Abstract
A new approach to assigning individuals to populations using genetic data is described. Most existing methods work by maximizing Hardy–Weinberg and linkage equilibrium within populations, neither of which will apply for many demographic histories. By including a demographic model, within a likelihood framework based on coalescent theory, we can jointly study demographic history and population assignment. Genealogies and population assignments are sampled from a posterior distribution using a general isolation-with-migration model for multiple populations. A measure of partition distance between assignments facilitates not only the summary of a posterior sample of assignments, but also the estimation of the posterior density for the demographic history. It is shown that joint estimates of assignment and demographic history are possible, including estimation of population phylogeny for samples from three populations. The new method is compared to results of a widely used assignment method, using simulated and published empirical data sets.
THE assignment of individuals to populations is a common population genetic application (Paetkau et al. 1995; Rannala and Mountain 1997; Pritchard et al. 2000; Dawson and Belkhir 2001; Corander et al. 2003; Baudouin et al. 2004; Guillot et al. 2005; François et al. 2006; Pella and Masuda 2006; Wu et al. 2006; Huelsenbeck and Andolfatto 2007; Zhang 2008; Reeves and Richards 2009). Most methods assume random mating within populations and free recombination between loci to find population assignments that minimize the amount of departure from Hardy–Weinberg equilibrium (HWE) and linkage equilibrium (LE) within populations. The population assignment with the highest likelihood, or highest posterior probability under Bayesian approaches, is taken as the assignment estimate. Generally the true allele frequencies are not known so that methods must either make use of estimated allele frequencies (e.g., Grant et al. 1980) or consider the range of possible allele frequencies (e.g., Pritchard et al. 2000).
Methods based on minimizing departure from HWE and LE typically offer little accommodation for the different potential causes of differentiation between populations. They allow populations to differ in allele frequencies, but they do so without including explicit evolutionary models of demography and mutation that cause such differences (Waples and Gaggiotti 2006; Listman et al. 2007). For example, it is possible that a method based on departures from HWE and LE will provide better estimates when divergence arises under an island model than when a similar amount of divergence arises under a phylogenetic branching model or vice versa. To assess such dependencies methods must be run on data simulated under various kinds of demographic histories. Without demographic history as a part of the model implemented in a method, such simulation studies tend to treat the computer program as a “black box” that can reveal interactions between demography and assignment only when observed in operation under different kinds of simulated histories (Evanno et al. 2005; Waples and Gaggiotti 2006; Chen et al. 2007; Fogelqvist et al. 2010).
In fact, likelihood-based methods for studying demographic histories offer a direct path to studying population assignment (e.g., Matz and Nielsen 2005; Nielsen and Matz 2006). For example, Kuhner et al. (1995) pioneered a Markov chain Monte Carlo (MCMC) approach (Metropolis et al. 1953; Hastings 1970) to sampling genealogies, which could be modified to include the population assignments of genes, at the terminal branches of genealogies, as free parameters. Here we take this general approach to study population assignment and demographic history by adapting an MCMC framework developed for the isolation-with-migration (IM) model for two or more populations (Hey and Nielsen 2007; Hey 2010b). The goals of this article are to describe a new method for estimating population assignment together with parameters of an IM model and to examine how well the method performs using simulated and real data and in comparison to another widely used assignment method that does not use an explicit divergence model
Models and Methods
The multipopulation IM model includes a population tree that is an ultrametric binary rooted tree (of populations) with a labeled history (Edwards 1970), in which nodes of the tree are ordered in time (Hey 2010b). A genealogy, represented by G, is also a bifurcating ultrametric tree that lies within the population tree and that describes, for all of the sampled gene copies of a locus, the times and topology of common ancestry and the times and directions of migration events. Individual loci have no recombination, but all of the loci, denoted by L, are assumed to be unlinked so that their corresponding genealogies are independent of each other. Every node of a genealogy is given the corresponding population label within which it falls in the population tree (see Figure 1).
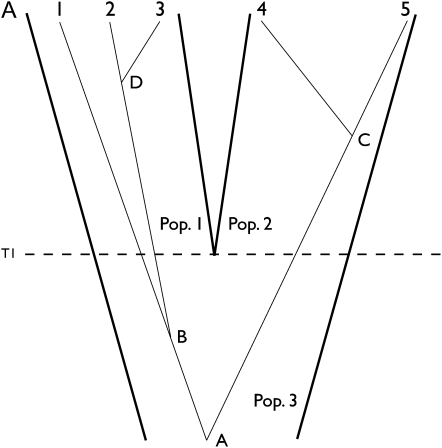
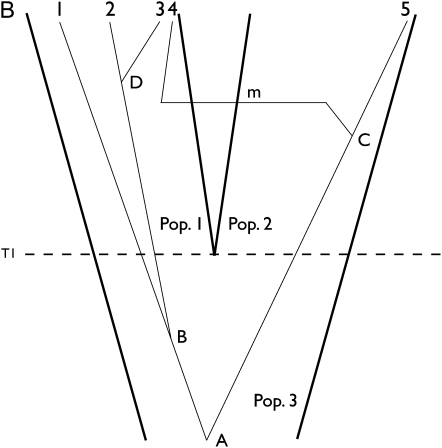
Figure 1 .
Assignment update for a genealogy with five gene copies overlaid upon a tree for two sampled populations. Population 3 splits into populations 1 and 2 at time T1. Two coalescent events A and B occur within population 3. Coalescent event D occurs within population 1 and event C within population 2. (A) A genealogy is drawn before reassigning gene 4 and (B) another after the reassignment. All four coalescent events remain unchanged when reassigning gene 4. The reassignment of gene 4 necessitates migration event m. The assignment of five individuals associated with the genealogy of A is (1, 1, 1, 2, 2), and that of B is (1, 1, 1, 1, 2).
Hey and Nielsen (2007) used MCMC to sample a multilocus genealogy G and splitting time t from the posterior distribution,
where f(X|G) is the likelihood of the data, π(G|t) and π(t) are prior distributions, and f(X) is the marginal likelihood. In the method of Hey and Nielsen (2007) the data, X, include gene copies from one or more loci that are already assigned to the populations from which they are sampled. We use f to denote likelihood and marginal-likelihood functions and π to designate prior and posterior distributions. It is possible to sample genealogies from the posterior distribution by having a closed-form expression for the prior distribution given by
where Θ is a vector of demographic parameters pertaining to population sizes and migration rates. Values of G drawn from the posterior distribution are used to estimate the posterior density of Θ given the data X,
| (1) |
where Ψ is the set of all possible genealogies, □ is the expectation over samples from π(G, t|X), and π(Θ|G, t) is the distribution of demographic parameters given genealogies and splitting times. Estimates of Θ are obtained by maximizing (1) over the range of parameter values (as specified by the prior distribution of Θ) using the posterior sample of G,
| (2) |
where J is the size of a posterior sample, π(Gj|tj, Θ) = π(Gj|Θ) is the probability of the jth posterior sample of genealogy given demographic parameters (Kingman 1982; Felsenstein 1992), and π(Θ|tj) is the prior of the demographic parameters. Typically in analyses of IM models the prior distribution on parameters is uniform over a specified range. In this case the posterior density is proportional to the likelihood over the range, permitting likelihood-ratio tests of nested demographic models (Hey and Nielsen 2007).
IM models with assignment
We consider data drawn from n individuals, with each individual having come from one of Kmax sampled populations. The genetic data without information on which populations individuals came from is denoted by Y. The genotype for individual i at locus l is a single-valued vector Yil for haploid data or a two-valued vector (Yil1, Yil2) for diploid data and the genotype for individual i, Yi, consists of L such vectors. Assignment A is a vector of size n with element Ai taking a value from 1 through Kmax. The number of sampled populations, K, can be less than Kmax because A can have zero individuals in some populations. The number of ways to assign n individuals to K populations is a Stirling number of the second kind (Bell 1934). All of the assignments are equally likely a priori. The posterior distribution of genealogy, splitting times, and assignment given data from unassigned individuals is
| (3) |
where f(Y|G) is the likelihood, π(G|t, A), π(t|A), and π(A) are priors, and f(Y) is the marginal likelihood. Note that because the likelihood f(Y|G) depends only on the topology and branch lengths of the genealogy, it does not depend on A.
MCMC with genealogies, splitting times, and assignment:
To estimate the posterior distribution (3) we employ an MCMC simulation in which we start with an initial value of (G(0), t(0), A(0)) and replace current values (G, t, A) by new values (G*, t*, A*) with acceptance probability
| (4) |
where q is a proposal density. An update of assignment requires relabeling branches on genealogies, which can entail changes to the structure of genealogies, including changes in coalescent times as well as migration times and directions thereof. Furthermore, when an individual is represented by multiple loci, an update of that individual’s assignment must be applied to the genealogies for all of the loci with genes sampled from that individual. Just as in the case of an update to a population splitting time, which applies to the genealogies for all of the loci in a study (Hey and Nielsen 2004), the acceptance rates of proposed assignment updates are expected to decrease as more loci are included in the study. Two update protocols for assignment were developed on the basis of previous methods for updating genealogies (Beerli and Felsenstein 1999; Nielsen and Matz 2006; Hey and Nielsen 2007). We first describe the approach of Nielsen and Matz followed by that of Beerli and Felsenstein.
Nielsen and Matz (2006) updating method:
Figure 1 shows a change of assignment on a genealogy in which five individuals are each represented by a single gene copy. Figure 1, A and B, shows a genealogy in a two-population model before and after gene copy 4 is reassigned from population 2 to 1. Prior to the reassignment migration events along the branch leading to the genes of the reassigned individual are erased, and new migration events are resimulated after the genes are reassigned (i.e., moved to a different population). Note that each internal node on the genealogy has a population designation, which depends upon the time of that node, the assignment of descendant gene copies, and migration events on branches leading to the descendant gene copies. When a sampled gene is reassigned to another population, its new label may become incompatible with the population label at the base of its branch. In Figure 1A the labels at nodes C and 4 are both population 2, but after gene 4 is moved to population 1 in Figure 1B its location is incompatible with the population label 2 on node C. This incompatibility is resolved by simulating migration events conditioned on the requirement that population labels at internal nodes be compatible with the population labels of sampled gene copies. With this type of update the probability of the data given the genealogy remains unchanged; i.e., f(Y|G*) = f(Y|G) because the update changes neither the topology nor the branch lengths of a genealogy. With a uniform prior for the splitting time and assignment, the acceptance probability of Equation 4 becomes
| (5) |
The Nielsen and Matz (2006) update is convenient because it does not require changing the tree topology and therefore does not require recalculation of the likelihood. However, this same feature, and the need to resimulate migration events to accommodate a fixed topology with a new assignment, leads to low acceptance rates when multiple loci must be updated simultaneously.
