Abstract
While it is well understood that the pace of evolution depends on the interplay between natural selection, random genetic drift, mutation, and gene flow, it is not always easy to disentangle the relative roles of these factors with data from natural populations. One popular approach to infer whether the observed degree of population differentiation has been influenced by local adaptation is the comparison of neutral marker gene differentiation (as reflected in FST) and quantitative trait divergence (as reflected in QST). However, this method may lead to compromised statistical power, because FST and QST are summary statistics which neglect information on specific pairs of populations, and because current multivariate tests of neutrality involve an averaging procedure over the traits. Further, most FST–QST comparisons actually replace QST by its expectation over the evolutionary process and are thus theoretically flawed. To overcome these caveats, we derived the statistical distribution of population means generated by random genetic drift and used the probability density of this distribution to test whether the observed pattern could be generated by drift alone. We show that our method can differentiate between genetic drift and selection as a cause of population differentiation even in cases with FST = QST and demonstrate with simulated data that it disentangles drift from selection more accurately than conventional FST–QST tests especially when data sets are small.
NULL models have an important role in many areas of biology (e.g., Whitlock and Phillips 2000; Hubbell 2001; Ohta 2002) and not least in studies of population differentiation. For instance, the expectation that genetic differentiation occurs most readily toward the direction of maximum additive genetic variance (Schluter 1996) has provided a useful null model for evaluating the role of the ancestral G matrix in the rate and direction of evolutionary transitions (e.g., McGuigan 2006). Likewise, while few evolutionary biologists would doubt the power of natural selection over genetic drift (Hohenlohe and Arnold 2008), comparisons of the index of quantitative genetic differentiation (QST) to the index of neutral genetic differentiation (FST) have provided a useful platform to identify traits and populations subject to directional selection (Merilä and Crnokrak 2001; McKay and Latta 2002; Leinonen et al. 2008).
The current neutrality tests of population differentiation make restrictive assumptions or suffer from various methodological or logical problems (e.g., O’Hara and Merilä 2005; Beaumont 2008; Whitlock 2008). To start with, the popular method of comparing the index of quantitative genetic differentiation (QST) to the index of neutral genetic differentiation (FST) does not necessarily provide practical means to analyze whether a small number of populations are locally adapted to their environments, as obtaining reliable estimates typically requires data from >10 populations in controlled environmental conditions (O’Hara and Merilä 2005; Martin et al. 2008). Multivariate extensions of FST–QST comparisons (Chenoweth and Blows 2008; Martin et al. 2008) are somewhat more powerful due to integration of information across multiple traits, but they also suffer from the same principal shortcomings. Second, while problems stemming from phylogenetic nonindependence in statistical testing are well recognized in interspecific comparative studies (Harvey and Pagel 1991; Martins and Hansen 1997; Martins 2000), in the context of intraspecific studies the phylogenetic nonindependence among populations is often not controlled for or even discussed (but see Edwards and Kot 1995).
Apart from the statistical issues, there are also more fundamental problems with FST–QST comparisons (e.g., Hendry 2002; Whitlock 2008). In the univariate case in which measurements are made on a single trait, QST is defined by
| (1) |
(Merilä and Crnokrak 2001; Whitlock 2008), where D is the additive variance among populations and G is the additive variance within populations, the latter averaged over the populations. A number of methods have been developed for accounting for the estimation error in QST (O’Hara and Merilä 2005), but an often ignored fact is that D and G are random variables due to the inherent randomness in the evolutionary process (to be called here process error to separate it from estimation error). However, in the context of FST–QST comparisons it has been pointed out only recently that QST is a random variable, which thus has a probability distribution around the expectation given by Equation 1 (Whitlock 2008; Whitlock and Guillaume 2009).
In the multivariate case in which measurements are made on a number of traits, QST is a matrix, defined, e.g., as
| (2) |
(Chenoweth and Blows 2008). An additional technical difficulty in conducting a multivariate FST–QST neutrality test is that one of the quantities (QST) is a matrix, while the other one (FST) is a scalar. Chenoweth and Blows (2008) suggested comparing FST either to the mean eigenvalue of QST or to the maximal eigenvalue of QST. While these are practical solutions that may well lead to correct conclusions, they lack a clear theoretical justification and do not fully utilize the multivariate information contained in QST. Martin et al. (2008) avoided the problem of comparing a matrix to a scalar by noting that in the multivariate case neutral dynamics lead to
| (3) |
This equation is consistent with QST = FST, but as both its sides are matrices, it contains additional degrees of freedom and thus additional statistical power. One can test (i) whether D and G are proportional and (ii) whether the proportionality constant equals 2FST/(1 − FST). As expected, a combination of such tests has more statistical power than a univariate test (Martin et al. 2008). However, even these approaches pool the data over the traits in the sense that they do not account for the full matrix structure in a single test. A further problem with Equation 3 is that it ignores the process error, and thus it strictly speaking holds only for the unknown expectations of the D and G matrices over the evolutionary process.
In this article, we propose an alternative approach to QST–FST comparisons to disentangle drift and selection. Our method is based on the simple observation that additive covariance among relatives, often expressed in the context of the so-called “animal model” by the equation
| (4) |
(Lynch and Walsh 1998), also holds at the population level. In the usual interpretation, ai is the genetic value of individual i and is the coancestry coefficient between individuals i and i′. As we show, Equation 4 also holds if we let ai be the mean genetic value in population i and be the mean coancestry between individuals belonging to populations i and i′. This population-level analog of Equation 4 is a statistical statement about the distribution of population means under neutrality, and it can thus be used to derive a statistical test asking whether the observed pattern can be explained by drift alone.
