Abstract
Thyroid hormone has pleiotropic effects on cochlear development, and genomic variation influences the severity of associated hearing deficits. DW/J-Pou1f1dw/dw mutant mice lack pituitary thyrotropin, which causes severe thyroid hormone deficiency and profound hearing impairment. To assess the genetic complexity of protective effects on hypothyroidism-induced hearing impairment, an F1 intercross was generated between DW/J-Pou1f1dw/+ carriers and an inbred strain with excellent hearing derived from Mus castaneus, CAST/EiJ. Approximately 24% of the (DW/J × CAST/EiJ) Pou1f1dw/dw F2 progeny had normal hearing. A genome scan revealed a locus on chromosome 2, named modifier of dw hearing, or Mdwh, that rescues hearing despite persistent hypothyroidism. This chromosomal region contains the modifier of tubby hearing 1 (Moth1) locus that encodes a protective allele of the microtubule-associated protein MTAP1A. DW/J-Pou1f1dw/+ carriers were crossed with the AKR strain, which also carries a protective allele of Mtap1a, and we found that AKR is not protective for hearing in the (DW/J × AKR) Pou1f1dw/dw F2 progeny. Thus, protective alleles of Mtap1a are not sufficient to rescue DW/J-Pou1f1dw/dw hearing. We expect that identification of protective modifiers will enhance our understanding of the mechanisms of hypothyroidism-induced hearing impairment.
THYROID hormone (TH) is an important regulator of many processes in mammalian development, including body growth (Cabello and Wrutniak 1989) and central nervous system maturation (Bernal 2005). Auditory function is particularly sensitive to the effects of TH, which is required for the complex development and physiology of the cochlea (Deol 1973; Uziel 1986; Sohmer and Freeman 1996). Mutations in several different genes can prevent or interfere with the TH-signaling pathway that is required for normal auditory development and function. In addition to gene mutations that cause hypothyroidism, mutations of genes encoding thyroid hormone receptors (Forrest et al. 1996; Rusch et al. 2001) and iodothyronine deiodinases (Ng et al. 2004, 2009) can diminish the developing cochlea’s response to TH and exhibit similar auditory phenotypes. Cases of congenital hypothyroidism (CH) are classified as primary if caused by thyroid gland dysfunction and as secondary if caused by pituitary gland abnormalities. Mouse models of primary hypothyroidism include the hypothyroid mutation (hyt) of the Tshr gene, which has residual activity and mild-to-negligible hearing deficit (O’Malley et al. 1995; Sprenkle et al. 2001; Karolyi et al. 2007), and the thyroid dyshormonogenesis mutation (thyd) of the Duox2 gene, which causes severe-to-profound deafness in mice (Johnson et al. 2007). Pax8 knockout mice are athyroid, exhibit ear abnormalities, and lack an auditory brainstem response (ABR) to sound. The utility of this model is limited because the mice survive only to postnatal day 21 (P21), and it is difficult to distinguish between the effects of Pax8 expression in the thyroid gland and the otic placode (Christ et al. 2004).
Hearing impairment has been examined in three mouse models of secondary hypothyroidism: the Snell dwarf (dw) mutation of the Pou1f1 gene (formerly Pit1), the Ames dwarf (df) mutation of the Prop1 gene, and a targeted knockout mutation of the Cga gene, which encodes an essential subunit of thyrotropin (Karolyi et al. 2007). Both Pou1f1 and Prop1 encode transcription factors in the same pathway of pituitary gland development. Mice with null mutations in Prop1, Pou1f1, or Cga lack pituitary thyrotropin and have no measurable thyroid hormone in the serum. In spite of these similarities, the hearing defects in DW/J-Pou1f1dw/dw and Cga mutant mice are profound, whereas DF/B-Prop1df/df mutants have only a mild hearing impairment. The Pou1f1dw and Prop1df mutations are on different strain backgrounds, and analysis of a small cross between DW/J-Pou1f1dw/+ and DF/B-Prop1df/+ mice showed that genetic background is likely responsible for the different hearing phenotypes (Karolyi et al. 2007).
Advances in understanding how TH signaling affects the development and function of the cochlea will help to illuminate the molecular mechanisms that underlie development of normal auditory functions. We chose the Pou1f1dw mutant mice because of the severity of their hearing impairment, their responsiveness to TH replacement, and their well-characterized cochlear pathologies (Mustapha et al. 2009). Detailed morphological, physiological, and gene expression analyses of Pou1f1dw mutants during the course of cochlear development revealed tectorial membrane abnormalities, loss of outer hair cells (OHCs) preceded by their compromised function [as evidenced by an absence of distortion product otoacoustic emissions (DPOAE) and cochlear microphonics], reduced endocochlear potential, and reduced expression of potassium channel proteins (KCNJ10 in the stria vascularis and KCNQ4 in OHCs), all of which are likely contributors to the severe hearing impairment of these mutant mice (Mustapha et al. 2009). Similar features, including lack of DPOAE, reduced endocochlear potential, and tectorial membrane abnormalities, are characteristic of the Cga knockout mice (Karolyi et al. 2007).
Although insights have been gained from studies of existing hypothyroid mouse models, our understanding of the molecular mechanisms underlying the hearing impairment associated with hypothyroidism is still incomplete. The observed influence of genetic background on the severity of hearing impairment in hypothyroid mice offers an opportunity to identify modifier genes and pathways that could enhance our understanding of these mechanisms. Here we describe results from a large linkage cross that was designed to map loci that modify the hearing of hypothyroid DW/J-Pou1f1 dwarf mice. Heterozygous DW/J-Pou1f1dw mice were intercrossed with the wild-derived, inbred strain CAST/EiJ, which have good hearing. F2 mutant progeny (dw/dw) from this intercross ranged from normal hearing to completely deaf, despite the fact that they were all profoundly hypothyroid and equally growth-impaired. By analysis of this cross, we identified a locus on chromosome (Chr) 2 that had a highly significant linkage association (LOD = 10) with the ABR threshold variation exhibited by the mutant intercross mice. This quantitative trait locus (QTL) was given the symbol Mdwh (modifier of dw hearing). The Mdwh interval contains Moth1, the modifier of tubby hearing, which varies among strains by the length of an alanine–proline amino acid repeat in microtubule-associated protein 1a (Mtap1a) (Ikeda et al. 2002). DW/J strain has a susceptible allele of Mtap1a, while CAST/EiJ and AKR strains have protective alleles. F2 dw/dw mutants born from an intercross of (AKR × DW/J) F1 mice are all deaf, suggesting that Mdwh is not Moth1, but represents a novel locus that protects against hearing impairment in hypothyroid Pou1f1dw mutants.