Beerli and Felsenstein (1999) updating method:
Beerli and Felsenstein (1999) devised an update in which a proposed genealogy is sampled from the prior distribution for genealogies, conditioned on the current values of the demographic parameters. We found that this method, adapted to accommodate assignment, performed better with data sets having multiple loci than did the Nielsen and Matz (2006) method and hereafter all results are based on the Beerli and Felsenstein (1999) method.
The Beerli and Felsenstein (1999) update begins by randomly selecting gene copies to be reassigned and proceeds by erasing the external branches for those gene copies, randomly selecting new population assignments, and simulating new branches. Beerli and Felsenstein called the new edge an “active lineage” and all the other lineages in the genealogy “inactive lineages”. The simulation of an active lineage can include adding migration events, and it ends in a coalescent event. When the active lineage coalesces at some time point, one of the inactive lineages is chosen as a new sister to the active lineage. To improve the acceptance rate of updates when the infinite sites mutation model (Kimura 1969) is used, we propose only genealogies that are compatible with the data under that model. When a finite sites mutation model is in use, the situation is more complex because selection of any inactive lineage as a sister to the active lineage will yield a tree that has a nonzero likelihood. We have not implemented the method for finite site mutation models.
The method of Beerli and Felsenstein requires the simulation of migration and coalescence using values of demographic parameters. However, unlike some MCMC-based genealogy samplers, the method of Hey and Nielsen (2007) does not include these parameters in the Markov chain, and so it was necessary to use values estimated from the current genealogy. Just as we estimate the posterior density for Θ using a larger sample of genealogies using (2), we can use the current genealogy Gj of a single locus to estimate Θ by maximizing
| (6) |
We describe one way to estimate Θ using a single genealogy in Appendix A. The acceptance probability αBF(G, t, A → G*, t*, A*) is given by
| (7) |
where f(Y|G*) ≠ f(Y|G), because of changes to the topology and branch lengths of the genealogy.
Implicit inference of the population tree:
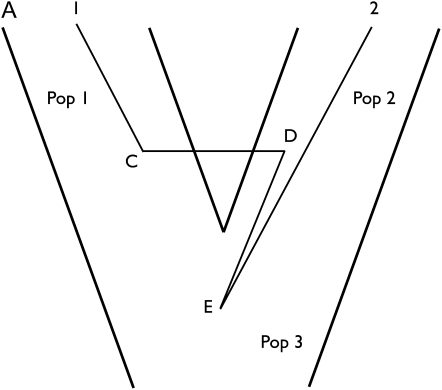
In the absence of a population tree, assignment information is limited to which individuals occur together in the same groups, and the actual population labels attached to groups are interchangeable. However, with a population tree, populations are not interchangeable but rather vary in how related they are to other populations, in which case assignment becomes a question not only of grouping individuals but also of where groups of individuals fall on the population tree; see also O’Meara (2010). Figure 2 shows an example in which sampling assignment, for populations on a tree, requires that we sample both the assignment and the population tree. The posterior distribution of genealogy, population tree, and assignment is
| (8) |
where τ is a population tree, which is taken to include the vector of splitting times t.
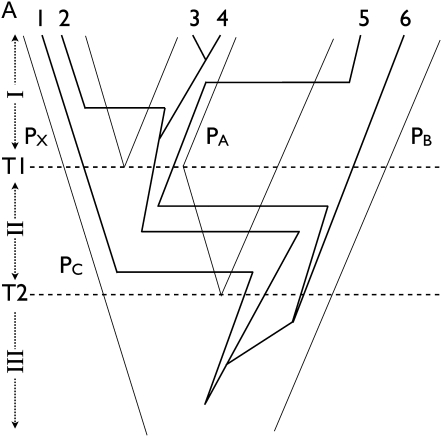
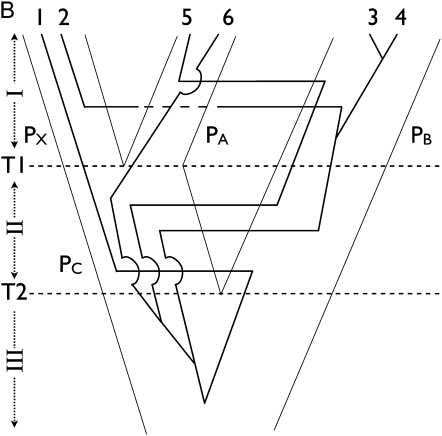
Figure 2 .
Three-point-turn update of the tree with three populations. (A) A tree is drawn with three populations (PX, PA, and PB). Splitting events T1 and T2 separate time in three periods, I, II, and III. A genealogy of six genes overlies the population tree and the actual assignment is (PX, PX, PA, PA, PB, PB). (B) We propose the population tree from the tree of A by swapping PA and PB only when there are no coalescent events during period II. Assignment of genes 1 and 2 remains unchanged while genes 5 and 6 are assigned to PA and genes 3 and 4 are assigned to PB. During period I, migration and coalescent events are labeled by swapping labels of PA and PB. During period II, events are labeled by swapping PC and PB only along the lineages that pass down from PA and PB and not from PX. The actual assignment changes to (PX, PX, PB, PB, PA, PA).
In trials using simulated data in a three-population IM model, neither the Nielsen and Matz (2006) nor the Beerli and Felsenstein (1999) protocols provided sufficient mixing of the MCMC simulation. Therefore we developed a method of updating the population tree and applied it to the case of three sampled populations. The acceptance probability with this update is
| (9) |
where the likelihood ratio disappears because f(Y|G*) and f(Y|G) cancel out. Figure 2 describes a population tree update, what we call the “three-point-turn”, in which a population branch first slides down along its sister branch to form a polytomy and then back up a different branch.
Summarizing assignment values sampled from the Markov chain
Huelsenbeck and Andolfatto (2007) introduced a method of summarizing a set of assignment values that are sampled from a posterior distribution. We employ this approach and adapt it to quantify assignment uncertainty and to estimate demographics jointly with assignment. Following Huelsenbeck and Andolfatto (2007) we make use of the concepts of “partition distance” between two assignments and of “mean assignment”. We explain these concepts in some detail because of their importance for jointly estimating population assignment and demographic history.
Partition distance:
The partition distance is the minimum number of individuals that must be removed to make two assignments equivalent (Almudevar and Field 1999; Gusfield 2002; Konovalov et al. 2005). For two assignments, with populations represented, respectively, we begin by relabeling each assignment using the restricted growth function (RGF) (e.g., Stanton and White 1986; Huelsenbeck and Andolfatto 2007). Under RGF indexing, individual assignments are numbered sequentially except that all individuals assigned to the same population are assigned the same label. For example, the RGF of assignment (3, 3, 1, 2) is (1, 1, 2, 3), where the first two individuals form a group, and each of the third and the fourth forms its own group. With the reindexed assignments A1 and A2, let d(A1, A2) be the number of individuals whose population assignment in A1 is different from what it is in A2, and let λ(A) be a relabeling in which the population labels in an assignment A are permuted. There are k! possible permutations of a set of k elements; e.g., six permutations of the ordered set {1, 2, 3} include (1, 2, 3), (1, 3, 2), (2, 1, 3), (2, 3, 1), (3, 1, 2), and (3, 2, 1). Because labels are ordered in an assignment, a relabeling operation λ(1, 2, 3) = (3, 2, 1) means that λ(1) = 3, λ(2) = 2, and λ(3) = 1. For the assignment with the most populations represented, there are s! possible relabeled assignments, where s = max(s1, s2), and s1 and s2 are the numbers of distinct populations in A1 and A2, respectively. For each possible relabeling of that assignment, we calculate d(A1, A2). The smallest of these values is the partition distance. For example, consider two assignments of (3, 1, 2) and (2, 2, 1). Their RGF values are (1, 2, 3) and (1, 1, 2), respectively. We choose the first one to relabel and fix the second. Among six possible assignments (1, 3, 2) differs from (1, 1, 2) in only one individual. The partition distance is 1.
When there are more than two populations, and they are connected by a population tree, not all labels are interchangeable. In this case we use the actual assignment samples without RGF conversion. For example, with three populations only two of six (3!) permutations are valid with a population tree of ((1, 2), 3) [i.e., (1, 2, 3) and (2, 1, 3)] because only these two permutations place population 3 as the outgroup to the sister pair of 1 and 2. If a binary tree of populations has Ks pairs of sister present-day populations, then there are only permutations that provide equivalent labelings. All of these labelings fall equivalently onto the same population tree. An example is shown in Table 1 for five assignment samples for a problem with six individuals and a population tree in which populations 1 and 2 are the most closely related: ((1 ,2), 3). We identify the partition distance that is constrained by the population tree, as the “tree-constrained partition distance”.
