Abstract
Meiotic recombination is a fundamental cellular mechanism in sexually reproducing organisms and its different forms, crossing over and gene conversion both play an important role in shaping genetic variation in populations. Here, we describe a coalescent-based full-likelihood Markov chain Monte Carlo (MCMC) method for jointly estimating the crossing-over, gene-conversion, and mean tract length parameters from population genomic data under a Bayesian framework. Although computationally more expensive than methods that use approximate likelihoods, the relative efficiency of our method is expected to be optimal in theory. Furthermore, it is also possible to obtain a posterior sample of genealogies for the data using this method. We first check the performance of the new method on simulated data and verify its correctness. We also extend the method for inference under models with variable gene-conversion and crossing-over rates and demonstrate its ability to identify recombination hotspots. Then, we apply the method to two empirical data sets that were sequenced in the telomeric regions of the X chromosome of Drosophila melanogaster. Our results indicate that gene conversion occurs more frequently than crossing over in the su-w and su-s gene sequences while the local rates of crossing over as inferred by our program are not low. The mean tract lengths for gene-conversion events are estimated to be ∼70 bp and 430 bp, respectively, for these data sets. Finally, we discuss ideas and optimizations for reducing the execution time of our algorithm.
CROSSING over and gene conversion refer to two different mechanisms of recombination by which homologous chromosomes exchange genetic material during meiosis. In many eukaryotes, recombination is an essential requirement for sexual reproduction because it maintains physical connections between homologous chromosomes and contributes to proper segregation at the end of the first meiotic division. As currently understood, recombination starts with the formation of a double-stranded break in the DNA and proceeds by a series of biochemical steps leading to its repair (e.g., see Szostak et al. 1983; Stahl 1994). This repair can result in either the reciprocal exchange of large chromosomal regions (called crossing over) or the exchange of short DNA tracts (called gene conversion). The stretch of DNA exchanged during a gene-conversion event is called a conversion tract. A crossing-over event involves a single breakpoint in the chromosome and regions beyond this breakpoint are swapped between homologs to create mosaic products. In contrast, a gene conversion creates a mosaic in which a short internal fragment comes from one homolog while the rest of the chromosome flanking this fragment comes from the other. The products are similar in length to the participating homologs.
Crossing over is the better studied mechanism of recombination and crossing-over rates are known to vary tremendously across the genome at all scales. In particular, sperm-typing experiments as well as population genetic analysis of human polymorphism data have provided considerable evidence for fine-scale rate variation along the genome (e.g., see Fullerton et al. 1994; Dunham et al. 1999; Jeffreys et al. 2001, 2005; Innan et al. 2003; Crawford et al. 2004; McVean et al. 2004; International Hapmap Consortium 2005; Myers et al. 2005; Fearnhead and Smith 2005; Tiemann Boege et al. 2006; Coop et al. 2008). Notably, the population genetic studies ignored the effects of gene conversion and assumed that crossing over was the only mechanism underlying all exchanges. In general, gene conversion has been harder to study due to lack of appropriate fine-scale data and powerful statistical tools. Sperm-typing studies at a few individual human loci have provided evidence of high rates of gene conversion relative to crossing over (Zangenberg et al. 1995; Jeffreys and May 2004) and observed tract lengths that appear to be in the range of 50–150 bp. Gene conversion has also been studied experimentally in yeast and fruit flies (Fogel et al. 1983; Hilliker et al. 1991, 1994; Paques and Haber 1999; Allers and Lichten 2001; Mancera et al. 2008) and tracts are estimated to be in the range of 350–2000 bp in these organisms. Nevertheless, the nature and extent of variation in gene-conversion rates along genomes remains by and large unknown.