Materials and Methods
Mice
Mice of the C3H/HeJ-Pou1f1dw-J/J, DW/J-Pou1f1dw, and CAST/EiJ inbred strains and their derivative F1 and F2 hybrids were housed in the Research Animal Facility of the Jackson laboratory. In parallel, stocks of DW/J-Pou1f1dw, CAST/EiJ, and AKR were purchased from the Jackson laboratory and bred at the University of Michigan to produce (DW/J-Pou1f1dw × CAST/EiJ) F2 hybrids and (DW/J-Pou1f1dw × AKR) F2 hybrids. All procedures were approved by the Institutional Animal Care and Use Committees of each institution. Both the Jackson laboratory and the University of Michigan are accredited by the American Association for the Accreditation of Laboratory Animal Care and are registered with the U.S. Department of Agriculture as research facilities. Genotyping processes for dw and dw-J alleles were previously described (Eicher and Beamer 1980; Karolyi et al. 2007).
Assessment of hearing by ABR threshold analysis
The ABR test was used to assess hearing thresholds of the mice. C3H/HeJ-Pou1f1dw-J/J, DW/J-Pou1f1dw, and their derivative F2 hybrids and 196 (DW/J-Pou1f1dw × CAST/EiJ) F2 hybrids were tested at the Jackson laboratory. At the University of Michigan 68 (DW/J-Pou1f1dw × CAST/EiJ) F2 hybrids and 62 (DW/J-Pou1f1dw × AKR) F2 hybrids were tested.
Similar ABR procedures were used at the Jackson laboratory and at the University of Michigan (Zheng et al. 1999; Karolyi et al. 2007). All animals were tested binaurally. The frequencies of tone bursts presented to the mice were click and 8, 16, 32 kHz at the Jackson laboratory and 10 and 20 kHz at the University of Michigan.
Linkage intercross mapping
Individual DNA samples from linkage intercross mice were genotyped by PCR amplification with primer pairs designed to amplify specific microsatellite markers purchased from Integrated DNA Technologies (Coralville, IA). PCR reactions and PCR product visualization methods were as previously described (Gagnon et al. 2006). ABR thresholds for click, 8, 16, and 32 kHz stimuli were evaluated as quantitative traits, and QTL linkage analysis was performed using the computer program Map Manager QTX (version b20), which uses a fast regression method to detect and localize quantitative trait loci within intervals defined by genetic markers and can perform pairwise locus analysis to search for QTL interactive effects. This program reports the resulting regression coefficients and a likelihood-ratio statistic that is based on natural logarithms but can be converted to conventional LOD scores by dividing by 4.61.
Candidate gene analysis
PCR for comparative DNA sequence analysis was performed according to the same conditions as described above for linkage mapping. DNA sequences of the PCR primers used to amplify the polymorphic repeat sequence GCTCCA of Mtap1a were 5′-TCTGGGACCTCACTCCTCTG (forward) and 5′-GTTTCTCCTGGGCCATTAGC (reverse). PCR products from genomic DNA were purified with the QIAquick PCR Purification Kit (Qiagen, Valencia, CA). DNA sequencing was performed using the same primers as for DNA amplification, then run on an Applied Biosystems 3700 DNA Sequencer with an optimized Big Dye Terminator Cycle Sequencing method.
Results
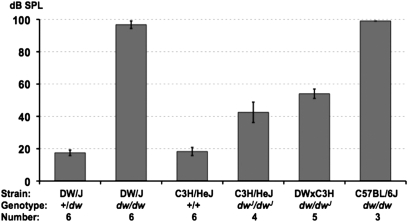
We reported strain background effects on hearing in hypothyroid mice that were produced from an intercross of DF/B-Prop1df and DW/J-Pou1f1dw strains (Karolyi et al. 2007). There were insufficient F2 progeny, however, to assess the heterogeneity of effects among them (six or fewer for each genotype group). The unique nature of each stock (DW/J and DF/B) and the fact that DF/B are not inbred compelled us to extend this analysis of modifier effects by comparing ABR thresholds of mice with different Pou1f1 mutant genotypes on multiple genetic backgrounds (Figure 1). As previously reported, Pou1f1dw/dw mice on the DW/J strain background are nearly deaf at 4 weeks of age, whereas Pou1f1dw/+ heterozygotes have normal hearing thresholds. On the C3H/HeJ strain background, mice homozygous for a different null mutation of Pou1f1, the dwJ allele, show a hearing impairment greater than that of the parental C3H/HeJ strain mice but not nearly as severe as that of dw mutants on the DW/J strain background. Both the dw and dwJ alleles are recessive, complete loss-of-function alleles that result from a missense mutation in the homeodomain that abolishes DNA binding and an intragenic rearrangement, respectively (Camper et al. 1990; Li et al. 1990). Compound heterozygote (DW/J × C3H/HeJ) F1 hybrids (dw/dwJ) exhibit a hearing impairment that is intermediate between that of DW/J-dw/dw and C3H/HeJ-dwJ/dwJ mutants. Mutant dw/dw mice of the B6.DW-Pou1f1dw/J congenic strain exhibit severe hearing impairment like that of dw/dw mutants on the DW/J strain background. The congenic strain was constructed by transferring the dw mutation from the DW/J strain onto an otherwise C57BL/6J strain background by multiple rounds of backcrossing and selection. All of these results support the idea that the genetic background affects the degree of hearing loss of hypothyroid Pou1f1 mutants.
Figure 1 .
Genetic background modifies severity of hearing impairment in Pou1f1dw/dw mice. Average 16-kHz ABR thresholds (with standard error bars) are shown for mice with different Pou1f1 genotypes on different strain backgrounds. Note that dwarf mutant mice on the DW/J and C57BL/6J strain backgrounds have a much more severe hearing impairment than mutant mice on the C3H/HeJ background (dB SPL = decibel sound pressure level).