Table 1 . Examples of relabeling and distances for assignment samples.
| Sample no. | Sampled assignment by individual | Relabeling island modela | Partition distanceb | Relabeling tree model (1, 2), 3)c | Tree-constrained distanced | ||
|---|---|---|---|---|---|---|---|
| 1 2 3 4 5 6 | 1 2 3 4 5 6 | Ae | Bf | Ae | Bf | ||
| 1 | 2 2 1 1 3 3 | 1 1 2 2 3 3 | 0 | 1 | 1 1 2 2 3 3 | 0 | 1 |
| 2 | 2 2 3 3 1 1 | 1 1 2 2 3 3 | 0 | 1 | 1 1 3 3 2 2 | 4 | 3 |
| 3 | 1 1 2 2 3 3 | 1 1 2 2 3 3 | 0 | 1 | 1 1 2 2 3 3 | 0 | 1 |
| 4 | 3 2 1 2 1 1 | 1 2 3 2 3 3 | 2 | 3 | 3 1 2 1 2 2 | 4 | 3 |
| 5 | 2 2 1 2 3 3 | 1 1 2 1 3 3 | 1 | 2 | 1 1 2 1 3 3 | 1 | 2 |
| Sumg | 3 | 8 | 9 | 10 | |||
| Squaredh | 5 | 16 | 33 | 24 | |||
Five sampled assignments are shown for six individuals from three populations, with each individual having a population value of 1, 2, or 3. The third column represents one possible relabeling of the assignment. The fourth column shows the partition distance (see text) between two candidate assignments, A and B. The fifth column represents one of two possible relabeled assignments by considering a particular population tree. The sixth column shows the tree-constrained distances between assignments A and B with the assignment of the fifth column.
Relabeled assignments, of six possible relabelings, under island population structure.
The minimum number of individuals that have to be removed from a relabeled assignment (among all six possible relabelings) to calculate the distance to assignment A or B (Almudevar and Field 1999).
Relabels of the sampled assignment that is constrained by the population tree such that only populations 1 and 2 form an equivalence class.
The minimum number of individuals required to make the relabel with the tree constraint equivalent to the true assignment A or B.
Assignment A: 1 1 2 2 3 3.
Assignment B: 1 1 2 2 2 3.
Sum of the five distances.
Sum of the five squared distances.
Determining the mean assignment:
The method of mean assignment developed by Huelsenbeck and Andolfatto (2007) is to summarize a posterior sample of assignment values by minimizing the squared partition distance to the sampled assignment values. To extend the idea of mean assignment to the case where we are given a population tree constraint, we define the tree-constrained mean assignment as the assignment that minimizes the tree-constrained partition distance to all sampled assignments. The tree-constrained mean assignment minimizes the tree-constrained partition distance, rather than the square of it, as used by Huelsenbeck and Andolfatto, because squared tree-constrained partition distances can be inconsistent with the partition distance. For example, using the total sample of size 5 in Table 1, the sum of the squared tree-constrained distances of assignment A is larger than the one using assignment B although assignment A is closer to the five samples than is assignment B.
Quantifying assignment uncertainty:
The unit of a partition distance is the number of individuals, and as such it is fairly easy to interpret a given value. For example, consider a case of samples from two populations for which the true assignment is known. Then a distance between a sampled assignment and the true value that is half the number of individuals in the study would suggest that the assignment was no better than random with respect to the true value (i.e., half the individuals are correctly assigned and half are not).
The partition distance also lends itself to quantifying each individual’s assignment uncertainty, which we define as the proportion of sampled assignment values in which that individual must be removed in the calculation of the distance between an assignment and the mean assignment. The more often an individual is removed in the procedure of computing assignment distance, the less confident we can be that the individual is assigned to the population in which it occurs in the mean assignment. Another useful measure of assignment uncertainty is the variance of a mean assignment, which is the mean squared partition distance between sampled assignment values and the mean assignment (Huelsenbeck and Andolfatto 2007). Similarly the standard deviation, in units of individuals, is the square root of the variance. This measure can serve as an indication of the number of individuals that are typically uncertain in their assignment.
Joint estimation of assignment and demographic parameters
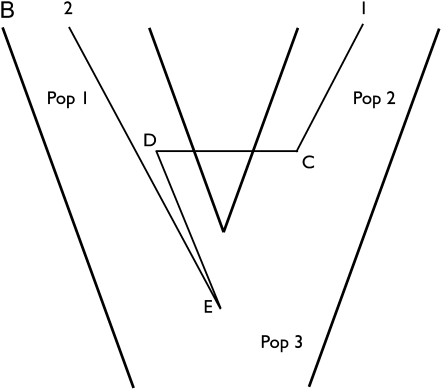
We developed two methods for jointly estimating assignment and population-specific demographic parameters. The first algorithm, joint demography and assignment (JDA), can be applied to an island model or a two-population model with a single splitting time. The second algorithm, joint demography and assignment with population tree (JDAP), can be applied when there are more than two populations on a population phylogeny. To understand these we first turn to the matter of “label switching”, in which the effective identity of populations changes with assignment (Stephens 2000). Consider a case of four individuals, with individuals 1 and 2 assigned to population A and individuals 3 and 4 assigned to population B. In a model with just two populations this assignment is equivalent to one in which individuals 3 and 4 are assigned to A and 1 and 2 are assigned to B. Failing to identify this equivalence will prevent the joint estimation of demographic parameters and assignment. Figure 3 shows a simple case with two gene copies in two populations. Note that in Figure 3, A and B are equivalent, but reversed with respect to population labels 1 and 2. If we simply use the two genealogies without any modification, population 1 is represented by gene 1 in Figure 3A and by gene 2 in Figure 3B. If gene 1 and gene 2 are sampled from two different populations, then we wish to estimate parameters that are associated with one population using genealogies in which that population contains either gene 1 or gene 2 but not a mixture of both. To accommodate these kinds of equivalencies, by following Stephens (2000) we relabel the genealogy in either Figure 3A or 3B by swapping the labels of populations 1 and 2 as well as the population labels for internal nodes C and D for this genealogy. The swapping operation on genes with labels 1 and 2 is denoted by ν(A) and the additional swapping of internal nodes is denoted by ν(G). Because the posterior distribution of assignment and genealogies is invariant to population label changes, under an island model and under a two-population IM model (see Appendix B), we can find the estimate and the list of permutations ν1, … , νJ that jointly maximizes (see Equations 1 and 2). This maximization is reminiscent of Algorithm 4.1 of Stephens (2000). Because this approach would be extremely slow due to the very large number of searches for the highest posterior probability, we use an approximation based on the mean assignment.
Figure 3 .
Label-switched genealogies. Genealogies of A and B are equivalent except that the labels are switched and the assignment of genealogy A is (1, 2), while that of genealogy B is (2, 1). The population labels at the two migration starting and ending points are also switched. The population label at the root node E remains unchanged because it is allotted to the ancestor population. Likelihoods of the two genealogies are equal, and their prior probabilities of the genealogies are also the same.
JDA algorithm under an island model:
Rather than maximizing the joint posterior density, for each of all possible permutations of sampled assignments we use the mean assignment and partition distance to approximate a list of permutations, which are then used to calculate the joint posterior density. The algorithm proceeds as follows:
Step 1: Find the mean assignment of a posterior sample of J assignments (A1, … , AJ).
Step 2: Identify the sequence of permutations {νj}j∈{1,…,J} such that for each Aj is the smallest among the possible K! relabelings.
Step 3: In Equation 2 replace π(Θ|Gj, t) with π(Θ|ν(Gj), t) to estimate the demographic parameters.
Note that we use the same prior for all population sizes or migration rates for populations that are a priori interchangeable, which renders the prior of the relabeled genealogy π(ν(G)) equal to the original prior π(G) (see Appendix B).
JDAP algorithm:
When sampling assignments from the posterior distribution we are also implicitly sampling the population tree that is associated with each of the assignments (see Equations 8 and 9). Demographic parameters can be meaningful only under a particular population tree. If two population trees are different, the ancestral population in one of the two population trees can lose its meaning in the other tree. Because population assignment and population tree are confounded, we first find disjoint sets of a posterior sample of population assignment on the basis of its proximity to population trees implied by population assignment. For a population tree with a considerable size of population assignment, we estimate demographic parameters that are pertinent to the population tree. For example, assignment sample 4 in Table 1 has only individual 1 in population 3 while assignment samples 1, 3, and 5 have individuals 5 and 6 assigned to population 3. Because population 3 is the outgroup in the population tree in the model ((1, 2), 3), these two groups of sampled assignments imply different phylogenies for the actual populations from which these samples came. Similarly the two groups of samples invoke different meanings for the demographic parameters associated with the outgroup population 3. Because the implied phylogeny and the meaning of the demographic parameters change with sampled assignment values, we use an algorithm for estimating the posterior density in which genealogy and assignment samples are grouped on the basis of which population tree they are most strongly associated with. To find the most close population tree of a population assignment we use the measure of tree-constrained partition distance and tree-constrained mean assignment. For three populations there are three groups of samples corresponding to the three rooted population trees. The algorithm begins with a categorization and proceeds as follows:
Step 1: Find the mean assignment of a posterior sample of J assignments (A1, … , AJ).
Step 2: Identify the sequence of permutations {νj}j∈{1,…,J} such that is the smallest among the K! possible relabelings for each Aj.
Step 3: Categorize each sampled assignment and genealogy, Aj and Gj, to groups on the basis of the corresponding population trees by using the sequence of permutations {νj}j∈{1,…,J}.
Step 4: That population tree that occurs most often in the categorized sample is considered to be that with the highest posterior probability. Only those assignments and genealogies that are associated with this tree are retained, and these are renumbered from 1 to Jc, where Jc is the total number of the retained values of A and G. The demographic parameters of the population tree are estimated using just the categorized subsample of genealogies because other genealogies may be related to disparate population trees. The remainder of the JDAP algorithm is similar to the JDA algorithm, but is adapted to the case of an IM model with a population tree.
Step 5: Find the tree-constrained mean assignment of the categorized subsample of assignments with the constraint of the best supported tree.
Step 6: Identify the sequence of permutations such that is the smallest among the possible relabelings for each Aj.
Step 7: Maximize Equation 2, with π(Θ|Gj, t) replaced by π(Θ|νj(Gj), t) to find the demographic parameter estimate that is associated with the population tree
We have not yet implemented the case where four or more populations are related by a binary species tree. In these cases the underlying topology of the population tree will also be variable.