We first review the fundamental theory of quantitative genetics from which Equation 4 and the expectation QST = FST can be derived. While this theory has been used and discussed in a large number of articles (see, e.g., Whitlock 1999; Merilä and Crnokrak 2001; McKay and Latta 2002) and textbooks (e.g., Lynch and Walsh 1998), we rederive the theory here to make our arguments transparent and to be specific about hidden assumptions, such as the exact nature of the process error. On the basis of the mathematical theory, we propose a new approach for testing the role of directional (or stabilizing) selection behind population divergence. In comparison to earlier methods, the method developed here (i) accounts for the inherent randomness in the evolutionary process, (ii) utilizes in the multivariate case the information on all traits without an averaging procedure, (iii) accounts for population-to-population–level coancestry coefficients and thus omits the averaging procedure used to compute FST, and (iv) accounts for population-to-population–level information on similarity in trait values and thus omits the averaging procedure used to compute QST. We illustrate with simulated data how the new approach can be used to reject the null hypothesis of divergence by random genetic drift even in cases where FST and QST are identical. Finally, we implement a Bayesian framework to connect our method to the kind of data typically used in an FST–QST test and compare its performance to the method of Martin et al. (2008). We demonstrate that contrary to earlier methods, our approach can disentangle drift from selection even for cases where data are available for a few populations only.
Population Divergence Under Neutral Genetic Drift
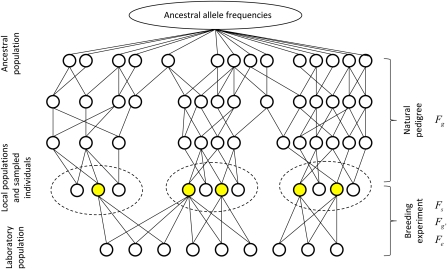
As illustrated in Figure 1, we consider a metapopulation consisting of a set of local populations, each local population X ∈ consisting of nX individuals. We assume that all local populations are derived from a common ancestral population, and our interest is in disentangling the roles of drift and selection as factors behind an observed genetic divergence among the local populations. We first derive the expectation under the null model, which assumes that evolution takes place from the ancestral population to the present generation under genetic drift only. We then develop a statistical framework that applies to typical study designs by assuming that a randomly selected subset of individuals is sampled from the local populations. This sample population is genotyped for neutral markers (to infer relatedness) and used in a “common garden breeding experiment” to measure the genetic component behind quantitative trait divergence, the offspring being called here the “laboratory population”.
Figure 1 .
Schematic illustration of divergence of local populations from a common ancestral population and of a “common garden” breeding experiment applied to the current generation. Dashed ellipses represent local populations, circles represent individuals, and lines connect parents and offspring. The four random processes refer to the flow of alleles through the natural pedigree (Fg) and through the breeding experiment (Fg′), the sampling of individuals to be used in the breeding experiment (Fs, sampled individuals shown by yellow), and the randomness associated to environmental effects for the individuals in the “laboratory” population (Fe).
Identity by descent, identity by state, coancestry, and FST
Relatedness among individuals can be measured through the identity by state, the realized identity by descent, or the probability of identity by descent, whose quantities have been estimated from neutral molecular marker data with a variety of statistical methods (Cockerham and Weir 1987; Weir and Hill 2002; Fernandez and Toro 2006; Kitakado et al. 2006; Anderson and Weir 2007; Kitada et al. 2007; Bink et al. 2008; Maenhout et al. 2009; Samanta et al. 2009). In this article we characterize relatedness through the coancestry coefficient , which is the probability that randomly chosen alleles from a given neutral locus of individuals i and i′ are identical by descent (IBD), i.e., that they are derived from the same allelic copy in the ancestral population. We denote average coancestry between populations X and Y by (see Table 1 for symbols used in this article)
| (5) |
where the superscript refers to population level. The average within-population relatedness in population X is denoted by We denote the average self-coancestry in population X by and the average self-coancestry among all populations by We note that if the mating structure within local populations is random, we have
Table 1 . Symbols used in this article.
| Type | Symbol | Explanation |
|---|---|---|
| Dimensions | No. of local populations | |
| nX | No. of individuals in population X | |
| No. of traits | ||
| nℒ | No. of loci | |
| Coancestry coefficients | Matrix θ, with elements θii′ | Coancestry coefficient = probability that randomly chosen alleles for individuals i and i′ are identical by descent for a neutral locus |
| Matrix , with elements | Population-to-population coancestry | |
| Average population-to-population coancestry | ||
| Within-population coancestry | ||
| Average within-population coancestry | ||
| Average self-coancestry in population X | ||
| Average self-coancestry | ||
| Distribution of alleles | xijku | Indicator function for the allele k in locus j of individual i being of the allelic type u |
| pju | The frequency of allele u in locus j in the ancestral population | |
| Genetic values | vjum | Additive value of allele u in locus j for trait m |
| Additive value of trait m for individual i | ||
| ai | Vector of additive values (of all traits) for individual i | |
| Mean additive value in population X | ||
| Mean additive value | ||
| Genetic variance–covariance matrices | Matrix of genetic variances and covariances in the ancestral population | |
| GX | Matrix of genetic variances and covariances in population X | |
| Matrix of genetic variances and covariances among populations |
The index of FST measures within-population relatedness compared to among-population relatedness, defined through identity by state (IBS) as
| (6) |
(Cockerham and Weir 1987). Here f1 is the IBS probability for two alleles sampled from two individuals belonging to the same population, whereas f2 is the IBS probability for two alleles sampled from two individuals belonging to different populations. The definition of Equation 6 is the most common basis for FST estimation methods in QST–FST comparisons (Leinonen et al. 2008). While FST is a widely used measure of population differentiation, recent studies have questioned its applicability, partly due to logical flaws and unintuitive interpretation in allele-frequency–based definitions of FST (Jost 2008) and partly due to the fact that it is often ignored that FST is a random variable (Whitlock 2008). Some of these problems can be avoided by defining FST through coancestry, i.e., the probability of identity by descent for neutral loci (Rousset 2004) as
| (7) |
where
| (8) |
and
| (9) |
are metapopulation-level averages of the relatedness coefficients. As shown in Supporting Information, File S1, the two definitions (Equations 6 and 7) coincide at the low mutation limit for neutral loci. As Equation 7 is defined through coancestry, it is a property of the underlying pedigree and thus independent of any specific marker locus.
