrodites were fixed and stained with DAPI to address the sterile phenotype. Sterile hermaphrodites had severely reduced germlines containing a small number of early germ cells and no evidence of undergoing a mitotic-to-meiotic transition (Figure 3). ok2759 males exhibited similarly reduced germlines indicating that, unlike the differential gamete phenotypes of emb-1(hc62ts) males and hermaphrodites, the germline proliferation aspect of the deletion phenotype is not sex specific (Figure 3, G and H).
Figure 3 .
emb-1 deletion homozygotes are sterile and have no gametes. Whole animal DAPI staining to visualize DNA. (A and B) N2 adult hermaphrodite. (C and D) ok2759 homozygous adult hermaphrodite. (E and F) ok2759/hT2; him-8 heterozygous male. (G and H) ok2759; him-8 homozygous adult male. Note that in the sterile ok2759 animals the gut nuclei are quite prominent, while they are normally obscured by the germline in wild-type animals. (I and J) Isolated gonads from apc-11(gk73) (I) and emb-1(ok2759) (J) deletion hermaphrodites, stained with DAPI (superscript D) plus inset of pHisH3(S10) (superscript H) immunostaining. In I, arrow points to the nucleus of a diakinetic oocyte. Bar in A, ∼100 μm (same scale for C, E, and G). Bar in B, ∼20 μm (same scale for D, F, and H).
To further evaluate the nature of this germline proliferation defect, we compared gonads from homozygous deletion mutants of apc-11 and emb-1, both derived from heterozygous parents. Isolated gonads of apc-11(gk37) homozygotes were larger and many produced at least a few normal sized oocytes (Figure 3I). However their mitotic zones were filled with large numbers of nuclei that were severely aneuploid and/or labeled with an antibody against the phosphorylated serine 10 form of histone H3 [pHisH3(S10)], an M-phase marker. In contrast, the gonads of emb-1(ok2759) homozygotes failed to completely develop. They exhibited little evidence of either spermatogenesis or oogenesis, and the arms remained linear and failed to reflex toward the dorsal side. Notably, the morphology of their germline nuclei revealed little evidence of aneuploidy and most gonads contained at most one to two nuclei that labeled positively for pHisH3(S10) (Figure 3J). At this point it remains unclear whether these differences reflect a distinct function for EMB-1 outside of the APC/C complex or merely a difference in the perdurance of the maternal product. Similarly, the observed germline proliferation defect could be due to cell nonautonomous effects from the somatic gonad, as both APC2 and CO9H10.7(APC17) may function in the migration of the somatic distal tip cell (Cram et al. 2006). However, given the well-established role of the APC/C in cell proliferation, we favor the more likely conclusion that the severe reduction of germline proliferation in ok2759 homozygotes reflects a requirement for zygotic emb-1 expression directly within the germ cells.
We previously have shown that adult ts APC/C mutants shifted to the nonpermissive temperature for short time periods (6 hr) resulted in defects in the germline mitotic divisions (Golden et al. 2000). When gonads were immunostained with the pHisH3(S10) antibody, they had elevated numbers of pHisH3 stained mitotic germ cells. We used this same assay to quantify increases in the number of M-phase nuclei within the gonads of emb-1 ts mutants. Although the number of pHisH3(S10) nuclei per gonad is highly variable (Table 4), we observed equivalent numbers of pHisH3(S10) nuclei in the gonads of wild-type (N2) and emb-1 heterozygotes and elevated numbers in emb-1 homozygotes and hemizygotes. Thus, reductions in EMB-1 expression in adult gonads result in an increase in germ cells in M phase, suggesting an extended pause in metaphase. Notably, emb-1 homozygotes had elevated numbers of pHisH3 staining germline nuclei even when maintained at the permissive temperature of 16°.
Table 4 . Number of M-phase nuclei in germline proliferative zone.
| Genotype | Growth conditions | N | pHisH3(S10) nuclei/arm |
|---|---|---|---|
| Wild type (N2) | 9 hr at 25° | 60 | 4.4 ± 3.5 |
| emb-1 (hc57ts) /+ | 9hr at 25° | 29 | 3.9 ± 1.9 |
| emb-1 (hc62ts) | Constant 16° | 52 | 6.4 ± 3.6 |
| emb-1 (hc62ts) | 6 hr at 25° | 36 | 6.2 ± 2.8 |
| emb-1 (hc57ts) | 9 hr at 25° | 68 | 8.7 ± 3.5 |
| emb-1 (hc57ts)/ok2759 | 9 hr at 25° | 32 | 8.0 ± 2.4 |
Strains and crosses were maintained at 16° before young adult hermaphrodites were shifted to 25° for the indicated duration. Animals were dissected to isolate gonadal arms, processed for immunofluorescence, and pHisH3(S10) positive nuclei were scored under epifluorescence.