Computer programs:
The IMa2 computer program (Hey 2010b) was modified to incorporate the methods described here, using a uniform prior on assignment. Data were simulated using a coalescent simulator program SIMDIV (Wang and Hey 2010) that generates data under IM models. These programs are available at http://genfaculty.rutgers.edu/hey/software. We also used the STRUCTURAMA program (Huelsenbeck and Andolfatto 2007) as a representative allele-based method. STRUCTURAMA implements a method that is similar to that developed by Pritchard et al. (2000), which is implemented in the STRUCTURE program. However STRUCTURAMA employs an analytic integration over allele frequencies, rather than Gibbs sampling of allele frequencies, and it does not require that the user specify the number of populations. STRUCTURAMA is also unique among population assignment programs in that it provides estimates of the mean assignment, which we use for comparison to the methods described here. We confirmed that for a fixed number of populations STRUCTURAMA uses a uniform prior on assignment by running the program for a fixed four-population model without data. The results revealed a uniform posterior probability of assignment, equal to the prior in the absence of data (results not shown).
Data
Four groups of simulated data sets were used to assess how well the new methods perform and to compare results for the new methods with a method that does not include a demographic model. We also analyzed and compared results for two real data sets drawn from the literature.
Simulated data
Simulated data sets were based on a two- or three-population IM model with parameters for sampled population and ancestral population sizes, splitting times and migration rates (Nielsen and Wakeley 2001; Hey and Nielsen 2004). Data were simulated under the infinite sites mutation model. MCMC conditions (in terms of burn-in duration, length of chain, heating scheme, and numbers of Metropolis-coupled chains) were determined on the basis of preliminary runs for representative simulated data sets to ensure samples of effectively independent genealogies and assignments. Results were compared to those obtained for the same data using the program STRUCTURAMA, which did not incorporate a mutation model or a demographic model. Because STRUCTURAMA required allelic data, it was necessary to convert simulated DNA sequences to alleles (i.e., each distinct sequence was given an allele designation). This conversion of sequences to alleles necessarily incurred loss of information. Because the new method uses the infinite sites model and does not require that sequence data be reduced to alleles, we expected that it would do a better job inferring assignment than one that does not.
Simulation set 1—varying the number of loci:
Data sets were simulated with varying numbers of loci with demographic parameters as follows: θ1 = θ2 = θA = 1, t = 0.1, and m12 = m21 = 0.1. A single gene copy was sampled from each of 20 individuals for each population, and the number of loci, L, ranged from 1 to 10. For each value of L, 50 independent data sets were generated. Prior distributions for parameters were uniform from zero to a maximal value: 10 for population mutation rates, 1 for population migration rates, and 3 for splitting time.
Simulation set 2—varying population size, migration rates, and splitting time:
Several values were considered for each of the parameters of the two-population IM model: population mutation rate θ’s were set to 0.5, 1.0, or 2.0 (in each case all three populations had the same size); population migration rates, m12 × θ and m21 × θ in both directions were set to 0.0, 0.1, or 1.0; and scaled splitting time, t/θ, was set to 0.05, 0.10, or 0.20. Twenty gene copies were sampled from each population, with four loci per data set; and 50 data sets were simulated for each of the 27 sets of parameter values. Parameter priors were the same as in simulation 1 except for the maximum of splitting time t being set to 10.
Simulation set 3—joint estimation of assignment and demographics:
To assess the quality of joint estimates of assignment and demography, data sets were simulated under models with recent divergence and/or high gene flow. Such models yield assignments with high uncertainty and it is in these types of situations where we want to see how well demography can be estimated together with assignment. Two models were used, one with no migration and splitting time set to t/θA = 0.05 and a second with migration rates of 0.5 in both directions and a splitting time of t/θA = 0.10. For both models population size parameters were set to unequal values (θ1 = 1, θ2 = 3, and θA = 2). Each data set consisted of 10 loci with 20 gene copies from each sampled population, and 50 independent data sets were generated for each set of parameter values.
Simulation set 4—implicit inference of population tree:
To assess how well sampled assignment values can reveal the population tree, we consider 12 three-population IM models. We assumed that all of the five populations (three present and two ancestral) share the same population size, with θ set to 1 in all of the simulations. The older splitting time was fixed to 0.5 in all of the 12 cases, while the most recent splitting time took on values of 0.1, 0.2, 0.3, and 0.4. Instead of just two migration rates (as is the case in a two-population IM model) a three-population model has eight migration rates (Hey 2010b). We considered three values for population migration rates (0, 0.1, and 1), in each case setting all of the migration rates to the same value. Upper bounds on the prior distributions were 3 for θ, 2 for t, and 2 for m. Each data set consisted of seven gene copies at each of four loci. We generated 30 replicates for each of the 12 parameter sets.
Real data sets
Two data sets drawn from the literature are described below and listed in Table 2 with the options used for the IMa2 and STRUCTURAMA programs. Unlike the simulation studies we do not know with certainty that the actual species assignments, in the original reference for each empirical data set that was used, are correct; and we acknowledge that apparently “incorrect” or uncertain assignment estimates could be due to mistaken assignments in the original study. Most of the loci in the data sets were diploid with two gene copies per locus sampled per individual. For the purposes of this paper the loci were treated as haploid with one gene copy sampled from each individual at each locus.
Table 2 . Runtime settings for IMa2 and STRUCTURAMA for the two real data sets: mouse (Geraldes et al. 2008) and chimpanzee (Fischer et al. 2006).
| Data | La | Kb | nc | Programd | me | Burn-inf | Total | Samples | hng | Bmaxh | θmaxi | Tmaxj | Mmaxk |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 7 | 2 | 113 | IM tree | IS | 1 × 105 | 1 × 106 | 5 × 105 | 40 | 0.85 | 10 | 5 | 0.5 |
| Mouse | 7 | 2 | 113 | ST | NA | 1 × 104 | 1 × 105 | 1 × 104 | 10 | NA | NA | NA | NA |
| Chimp | 10 | 4 | 39 | IM island | IS | 5 × 104 | 2 × 106 | 4 × 104 | 150 | 0.50 | 5 | NA | 1 |
| Chimp | 10 | 4 | 39 | ST | NA | 1 × 104 | 1 × 105 | 1 × 104 | 100 | NA | NA | NA | NA |
| Chimp | 9 | 2 | 20 | ST | NA | 1 × 104 | 1 × 105 | 1 × 104 | 10 | NA | NA | NA | NA |
| Chimp | 9 | 3 | 30 | IM tree | IS | 1 × 105 | 2 × 106 | 2 × 104 | 150 | 0.10 | 2 | 1.5 | 0.0001 |
Number of loci.
Number of populations: IMa2 was run with either a tree or an island structure of K populations, and STRUCTURAMA was run with a fixed number of K populations.
Number of individuals.
IM tree, IMa2 with a population tree; IM island, IMa2 with an island; ST, STRUCTURAMA.
Mutation model for IMa2. IS, infinite sites model; NA, not applicable.
Burn-in, total, and samples are in generations. After steps of burn-in, we take samples from total generations; one generation for IMa2 tries to update the population label of a single individual whereas one generation for STRUCTURAMA tries to update the labels of multiple individuals.
The number of Metropolis-coupled chains (Geyer 1991).
The heating level of the most heated chain.
Maximum of uniform prior of population size.
Maximum of uniform prior of splitting time.
Maximum of uniform prior of migration rates.
Mouse data set:
Geraldes et al. (2008) studied the divergence of mouse species using eight loci. We focused on samples for Mus domesticus and M. castaneus with seven of the loci, excluding the mitochondrial control region locus because of lack of individual information for these species at this locus. The total number of individuals was 113: 55 from M. domesticus and 58 from M. castaneus. A two-population IM model with the infinite sites mutation model was used.
Chimpanzee data set:
Fischer et al. (2006) studied divergence among chimpanzee taxa, including West (Pan troglodytes verus), East (P. t. schweinfurthii), and Central African common chimpanzees (P. t. troglodytes) and bonobos (P. paniscus). The population tree of the four populations was estimated as ((Eastern, Central), Western), bonobo) (Becquet et al. 2007; Caswell et al. 2008). We used a partial data set of 10 loci under the infinite sites mutation model. The total number of individuals was 39: 9 from bonobos and 10 from each of the chimpanzee populations. Analyses were conducted under a four-population islands model as well as an IM tree model of the three populations of common chimpanzee. In the three-population analysis, without the bonobo, 1 of the 10 loci was not variable and so was excluded.
Results
Simulation study
Simulation set 1:
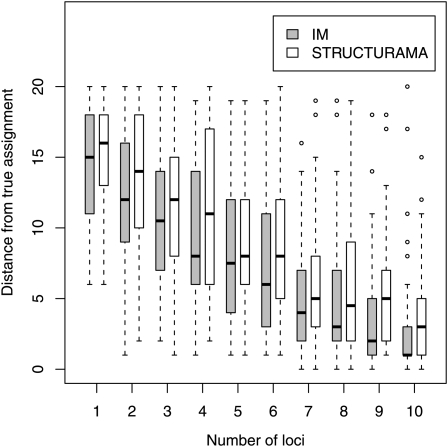
The effect of varying the number of loci is shown in Figure 4. With 20 individuals from each population the largest possible partition distance is 20. Although results for both methods show very little resolution for assignment with small numbers of loci, the trends are useful for seeing how the methods compare as the number of loci is increased. For both IMa2 and STRUCTURAMA the distances from the true value become smaller with more loci, and both programs start recovering the true assignment (i.e., some data sets had a mean assignment equal to the true value) with data sets of seven loci. The two programs performed qualitatively similarly, with the median distances from the true value for IMa2 runs consistently less than those for STRUCTURAMA runs.
Figure 4 .
The result of simulation 1 for assessing the effect of numbers of loci on accuracy of assignment. For each simulated data set the distance of the mean assignment from the true assignment was recorded. Each box plot includes the median of the 50 distances and is as large as the interquartile range or the first to the third quartile. The whiskers extend to the most extreme data point but not more than 1.5 times the interquartile range.
Simulation set 2:
Table 3 shows the effect of varying the parameters of a two-population IM model for a model with four loci. Assignment accuracy decreased for more recent divergence times, larger migration rates, and smaller present-day population sizes. To compare the assignment performance of the two methods we counted the number of data sets in which IMa2 inferred assignments better than, worse than, or as good as STRUCTURAMA. One method performed no better than the other in the cases of θ = 0.5 or 1.0 and those of t/θ = 0.05 or 0.10. IMa2 outperformed STRUCTURAMA in the cases of θ = 2 and those of t/θ = 0.2.