Quantitative traits and the G matrix
We consider a set of quantitative traits affected by a set of nℒ loci (or genes) and denote by xijku the indicator function for the allele k (we consider here diploidic individuals, so k = 1, 2) in locus j of individual i being of the allelic type u. We consider only additive genetic effects and denote the additive value of allele u in locus j for trait m by vjum so that the additive value of trait m for individual i is We denote the vector of additive values (of all traits) for individual i by ai and the matrix consisting of additive vectors for all individuals by A = (ai)i.
We denote the frequency of the allele u in locus j in the ancestral population by pju and consider this as a fixed parameter for the development of the theory. We next consider a random process Fg which describes the flow of alleles from the ancestral population to the local populations. To create a random sample (i.e., realization of the xijku values) of this process, the first step is to randomize the alleles to the individuals forming the ancestral population with the multinomial distribution with parameter pju, independently for each allele, each locus, and each individual. In other words, the individuals in the ancestral generation are assumed to be unrelated and noninbred. The second step is to mimic neutral drift by randomizing the alleles to the forthcoming generations by deriving for each locus one allele from each parent. This is done independently for each locus, so no linkage is assumed.
The process Fg affects only the flow of alleles, not the pedigree structure, which is considered here to be fixed. We note that even though natural pedigrees are surely affected by selection on a number of traits, the flow of neutral alleles (and thus the process Fg) can be modeled by considering the pedigree as fixed. We denote expected values, variances, and covariances over Fg by E[⋅], Var[⋅], and Cov[⋅]. In neutral loci, the expected allele frequency for all individuals equals that in the ancestral generation, E[xijku] = pju.
We denote the additive genetic variance–covariance matrix of the ancestral population by to separate it from the mean additive genetic variance–covariance matrix averaged over the local populations, the latter denoted by G throughout this article. We define as the covariance (over Fg) in additive value over the individuals belonging to the ancestral generation. As shown in File S1, can be written with the help of the ancestral allele frequencies and the additive values of the alleles as
| (10) |
where δu,u′ is Kronecker’s delta, with δu,u = 1 and δu,u′ = 0 for u ≠ u′. We confirm in File S1 that with this definition, the phenotypic covariance among relatives equals the assumption of the animal model (Lynch and Walsh 1998),
| (11) |
We note that Equation 11 does not assume any specific distributional form for the additive values. If following the usual convention in quantitative genetics, i.e., that of multinormally distributed traits, the above leads to
| (12) |
where μ is the expected mean determined by the allele frequencies in the ancestral generation, n is the total number of individuals, In is the n × n identity matrix, θ is the coancestry matrix with elements , and ⊗ denotes the Kronecker product.
One can define the additive genetic variance–covariance matrix with respect to any reference (or base) population instead of the ancestral population. We denote by GX the additive variance–covariance matrix with the current local population X considered as the reference population. As described above, GX depends on the additive values of the alleles and on the allele frequencies in the population X. While the additive values of alleles (vjum) are here considered fixed parameters and thus independent of the population, the allele frequencies in population X depend on the process Fg, and thus GX is a random variable. The expectation of GX is (see File S1)
| (13) |
which is small in populations in which the individuals are closely related to each other ( large). Equation 13 has been written in the form E[GX] = (1 − F)G, where F is an “inbreeding coefficient” (Lande 1980; Wright 1951), the relevant F statistic here being the average coancestry coefficient among individuals in population X. Quoting Phillips et al. (2001, p. 1137) on Equation 13, “this result, if true, is important, because it implies that drift would not affect the trajectory of evolution but merely the pace at which evolution would proceed.” Defining as the average of the population-specific additive genetic variance–covariance matrices, by the above it holds that E[G] = (1 − θ). The distribution of GX around the expectation, generated by the random drift process Fg, remains partly an open problem (Phillips et al. 2001).
Additive genetic variation among populations
We denote by
| (14) |
the mean additive value in population X and by
| (15) |
the mean of the means over the populations. Additive variance–covariance among the populations can be measured by With respect to the process Fg, D is a random variable, the expectation of which is given by (see File S1)
| (16) |
Thus, the variance among the additive means of the local populations is large if the populations contain closely related individuals (θ large) and if the populations are not much related to each other (θ small). Also this result was suggested already by Wright (1951). Phillips et al. (2001, p. 1138) expressed it as “the amount of genetic variance is expected to be 2F times the genetic variance of the base population.” Comparing to Equation 16, in this case the relevant F statistic is θ − θ, i.e., the average coancestry among the populations minus the average coancestry within the populations. Utilizing Equation 7, Equation 16 can be equivalently written as
| (17) |
which was the starting point of Martin et al. (2008).
Uncovering the Signature of Selection
As discussed in the Introduction, there are several caveats with most QST–FST tests. First, these seldom account for the process error, i.e., randomness generated by Fg, and thus they may potentially result in flawed judgment. Second, QST and FST are summary statistics over the populations, and they thus neglect any information at the population-to-population level. Third, the current multivariate tests of neutrality (Chenoweth and Blows 2008; Martin et al. 2008) involve an averaging procedure over the traits. To overcome these caveats, we next propose an alternative approach for pinpointing the role of selection in population divergence.
Our reasoning is based on the observation that the covariance in population mean additive values is given by (see File S1)
| (18) |
Denoting the matrix of population-level effects by and assuming normally distributed traits, we obtain
| (19) |
where the matrix θ consists of the elements Note that these equations are the population-level analogs of the individual-level Equations 11 and 12.