Characterizing the basic parameters of gene conversion (i.e., rates and tract lengths) within organisms is useful for a variety of reasons. First, it helps to better explain the patterns of linkage disequilibrium observed in single-nucleotide polymorphism (SNP) data (Wall and Pritchard 2003; Padhukasahasram et al. 2004). Second, algorithms for association mapping (e.g., Liu et al. 2001; Morris et al. 2002; Niu et al. 2002; Carlson et al. 2004) and for detecting natural selection (e.g., Voight et al. 2006) make assumptions about the local recombination parameters and conversion estimates may help fine-tune such methods. Finally, meiotic gene conversion is a fundamental biological mechanism and its relationship to crossing over is not yet fully understood and is an important open question. The emergence of dense SNP data sets from next generation sequencing technologies in the coming years presents a major opportunity to accurately quantify recombination rates from a population genetics standpoint. Thus, population genetic analysis of gene-conversion and crossing-over rates can eventually shed light on the relationship between these two processes and can supplement experimental approaches in answering this question (see previous studies Langley et al. 2000; Allers and Lichten 2001; Andolfatto and Wall 2003; Padhukasahasram et al. 2006; Mancera et al. 2008).
Currently, several statistical methods exist that are designed to jointly estimate the crossing-over and gene-conversion rates from population genetic data. Methods developed by Frisse et al. (2001), Ptak et al. (2004), and Wall (2004) generalize the composite-likelihood approach first proposed in Hudson (2001). These approaches divide the data into small subsets (pairs or triplets of segregating sites), calculate likelihoods for these subsets, and multiply them together. The likelihood thus obtained is called the composite likelihood. Composite-likelihood methods use precomputed likelihood lookup tables for all the possible configurations of the subsets and are typically fast. Because the subsets are not independent of one another, they do not calculate the true likelihood of the data. Therefore, correct confidence intervals can be obtained only by using simulations.
Padhukasahasram et al. (2006) describe a rejection-sampling method that simultaneously utilizes informative long-range and short-range summary statistics to infer the recombination parameters. This approach uses only part of the information available in the data for the sake of computational efficiency. Confidence intervals may be directly calculated from the likelihood surface in this method.
The approximate-likelihood method for estimating crossing-over rates [called product of approximate conditionals (PAC)] that was proposed in Li and Stephens (2003) has also been extended by several recent studies to include gene conversion. Briefly, this method infers recombination parameters under a heuristic model and is computationally efficient. However, inference is currently restricted to the constant population size Wright–Fisher model only. Hellenthal (2006) used a PAC model where the conversion tract can include at most one marker. Gay et al. (2007) improved on that work to allow for arbitrary gene-conversion tract lengths and this method can be used for coestimating crossing-over and gene-conversion rates as well as tract lengths. One simplification in their model was that they disallowed overlapping gene-conversion events. Yin et al. (2009) further generalized this work to allow for overlapping events and in theory this method is expected to perform at least as well as the method of Gay et al. (2007). Simulations indicate that this generalization (at least for a subset of parameters) leads to a more accurate PAC-based method for jointly estimating all three recombination parameters.
In this article, we extend the Bayesian Markov chain Monte Carlo (MCMC) method (originally developed to infer crossing over exclusively) of Wang and Rannala (2008) to jointly estimate the population crossing-over rate, the population gene-conversion rate, and the mean conversion tract length from SNP data. We first check the performance of the new method on simulated data and verify its correctness. In addition, we extend the method for inference under models with variable gene-conversion and crossing-over rates and demonstrate its ability to identify recombination hotspots. Next, we apply the method to two empirical data sets that were sequenced in the telomeric regions of the X chromosome of Drosophila melanogaster. Because the new method generates a posterior sample of genealogies consistent with the observed data, it is possible to obtain the distribution of gene-conversion breakpoints for a data set. Comparison of this distribution with a prior using a Bayes factor may be informative about the locations of conversion breakpoints in the history of the sample. We also calculate the Bayes factors for gene-conversion initiation points for these two Drosophila data sets. Our analysis suggests that gene conversion occurs more frequently than crossing over in the su-w and su-s gene sequences while the local rates of crossing over as inferred by our program are not low. The estimated mean tract lengths for these regions are ∼70 bp and 430 bp, respectively. Furthermore, plots of the logarithm of Bayes factors for conversion start points do not indicate any strong deviations from a prior distribution expected under a model with uniform recombination rates. Finally, we discuss ideas and optimizations for improving the run-time efficiency of our algorithm.