Discussion
In this report, we demonstrate that maternal expression of the emb-1 gene in C. elegans oocytes is essential for progression beyond metaphase of meiosis I, while zygotic expression of emb-1 is essential for mitotic germline proliferation. Our unpublished observations also suggest that EMB-1 is required for the late developmental events of male tail and hermaphrodite vulva formation. While previous reports had noted the similarity of emb-1’s meiotic arrest phenotype to that of known subunits of the APC/C subunit (Golden et al. 2000), this study extends these findings to show that the emb-1(hc62ts) phenotype is enhanced or suppressed in double mutant combinations with mutations in either APC/C subunits or known APC/C suppressors. These studies also reveal a role for EMB-1 in the mitotic divisions of early embryos and suggest that EMB-1 may be functioning as a novel subunit of the APC/C.
Our results reveal that emb-1 encodes a novel 81-residue protein with no recognizable protein motifs. Though a few groups have recently identified the human APC16 subunit (Hubner et al. 2010; Hutchins et al. 2010; Kops et al. 2010; Ohta et al. 2010), only one has identified K10D2.4 as the C. elegans ortholog (Kops et al. 2010). Kops et al. (2010) identified the APC16 ortholog from 27 different metazoan species, but did not identify orthologs in S. pombe or S. cerevisiae. Sequence similarity was found in four patches called AH1–4 (for APC homology) with the most significant similarity residing in AH4. Though the C. elegans homology in this region was the weakest of the 27 species, the authors did identify K10D2.4 as the C. elegans gene with the most similarity.
Our results indicate that emb-1 is essential for both oocyte meiosis and germline development. Depletion of EMB-1 either in a temperature-sensitive mutant or in RNAi mediated knockdowns results in embryos that arrest at metaphase of meiosis I. This phenotype is consistent with that of known APC/C subunits in C. elegans and distinct from that of meiotic-specific activators in other organisms, which arrest after metaphase I. Analysis of mutants homozygous for the deletion ok2759 revealed an additional zygotic requirement for the emb-1 gene; ok2759 animals are sterile and exhibit defects in the mitotic proliferation of germ cells combined with a failure in gametogenesis. Our current results leave open the intriguing question of whether or not emb-1 functions in a gamete-specific fashion. We have been unable to detect spermatogenesis-associated meiotic arrest defects in either emb-1(hc62ts) homozygotes or in emb-1(hc62ts)/deletion males. Conversely, in studying ts alleles of other APC/C subunits, only a subset of alleles exhibit defects in spermatocyte meiosis, while all except one exhibit defects in oocyte meiosis (P. L. Sadler, D. Fox, A. Pletcher, and D. Shakes, unpublished observations; Tarailo et al. 2007). Recent work indicates that operon-encoded genes are expressed in a germline-intrinsic and/or oogenesis-specific pattern (Reinke and Cutter 2009), but the third gene of this operon (cid-1) is expressed in both male and female germlines (van Wolfswinkel et al. 2009).
The idea that emb-1 encodes a novel subunit of the APC/C is suggested not only by the phenotype of emb-1 homozygotes but also by the phenotype of emb-1 in combination with known APC/C subunits and their suppressors. The human APC16 subunit was recently identified in a number of proteomics/biochemistry studies (Hubner et al. 2010; Hutchins et al. 2010; Kops et al. 2010; Ohta et al. 2010). By homology, Kops et al. (2010) identified K10D2.4 as a potential C. elegans ortholog and showed by RNAi that depleted embryos arrested as one-cell embryos (Kops et al. 2010). Green et al. (2011) subsequently demonstrated that K10D2.4 biochemically associates with numerous C. elegans APC/C subunits in immunoprecipitation assays. These studies, together with our own molecular identification of emb-1 as K10D2.4 and evidence of genetic interactions with known APC/C subunits and suppressors, provide multiple lines of evidence that the EMB-1 protein functions as a component of the C. elegans APC/C.