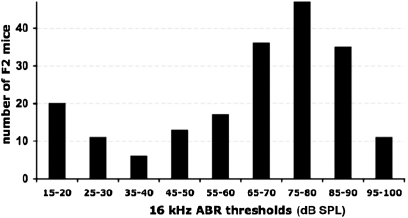
To investigate the genetic complexity of the strain-related hearing loss variation of Pou1f1dw mutant mice and to map the putative modifier genes, we produced a linkage intercross between (DW/J-Pou1f1dw × CAST/EiJ) F1 hybrids. We chose the CAST/EiJ strain because mice of this strain retain normal hearing ability at advanced ages and because of the increased likelihood of genetic marker polymorphisms with the DW/J strain (Zheng et al. 1999). Pou1f1 mutant (dw/dw) F2 progeny from this intercross represented ∼25% of the total, as expected, and these mutants were tested for hearing and analyzed for genetic linkage. ABR thresholds for click, 8-, 16-, and 32-kHz stimuli were measured at 1–2 months of age (average 40 days) for each of 196 F2 mutant mice. The dw/dw F2 mice exhibited a wide range of ABR thresholds that were distributed in a skewed bimodal fashion, suggesting a major influence of one or a few genes (Figure 2). A bell-shaped distribution with a single mode would have suggested that many genes with small effects contribute to the hearing differences among the mutant mice. A substantial portion—16% (N = 31)—of the mutant F2 mice exhibited normal hearing thresholds (<35 dB SPL at 16 kHz). The body weights of the mutant dw/dw F2 mice were about threefold smaller than normal mice, consistent with profound hypothyroidism and growth deficiency. There was no significant correlation between body weight and ABR thresholds (Table 1). This suggests that the rescue of hearing does not occur concomitantly with a growth rescue.
Figure 2 .
Degree of hearing impairment varies among F2 dw/dw progeny of the (DW-Pou1f1dw/+ × CAST) intercross. A frequency distribution is shown for 196 dw/dw F2 mice sorted according to their 16-kHz ABR thresholds. All mice were tested at 4–8 weeks of age. Note the bimodal shape of the distribution with peaks at 15–20 and 75–80 dB SPL.
Table 1 . Body size of dwarf intercross mice is not affected by strain background.
| ABR threshold (dB SPL) | Count | Average weight (g) | Variance | ||
|---|---|---|---|---|---|
| 15–25 | 23 | 7.8 | 0.6 | ||
| 30–50 | 27 | 8.0 | 1.0 | ||
| 55–60 | 17 | 7.8 | 0.6 | ||
| 65–70 | 36 | 7.8 | 0.8 | ||
| 75–80 | 47 | 7.6 | 0.4 | ||
| 85–90 | 35 | 7.7 | 0.6 | ||
| 95–100 | 11 | 7.5 | 0.3 | ||
| ANOVA | |||||
| Source of variation | SS | d.f. | MS | F-value | P-value |
| Between | 3.2 | 6 | 0.53 | ||
| Within | 119 | 189 | 0.63 | 0.8 | 0.54 |
| Total | 122.2 | 195 | |||
ANOVA confirms that the similar body weights of dw/dw F2 mice from the (DW-Pou1f1dw × CAST/EiJ) intercross do not correspond with their highly variable ABR thresholds.
Secretory otitis media can affect the hearing functions in both humans and mice, mostly at low frequencies (Hardisty et al. 2003; Bluestone and Klein 2007). A study of hearing loss in children with sporadic CH showed no increased susceptibility for middle ear effusion (Vanderschueren-Lodeweyckx et al. 1983). In mice of the DW/J background, we observed mild otitis media with a similar frequency in Pou1f1dw/dw mutants (8/11) and in wild types (5/11) (P > 0.05; our unpublished data). All mutant Pou1f1dw/dw mice on the DW strain background exhibit profound hearing impairment, even those without any sign of otitis media. The presence or absence of otitis media, therefore, cannot explain the variable hearing loss observed in Pou1f1dw/dw mice of the (DW/J-Pou1f1dw × CAST/EiJ) F2 intercross.
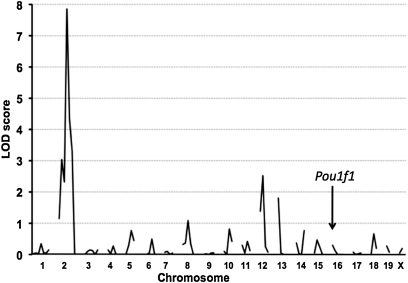
As a first approach for mapping modifier loci for hearing, we performed a genome-wide scan for linkage by limiting our analysis to F2 mice with the most extreme ABR thresholds, 30 with the lowest (15–30 dB SPL), and 11 with the highest (95–100 dB SPL) thresholds among 192 dw/dw F2 mutants generated at the Jackson laboratory. Selecting mice with extreme phenotypes maximizes information for more efficient linkage scanning without sacrificing detection capability (Darvasi 1997). We typed 90 microsatellite markers spaced at 15-cM intervals on all 20 mouse chromosomes, which covered >90% of the genome. We used the Map Manager QTX computer program (Manly et al. 2001) to analyze QTL associated with ABR threshold variation. We evaluated intercross linkage using a free model (2 d.f.) without prior assumptions of dominant, recessive, or additive effects. The LOD scores for associations of 16-kHz thresholds with the 90 markers are shown in Figure 3. With this model, a LOD score of 4.3 is considered a significant intercross linkage value, corresponding to a genome-wide type I error of 0.05 (Lander and Kruglyak 1995). The highest and only statistically significant linkage associations were found with markers on Chr 2, and additional Chr 2 markers were then analyzed (Figure 4A). The maximum LOD score was 7.9 for the linkage associations of D2Mit134 and D2Mit304 with 16-kHz ABR thresholds (LOD scores were 7.0, 6.7, and 7.4 for click, 8-, and 32-kHz thresholds, respectively). Given that a LOD score of 4.3 is considered significant, the Chr 2 linkage with a LOD score of 7.9 validated designation of this new QTL as “modifier of dw hearing” (Mdwh).
Figure 3 .
Genome-wide linkage analysis of (DW-Pou1f1dw/+ × CAST) F2 mice identifies a hearing modifier locus on Chr 2. The 41 dw/dw F2 mice with the most extreme ABR thresholds were analyzed, and LOD scores are shown for the linkage associations of these threshold values with each of 90 chromosomal markers. Only markers on Chr 2 showed statistically significant linkage. The Pou1f1 gene is located on Chr 16.