Table 3 . The result of simulation 2 for assessing population assignment inference with the two-population IM model by varying population sizes, splitting time, and migration rates.
| Summaryd | Comparison | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| θa | t/θb | θ × mc | IM | ST | IMe | STf | Tidedg | ||
| 0.5 | 0.05 | 0 | 15.5 | (8, 20) | 15.0 | (8, 20) | 19 | 18 | 13 |
| 0.5 | 0.05 | 0.1 | 14.0 | (7, 20) | 14.0 | (7, 20) | 21 | 21 | 8 |
| 0.5 | 0.05 | 1 | 15.5 | (9, 20) | 16.0 | (9, 19) | 21 | 19 | 10 |
| 0.5 | 0.10 | 0 | 12.0 | (2, 19) | 11.5 | (2, 20) | 25 | 16 | 9 |
| 0.5 | 0.10 | 0.1 | 10.5 | (3, 19) | 10.5 | (3, 20) | 22 | 22 | 6 |
| 0.5 | 0.10 | 1 | 14.5 | (5, 20) | 14.0 | (4, 20) | 18 | 19 | 13 |
| 0.5 | 0.20 | 0 | 6.0 | (0, 15) | 7.0 | (0, 17) | 26 | 7 | 17 |
| 0.5 | 0.20 | 0.1 | 5.0 | (0, 17) | 6.0 | (0, 19) | 28 | 9 | 13 |
| 0.5 | 0.20 | 1 | 11.0 | (1, 20) | 10.5 | (2, 19) | 24 | 15 | 11 |
| 1 | 0.05 | 0 | 13.5 | (6, 20) | 14.0 | (5, 20) | 19 | 21 | 10 |
| 1 | 0.05 | 0.1 | 15.5 | (7, 20) | 16.0 | (9, 20) | 24 | 17 | 9 |
| 1 | 0.05 | 1 | 14.0 | (9, 20) | 15.0 | (7, 19) | 25 | 16 | 9 |
| 1 | 0.10 | 0 | 7.0 | (2, 20) | 8.5 | (2, 18) | 27 | 15 | 8 |
| 1 | 0.10 | 0.1 | 8.5 | (1, 20) | 10.0 | (2, 19) | 27 | 16 | 7 |
| 1 | 0.10 | 1 | 11.0 | (4, 20) | 11.5 | (4, 18) | 26 | 20 | 4 |
| 1 | 0.20 | 0 | 3.0 | (0, 14) | 4.0 | (0, 17) | 28 | 5 | 17 |
| 1 | 0.20 | 0.1 | 3.0 | (0, 17) | 5.0 | (0, 18) | 36 | 7 | 7 |
| 1 | 0.20 | 1 | 6.0 | (1, 20) | 7.0 | (1, 18) | 32 | 8 | 10 |
| 2 | 0.05 | 0 | 9.5 | (5, 19) | 12.0 | (4, 19) | 32 | 9 | 9 |
| 2 | 0.05 | 0.1 | 12.0 | (3, 19) | 13.0 | (4, 20) | 25 | 20 | 5 |
| 2 | 0.05 | 1 | 13.0 | (6, 19) | 13.5 | (5, 19) | 24 | 22 | 4 |
| 2 | 0.10 | 0 | 5.0 | (0, 18) | 7.0 | (1, 18) | 34 | 10 | 6 |
| 2 | 0.10 | 0.1 | 5.0 | (1, 17) | 6.5 | (2, 17) | 35 | 9 | 6 |
| 2 | 0.10 | 1 | 7.0 | (1, 19) | 8.0 | (2, 20) | 33 | 8 | 9 |
| 2 | 0.20 | 0 | 0.0 | (0, 5) | 2.0 | (0, 7) | 33 | 1 | 16 |
| 2 | 0.20 | 0.1 | 1.0 | (0, 7) | 2.0 | (0, 11) | 22 | 5 | 23 |
| 2 | 0.20 | 1 | 3.0 | (0, 14) | 5.0 | (0, 17) | 34 | 3 | 13 |
Fifty data sets were simulated for each row.
Population sizes, 2Nμ.
Splitting time scaled by θ, and the splitting time itself is the product of time in generations and mutation rate per generation.
Migration rate scaled by θ.
Summary of the 50 distances from true to mean assignments using IMa2 (IM) and STRUCTURAMA (ST): The median and two quantiles (2.5% and 97.5%), within parentheses and separated by a comma, are shown.
The number of cases in which the distance from IMa2 is shorter than that from STRUCTURAMA.
The number of cases in which the distance from IMa2 is longer than that from STRUCTURAMA.
The number of cases in which the two distances are equal.
Simulation set 3:
Table 4 shows how well the method performed for the joint estimation of assignment and demography. For t = 0.05 and no migration, the distance between the mean and true assignments was 13.68, which was quite large for a total sample of 40 gene copies per locus. Under this model the estimates of t tended to be much lower when assignment was variable than when it was fixed. Also under this model, the upper 95% quantile for θ1 was much higher (near the upper bound for this parameter) when assignment was variable. The same patterns for t and θ1 were seen for the second model in which the splitting time was greater (t = 0.1) but there was substantial gene flow. Under this model the distance between the mean and true assignments was 6.64. In both of these models, regardless of whether assignment was fixed or variable, the estimated posterior distributions for migration rates were nearly flat and the population size estimates had a substantial bias, with estimated values tending to be between 65% and 85% of the true value. In short, the conditions of low divergence and modest data set size that made assignment difficult to estimate were also those in which it was difficult to estimate demographic parameters.
Table 4 . The result of simulation 3 for assessing joint estimation of assignment and demographics.
| ta (m12b, m21c) assignmentd | 0.05 fixed | (0, 0) variable | 0.10 fixed | (0.5, 0.5) variable |
|---|---|---|---|---|
| Distancee(0) | NA | 13.68 | NA | 6.64 |
| (5.225, 19.775) | (1.225, 17) | |||
| Splitting timef | 0.05 | 0.014 | 0.074 | 0.03 |
| (0.006, 0.122) | (0.002, 0.102) | (0.022, 0.198) | (0.002, 0.178) | |
| θ1(1.0)g | 0.845 | 0.755 | 0.745 | 0.695 |
| (0.255, 4.665) | (0.385, 9.675) | (0.315, 2.535) | (0.245, 9.475) | |
| θ2(3.0)h | 1.665 | 1.245 | 1.945 | 1.525 |
| (0.695, 9.615) | (0.555, 9.695) | (0.815, 9.405) | (0.595, 9.645) | |
| θA(2.0)i | 1.475 | 1.565 | 1.515 | 1.555 |
| (0.925, 2.555) | (0.915, 2.515) | (0.935, 2.635) | (0.985, 2.605) |
For each simulation set the estimates with 2.5% and 97.5% quantile values are given for partition distances and demographic parameter estimates from 50 independent simulations.
Splitting time of the two-population IM model. The three true values of splitting time are shown in the column headings.
True migration rate from population 1 to 2.
True migration rate from population 2 to 1.
Assignment is fixed or variable in the model.
Distances between the mean assignment and the true assignment.
Splitting time estimates.
Size of population 1: The true value is 1.0.
Size of population 2: The true value is 3.0.
Size of the ancestor of populations 1 and 2: The true value is 2.0.
Simulation set 4:
Table 5 shows results for inferring the tree with three populations, using the three-point-turn update, for different migration rates and splitting times. As expected, the probabilities for the correct tree were highest for lower migration rates and when the time between the two splitting events was greater.
Table 5 . The result of simulation 4 for assessing the inference of the tree with three populations.
| ma | tb | Tree 1c | Tree 2d | Tree 3d |
|---|---|---|---|---|
| 0 | 0.1 | 0.7282 (0.2082) | 0.1315 (0.1058) | 0.1390 (0.1077) |
| 0 | 0.2 | 0.5451 (0.1646) | 0.2200 (0.0854) | 0.2334 (0.1051) |
| 0 | 0.3 | 0.4356 (0.1941) | 0.2875 (0.1249) | 0.2755 (0.1062) |
| 0 | 0.4 | 0.3793 (0.1604) | 0.3123 (0.1117) | 0.3070 (0.1071) |
| 0.1 | 0.1 | 0.7217 (0.1731) | 0.1359 (0.0876) | 0.1411 (0.0908) |
| 0.1 | 0.2 | 0.5386 (0.2014) | 0.2462 (0.1252) | 0.2139 (0.0910) |
| 0.1 | 0.3 | 0.4480 (0.1203) | 0.2874 (0.1044) | 0.2634 (0.0547) |
| 0.1 | 0.4 | 0.4177 (0.1569) | 0.2981 (0.1024) | 0.2828 (0.1167) |
| 1 | 0.1 | 0.5662 (0.1972) | 0.2249 (0.1430) | 0.2076 (0.1309) |
| 1 | 0.2 | 0.4150 (0.1547) | 0.2843 (0.1205) | 0.2994 (0.1208) |
| 1 | 0.3 | 0.3948 (0.1211) | 0.3044 (0.1060) | 0.2993 (0.1223) |
| 1 | 0.4 | 0.3341 (0.1225) | 0.3756 (0.1226) | 0.2889 (0.0947) |
The proportions of one of three trees in posterior samples are shown with their standard deviation in parentheses.
Migration rates between all of the pairs of populations in which migration events are possible under each population tree.
The time at which the two sister populations emerge from their common ancestor. The time at which the ancestor of all of the three populations splits is 0.5.
The true population tree.
The false population trees.
Real data analysis
Mouse data set of Geraldes et al. (2008):
Both IMa2 and STRUCTURAMA estimated the true assignment correctly. The average squared distance of the assignment to posterior samples for IMa2 was 0.6. The average squared distance using STRUCTURAMA was considerably higher at 9, which was equivalent to a standard deviation of three individuals having assignment different from the mean assignment. In a model in which population assignment was an unknown, the estimated demographic parameter values were similar to those obtained in a model in which population assignment was fixed (Table 6). This demonstrated that assignment could be studied jointly with demography, but was not surprising in this case because population assignment was estimated correctly with low uncertainty. Population size of M. domesticus was estimated to be smaller than half the population size of M. castaneus. Their ancestor was estimated to be smaller in population size than M. castaneus and larger than M. domesticus. The migration rate was estimated to be nonzero in the direction of M. domesticus to M. castaneus. These patterns were very similar to those reported by Geraldes et al. (2008) for the full data set for these species.