The above equations (Equation 18 or Equation 19) contain all the information used in a QST–FST test. First, the value of FST is a summary statistic of the matrix θ. Second, the mean additive genetic variance–covariance matrix over the local populations (G) can be computed from and θ. Third, the among-population variance D can be computed from the matrix A. However, unlike most QST–FST tests, Equations 18 and 19 give the neutral expectation of population divergence separately for all population pairs and for all traits, and thus they do not involve any averaging procedures over the coancestry or over the traits. Further, the distributional form of Equation 19 accounts for the process error generated by Fg.
Patterns of population divergence under drift and selection
The above theory assumes that evolution is influenced by neutral genetic drift only. We next discuss what kind of signatures of selection may be found from a pattern of population divergence if the traits have been under local adaptation. To simplify the discussion, we first ignore estimation error and thus assume that the following quantities are known exactly: mean additive values for all local populations (A), their common expected mean (μ), population-level coancestry matrix (θ), and the additive variance–covariance matrix of the ancestral population (). In the next section, we account for estimation error by illustrating with simulated data how the framework can be applied to the kinds of data that are typically used in QST–FST tests.
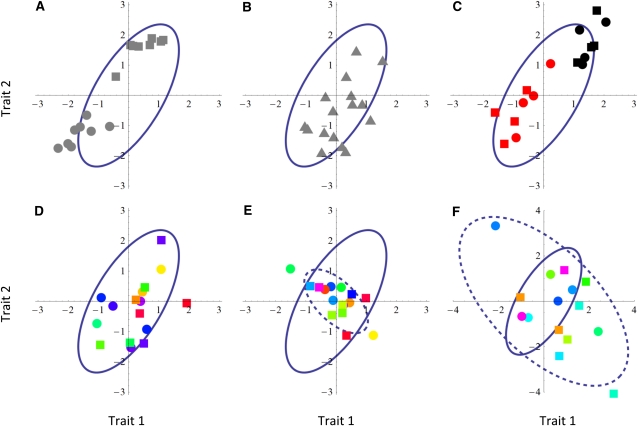
Figure 2 depicts patterns of population divergence under different scenarios of drift and local adaptation. In Figure 2, A and B, the traits under consideration are selectively neutral and thus the populations have diverged due to drift only (Equation 19). In Figure 2A, the set of local populations, referred to as (a), includes two clusters of closely related (in the sense of coancestry) local populations (the circles and the squares). In Figure 2B, the set of local populations, referred to as (b), is homogeneous so that all local populations (the triangles) are equally related to each other by coancestry. The ancestral matrix is represented by the continuous ellipse (variance–covariance matrices can be visualized by ellipsoids; see, e.g., Ovaskainen et al. 2008), and it is assumed to be identical in Figure 2, A and B. As expected under drift (Equation 19), in Figure 2A the two sets of local populations form distinct groups also in terms of the quantitative traits, whereas in Figure 2B the local populations are spread more evenly around the common expectation. Note that we have organized the matrices θ(a) and θ(b) so that FST is the same for both population structures.
Figure 2 .
Patterns of population divergence for = 2 traits under drift (A and B) and different kinds of selection scenarios (C–F). The symbols represent the distribution of population means (A) around their common expectation, with a total of n = 16 populations. Circles and squares represent two lines of populations that have diverged from each other ( if X is represented by a circle and Y by a square, and if both X and Y are shown by the same symbol). The triangles represent a set of populations that have diverged from a common ancestor equally long ago ( for all population pairs). Both population structures a and b correspond to FST = 0.37. In c, the black and red colors correspond to two different environments, with within a color and between the colors. In D–F, the mixture of colors refers to random variation in the selection gradient, implemented as ϕ = 0.5In . The solid ellipses show the additive genetic variance–covariance matrix of the common ancestral population () and the dashed ellipses show the variance–covariance matrix Vs of selection (identical to in C and D). Parameters: = (1, 0.9; 0.9, 2), Vs = 0.4V. (E), Vs = 5V. (F), with V = (1, −0.6; −0.6, 1). Note that F is shown in a different scale from the other panels.
The colors of the symbols represent variation in environmental conditions, which impose differential selective pressures for the traits under consideration in Figure 2, C–F). We assume that the populations inhabit either two kinds of environments (red and black, Figure 2C) or many kinds of environments (colors in Figure 2, D–F). To generate data dominated by selection, we replaced Equation 19 by
| (20) |
where ϕ is a correlation matrix of environmental similarity, and VS is a variance–covariance matrix describing the space of trait combinations favored by selection. While Equation 19 assumes that genetic drift is the only factor shaping population divergence, Equation 20 makes the opposite assumption that all local populations have become locally adapted to their environments, and thus the link between coancestry and similarity in quantitative traits is fully uncoupled. The matrix VS is represented by a dashed ellipse in Figure 2, E and F, whereas it is assumed to be identical to in Figure 2, C and D. Consequently, in Figure 2, A–D, Equation 17 holds exactly, and hence any FST–QST comparison (univariate or multivariate, such as implemented in Martin et al. 2008) would fail to recover the signature of selection present in Figure 2, C and D. However, by visual inspection Figure 2A and Figure 2, C and D, clearly differ from each other, as in Figure 2A the related populations (squares and circles) are grouped close to each other, whereas in Figure 2, C and D, this is not the case. Thus, the signature of selection in Figure 2, C and D, is that the observed pattern does not show the expected feature under genetic drift, namely that closely related populations should be located close to each other in the trait space. Figure 2E represents stabilizing selection, in which the populations have diverged less than expected by drift (QST < FST), whereas in Figure 2F directional selection has made the populations diverge more than expected by drift alone (QST > FST). Further, in Figure 2, E and F, we have assumed that the direction of the is not proportional to VS, so the signature of selection involves a multivariate component.