Acknowledgments
We acknowledge Yuji Kohara (National Institute of Genetics, Mishima, Japan) for generously providing us with the cDNA clone for emb-1. We are also grateful to Shohei Mitani (Tokyo Women’s Medical University School of Medicine) for generating the cid-1 deletion alleles, the C. elegans Gene Knockout Consortium (Oklahoma Medical Research Foundation), and the C. elegans Reverse Genetics Core Facility (Vancouver) for isolating the ok2757 and ok2759 deletion alleles of emb-1. We thank Colin Thacker (University of Utah) for generating our transgenic lines, the Caenorhabditis Genetics Center for providing numerous strains for mapping, James LaRue (College of William and Mary) for preliminary studies of emb-1 doubles, and Kevin O’Connell for critical comments on this manuscript. We are also grateful for the discussions and suggestions from members of our laboratories and the Baltimore Worm Club. This research was supported, in part, by National Institutes of Health (NIH) grant R15GM60359 (D.C.S.), Jeffress Memorial Trust Grant (J-840; D.C.S.), and the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (A.G.).
Literature Cited
- Asakawa H., Kitamura K., Shimoda C., 2001. A novel Cdc20-related WD-repeat protein, Fzr1, is required for spore formation in Schizosaccharomyces pombe. Mol. Genet. Genomics 265: 424–435 [DOI] [PubMed] [Google Scholar]
- Blanco M. A., Pelloquin L., Moreno S., 2001. Fission yeast mfr1 activates APC and coordinates meiotic nuclear division with sporulation. J. Cell Sci. 114: 2135–2143 [DOI] [PubMed] [Google Scholar]
- Blumenthal T., 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11: 132–136 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K. F., Mallory M. J., Egeland D. B., Jarnik M., Strich R., 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. USA 97: 14548–14553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram E. J., Shang H., Schwarzbauer J. E., 2006. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J. Cell Sci. 119: 4811–4818 [DOI] [PubMed] [Google Scholar]
- Davis E. S., Wille L., Chestnut B. A., Sadler P. L., Shakes D. C., et al. , 2002. Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160: 805–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A. E., Park J. S., Inoue I., Tachikawa H., Neiman A. M., 2009. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell 20: 134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Bogdanova A., Habermann B., Zachariae W., Ahringer J., 2007. Identification of the C. elegans anaphase promoting complex subunit Cdc26 by phenotypic profiling and functional rescue in yeast. BMC Dev. Biol. 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P., Herzog F., Gieffers C., Sander B., Riedel D., et al. , 2005. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol. Cell 20: 867–879 [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Furuta T., Tuck S., Kirchner J., Koch B., Auty R., et al. , 2000. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11: 1401–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A., Sadler P. L., Wallenfang M. R., Schumacher J. M., Hamill D. R., et al. , 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. A., Kao H. L., Audhya A., Arur S., Mayers J. R., et al. , 2011. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145: 470–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ganoth D., Sudakin V., Dahan A., Cohen L. H., et al. , 1994. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein-kinase Cdc2. J. Biol. Chem. 269: 4940–4946 [PubMed] [Google Scholar]
- Herzog F., Primorac I., Dube P., Lenart P., Sander B., et al. , 2009. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323: 1477–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., et al. , 2010. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189: 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins J. R., Toyoda Y., Hegemann B., Poser I., Heriche J. K., et al. , 2010. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321 [DOI] [PubMed] [Google Scholar]
- King R. W., Peters J. M., Tugendreich S., Rolfe M., Hieter P., et al. , 1995. A 20s complex containing Cdc27 and Cdc16 catalyzes the mitosis-specific conjugation of ubiquitin to Cyclin-B. Cell 81: 279–288 [DOI] [PubMed] [Google Scholar]
- Kops G. J., van der Voet M., Manak M. S., van Osch M. H., Naini S. M., et al. , 2010. APC16 is a conserved subunit of the anaphase-promoting complex/cyclosome. J. Cell Sci. 123: 1623–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., York J. P., Zhang P., 2007. Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol. Cell. Biol. 27: 3481–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca T., Castro A., Martinez A. M., Vigneron S., Morin N., et al. , 1998. Fizzy is required for activation of the APC/cyclosome in Xenopus egg extracts. EMBO J. 17: 3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa J., Schierenberg E., Miwa S., von Ehrenstein G., 1980. Genetics and mode of expression of temperature-sensitive mutations arresting embryonic development in Caenorhabditis elegans. Dev. Biol. 76: 160–174 [DOI] [PubMed] [Google Scholar]