Table 6 . Demographic parameter estimates and their 95% confidence intervals in parentheses with assignment fixed to the true and assignment being a random variable using the Geraldes data set.
| Assignment | θ1a | θ2b | θAc | m12d | m21e | tf |
|---|---|---|---|---|---|---|
| Fixed | 1.105 | 3.475 | 2.865 | 0.00025g | 0.04625 | 1.438 |
| (0.725, 1.675) | (2.545, 5.015) | (0.525, 7.975) | (0.00125, 0.1787) | (0.00725, 0.2072) | (1.048, 4.577) | |
| Variable | 1.095 | 3.515 | 2.905 | 0.00025g | 0.03025 | 1.518 |
| (0.7450, 1.675) | (2.585, 5.035) | (0.555, 8.045) | (0.00175, 0.2422) | (0.00325, 0.1862) | (1.032, 4.433) |
Population size of M. domesticus.
Population size of M. castaneus.
The common ancestor’s population size.
Migration rate into M. domesticus from M. castaneus.
Migration rate into M. castaneus from M. domesticus.
Time at which the common ancestor of the two populations split.
Corresponds to the first bin of the histogram, which represents zero.
Chimpanzee data of Fischer et al. (2006) under an island model of population structure:
In Table 7 the column labeled “IMa2” shows the mean assignment for these data under an island model for four populations. While most individuals were assigned to their reported subspecies, IMa2 assigned East African common chimpanzee individuals Alley and Sultana to the same population as all of the Central African chimpanzee individuals. The average squared distance from the mean assignment under IMa2 was 11, corresponding to a standard deviation of 3.31 individuals. Individual assignment uncertainties for the East and Central African chimpanzees were larger than those for the bonobo and West African populations, with bonobos having the smallest assignment uncertainties. Three chimpanzees that were of higher individual assignment uncertainty were Alley, Judy, and Sultana among which Alley and Sultana were incorrectly assigned. STRUCTURAMA was also run on the chimpanzee data, with a setting of a maximum of four populations. Interestingly only three populations were inferred, with all of the Central and East African individuals estimated to have come from a single population (Table 7). The average squared distance of the mean assignment was 3. Individual assignment uncertainties for bonobo and Central and West African populations were smaller than those for East African populations. The four chimpanzee populations had estimated divergence times that varied over approximately an order of magnitude (Won and Hey 2005; Becquet et al. 2007; Caswell et al. 2008; Hey 2010a). To see whether the inclusion of the most divergent populations might have contributed to the failure by STRUCTURAMA to resolve four populations, a two-population model with STRUCTURAMA was run with only the 20 chimpanzees of the Central and East African populations. In this case [Table 7, column ST(2)] two populations were inferred rather than a single population. Three East African chimpanzees, Alley, Judy, and Sultana, were assigned to Central African chimpanzees, and Chiquita and Clara from the Central African population were assigned to the East African population. The average squared distance of the assignment to the posterior sample was 32. Individual assignment uncertainties were rather large. For the case of a four-population island model the population size estimates obtained jointly with assignment were in three cases about one-third lower than the values estimated under a fixed true assignment (Table 8). Central African chimpanzee population size was estimated to be larger than that estimated from the fixed assignment presumably because IMa2 assigned a few more individuals of the East African population to the Central African population. However, the order of magnitude of population size estimates remained unchanged: Central African, East African, bonobo, and West African populations in decreasing order of estimated value.
Table 7 . Population assignment of chimpanzees (three subspecies) and Bonobos.
| Individual | Referencea | IMa2b | STc | ST(2)d | IMa2(3)e | ||||
|---|---|---|---|---|---|---|---|---|---|
| Bono | 1 | 1 | 0.000000 | 1 | 0.000900 | ||||
| Catherine | 1 | 1 | 0.000800 | 1 | 0.000700 | ||||
| Joey | 1 | 1 | 0.000850 | 1 | 0.000300 | ||||
| Kombate | 1 | 1 | 0.000000 | 1 | 0.000300 | ||||
| Kosana | 1 | 1 | 0.000050 | 1 | 0.003000 | ||||
| Sandy | 1 | 1 | 0.000050 | 1 | 0.000100 | ||||
| Ulindi | 1 | 1 | 0.000800 | 1 | 0.000200 | ||||
| Yasa | 1 | 1 | 0.000750 | 1 | 0.000400 | ||||
| Zorba | 1 | 1 | 0.000050 | 1 | 0.000800 | ||||
| Akila | 2 | 2 | 0.040275 | 2 | 0.012300 | 1 | 0.151200 | 1 | 0.043350 |
| Alley | 2 | 3 | 0.318325 | 2 | 0.021200 | 2 | 0.335100 | 2 | 0.243950 |
| Amizero | 2 | 2 | 0.025200 | 2 | 0.005500 | 1 | 0.098000 | 1 | 0.028850 |
| Annie | 2 | 2 | 0.032100 | 2 | 0.024200 | 1 | 0.096300 | 1 | 0.030200 |
| Eva | 2 | 2 | 0.083275 | 2 | 0.003200 | 1 | 0.319600 | 1 | 0.162800 |
| Judy | 2 | 2 | 0.567625 | 2 | 0.042100 | 2 | 0.313800 | 2 | 0.432500 |
| Mary | 2 | 2 | 0.082400 | 2 | 0.002300 | 1 | 0.324100 | 1 | 0.156850 |
| Mimi | 2 | 2 | 0.032325 | 2 | 0.003400 | 1 | 0.170000 | 1 | 0.040700 |
| Mzee | 2 | 2 | 0.084200 | 2 | 0.001800 | 1 | 0.319900 | 1 | 0.159950 |
| Sultana | 2 | 3 | 0.434900 | 2 | 0.044700 | 2 | 0.316900 | 1 | 0.561900 |
| Chiquita | 3 | 3 | 0.062750 | 2 | 0.046700 | 1 | 0.388700 | 2 | 0.136050 |
| Clara | 3 | 3 | 0.055550 | 2 | 0.053100 | 1 | 0.435000 | 2 | 0.123300 |
| Dodo | 3 | 3 | 0.015375 | 2 | 0.042000 | 2 | 0.170000 | 2 | 0.103700 |
| Henri | 3 | 3 | 0.009850 | 2 | 0.054500 | 2 | 0.141000 | 2 | 0.076850 |
| Ivindo | 3 | 3 | 0.019525 | 2 | 0.055900 | 2 | 0.166200 | 2 | 0.089650 |
| Makata | 3 | 3 | 0.082000 | 3 | 0.568300 | 2 | 0.236200 | 2 | 0.135800 |
| Masuku | 3 | 3 | 0.027925 | 2 | 0.043900 | 2 | 0.183500 | 2 | 0.108750 |
| Moanda | 3 | 3 | 0.102475 | 2 | 0.108100 | 2 | 0.483100 | 2 | 0.270950 |
| Noemie | 3 | 3 | 0.016125 | 2 | 0.043500 | 2 | 0.167600 | 2 | 0.093750 |
| Ntoum | 3 | 3 | 0.337625 | 2 | 0.081600 | 2 | 0.210400 | 2 | 0.260200 |
| Frits | 4 | 4 | 0.004050 | 3 | 0.003200 | 3 | 0.000550 | ||
| Hilko | 4 | 4 | 0.003575 | 3 | 0.003100 | 3 | 0.000750 | ||
| Louise | 4 | 4 | 0.216425 | 3 | 0.021300 | 3 | 0.000250 | ||
| Marco | 4 | 4 | 0.003525 | 3 | 0.000800 | 3 | 0.000500 | ||
| Oscar | 4 | 4 | 0.003500 | 3 | 0.000600 | 3 | 0.000500 | ||
| Regina | 4 | 4 | 0.003500 | 3 | 0.000400 | 3 | 0.000350 | ||
| Socrates | 4 | 4 | 0.005650 | 3 | 0.000900 | 3 | 0.000250 | ||
| Sonja | 4 | 4 | 0.003575 | 3 | 0.000600 | 3 | 0.000350 | ||
| Yoran | 4 | 4 | 0.003925 | 3 | 0.000600 | 3 | 0.000350 | ||
| Yvonne | 4 | 4 | 0.056800 | 3 | 0.006600 | 3 | 0.000650 | ||
Open cells indicate individuals that are not included in the analyses.
Reported assignment (Fischer et al. 2006) 1 for bonobos, 2 for East African, 3 for Central African, and 4 for West African chimpanzees.
Mean assignment and uncertainty inferred using IMa2 with the island model. We compare each of the posterior sampled assignments with the mean assignment to compute the partition distance. The partition distance is the number of individuals that have to be removed so that the two assignments are equivalent. The assignment uncertainty of an individual is the proportion of sampled assignments in which the individual is removed.
Mean assignments and uncertainty using STRUCTURAMA.
Mean assignments and uncertainty using STRUCTURAMA with East and Central African chimpanzees.
Mean assignments and uncertainty inferred using IMa2 with the tree model.
Table 8 . Population sizes and their 95% confidence intervals in parentheses under a four-population island model with assignment fixed to the true and assignment being a random variable using Fischer’s data set.
| Assignment | θ1a | θ2b | θ3c | θ4d |
|---|---|---|---|---|
| Fixed | 0.1275 | 0.1775 | 0.4275 | 0.1175 |
| (0.0575, 0.3325) | (0.0875, 0.4225) | (0.2475, 0.8075) | (0.0425, 0.3075) | |
| Variable | 0.0925 | 0.1075 | 0.4425 | 0.0875 |
| (0.0425, 0.2625) | (0.0475, 0.5775) | (0.2075, 0.9875) | (0.0375, 0.2575) |
Population size of bonobos.