A challenge with the above line of reasoning is to turn it into a statistical test, as it is difficult to develop natural metrics for patterns. However, as the neutral expectation (Equation 19) is expressed in terms of a probability distribution, one very natural measure for deviation from the theoretical expectation is the likelihood of the observed pattern. To formalize this, we let f be the probability density of the distribution of Equation 19 under population relatedness θ and A be the realized pattern of population divergence. We define the signature of selection as S = Pr(f(A) < f(AR)), whose probability is computed over random realizations AR of Equation 19 (10,000 replicate draws in the examples below). A value with S ≈ 1 indicates that the observed pattern A is unlikely under neutrality, whereas a typical neutral pattern should give S ≈ 0.5 In some cases, such as under strong stabilizing selection, one may also expect the value probability density to be higher for A than for a random realization; thus S ≈ 0 also indicates a departure from neutrality.
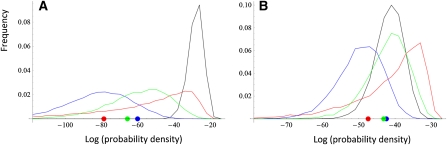
Figure 3 illustrates how the test statistic S behaves in the examples of Figure 2. For example, the red dot in Figure 3A measures the likelihood of observing the pattern in Figure 2C under neutrality and coancestry matrix corresponding to population (a). To ask whether this is an unusual value under the assumption of drift only, we compare it to the theoretical expectation (the black line in Figure 3A), which is obtained by random draws from Equation 19. As the red dot is clearly outside the central probability mass of the theoretical expectation (S > 0.99), we may reject the null hypothesis of drift with high confidence. Similarly, the patterns in Figure 2, D–F, are clearly distinguishable from the neutral expectation (Figure 3A). Conducting the above test with the coancestry matrix corresponding to population (b) leads to a somewhat different result, as now only the pattern A(f) could be safely distinguished from drift using the test above (Figure 3B).
Figure 3 .
Statistical power for disentangling drift and selection under the scenarios shown in Figure 2. A and B correspond to coancestry matrices used in Figure 2, A and B, respectively. The black lines show the theoretical distribution f(A) under drift, generated by 10,000 randomizations of Equation 19. The red, blue, and green dots correspond to the patterns shown in Figure 2, C–E, respectively. Regarding the pattern in Figure 2F, log(f(a)(A(f))) = −365 and log(f(b)(A(f))) = −167; hence these values are outside of the ranges of the panels. The red, blue, and green lines show the distribution of values of which the same colored dots are examples, based on 10,000 replicates simulated using the models in Figure 2. Almost the entire distribution corresponding to the scenario in Figure 2F is outside the range shown in these figures, the maximum values among the replicates being log(f(a)(A(f))) = −129 and log(f(b)(A(f))) = −68 .
Repeating the above tests for a number of replicate simulations of the processes used to generate the patterns in Figure 2, C–F, we note that the above test needs to be considered in the two-sided sense. For example, in Figure 3B there are cases in which the observed value of f(b)(A(c)) is much higher than 99% of the values under the theoretical expectation. This is expected to happen, e.g., in the case of stabilizing selection, as the probability density is maximized at the origin, i.e., if the population means are more close to that of the ancestral population than expected under drift.
A Statistical Framework for Testing for Neutrality
To make our approach practical, the quantities θ, μ, A, and need to be estimated from data. To estimate θ, we have extended the F-model, which models neutral drift through the Dirichlet distribution (Gaggiotti and Foll 2010) to allow for an arbitrary level of gene flow among the populations (see File S2 for details). Here we combine the estimation of θ with the estimation of μ, A, and in a single Bayesian model.
Sample population, breeding design, and laboratory population
In the context of FST–QST comparisons, the additive genetic variance–covariance matrix G is usually estimated for the current populations, either assuming a common G matrix for all populations (e.g., Chenoweth and Blows 2008) or estimating population-specific G matrices (e.g., Cano et al. 2004). Our aim is to use data on all populations simultaneously to estimate the ancestral variance–covariance matrix . As depicted in Figure 1, we assume the usual situation in which a common garden breeding design is used to separate the genetic and environmental components. Some individuals are sampled from the local populations, and these individuals are crossed (typically within the local populations but for our purposes equally well also between the local populations) to produce the laboratory population. For simplicity we assume here only one generation for the breeding design. We extend the definition of the coancestry coefficient θii′ for the laboratory individuals and recall that for θii′ the ancestral population is considered as the reference population. We also need such coancestry coefficients among the laboratory individuals for which the present population is considered as the reference population. This coancestry matrix, denoted by θℬ (with elements ), measures the amount of relatedness generated by the breeding design, the individuals being full-sibs, half-sibs, or unrelated.
We denote by zi the vector of measured trait values of individual i in the laboratory population and assume the animal model (Lynch and Walsh 1998),
| (21) |
where μi contains the overall mean and possible individual-level covariates such as the effect of sex, ai is the additive genetic value, and ei is the environmental effect, which we assume to be multinormally distributed with mean zero and covariance–variance matrix VE. We decompose the additive genetic effects further as
| (22) |
where pi is a population-level effect and si an individual-level effect. The population-level effect for laboratory-individual i is defined as
| (23) |
where S(i) and D(i) denote the local populations from which the sire s(i) and the dam d(i) of the individual i originate, respectively.