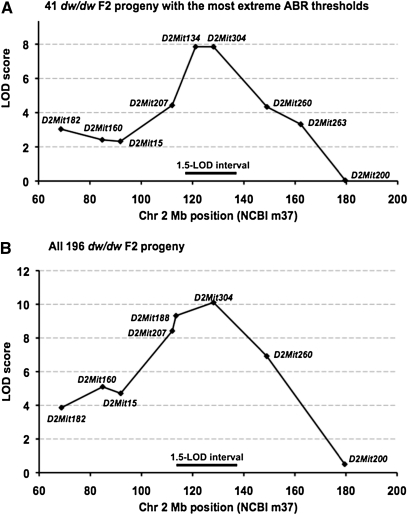
Figure 4 .
Additional mice and markers refined the map position of Mdwh on Chr 2. (A) Map position as determined from the 41 dw/dw F2 mice with the most extreme thresholds. The peak LOD scores (7.9) are for D2Mit134 at the 121-Mb position and D2Mit304 at the 128-Mb position. (B) Map position as determined from all 196 dw/dw F2 mice. The peak LOD score (10.1) is for D2Mit304 at the 128-Mb position. The 1.5-LOD support intervals for both analyses show that Mdwh is most likely located within the 118- to 138-Mb region of Chr 2.
To confirm the significance of this linkage and to clarify the additive and dominance components of the phenotypic variance, we extended the linkage studies by genotyping the remaining 155 dw/dw F2 mice from the intercross for Chr 2 markers surrounding the Mdwh locus. Identical most likely map positions for the Mdwh QTL were determined from all 196 Pou1f1dw/dw F2 mice and the 41 dw/dw mice with the most extreme hearing phenotypes (Figure 4). Both analyses gave peak associations with D2Mit304 (which was nonrecombinant with D2Mit134 in the 41 extreme mice). The 1.5-LOD support interval, which provides 95% confidence of coverage (Dupuis and Siegmund 1999), for both analyses is the 118- to 138-Mb region of Chr 2 (National Center for Biotechnology Information Build m37). Because of the increased sample size, the LOD scores for linkage associations were higher when all 196 mice were analyzed, with a peak score of 10.1 for marker D2Mit304 (Figure 4B).
The effects of Mdwh on the hearing thresholds of dwarf intercross mice are highly significant and vary little in response to the different auditory stimuli (LOD 9.0–10.1; Table 2). The LOD scores shown in Figure 4 and Table 2 are for an additive inheritance model. These values were essentially the same as those calculated for a free model (no inheritance assumptions) and larger than those of recessive and dominant models, indicating that the effects of Mdwh are primarily additive.
Table 2 . Mdwh locus has a large effect on ABR thresholds of intercross mice.
| ABR thresholds | ||||
|---|---|---|---|---|
| Genotype | Click | 8 kHz | 16 kHz | 32 kHz |
| DD, N = 50 | 97.9 (8.1) | 95.9 (8.2) | 82.1 (12.6) | 97.9 (7.5) |
| DC, N = 96 | 84.8 (16.7) | 81.6 (16.9) | 64.3 (20.8) | 87.1 (15.6) |
| CC, N = 44 | 72.8 (23.7) | 72.4 (21.5) | 51.6 (27.1) | 75.2 (22.4) |
| LOD score | 10.1 | 9.5 | 9.9 | 9 |
| % variation | 22 | 20 | 21 | 20 |
Average ABR thresholds of dw/dw F2 mice from the (DW-Pou1f1dw × CAST/EiJ) intercross are shown sorted by their D2Mit304 (Mdwh) genotypes. LOD scores of linkage associations are shown for an additive inheritance model with 1 d.f., and estimates are given for the percentage of total ABR variation that can be explained by the Mdwh locus. Genotype designations: DD, homozygous for DW allele; DC, heterozygous for DW and CAST alleles; CC, homozygous for CAST allele. Mice were tested at 4–8 weeks of age. Standard deviations are in parentheses.
The genetic interval containing Mdwh was estimated to span ∼20 Mb between the 118- and 138-Mb positions of Chr 2 (Figure 4). An interesting candidate gene within this interval is Mtap1a, the gene encoding microtubule-associated protein 1 A. Mtap1a, located at 121.1 Mb on Chr 2, is particularly intriguing because it modifies the hearing of mice with the tubby (tub) mutation in a strain-specific manner (Ikeda et al. 2002). The wild-type Mtap1a alleles from strains AKR/J, CAST/EiJ, and 129P2/OlaHsd protect tubby mice from hearing deficits, whereas a sequence variant in the C57BL/6J strain confers susceptibility to hearing impairment. We examined the Mtap1 gene in the DW/J strain and found that it is the same allelic form as that of the C57BL/6J strain (Figure 5), consistent with the possibility that this allele confers hearing loss susceptibility to Pou1f1dw/dw mutants as it does for tub/tub mutant mice. Both DW/J-Pou1f1dw/J and C57BL/6J strains contain five repeats of a 6-bp sequence in the large exon 5 of Mtap1a, whereas all other strains that we examined (AKR/J, CAST/EiJ, 129P2/OlaHsd, C3H/HeJ, BALB/cByJ, and DF/B-Prop1df/J) contain 15 repeats of this sequence.
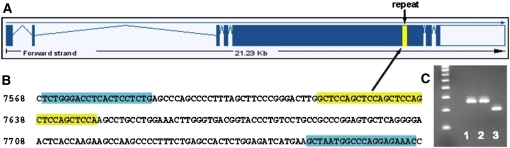
Figure 5 .
Mtap1a is a candidate gene for Mdwh. (A) Genetic structure of Mtap1a showing the position of a polymorphic repeat sequence (vertical yellow bar) in the large exon 5. (B) Primers (highlighted in blue) were designed to amplify the GCTCCA repeat sequence (highlighted in yellow; from C57BL/6J DNA), which encodes an alanine–proline repeat. (C) Sequencing of the 268-bp PCR amplification products from CAST/EiJ (lane 1) and C3H/HeJ (lane 2) showed that these strains have 15 repeats of the 6-bp sequence, whereas the 208-bp PCR product from the DW strain (lane 3) showed only 5 repeats.