Population size of East African Chimpanzees.
Population size of Central African Chimpanzees.
Population size of West African Chimpanzees.
Chimpanzee data set of Fischer et al. (2006) using IM with a population tree:
With data from three subspecies of common chimpanzees (i.e., without using the Bonobo data) we used the JDAP algorithm to jointly estimate assignment and the population tree. The resulting mean assignment [see the IMa2(3) column in Table 7] was correct with the exceptions that it allotted Alley and Judy to the Central African population and Sultana to the East African population. These same individuals were also uncertain in their assignment under the island model. We also used the total posterior sample of assignments to find the tree-constrained mean assignment of which was the same as the mean assignment of except that Sultana along with Alley and Judy was misassigned to the Central African population (data not shown). Individual assignment uncertainties of West African chimpanzees were relatively small, and those of East and Central African chimpanzees were large. The true population tree was most supported by the posterior sample of assignments among the three population trees that were equally likely a priori. We used each of the three sets of genealogy samples to estimate demographic parameters for each corresponding population tree (Table 9). The two splitting events were farther apart in the true population tree than in the other wrong trees. East African population size was underestimated, and Central African population size was overestimated presumably because the three individuals, Alley, Judy, and Sultana, of the East African population were uncertain in their individual assignment.
Table 9 . Population size and splitting time estimates and their 95% confidence intervals in parentheses under the tree model using the three chimpanzee populations.
| Tree | Aa | Counts (%) | θ1b | θ2c | θ3d | θ4e | θ5f | t1g | t2h |
|---|---|---|---|---|---|---|---|---|---|
| (1, 2), 3) | F | NA | 0.2430 | 0.7630 | 0.143 | 0.571 | 0.207 | 0.08175 | 0.1703 |
| (0.091, 0.925) | (0.325, 1.931) | (0.069, 0.461) | (0.183, 1.939) | (0.021, 0.889) | (0.02625, 0.2003) | (0.09075, 0.4627) | |||
| (1, 2), 3) | V | 14,706 | 0.1610 | 1.101 | 0.1470 | 0.5570 | 0.2250 | 0.06225 | 0.1598 |
| (73.5) | (0.063, 1.721) | (0.357, 1.957) | (0.061, 0.443) | (0.159, 1.931) | (0.027, 0.851) | (0.00825, 0.2003) | (0.08025, 0.4148) | ||
| (1, 3), 2) | V | 3,141 | 0.2030 | 1.405 | 0.1030 | 0.089 | 0.2910 | 0.09225 | 0.1462 |
| (15.7) | (0.087, 1.587) | (0.527, 1.963) | (0.041, 0.747) | (0.065, 1.945) | (0.071, 0.739) | (0.03975, 0.2303) | (0.06375, 0.2873) | ||
| (2, 3), 1) | V | 2,153 | 0.2490 | 1.289 | 0.1090 | 0.8310 | 0.2170 | 0.1118 | 0.1177 |
| (10.8) | (0.125, 1.451) | (0.515, 1.961) | (0.051, 0.331) | (0.129, 1.951) | (0.051, 0.751) | (0.04575, 0.2437) | (0.06975,0.3008) |
F, fixed assignment; V, variable assignment.
Population size of East African Chimpanzees.
Population size of Central African Chimpanzees.
Population size of West African Chimpanzees.
Population size of ancestor of two ingroup populations.
Population size of the ancestor of all three populations.
Time at which the common ancestor of two ingroup populations split.
Time at which the common ancestor of the three chimpanzee populations split.
Discussion
Investigators in many contexts are often faced with the situation of having data that have multiple sources of variation. Depending on the state of theory in the particular field, it may be possible for these sources of variation to be modeled with parameters. But without a way to simultaneously consider the different causes of variation, analyses may need to be done in a piecemeal fashion. For example, an analysis may begin with the estimation of just one part of the full model and then proceed by plugging this estimate into a second analysis to address a different part of the model. In this way a series of conditional estimates can be obtained, all but the first of which are at added risk of error because of conditioning on previous estimates. In just this way it is common for investigators to first estimate population assignment, using a method that maximizes Hardy–Weinberg and linkage equilibria, and then use these estimates to conduct additional analyses (e.g., Sacks et al. 2004; Coulon et al. 2006; Bergl and Vigilant 2007).
The methods described here permit investigators to avoid a series of separate but interdependent analyses and to jointly study population assignment and demographic history. In addition to estimated assignments and estimates of demographic parameters, investigators can ask how uncertainty in assignment varies across the demographic history that has been estimated. It is expected that recently formed populations may be estimated with less certainty; but by studying the age of population formation together with assignment, this interaction can be examined directly. Similarly populations that have been exchanging genes may give rise to data that are not easily assigned, but nevertheless it is possible to study assignment and gene exchange together.
The isolation-with-migration model is flexible with regard to mutation models and demographic histories, particularly when multiple sampled populations are included. However, the general approach that we describe, of including population assignment as part of the genealogy in an MCMC analysis, is one that could be adapted to other kinds of demographic models. Recently, Yang and Rannala (2010) described a method for jointly estimating phylogeny, assignment, and population size parameters. Their method assumes that there has been no migration, unlike the applications described here, but it can be applied to data sets with more populations and a larger phylogeny (with the aid of a user-supplied guide tree) than described here (Yang and Rannala 2010).
These methods are also flexible in that it is straightforward to include both data from individuals with unknown assignment and data from individuals whose population assignment status is known. In this way the general assignment problem can be seen to include both the classification of individuals to previously identified populations (a procedure that is in some contexts known as an assignment test) and the discovery of previously unidentified populations. At one extreme we might have a DNA barcoding problem, where an assignment of a single individual is to be determined against a backdrop of previously assigned data (Matz and Nielsen 2005; Nielsen and Matz 2006), and at the other extreme an investigator might have little previous knowledge of how many populations exist.
Our studies with simulated and real data show that it is possible to estimate assignment together with demographic history and that assignment estimates are improved when they are obtained under an IM model, relative to the case without. However, it is important to point out that the comparisons between IMa2 and STRUCTURAMA, while they represent a contrast of with and without a demographic model, also include the contrast of with and without a mutation model. STRUCTURAMA, like most other population assignment methods, uses an allelic model of variation, whereas the simulated and real data in the present studies were based on mutation models for DNA sequence variation. To run STRUCTURAMA, or any similar allele-based method, requires pruning the data so that each distinct DNA haplotype is reduced to an allelic representation. Thus we do not know how much of the difference in assignment estimation between STRUCTURAMA and IMa2 is due to the inclusion of a demographic model in the latter and how much is due to the inclusion of a mutation model.
Assignment and phylogeny
In any demographic model with more than two populations, and that includes a phylogenetic component, the estimation of assignment entails the estimation of the population phylogeny. In this article we studied this issue in detail for the case of three-population models. This required that we extend the ideas of partition distance (Almudevar and Field 1999; Gusfield 2002; Konovalov et al. 2005) and mean assignment (Huelsenbeck and Andolfatto 2007) to tree-constrained partition distance and tree-constrained mean assignment. We are then able to use tree-constrained partition distances in the JDAP algorithm to jointly estimate assignment, demographic history, and population phylogeny. When we applied these methods to three subspecies of chimpanzees, they returned the true phylogenetic tree and simultaneously provided population assignments that were more accurate, and that came with less uncertainty, than those found using the STRUCTURAMA program.
Limitations
A major limitation of the methods presented here is that they are computationally quite slow, particularly when compared to the speed of programs implementing allele-based methods. The run time for the two real data sets ranged from 40 to 129 hr, many times that required by STRUCTURAMA for the same data. When included as part of the Markov chain simulation, population assignment takes time to update and can significantly impede the mixing of the Markov chain. The mixing issue can be significantly mitigated by the addition of Metropolis-coupled chains, but the resulting analysis can be quite slow. It is for this reason that the analyses described here are for modestly sized data sets.
The assumptions of the IM models adapted here all stem from those adopted by Nielsen and Wakeley (2001) in their original MCMC method for this model. In particular these include the assumptions of selective neutrality, free recombination within loci, and zero recombination within loci. Allele-based methods that minimize Hardy–Weinberg also, at least implicitly, assume that selection has not been a factor causing a departure from Hardy–Weinberg. They do not require an assumption of no recombination within loci, because in an allelic context intragenic recombination is just an additional source of new alleles, and they can be extended to handle cases of restricted recombination between loci (Falush et al. 2003). Strasburg and Rieseberg (2010) examined the case of a failure of the assumption of zero recombination in IM analyses using the IMa program (Hey and Nielsen 2007) and found that the method was fairly robust to modest levels of recombination when data are reduced to nonrecombining blocks, as determined by the four-gamete test (Hudson and Kaplan 1985).
Acknowledgments
We thank Yong Wang, Jongwoo Jung, Kasper Munch, and Rasmus Nielsen for helpful discussions and Armando Geraldes and Michael Nachman for the mouse data set. This research was supported by grants to J.H. from the National Institutes of Health (GM078204) and the National Science Foundation (DEB-0949561).
Appendix A
Genealogy Updating Using Estimates of the Population Mutation and Migration Parameters From the Current Genealogy
The probability of a genealogy, given parameter values under a two-population IM model, is given in Equation 15 of Hey and Nielsen (2007) and in (A1) of the supporting information of that article. This probability is a function of the numbers of a series of terms calculated from the genealogy, including coalescent events (c1, c2, cA), total coalescent rates (f1, f2, fA), numbers of migration events (w1, w2), and total migration rates (g1, g2), where the subscripts refer to the corresponding population (A refers to the ancestral population). By taking the derivative of this function with respect to a parameter, we find the individual maximum-likelihood estimates for each parameter associated with that genealogy. The estimate of θ1 is derived as
and for the parameter m12 as
A nonzero value of 1.0 is used for mmin because a value of zero violates the MCMC criterion that an update to the genealogy be reversible.