The data matrix Z = (zi)i arising from the breeding design involves three additional components of uncertainty on top of the flow of alleles from the ancestral generation to the local populations: the sampling of individuals to be used as parents (we denote the sampling process by Fs), the flow of alleles from the sampled individuals to the laboratory population (denoted by Fg′), and the assignment of the environmental effects to the laboratory individuals (denoted by Fe). The measured data Z are conditional on the pedigree, the allelic frequencies in the ancestral generation, and the breeding design, one realization of the random process F consisting of all the four components, F = (Fg, Fs, Fg′, Fe). In File S1, we first treat the effects of these four random processes separately and then consider their joint effect, to show that the observed data Z are distributed (over F) as
| (24) |
| (25) |
| (26) |
Here A, S, and E are independent of each other. The matrices S and E consist of the elements S = (si)i, E = (ei)i, and the elements of the matrix M(θℬ, θ, θ) are given by
| (27) |
We note that the covariance in the individual-level effects S is typically estimated as 2G⊗ θℬ, where if i and j are full-sibs, if i and j are half-sibs, and otherwise These numbers are recovered if the individuals in the field populations are unrelated and noninbred , i.e., if considering the field population as the base population. Thus the somewhat different form above follows as we refer to the ancestral matrix, not the average G over the local populations. The main reason for doing so is that, as discussed above, G is a random variable whereas does not depend on the random process F, and thus the inference based on θ, θ, μ, A, and can be done without resorting to expectations.
Importantly, in terms of the coancestry coefficients, Equation 27 refers only to population-level averages, not to the coancestry coefficients among the particular individuals sampled for the breeding experiment. This makes the use of Equations 24–27 practical, as θ and θ can be measured for a different set of individuals from the ones used for the breeding experiment, assuming that both represent random samples of the respective local populations.
To estimate the parameters, we combined a Bayesian framework of the animal model (Sorensen and Gianola 2002; Ovaskainen et al. 2008) with the extended F-model to estimate all model parameters (θ, μ, A, and ) simultaneously from the neutral molecular data and the breeding experiment data. The details of the estimation scheme as well as the assumed prior distributions are presented in File S2. We note that our method is conservative because we fitted data to a model that assumes neutrality and thus, with limited data the parameter estimates are biased toward the neutral case.
Testing the approach with simulated data
To test the approach, we generated simulated data sets with or without local adaptation and used the above framework to attempt to uncover the signal of selection from these data. The data sets were constructed with an individual-based simulation model in which we assumed nonoverlapping generations. In the baseline scenario (scenario I), we assumed that eight small populations (with 20 individuals in each) had diverged from a common ancestral population 30 generations ago, with a small amount of gene flow (per capita probability of dispersal 0.01) and a shared pool of migrants. Scenario II differs from the baseline scenario by having a 10 times longer time since population divergence, i.e., the simulations lasting for 300 generations. Scenario III differs from the baseline scenario by having asymmetric gene flow, generated by a stepping-stone model where migrants from population 1 (8) always disperse to population 2 (7), whereas migrants from populations 2–7 disperse into either of their neighboring populations by probability 0.5. For each of these scenarios, we generated 100 replicate data sets where the populations were influenced by local adaptation and 100 data sets where they were not influenced by local adaptation. In the former case, we assumed that the reproductive success of the individuals depended on the match between their phenotype and an optimum set by the environment. The data consist of neutral molecular marker data (on 32 loci, with five allelic variants in each) measured for the present generation and a breeding experiment in which five sets of parents were sampled from each local population, five full-sibs per pair were raised in a common environment, and two traits were measured for the offspring. Details of the simulation scheme and the assumed parameter values are given in File S3.
To examine the effect of sample size, we subsampled the data to include two, four, or eight populations and measurements on either one or two of the traits. We applied the statistical framework developed above to each of the 3600 data sets (3 × 2 scenarios × 100 replicates × 6 sample sizes) to ask whether the data showed a signal of selection. We also applied to the same data the two-step test of Martin et al. (2008), which asks (i) whether the matrices D and G are proportional and (ii) whether the proportionality constant equals 2FST/(1 − FST). For details on the implementation of these tests, see Martin et al. (2008) and File S4.
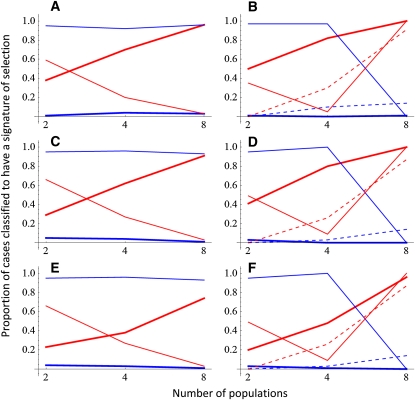
Due to the conservative nature of our test (see above), we used S > 0.8 as the criterion for a signal of selection, whose value still resulted in a very low frequency of false rejection of the null hypothesis (thick blue line in Figure 4). As expected, the statistical power of our method for detecting the signal of selection increased with the size of the data set, both in terms of the number of populations and in terms of the number of traits measured (thick lines in Figure 4). The proportionality test of Martin et al. (2008), which can be applied only in the case with two or more traits, also had some resolution to disentangle cases with and without selection (the dashed red line is higher than the dashed blue line in Figure 4B). However, this test had a higher misclassification rate than our method, both in terms of classifying data influenced by selection as neutral data and in terms of classifying neutral data as data influenced by selection (Figure 4B). The proportionality constant test performed well only in the case with two traits and eight populations (Figure 4B). In all other cases, i.e., for very small data sets, it performed very badly, finding a signature of selection on neutral data more often than from data influenced by selection. The counterintuitive behavior is at least partly explained by the fact that due to the small number of degrees of freedom the confidence interval for the proportionality constant included negative values in these cases. Note that the simulation results here are conditional to the case study simulated and are aimed to illustrate the influence of sample size on the probability of observing the signature of selection. Thus, they should not be interpreted as general power tests.
Figure 4 .
Performance of neutrality tests with simulated data that were generated with a scenario influenced by selection (red lines) or not influenced by selection (blue lines). The lines show the fraction of cases, of the 100 replicate data sets, in which the three statistical tests recovered evidence for population differentiation by selection. The thick line corresponds to the test developed in this article (with S > 0.8 as the criterion for a signal of selection), the thin solid line to the proportionality constant test of Martin et al. (2008) (with nonoverlapping confidence intervals as the criteria for a signal of selection), and the thin dashed line to the matrix proportionality test of Martin et al. (2008) (with P < 0.05 as the criterion for a signal of selection, applicable only for two or more traits). A and B depict scenario I, C and D show scenario II, and E and F show scenario III. A, C, and E and B, D, and F correspond to data sets with one and two traits, respectively.