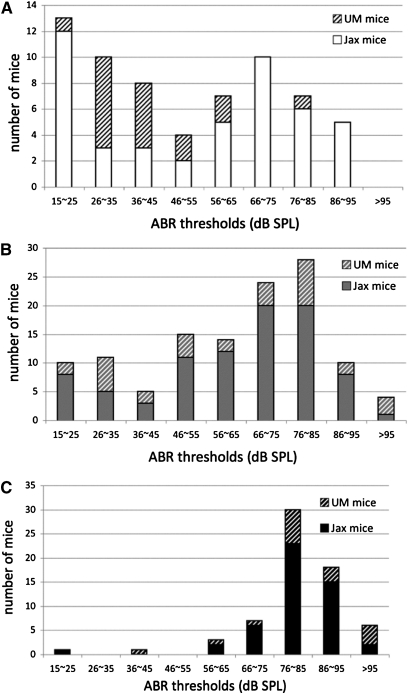
To assess the likelihood that the Moth1 allele of Mtap1a is identical to Mdwh, we created F1 intercrosses between DW/J-Pou1f1dw/+ and each of the two strains CAST/EiJ and AKR/J at the University of Michigan and assessed the segregation of non-DW/J Mtap1a alleles with the ability of F2 Pou1f1dw/dw mutants to hear. Sixty-eight Pou1f1dw/dw (DW/J-Pou1f1+/dw × CAST/EiJ) F2 mice were generated at the University of Michigan. Sixteen of them (23.5%) had ABR thresholds <35 dB SPL, which we designated as normal hearing. In both cohorts of (DW/J-Pou1f1+/dw × CAST/EiJ) F2 dw/dw mice generated at the Jackson laboratory and the University of Michigan, there is an enrichment of CAST/EiJ Mtap1a alleles among the hearing F2 mutants, but homozygosity for CAST/EiJ alleles is not sufficient for normal hearing ability (Figure 6A). Two F2 Pou1f1dw/dw mice homozygous for DW/J Mtap1a alleles also exhibited normal hearing ability (one is 20 dB SPL and one is ∼37 dB SPL) (Figure 6C), suggesting that CAST/EiJ Mtap1a alleles are not necessary for hearing either. ABR thresholds of mice heterozygous for Mtap1a alleles (DC in Table 2) are distributed through the range represented by the homozygous alleles (DD or CC), indicating the contribution of additional genes (Figure 6B).
Figure 6 .
The Mtap1a allele correlates with, but is neither necessary nor sufficient for, protection of hearing in Pou1f1dw mice. Two cohorts of dw/dw F2 (DW/J-Pou1f1dw/+ × CAST/EiJ) mice, with a total number of 251, were tested for their hearing sensitivity and genotyped for sequence variants of GCTCCA repeats in the Mtap1a gene. Mice were grouped by their Mtap1a genotypes, and we plotted the number of mice with ABR thresholds in each 20-kHz interval. Within each bar, the solid or open portion represents the mice generated at the Jackson laboratory (Jax mice); the hatched portion represents the mice generated at the University of Michigan (UM mice). (A) The numbers of mice homozygous for CAST/EiJ Mtap1a alleles at each ABR threshold interval. (B) The numbers of mice heterozygous for CAST/EiJ and DW/J Mtap1a alleles at each ABR threshold interval. (C) The numbers of mice homozygous for DW/J Mtap1a alleles at each ABR threshold interval.
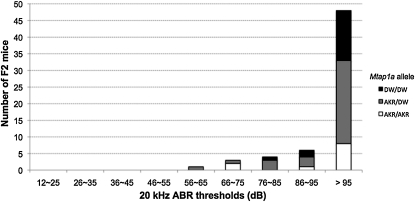
Sixty-two F2 Pou1f1dw/dw mice were collected from an intercross between (DW/J-Pou1f1dw/+ × AKR/J) F1 mice, and none of the progeny had normal hearing ability (Figure 7). We confirmed that 9 of 62 F2 Pou1f1dw/dw (DW/J-Pou1f1dw/+ × AKR/J) mice were homozyous for alleles of Mtap1a that protect against deafness of Tubby mutants, despite the lack of protection against dw hearing impairment. Thus, the AKR/J genetic background cannot protect against hypothyroidism-induced hearing loss in Pou1f1dw/dw mutants even though AKR/J mice have the same Mtap1a allele as CAST/EiJ.
Figure 7 .
AKR genetic background is not protective against hypothyroidism for the hearing in DW/J-Pou1f1dw/dw mice. Sixty-two dw/dw F2 (DW/J-Pou1f1dw/+ × AKR) mice were tested for their hearing sensitivity and genotyped for sequence variants of GCTCCA repeats in the Mtap1a gene. None of the mice showed ABR thresholds <55 dB SPL.
The Mouse Diversity Genotyping Array (Yang et al. 2009) was used to characterize the genetic background of the DW/J-Pou1f1dw strain and compare it to 72 inbred strains (SNP data from Mouse Diversity Array for the DW/J-Pou1f1dw strain can be found on the Mouse Phenome Database website at http://phenome.jax.org; MPD#: TJL4). In the Mdwh critical region (Chr 2: 118–138 Mb), the DW/J-Pou1f1dw strain is more similar to C57 substrains than to any of the other inbred strains used in the Mouse Diversity Genotyping Array. There are only 34 mismatches of 4508 SNPs analyzed between the DW/J-Pou1f1dw and C57BL/6J strains. Almost all of these apparent mismatches occur in cases where the allele of one or the other strain was uncertain. There is only one SNP that is a true mismatch: C/T. The strain distribution pattern (SDP) is the pattern of allelic similarities and differences among strains at a locus (Yalcin et al. 2004). The most consistent SDP between the susceptible (DW/J-Pou1f1dw, C57BL/6J, AKR) and protective (Cast/EiJ, C3H/HeJ) strains for hypothyroidism-induced deafness is the 124- to 131-Mb region of Chr 2, suggesting that Mdwh is in this interval (Table 3).
Table 3 . Comparing SNPs of DW-Pou1f1dw, C57B6/J, and AKR strains with SNPs of C3H and CAST/EiJ in the Chr 2 candidate region for the Mdwh QTL (118–138 Mb).
| Mb region | No. of SNPs with appropriate SDP |
|---|---|
| 118.0–118.9 | 6 (179) |
| 119.0–119.9 | 1 (185) |
| 120.0–120.9 | 2 (184) |
| 121.0–121.9 | 4 (166) |
| 122.0–122.9 | 1 (142) |
| 123.0–123.9 | 0 (189) |
| 124.0–124.9 | 14 (220) |
| 125.0–125.9 | 46 (209) |
| 126.0–126.9 | 58 (187) |
| 127.0–127.9 | 74 (243) |
| 128.0–128.9 | 46 (301) |
| 129.0–129.9 | 9 (333) |
| 130.0–130.9 | 34 (219) |
| 131.0–131.9 | 2 (235) |
| 132.0–132.9 | 6 (211) |
| 133.0–133.9 | 3 (190) |
| 134.0–134.9 | 0 (222) |
| 135.0–135.9 | 0 (196) |
| 136.0–136.9 | 0 (148) |
| 137.0–138.0 | 0 (175) |
Numbers in italics indicate the region with a consistent SDP. Numbers in parentheses are the numbers of SNPs that have allele information in each of the five mouse strains.