With values for the demographic parameters, we used expressions (3) and (7) of Beerli and Felsenstein (1999) to simulate the time to the next event in the genealogy. We choose an external branch of a gene tree and label the tip of the branch to other populations at random. We detach the branch from the genealogy. We divide the time duration from the present to the root of the genealogy by events including coalescent, migration, and population splits. For each time interval starting at the present we sample the next event and time using expression (3) of Beerli and Felsenstein (1999). If the time sampled is larger than the current time interval, we skip the current time interval and move to the next time interval. The simulation of events ends in a coalescent event. If the simulation does not complete even in the last time interval before the time at the root, we use expression (7) of Beerli and Felsenstein (1999) to sample the next event and time. One difference between the case of Beerli and Felsenstein (1999) and that of ours is that we need to consider population splitting events. In other respects the procedure follows Beerli and Felsenstein (1999).
Appendix B
Population Relabeling Preserves the Posterior Distribution of Genealogy and Splitting Time
We wish to show that permutations of assignment labels change neither the prior distribution nor the posterior distribution of a genealogy under a two-population IM model. The following derivation is based on Equation A1 in the supporting information to Hey and Nielsen (2007) that describes the terms in the probability of genealogy and population splitting time given model parameters π(G, t|Θ), where Θ = {θ1, θ2, θA, m1, m2}. For calculating the prior probability, all of the information in a genealogy in an IM model can be described as a vector with elements being numbers of coalescent events (c), total coalescent rates (f), numbers of migration events (w), and total migration rates (g) or G = (c1, f1, w1, g1, c2, f2, w2, g2, cA, fA) (Hey and Nielsen 2007) (see Appendix A). We relabel the genealogy by swapping the terms for populations 1 and 2,
where ν is an operator that relabels the ordered list of numbers that represents the given genealogy. The probability of genealogy given model parameters π(G|Θ) may differ from that of the relabeled genealogy, π(ν(G)|Θ), given the same model parameters. However, if the prior probabilities for the parameters for population 1 are the same as those for population 2 [i.e., π(θ1) = π(θ2) and π(m1) = π(m2)], then the prior of the genealogy is equal to the prior of the relabeled genealogy:
Turning to the posterior density, we note that the likelihood does not depend on assignment, f(X|G) = f(X|ν(G)), and therefore the posterior distribution of the genealogy given the data is invariant to label permutation:
Although we assume the two-population IM model, the same argument applies to the multipopulation island model. In the case of a three-population IM model there are three possible labeled tree topologies, but the symmetry applies only to the two sister populations. In general for trees with multiple populations we can relabel genealogies only via swapping for populations that are each other’s sisters.
Literature Cited
- Almudevar A., Field C., 1999. Estimation of single-generation sibling relationships based on dna markers. J. Agric. Biol. Environ. Stat. 4: 136–165 [Google Scholar]
- Baudouin L., Piry S., Cornuet J. M., 2004. Analytical Bayesian approach for assigning individuals to populations. J. Hered. 95: 217–224 [DOI] [PubMed] [Google Scholar]
- Becquet C., Patterson N., Stone A. C., Przeworski M., Reich D., 2007. Genetic structure of chimpanzee populations. PLoS Genet. 3: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P., Felsenstein J., 1999. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152: 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E. T., 1934. Exponential numbers. Am. Math. Mon. 41: 411–419 [Google Scholar]
- Bergl R. A., Vigilant L., 2007. Genetic analysis reveals population structure and recent migration within the highly fragmented range of the cross river gorilla (gorilla gorilla diehli). Mol. Ecol. 16: 501–516 [DOI] [PubMed] [Google Scholar]
- Caswell J. L., Mallick S., Richter D. J., Neubauer J., Schirmer C., et al. , 2008. Analysis of chimpanzee history based on genome sequence alignments. PLoS Genet. 4: e1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Durand E., Forbes F., Francois O., 2007. Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol. Ecol. Notes 7: 747–756 [Google Scholar]
- Corander J., Waldmann P., Sillanpää M. J., 2003. Bayesian analysis of genetic differentiation between populations. Genetics 163: 367–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A., Guillot G., Cosson J.-F., Angibault J. M. A., Aulagnier S., et al. , 2006. Genetic structure is influenced by landscape features: empirical evidence from a roe deer population. Mol. Ecol. 15: 1669–1679 [DOI] [PubMed] [Google Scholar]
- Dawson K. J., Belkhir K., 2001. A Bayesian approach to the identification of panmictic populations and the assignment of individuals. Genet. Res. 78: 59–77 [DOI] [PubMed] [Google Scholar]
- Edwards A. W. F., 1970. Estimation of the branch points of a branching diffusion process. J. R. Stat. Soc. B 32: 155–174 [Google Scholar]
- Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1992. Estimating effective population size from samples of sequences: a bootstrap Monte Carlo integration method. Genet. Res. 60: 209–220 [DOI] [PubMed] [Google Scholar]
- Fischer A., Pollack J., Thalmann O., Nickel B., Pääbo S., 2006. Demographic history and genetic differentiation in apes. Curr. Biol. 16: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Fogelqvist J., Niittyvuopio A., Ågren J., Savolainen O., Lascoux M., 2010. Cryptic population genetic structure: the number of inferred clusters depends on sample size. Mol. Ecol. Resour. 10: 314–323 [DOI] [PubMed] [Google Scholar]
- François O., Ancelet S., Guillot G., 2006. Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics 174: 805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A., Basset P., Gibson B., Smith K. L., Harr B., et al. , 2008. Inferring the history of speciation in house mice from autosomal, x-linked, y-linked and mitochondrial genes. Mol. Ecol. 17: 5349–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer C. J., 1991. Markov chain Monte Carlo maximum likelihood. Comp. Sci. Stat. 23: 156–163 [Google Scholar]
- Grant W. S., Milner G. B., Krasnowski P., Utter F. M., 1980. Use of biochemical genetic variants for identification of sockeye salmon (Oncorhynchus nerka) stocks in Cook Inlet, Alaska. Can. J. Fish. Aquat. Sci. 37: 1236–1247 [Google Scholar]
- Guillot G., Estoup A., Mortier F., Cosson J. F., 2005. A spatial statistical model for landscape genetics. Genetics 170: 1261–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusfield D., 2002. Partition-distance: a problem and class of perfect graphs arising in clustering. Inf. Process. Lett. 82: 159–164 [Google Scholar]
- Hastings W. K., 1970. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57: 97–109 [Google Scholar]
- Hey J., 2010a The divergence of chimpanzee species and subspecies as revealed in multi-population isolation-with-migration analyses. Mol. Biol. Evol. 27: 921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J., 2010b Isolation with migration models for more than two species. Mol. Biol. Evol. 27: 905–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J., Nielsen R., 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J., Nielsen R., 2007. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc. Natl. Acad. Sci. USA 104: 2785–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Andolfatto P., 2007. Inference of population structure under a Dirichlet process model. Genetics 175: 1787–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1969. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics 61: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J., 1982. The coalescent. Stoch. Proc. Appl. 13: 235–248 [Google Scholar]
- Konovalov D. A., Litow B., Bajema N., 2005. Partition-distance via the assignment problem. Bioinformatics 21: 2463–2468 [DOI] [PubMed] [Google Scholar]
- Kuhner M. K., Yamato J., Felsenstein J., 1995. Estimating effective population size and mutation rate from sequence data using Metropolis-Hastings sampling. Genetics 140: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listman J. B., Malison R. T., Sughondhabirom A., Yang B.-Z., Raaum R. L., et al. , 2007. Demographic changes and marker properties affect detection of human population differentiation. BMC Genet. 8: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz M. V., Nielsen R., 2005. A likelihood ratio test for species membership based on DNA sequence data. Philos. Trans. R. Soc. B 360: 1969–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metropolis N., Rosenbluth A. W., Rosenbluth M. N., Teller A. H., 1953. Equation of state calculations by fast computing machines. J. Chem. Phys. 21: 1087–1091 [Google Scholar]
- Nielsen R., Matz M., 2006. Statistical approaches for DNA barcoding. Syst. Biol. 55: 162–169 [DOI] [PubMed] [Google Scholar]
- Nielsen R., Wakeley J., 2001. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara B., 2010. New heuristic methods for joint species delimitation and species tree inference. Syst. Biol. 55: 162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetkau D., Calvert W., Stirling I., Strobeck C., 1995. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 4: 347–354 [DOI] [PubMed] [Google Scholar]
- Pella J., Masuda M., 2006. The Gibbs and split-merge sampler for population mixture analysis from genetic data with incomplete baselines. Can. J. Fish. Aquat. Sci. 63: 576–596 [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B., Mountain J. L., 1997. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 94: 9197–9201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P. A., Richards C. M., 2009. Accurate inference of subtle population structure (and other genetic discontinuities) using principal coordinates. PLoS ONE 4: e4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks B. N., Brown S. K., Ernest H. B., 2004. Population structure of California coyotes corresponds to habitat-specific breaks and illuminates species history. Mol. Ecol. 13: 1265–1275 [DOI] [PubMed] [Google Scholar]
- Stanton D. W., White D. E., 1986. Constructive Combinatorics. Springer-Verlag, Berlin/Heidelberg, Germany/New York [Google Scholar]
- Stephens M., 2000. Dealing with label-switching in mixture models. J. R. Stat. Soc. B Met. 62: 795–809 [Google Scholar]
- Strasburg J. L., Rieseberg L. H., 2010. How robust are “isolation with migration” analyses to violations of the IM model? A simulation study. Mol. Biol. Evol. 27: 297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hey J., 2010. Estimating divergence parameters with small samples from a large number of loci. Genetics 184: 363–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R. S., Gaggiotti O., 2006. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 15: 1419–1439 [DOI] [PubMed] [Google Scholar]
- Won Y.-J., Hey J., 2005. Divergence population genetics of chimpanzees. Mol. Biol. Evol. 22: 297–307 [DOI] [PubMed] [Google Scholar]
- Wu B., Liu N., Zhao H., 2006. PSMIX: an R package for population structure inference via maximum likelihood method. BMC Bioinformatics 7: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Rannala B., 2010. Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci. USA 107: 9264–9269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., 2008. Tree-guided Bayesian inference of population structures. Bioinformatics 24: 965–971 [DOI] [PubMed] [Google Scholar]