Discussion
In this article, we have set the theoretical background for moving forward from QST–FST comparisons to more refined tests of population divergence. With the theoretical examples above, we have shown that it is sometimes possible to reject the null hypothesis of drift even if the overall pattern of population divergence would match exactly to the expectation under drift according to conventional FST–QST comparisons (e.g., Merilä and Crnokrak 2001; Martin et al. 2008; and Equation 17 in this article). This can occur because the averaging nature of FST leads to reduced statistical power for picking up a signal of selection compared to inference based on the full θ matrix, and the same consideration is true also in the case of QST. To connect the theory to data, we derived the statistical distribution of typical breeding experiment data under the assumption of drift (Equations 20–25) and developed a Bayesian estimating scheme. With simulated data, we have shown that our method can find a signature of selection even with very limited data on few populations, whereas the method of Martin et al. (2008) that requires the estimation of the D matrix fails to do so. Further, our method is less likely to lead to false positive results than that of Martin et al. (2008).
Similar to earlier methods, our approach to differentiate effects of drift from natural selection is based on number of simplifying assumptions (cf. Merilä and Crnokrak 2001; Whitlock 2008). First, we have assumed that the nonadditive (dominance and epistatic) effects of quantitative trait divergence are negligible. Earlier studies suggested that dominance effects are not likely to strongly influence QST estimates, and, if occurring, they are likely deflate rather than inflate the estimates (Whitlock 1999; Goudet and Martin 2007). However, the issue is still debated (Lopez-Fanjul et al. 2003, 2007), and the potential influence of nonadditive effects and a way to account for these effects on our approach need to be evaluated in future studies. Second, our approach assumes that mutational effects are negligible or, at least, that their influence on quantitative traits and neutral genetic variability is similar. Mutational effects are also ignored in conventional FST–QST comparisons (cf. Merilä and Crnokrak 2001; Whitlock 2008). The effect of mutations is to increase levels of genetic variation within populations and thus to lower the QST and FST, biasing FST–QST comparisons if the mutation rates are dissimilar in neutral marker loci and quantitative trait loci (Edelaar and Björklund 2011). Similar to mutations, also genotyping errors (e.g., Bonin et al. 2004) might influence our—as well as the conventional—approaches aiming to disentangle effects of drift and selection.
Phenotypic similarity among populations can occur due to adaptation to a shared selective regime or due to a shared phylogenetic history (Harvey and Pagel 1991). To this end, our approach could be taken a step further by accounting for information on environmental similarity. In our treatment we did not use the information in Figure 2C that the populations group in the trait space according to their local environments, shown by the red and black colors. If one would a priori hypothesize that the selective pressures are different in two environments [say, sea and freshwater populations of sticklebacks (Bell and Foster 1994)], then the grouping of the colors—independently of phylogenetic history—would represent strong additional evidence for selection. We leave the challenge of turning this statement into an appropriate statistical test for the future.
In conclusion, we expect that the theoretical and statistical frameworks set here will provide a basis for a sound and flexible tool for differentiating between adaptive and neutral explanations for observed patterns of population differentiation in the wild. In contrast to earlier similar approaches, our method deals with nonindependence among local populations, because the historical relationships among populations are explicitly part of the testing procedure. The issue of phylogenetic nonindependence was formerly considered mainly in the context of interspecific comparative studies (e.g., Harvey and Pagel 1991), but seldom in the context of intraspecific comparisons (but see Edwards and Kot 1995). To this end, our method should be a more rigorous way of analyzing intraspecific differentiation than those that assume populations to be phylogenetically independent, such as the methods based on simple comparisons of G-matrix statistics (e.g., Schluter 1996).
Acknowledgments
We thank Juho Pennanen for useful comments on earlier version of this article. Our research was supported by the Academy of Finland (grants 124242 and 213457 to O.O. and 213491, 129662, 118673, and 134728 to J.M.) and the European Research Council (Starting Grant 205905 to O.O.).