Discussion
TH deficiency can have major effects on neuronal development, body growth, and cochlear development resulting in cretinism, dwarfism, and deafness if not treated promptly. It is intriguing that the consequences of severe thyroid hormone deficiency can be highly variable in both humans and mice. Mutations in the pituitary transcription factor Pou1f1 result in severe hypothyroidism, dwarfism, and profound deafness on the DW/J genetic background, but the hearing deficit is milder when crossed to the normal-hearing, but genetically undefined, DF/B strain. Because the critical period for TH replacement is late gestation and early neonatal life, we assessed the contribution of the maternal environment to the differential hearing impairments between DW/J and DF/B strains. Our results prove that this particular genetic background effect is intrinsic to the fetus, rather than an influence of maternal TH during gestation or lactation (Q. Fang, unpublished data).
In this study, we examine the effects of the CAST/EiJ genetic background on the hearing of Pou1f1 mutants and show that strain-specific alleles of the Mdwh locus on Chr 2 have a major influence on the hearing of hypothyroid Pou1f1 dwarf mice (dw/dw). This locus showed a highly significant association (LOD score of 10) with hearing ability and could explain ∼20% of the ABR threshold variation of dwarf F2 mice from a DW/J-Pou1f1dw × CAST/EiJ intercross. Given the pleiotropic effects of TH on the inner ear development, it is very surprising that the Mdwh appears simple and genetically tractable.
The Mdwh modifier locus does not appear to affect the basal serum level of TH in Pou1f1 mutant mice, which lack thyroid stimulating hormone (TSH). If the modifier had a general effect on TH levels, then the dwarf mice with normal hearing would be expected to be larger than the deaf dwarfs. We have detected such generalized effects in other models with partial restoration of TSH production (Cushman et al. 2001). Because the weights of hearing and deaf dwarfs are indistinguishable and only ABR thresholds are variable (Table 1), it is likely that Mdwh is modifying processes that affect cochlear development or function even in the absence of systemic TH stimulation. Although we cannot rule out a local effect on cochlear TH production or transport, there are no known TH transporters or deiodinases in the critical region (Mouse Genome Informatics: http://www.informatics.jax.org).
The Mdwh gene may modify the degree of hearing impairment due to hypothyroidism by altering the basal expression levels of critical gene(s) without enhancing TH production. This model is drawn from a similar case of genetic background effects in which the survival of Prop1 mutant newborn mice correlated with higher basal levels of surfactant B gene expression in the lung at birth compared to mutants that exhibited lethality (Nasonkin et al. 2004). The transcription factor Nkx2.1 is necessary for surfactant B expression in the lung, and Nkx2.1 transcription is upregulated by TH. The mutant mice that were cyanotic and died had lower basal levels of Nkx2.1 expression than those that were viable, suggesting that genetic background affected the basal level of Nkx2.1 expression in the absence of TH. In the case of Mdwh, the DW/J hearing loss susceptibility allele may have a less active basal state (in the absence of TH) than the CAST/EiJ protective allele. If cochlear development and function were sensitive either directly or indirectly to slight variations in the level of Mdwh gene activity, then the lower basal levels in hypothyroid F2 mice with the DW/J allele could explain their more severe hearing impairment, whereas the higher basal level of Mdwh gene activity associated with the CAST/EiJ allele would protect from severe hearing loss and could explain the additive nature of Mdwh allele effects on ABR thresholds.
We considered the Mtap1a gene as a particularly intriguing candidate gene for Mdwh. It maps within the 20-Mb candidate interval on Chr 2 and is the gene responsible for the strain-specific modification of hearing in tubby (tub) mutant mice (Ikeda et al. 2002). The underlying mechanism proposed for this modification is an altered protein–protein interaction between MTAP1A (a microtubule-associated protein) and a protein localized to the post-synaptic density in hair cells, PSD95, a protein critical for the cyto-architecture at the synapse. Another hearing-related QTL on Chr 2, which could be equivalent to Mtap1a or Mdwh, modifies the degree of hearing loss in Beethoven mutant mice, which carry a mutation in Tmc1 (Noguchi et al. 2006). Intriguingly, the phenotype of these mice is outer hair cell loss and reduced DPOAE, the same as that observed in Pou1f1dw mutants (Mustapha et al. 2009).
Evidence implicating Mtap1a as the gene underlying the modifier effects of the Mdwh locus includes the following. First, the DW/J strain has the same Mtap1a allele as C57BL/6J (Figure 5), which was shown to confer susceptibility to hearing loss in tub mutant mice (Ikeda et al. 2002). Both DW/J and C57BL/6J strain backgrounds confer an increased susceptibility to hearing impairment in Pou1f1dw mutant mice (Figure 1), and both have the same Mtap1a allele. Second, the expression of Mtap1a is altered by TH in the cerebellum of hypothyroid rats (Benjamin et al. 1988) and in the sensory motor cortex of hypothyroid (hyt) mutant mice (Biesiada et al. 1996). Third, MTAP1A interacts directly with the calcium-activated potassium channel BKCa (Park et al. 2004), and defects in Kcnma1, which encodes one of the BKCa subunits, cause progressive hearing loss and absence of KCNQ4 expression (Ruttiger et al. 2004). Fourth, KCNQ4 is regulated by the thyroid hormone receptors TRα1 and TRβ in outer hair cells (Winter et al. 2006), and its expression is reduced in Pou1f1dw mutant outer hair cells (Mustapha et al. 2009). Finally, recent studies demonstrate that TH is essential for morphological and functional maturation of inner hair cell (IHC) ribbon synapses (Brandt et al. 2007; Sendin et al. 2007). These studies suggest that presynaptic dysfunction of IHCs, which could be proven by demonstrating reduced exocytosis efficiency, is a mechanism in congenital hypothyroid deafness, and MTAP1A has been proposed to be involved in synaptic function (Ikeda et al. 2002). Although these lines of evidence seem compelling when taken together, our study on segregation of CAST/EiJ and AKR/J Mtap1a alleles in F2 Pou1f1dw mutants showed that CAST/EiJ and AKR/J Mtap1a alleles are neither sufficient nor necessary for normal hearing abilities in those mice. We observed the same result in F2 mutants from an intercross between the DW/J strain and the 129P2/OlaHsd strain (129P2), which contains the same Mtap1a allele as CAST/EiJ and AKR/J (data not shown). We conclude that Mdwh is not Mtap1a, that Mtap1a is not the sole component of Mdwh, or that additional modifiers obscure the effects of Mtap1 on hearing.