Materials and Methods
Our recombination inference method is based on the retrospective coalescent framework in which the genealogy of a sample of sequences is approximated by a graph called the ancestral recombination graph (ARG) (Kingman 1982; Hudson 1983; Griffiths and Marjoram 1996). In particular, the method uses the coalescent with gene conversion as described in Wiuf and Hein (2000). In this model, conversion tract lengths are assumed to be geometrically distributed (Hilliker et al. 1994). The distribution of tract length (Z) given that a gene-conversion event has occurred, i.e., P(Z = z|gene conversion), is thus equal to q(1 − q)z–1, where q denotes the reciprocal of the mean conversion tract length (m). Consider a sequence of length L + 1 bp. A gene-conversion event can initiate between any two adjacent base pairs along the sequence. Without loss of generality, the model also assumes that a tract will always be to the right of the initiation position. Thus, if a conversion event initiates at gap S, S = 1, 2, 3, . . . , L, then the end point is S + z. Gene-conversion events that initiate outside the sequence on the left and terminate within the sequence must also be included in our analysis. The probability that the tract length is greater than km, where m is the mean conversion tract length and k is a positive integer, becomes negligible as k grows. So, only a finite sequence (say 50m) on the left of the sequence of interest needs to be taken into account for the latter kind of events and the rest of the chromosome can be ignored.
Let ρ denote the population crossing-over rate, γ the population gene-conversion rate, and θ the population mutation parameter. Let X denote a sample of haplotypes or genotypes and GS denote a collection of correlated trees (i.e., an ARG) at the marker locations consistent with X. We are interested in sampling from the following posterior probability density:
| (1) |
The posterior distribution of recombination parameters can be numerically evaluated by using a reversible-jump MCMC (RJMCMC) scheme. In particular, a Metropolis–Hastings (MH) algorithm is used to estimate the parameters described in Equation 1. The MH algorithm has two steps: (i) the proposal step in which potential new parameter values are simulated from the proposal density Q(φ′|φ) and (ii) the acceptance step in which the proposed values are accepted with probability α or rejected with probability 1 – α. If accepted, φ′ becomes the current state in the chain; otherwise the chain remains at φ and φ′ is discarded. The acceptance probability for a parameter is given by
| (2) |
where f denotes the prior density and l denotes the likelihood. For each node in the ARG, we store an array of size equal to the number of markers and keep track of marker positions that are ancestral to the sample. We call this the ancestry vector [also the Marker Ancestry (MA) vector, see Wang and Rannala 2008]. The distance between the start and the end of the ancestral locations in this vector is referred to as the length of the ancestry vector. Our joint estimation program begins with a binary tree that is consistent with the observed data and then proceeds according to the following proposal moves.
Adding (or removing) a pair of crossing-over and coalescence nodes
We generate two uniform random variables u and v from the interval (0, tH), where tH denotes the height of the genealogy. The smaller of the two becomes the time for inserting a new crossing-over node and the larger is the time corresponding to the coalescence node. The remainder of this move is identical to the one in Wang and Rannala (2008) except that the probability of acceptance becomes
where the proposal ratio is
GS denotes the current genealogy and denotes the newly proposed genealogy. npair here refers to the number of possible pairs of crossing-over and coalescence nodes that can be deleted from . and refer to the number of eligible branches at the times at which new crossing-over and coalescence nodes will be inserted where eligible branches must have ancestry vector length >1 when inserting crossing-over nodes. l2 − l1 refers to the distance between the start and the end of the ancestral segment in a branch.
The probability of adding a pair of nodes to move from GS to G′S is given by
The first term denotes the probability of proposing the ages of the two nodes, the second and third terms denote the probability that the two nodes are inserted into the particular eligible branches at their respective times, and the last term denotes the probability of proposing a recombination breakpoint in the crossing-over node that is inserted. Note that when deleting a pair of crossing-over and coalescence nodes, we randomly choose a pair to delete. Thus, in the proposal ratio 1/npair denotes the probability that the same pair of crossing-over and coalescent nodes that were added are chosen for deletion, to move back from to GS.