- Ohi M. D., Feoktistova A., Ren L., Yip C., Cheng Y., et al. , 2007. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol. Cell 28: 871–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Bukowski-Wills J. C., Sanchez-Pulido L., Alves Fde L., Wood L., et al. , 2010. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142: 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore L. A., Booth C. R., Venien-Bryan C., Ludtke S. J., Fioretto C., et al. , 2005. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol. Cell 20: 855–866 [DOI] [PubMed] [Google Scholar]
- Pesin J. A., Orr-Weaver T. L., 2008. Regulation of APC/C activators in mitosis and meiosis. Annu. Rev. Cell Dev. Biol. 24: 475–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M., 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Yu H., 2007. KEN-box-dependent degradation of the Bub1 spindle checkpoint kinase by the anaphase-promoting complex/cyclosome. J. Biol. Chem. 282: 3672–3679 [DOI] [PubMed] [Google Scholar]
- Reinke V., Cutter A. D., 2009. Germline expression influences operon organization in the Caenorhabditis elegans genome. Genetics 181: 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenberg E., Miwa J., von Ehrenstein G., 1980. Cell lineages and developmental defects of temperature-sensitive embryonic arrest mutants in Caenorhabditis elegans. Dev. Biol. 76: 141–159 [DOI] [PubMed] [Google Scholar]
- Schwab M., Lutum A. S., Seufert W., 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90: 683–693 [DOI] [PubMed] [Google Scholar]
- Seidl M. F., Schultz J., 2009. Evolutionary flexibility of protein complexes. BMC Evol. Biol. 9: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Sadler P. L., Schumacher J. M., Abdolrasulnia M., Golden A., 2003. Developmental defects observed in hypomorphic anaphase-promoting complex mutants are linked to cell cycle abnormalities. Development 130: 1605–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Lavy K. J., Oren Y. S., Feine O., Sajman J., Listovsky T., et al. , 2010. Fifteen years of APC/cyclosome: a short and impressive biography. Biochem. Soc. Trans. 38: 78–82 [DOI] [PubMed] [Google Scholar]
- Sonnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., et al. , 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469 [DOI] [PubMed] [Google Scholar]
- Stein K. K., Davis E. S., Hays T., Golden A., 2007. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics 175: 107–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K. K., Nesmith J. E., Ross B. D., Golden A., 2010. Functional redundancy of paralogs of an anaphase promoting complex/cyclosome subunit in Caenorhabditis elegans meiosis. Genetics 186: 1285–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel M. L., Cheng K. C., Seydoux G., 2007. Regulation of MBK-2/Dyrk kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr. Biol. 17: 1545–1554 [DOI] [PubMed] [Google Scholar]
- Sudakin V., Ganoth D., Dahan A., Heller H., Hershko J., et al. , 1995. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell 6: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo M., Kitagawa R., Rose A. M., 2007. Suppressors of spindle checkpoint defect (such) mutants identify new mdf-1/MAD1 interactors in Caenorhabditis elegans. Genetics 175: 1665–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton B. R., Toczyski D. P., 2006. Precise destruction: an emerging picture of the APC. Genes Dev. 20: 3069–3078 [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112 [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Claycomb J. M., Batista P. J., Mello C. C., Berezikov E., et al. , 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148 [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A., 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463 [DOI] [PubMed] [Google Scholar]
- Wasch R., Robbins J. A., Cross F. R., 2010. The emerging role of APC/CCdh1 in controlling differentiation, genomic stability and tumor suppression. Oncogene 29: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. Y., McNally K., McNally F. J., 2003. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C. elegans. Dev. Biol. 260: 245–259 [DOI] [PubMed] [Google Scholar]
- Yoon H. J., Feoktistova A., Wolfe B. A., Jennings J. L., Link A. J., et al. , 2002. Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr. Biol. 12: 2048–2054 [DOI] [PubMed] [Google Scholar]
- Zachariae W., Nasmyth K., 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13: 2039–2058 [DOI] [PubMed] [Google Scholar]
- Zachariae W., Shin T. H., Galova M., Obermaier B., Nasmyth K., 1996. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 274: 1201–1204 [DOI] [PubMed] [Google Scholar]
- Zachariae W., Shevchenko A., Andrews P. D., Ciosk R., Galova M., et al. , 1998. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science 279: 1216–1219 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Ching Y. P., Ng R. W., Jin D. Y., 2002. The APC regulator CDH1 is essential for the progression of embryonic cell cycles in Xenopus. Biochem. Biophys. Res. Commun. 294: 120–126 [DOI] [PubMed] [Google Scholar]
- Zich J., Hardwick K. G., 2010. Getting down to the phosphorylated 'nuts and bolts' of spindle checkpoint signalling. Trends Biochem. Sci. 35: 18–27 [DOI] [PubMed] [Google Scholar]