Given the pleiotropic effects that the thyroid hormone has on cochlear development (Mustapha et al. 2009), Mdwh could consist of multiple genes, possibly including Mtap1a, whose combined actions protect against hypothyroidism-induced hearing impairment in Pou1f1dw mutants. The information gleaned by genotyping the DW/J-Pou1f1dw genome for >570,000 SNPs in the Mouse Diversity Genotyping Array will facilitate the search for candidate genes within the Mdwh locus as well as assess candidate regions of additional modifier QTL on other chromosomes. Future directions to identify the protective genes could include refining the critical interval by generating more animals and genotyping more markers on Chr 2 and by creating a congenic mouse line with CAST/EiJ alleles of Mdwh and DW/J alleles at other loci to determine if the CAST/EiJ Mdwh locus is sufficient to protect against hearing loss. Another strategy would be to directly sequence the candidate genes within the Mdwh locus from the susceptible DW/J and/or C57BL/6J strains and compare them with the C3H/HeJ, CAST/EiJ, and other inbred strain sequences generated by the Mouse Genomes Project (http://www.sanger.ac.uk/resources/mouse/genomes/). This strategy was successful in identifying two other modifier loci (Buchner et al. 2003; Amendola et al. 2010). There are >1000 genes and >50,000 SNPs between C57BL/6J and CAST/EiJ within the Mdwh critical region, but no obvious nonsense SNPs are evident. Thus, genetic studies to narrow the interval are the next logical step for identification of the Mdwh modifier gene(s).
In conclusion, identifying the gene(s) that modify the expression of mutant auditory phenotypes related to congenital hypothyroidism will be important for determining the molecular mechanisms that underlie the influence of TH on cochlear development and function. In addition, it is likely that a mechanistic understanding of modifier gene action will provide a conceptual framework for understanding the multi-factorial basis for hearing loss.
Acknowledgments
We thank the following individuals for their important contributions to this work: Mirna Mustapha and Thomas Glaser for discussion and suggestions, Michelle Fleming and Emily Maclary for SNP analysis, Jennifer Benson for auditory brainstem response tests at the University of Michigan, and Sandra Gray for mouse husbandry at the Jackson laboratory. Funding for this study came from March of Dimes grant 6-FY08-262 (S.A.C.), National Institutes of Health/National Institute on Deafness and Other Communication Disorders grants DC04301 (K.R.J.) and P30 DC05188 (D.F.D.), a University of Michigan Barbour Fellowship (Q.F.), and a Rackham Graduate School Summer Grant (Q.F.).
Literature Cited
- Amendola E., Sanges R., Galvan A., Dathan N., Manenti G., et al. , 2010. A locus on mouse chromosome 2 is involved in susceptibility to congenital hypothyroidism and contains an essential gene expressed in thyroid. Endocrinology 151: 1948–1958 [DOI] [PubMed] [Google Scholar]
- Benjamin S., Cambray-Deakin M. A., Burgoyne R. D., 1988. Effect of hypothyroidism on the expression of three microtubule-associated proteins (1A, 1B and 2) in developing rat cerebellum. Neuroscience 27: 931–939 [DOI] [PubMed] [Google Scholar]
- Bernal J., 2005. Thyroid hormones and brain development. Vitam. Horm. 71: 95–122 [DOI] [PubMed] [Google Scholar]
- Biesiada E., Adams P. M., Shanklin D. R., Bloom G. S., Stein S. A., 1996. Biology of the congenitally hypothyroid hyt/hyt mouse. Adv. Neuroimmunol. 6: 309–346 [DOI] [PubMed] [Google Scholar]
- Bluestone C. D., Klein J. O., 2007. Otitis Media in Infants and Children. BC Decker, Hamilton, Ontario, Canada [Google Scholar]
- Brandt N., Kuhn S., Munkner S., Braig C., Winter H., et al. , 2007. Thyroid hormone deficiency affects postnatal spiking activity and expression of Ca2+ and K+ channels in rodent inner hair cells. J. Neurosci. 27: 3174–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner D. A., Trudeau M., Meisler M. H., 2003. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301: 967–969 [DOI] [PubMed] [Google Scholar]
- Cabello G., Wrutniak C., 1989. Thyroid hormone and growth: relationships with growth hormone effects and regulation. Reprod. Nutr. Dev. 29: 387–402 [DOI] [PubMed] [Google Scholar]
- Camper S. A., Saunders T. L., Katz R. W., Reeves R. H., 1990. The Pit-1 transcription factor gene is a candidate for the murine Snell dwarf mutation. Genomics 8: 586–590 [DOI] [PubMed] [Google Scholar]
- Christ S., Biebel U. W., Hoidis S., Friedrichsen S., Bauer K., et al. , 2004. Hearing loss in athyroid pax8 knockout mice and effects of thyroxine substitution. Audiol. Neurootol. 9: 88–106 [DOI] [PubMed] [Google Scholar]
- Cushman L. J., Watkins-Chow D. E., Brinkmeier M. L., Raetzman L. T., Radak A. L., et al. , 2001. Persistent Prop1 expression delays gonadotrope differentiation and enhances pituitary tumor susceptibility. Hum. Mol. Genet. 10: 1141–1153 [DOI] [PubMed] [Google Scholar]
- Darvasi A., 1997. The effect of selective genotyping on QTL mapping accuracy. Mamm. Genome 8: 67–68 [DOI] [PubMed] [Google Scholar]
- Deol M. S., 1973. Congenital deafness and hypothyroidism. Lancet 2: 105–106 [DOI] [PubMed] [Google Scholar]
- Dupuis J., Siegmund D., 1999. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics 151: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher E. M., Beamer W. G., 1980. New mouse dw allele: genetic location and effects on lifespan and growth hormone levels. J. Hered. 71: 187–190 [DOI] [PubMed] [Google Scholar]
- Forrest D., Erway L. C., Ng L., Altschuler R., Curran T., 1996. Thyroid hormone receptor beta is essential for development of auditory function. Nat. Genet. 13: 354–357 [DOI] [PubMed] [Google Scholar]