Calculating the likelihood of data given a genealogy GS:
l(X|GS, θ) denotes the likelihood of observing a set of haplotypes/genotypes given an ARG and the population mutation rate. Given the genealogical tree τi for a marker site i, and conditional on one or more mutations having occurred, the likelihood of a marker tree is given by
where l indicates a branch in τi with length tl, connecting and , with being the ancestral allele of in that branch. The likelihood is calculated across all branches in τi and I denotes the indicator function. Parameter Ti represents the sum of the branch lengths in τi and πj is the frequency of allele j at stationarity. l(X|GS, θ) is the product of the likelihoods across all the marker trees.
Adding (or removing) a pair of gene-conversion and coalescence nodes
As before, we generate two uniform random variables u and v from the interval (0, tH), where tH denotes the height of the genealogy. The smaller of the two becomes the time for inserting a new gene-conversion node and the larger is the time corresponding to the coalescence node. Note that we are interested only in gene-conversion events that can potentially change the genealogy of the sample at the marker locations and thus the sample configuration.
Let l2 − l1 denote the distance between the start and the end of the ancestral segment in a branch of the graph. We propose a gap position (g) for initiation of gene conversion nonuniformly from a region of length l2 − l1 + 50m, where the sequence of length 50m lies to the immediate left of the start of the ancestral region in the MA vector and m denotes the mean conversion tract length.
The probability that a gap location is proposed is proportional to the chance that the tract length is greater than or equal to the distance (k) from the nearest MA vector position on the right of that gap location. The chance that the tract length is ≥k is given by
After choosing the initiation gap, we propose a tract length (z) according to a truncated geometric distribution conditional on the tract length greater than or equal to the distance from the nearest MA vector position on the right, so that only tracts that can affect the genealogy of the sample at markers are included. When the start of the gene-conversion tract is to the left of the start of the MA vector, we include the additional condition that the end of the tract is always to the left of the end of the MA vector. Thus, we exclude tracts ≥l2 – l1 + k for such cases because such tracts do not affect the sample configuration (the probability of picking such a gap is proportional to ). The probability of acceptance is equal to
where the proposal ratio is
The terms are the same as before except that p(g) denotes the chance that a gap location is proposed. Let pj denote the probability that a gene-conversion tract that initiates at the jth base pair along the sequence includes some but not all ancestry vector positions. Then, . d(z) represents the density of the truncated geometric distribution for tract length at the chosen gap location; i.e., P(Z = z|Tract includes some but not all MA vector positions. Note that the proposal distributions for both initiation points and tract lengths are identical to the prior distributions when gene-conversion rates are assumed to be uniform along the sequence. npair refers to the number of possible pairs of gene-conversion and coalescence nodes that can be deleted. and refer to the number of eligible branches at the times at which new gene-conversion and coalescence nodes will be inserted, where eligible branches must have ancestry vector length >1 when inserting gene-conversion nodes.
Local topology rearrangements
This move consists of moving either a recombination or a coalescence node to a new location in the graph. It is identical to what is described in Wang and Rannala (2008) except that the conditional probability terms will include parameters ρ, γ, and m instead of ρ alone.
Propose new waiting times between events in the graph
We propose a new graph with new waiting times between the successive (recombination or coalescence) events. The topology of the graph is left unchanged in this new move. The waiting times to the next event are proposed from the prior distribution expected under coalescent theory on the basis of the number of lineages and the ancestry vectors after the previous event. Note that this proposal is independent of the current waiting times. The probability of acceptance is given by
Let k denote the number of lineages after the previous event. Under coalescent theory, the rate of coalescence is 0.5k(k – 1), the rate of crossing over is 0.5kρ, the rate of gene conversion is 0.5kγ, and the waiting time to the next event is exponentially distributed with an overall rate of [0.5k(k – 1) + 0.5kρ + 0.5kγ]. However, because here we are interested only in informative gene-conversion and crossing-over events, we have to make some modifications when calculating the rates. Let fi denote the fraction of the sequence included within the start and the end of the ancestry vector. Then, the rate of informative crossing-over events for the ith lineage is given by ρfi and the overall rate of crossing over becomes Similarly, let pij denote the probability that a gene-conversion tract that initiates at the jth basepair along the sequence in lineage i includes some but not all ancestry vector positions at that lineage. Let γ/L denote the gene-conversion rate per base pair. Then, the rate of informative gene conversions is given by where l1(i) and l2(i) denote the start and the end of the ancestry vector for the ith lineage. Thus, the waiting times to informative events are exponentially distributed with a rate of [0.5k(k – 1) + 0.5ρt + 0.5γt]. The overall probability of a proposal is equal to the product of the exponential density functions of successive events. Note that since we propose the new waiting times from the coalescent prior,
Coalescent prior density for a genealogy:
To calculate the coalescent prior probability for a genealogy GS, i.e., f(GS|ρ, γ, m), we calculate the product of three quantities at each event and multiply them together:
Density of the waiting time for that event: The time is exponentially distributed with a rate of [0.5k(k – 1) + 0.5ρt + 0.5γt].