Literature Cited
- Anderson A. D., Weir B. S., 2007. A maximum-likelihood method for the estimation of pairwise relatedness in structured populations. Genetics 176: 421–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont M. A., 2008. Selection and sticklebacks. Mol. Ecol. 17: 3425–3427 [DOI] [PubMed] [Google Scholar]
- Bell M. A., Foster S. A. (Editors), 1994. The Evolutionary Biology of the Threespine Stickleback. Oxford University Press, Oxford [Google Scholar]
- Bink M., Anderson A. D., van de Weg W. E., Thompson E. A., 2008. Comparison of marker-based pairwise relatedness estimators on a pedigreed plant population. Theor. Appl. Genet. 117: 843–855 [DOI] [PubMed] [Google Scholar]
- Bonin A., Bellemain E., Eidesen P. B., Pompanon F., Brochmann C., et al. , 2004. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 13: 3261–3273 [DOI] [PubMed] [Google Scholar]
- Cano J. M., Laurila A., Palo J., Merilä J., 2004. Population differentiation in G matrix structure due to natural selection in Rana temporaria. Evolution 58: 2013–2020 [DOI] [PubMed] [Google Scholar]
- Chenoweth S. F., Blows M. W., 2008. Q(st) meets the G matrix: the dimensionality of adaptive divergence in multiple correlated quantitative traits. Evolution 62: 1437–1449 [DOI] [PubMed] [Google Scholar]
- Cockerham C. C., Weir B. S., 1987. Correlations, descent measures—drift with migration and mutation. Proc. Natl. Acad. Sci. USA 84: 8512–8514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelaar P. I. M., Björklund M., 2011. If FST does not measure neutral genetic differentiation, then comparing it with QST is misleading. Or is it? Mol. Ecol. 20: 1805–1812 [DOI] [PubMed] [Google Scholar]
- Edwards S. V., Kot M., 1995. Comparative methods at the species level: geographic variation in morphology and group size in grey-crowned babblers (Pomatostomus temporalis). Evolution 49: 1134–1146 [DOI] [PubMed] [Google Scholar]
- Fernandez J., Toro M. A., 2006. A new method to estimate relatedness from molecular markers. Mol. Ecol. 15: 1657–1667 [DOI] [PubMed] [Google Scholar]
- Gaggiotti O. E., Foll M., 2010. Quantifying population structure using the F-model. Mol. Ecol. Res. 10: 821–830 [DOI] [PubMed] [Google Scholar]
- Goudet J., Martin G., 2007. Under neutrality, Q(ST)<= F-ST when there is dominance in an island model. Genetics 176: 1371–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey P. H., Pagel M. D., 1991. The Comparative Method in Evolutionary Biology. Oxford University Press, Oxford [Google Scholar]
- Hendry A. P., 2002. Q(ST)>=not equal < F-ST? Trends Ecol. Evol. 17: 502 [Google Scholar]
- Hohenlohe P. A., Arnold S. J., 2008. MIPoD: a hypothesis-testing framework for microevolutionary inference from patterns of divergence. Am. Nat. 171: 366–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell S. P., 2001. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton University Press, Princeton, NJ [Google Scholar]
- Jost L., 2008. G(ST) and its relatives do not measure differentiation. Mol. Ecol. 17: 4015–4026 [DOI] [PubMed] [Google Scholar]
- Kitada S., Kitakado T., Kishino H., 2007. Empirical Bayes inference of pairwise F-sT and its distribution in the genome. Genetics 177: 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakado T., Kitada S., Kishino H., Skaug H. J., 2006. An integrated-likelihood method for estimating genetic differentiation between populations. Genetics 173: 2073–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1980. Genetic variation and phenotypic evolution during allopatric speciation. Am. Nat. 116: 463–479 [Google Scholar]
- Leinonen T., O’Hara R. B., Cano J. M., Merila J., 2008. Comparative studies of quantitative trait and neutral marker divergence: a meta-analysis. J. Evol. Biol. 21: 1–17 [DOI] [PubMed] [Google Scholar]
- Lopez-Fanjul C., Fernandez A., Toro M. A., 2003. The effect of neutral nonadditive gene action on the quantitative index of population divergence. Genetics 164: 1627–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Fanjul C., Fernandez A., Toro M. A., 2007. The effect of dominance on the use of the Q(ST)-F-ST contrast to detect natural selection on quantitative traits. Genetics 176: 725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Maenhout S., De Baets B., Haesaert G., 2009. CoCoa: a software tool for estimating the coefficient of coancestry from multilocus genotype data. Bioinformatics 25: 2753–2754 [DOI] [PubMed] [Google Scholar]
- Martin G., Chapuis E., Goudet J., 2008. Multivariate Qst-Fst comparisons: a neutrality test for the evolution of the G matrix in structured populations. Genetics 180: 2135–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins E. P., 2000. Adaptation and the comparative method. Trends Ecol. Evol. 15: 296–299 [DOI] [PubMed] [Google Scholar]
- Martins E. P., Hansen T. F., 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149: 646–667 [Google Scholar]
- McGuigan K., 2006. Studying phenotypic evolution using multivariate quantitative genetics. Mol. Ecol. 15: 883–896 [DOI] [PubMed] [Google Scholar]
- McKay J. K., Latta R. G., 2002. Adaptive population divergence: markers, QTL and traits. Trends Ecol. Evol. 17: 285–291 [Google Scholar]
- Merilä J., Crnokrak P., 2001. Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 14: 892–903 [Google Scholar]
- O’Hara R. B., Merilä J., 2005. Bias and precision in Q(ST) estimates: problems and some solutions. Genetics 171: 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., 2002. Near-neutrality in evolution of genes and gene regulation. Proc. Natl. Acad. Sci. USA 99: 16134–16137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovaskainen O., Cano J. M., Merilä J., 2008. A Bayesian framework for comparative quantitative genetics. Proc. R. Soc. Lond. B Biol. Sci. 275: 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P. C., Whitlock M. C., Fowler K., 2001. Inbreeding changes the shape of the genetic covariance matrix in Drosophila melanogaster. Genetics 158: 1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F., 2004. Genetic Structure and Selection in Subdivided Populations. Princeton University Press, Princeton, NJ [Google Scholar]
- Samanta S., Li Y. J., Weir B. S., 2009. Drawing inferences about the coancestry coefficient. Theor. Popul. Biol. 75: 312–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D., 1996. Adaptive radiation along genetic lines of least resistance. Evolution 50: 1766–1774 [DOI] [PubMed] [Google Scholar]
- Sorensen D., Gianola D., 2002. Likelihood, Bayesian, and MCMC Methods in Quantitative Genetics. Springer-Verlag, New York [Google Scholar]
- Weir B. S., Hill W. G., 2002. Estimating F-statistics. Annu. Rev. Genet. 36: 721–750 [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., 1999. Neutral additive genetic variance in a metapopulation. Genet. Res. 74: 215–221 [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., 2008. Evolutionary inference from Q(ST). Mol. Ecol. 17: 1885–1896 [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., Guillaume F., 2009. Testing for spatially divergent selection: comparing QST to FST. Genetics 183: 1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M. C., Phillips P. C., 2000. The exquisite corpse: a shifting view of the shifting balance. Trends Ecol. Evol. 15: 347–348 [DOI] [PubMed] [Google Scholar]
- Wright S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354 [DOI] [PubMed] [Google Scholar]