- Gagnon L. H., Longo-Guess C. M., Berryman M., Shin J. B., Saylor K. W., et al. , 2006. The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J. Neurosci. 26: 10188–10198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisty R. E., Erven A., Logan K., Morse S., Guionaud S., et al. , 2003. The deaf mouse mutant Jeff (Jf) is a single gene model of otitis media. J. Assoc. Res. Otolaryngol. 4: 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A., Zheng Q. Y., Zuberi A. R., Johnson K. R., Naggert J. K., et al. , 2002. Microtubule-associated protein 1A is a modifier of tubby hearing (moth1). Nat. Genet. 30: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. R., Marden C. C., Ward-Bailey P., Gagnon L. H., Bronson R. T., et al. , 2007. Congenital hypothyroidism, dwarfism, and hearing impairment caused by a missense mutation in the mouse dual oxidase 2 gene, duox2. Mol. Endocrinol. 21: 1593–1602 [DOI] [PubMed] [Google Scholar]
- Karolyi I. J., Dootz G. A., Halsey K., Beyer L., Probst F. J., et al. , 2007. Dietary thyroid hormone replacement ameliorates hearing deficits in hypothyroid mice. Mamm. Genome 18: 596–608 [DOI] [PubMed] [Google Scholar]
- Lander E., Kruglyak L., 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247 [DOI] [PubMed] [Google Scholar]
- Li S., Crenshaw E. B., III, Rawson E. J., Simmons D. M., Swanson L. W., et al. , 1990. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature 347: 528–533 [DOI] [PubMed] [Google Scholar]
- Manly K. F., Cudmore R. H., Jr, Meer J. M., 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930–932 [DOI] [PubMed] [Google Scholar]
- Mustapha M., Fang Q., Gong T. W., Dolan D. F., Raphael Y., et al. , 2009. Deafness and permanently reduced potassium channel gene expression and function in hypothyroid Pit1dw mutants. J. Neurosci. 29: 1212–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasonkin I. O., Ward R. D., Raetzman L. T., Seasholtz A. F., Saunders T. L., et al. , 2004. Pituitary hypoplasia and respiratory distress syndrome in Prop1 knockout mice. Hum. Mol. Genet. 13: 2727–2735 [DOI] [PubMed] [Google Scholar]
- Ng L., Goodyear R. J., Woods C. A., Schneider M. J., Diamond E., et al. , 2004. Hearing loss and retarded cochlear development in mice lacking type 2 iodothyronine deiodinase. Proc. Natl. Acad. Sci. USA 101: 3474–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L., Hernandez A., He W., Ren T., Srinivas M., et al. , 2009. A protective role for type 3 deiodinase, a thyroid hormone-inactivating enzyme, in cochlear development and auditory function. Endocrinology 150: 1952–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi Y., Kurima K., Makishima T., de Angelis M. H., Fuchs H., et al. , 2006. Multiple quantitative trait loci modify cochlear hair cell degeneration in the Beethoven (Tmc1Bth) mouse model of progressive hearing loss DFNA36. Genetics 173: 2111–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley B. W., Jr, Li D., Turner D. S., 1995. Hearing loss and cochlear abnormalities in the congenital hypothyroid (hyt/hyt) mouse. Hear. Res. 88: 181–189 [DOI] [PubMed] [Google Scholar]
- Park S. M., Liu G., Kubal A., Fury M., Cao L., et al. , 2004. Direct interaction between BKCa potassium channel and microtubule-associated protein 1A. FEBS Lett. 570: 143–148 [DOI] [PubMed] [Google Scholar]
- Rusch A., Ng L., Goodyear R., Oliver D., Lisoukov I., et al. , 2001. Retardation of cochlear maturation and impaired hair cell function caused by deletion of all known thyroid hormone receptors. J. Neurosci. 21: 9792–9800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttiger L., Sausbier M., Zimmermann U., Winter H., Braig C., et al. , 2004. Deletion of the Ca2+-activated potassium (BK) alpha-subunit but not the BKbeta1-subunit leads to progressive hearing loss. Proc. Natl. Acad. Sci. USA 101: 12922–12927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendin G., Bulankina A. V., Riedel D., Moser T., 2007. Maturation of ribbon synapses in hair cells is driven by thyroid hormone. J. Neurosci. 27: 3163–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohmer H., Freeman S., 1996. The importance of thyroid hormone for auditory development in the fetus and neonate. Audiol. Neurootol. 1: 137–147 [DOI] [PubMed] [Google Scholar]
- Sprenkle P. M., McGee J., Bertoni J. M., Walsh E. J., 2001. Consequences of hypothyroidism on auditory system function in Tshr mutant (hyt) mice. J. Assoc. Res. Otolaryngol. 2: 312–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel A., 1986. Periods of sensitivity to thyroid hormone during the development of the organ of Corti. Acta Otolaryngol. Suppl. 429: 23–27 [DOI] [PubMed] [Google Scholar]
- Vanderschueren-Lodeweyckx M., Debruyne F., Dooms L., Eggermont E., Eeckels R., 1983. Sensorineural hearing loss in sporadic congenital hypothyroidism. Arch. Dis. Child. 58: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H., Braig C., Zimmermann U., Geisler H. S., Franzer J. T., et al. , 2006. Thyroid hormone receptors TRalpha1 and TRbeta differentially regulate gene expression of Kcnq4 and prestin during final differentiation of outer hair cells. J. Cell Sci. 119: 2975–2984 [DOI] [PubMed] [Google Scholar]
- Yalcin B., Fullerton J., Miller S., Keays D. A., Brady S., et al. , 2004. Unexpected complexity in the haplotypes of commonly used inbred strains of laboratory mice. Proc. Natl. Acad. Sci. USA 101: 9734–9739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ding Y., Hutchins L. N., Szatkiewicz J., Bell T. A., et al. , 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods 6: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. Y., Johnson K. R., Erway L. C., 1999. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear. Res. 130: 94–107 [DOI] [PMC free article] [PubMed] [Google Scholar]