- The probability that the event is a coalescence or a gene conversion or a crossing over:
The chance is calculated that (a) a particular pair of lineages will be picked to coalesce; i.e., 2/k(k – 1) if the event is coalescence. (b) If the event is crossing over, the chance that a particular lineage will be picked for crossing over is multiplied by the chance that its breakpoint will be picked. (c) If the event is gene conversion, the chance that the particular lineage is picked for gene conversion is multiplied by the chance that its initiation point is chosen multiplied by the chance that the tract length is chosen.
Propose a new breakpoint (and tract length) for a recombination node
In this move, we pick a crossing-over or gene-conversion node at random and propose a new breakpoint (and tract length) for that node. We also randomly propose a new direction for how the products of recombination get distributed (i.e., which part goes to the left branch/right branch of the recombination node). We then update the ancestry vectors of the subsequent nodes in the graph on the basis of the modified node. The probability of acceptance as before is equal to
If the chosen node is a crossing-over node, we propose a new breakpoint uniformly between the start and the end of the ancestry vector positions and the proposal ratio for the node is equal to 1. The proposal ratio for a new breakpoint and tract length for a gene-conversion node is given by
Here, p(g) denotes the probability that a breakpoint location is proposed in a gene-conversion node. Let pj denote the probability that a gene-conversion tract that initiates at the jth basepair along the sequence includes some but not all ancestry vector positions. Then, , where l1 and l2 denote the start and the end of the ancestry vector, respectively. d(z) is the density of the truncated geometric distribution for tract length at the chosen gap location; i.e., P(Z = z| Tract includes some but not all MA vector positions.
Modify ancestral states, haplotype phases, and missing data
These steps are identical to those found in Wang and Rannala (2008) because the likelihood of the data given the genealogy trees and θ, i.e., l(X|GS, θ), is the same as described there.
Propose new values for the parameters ρ, γ, m, and θ
New ρ is proposed according to a sliding window with reflecting boundary at 0. The acceptance probability is given by
New γ is proposed according to a sliding window with reflecting boundary at 0. The acceptance probability is given by
New m is proposed according to a sliding window with reflecting boundaries at 1 and at 2000 or 3000 bp. The acceptance probability is given by
New θ is proposed according to a sliding window with reflecting boundary at 0. The acceptance probability is given by
Variable recombination rate model
We also use a variable recombination rate model in our analysis that includes background rate variation and hotspots. In this model, the background crossing-over rate follows a gamma distribution with shape and scale parameters and recombination hotspots that arise according to a Markov process. The waiting distance until the occurrence or loss of a crossing-over hotspot is exponentially distributed while the intensity of a hotspot follows a log-normal distribution. More details about this crossing-over model can be found in Wang and Rannala (2009). In our extension of the Wang and Rannala model, we assume that gene-conversion and crossing-over rates vary in an identical pattern such that their ratio (f) remains uniform along the sequence. Thus, all crossing-over hotspots are also gene-conversion hotspots.
Results
Checking the MCMC program
To check the correctness of our MCMC algorithm, we first examined the stationary distribution of several genealogy-based summary statistics when the chain is run with no data (i.e., with the likelihood ratio set to 1). We fixed the values of ρ, γ, and m when running the chain and then compared the distribution of the number of crossing-over (CO) events, the number of gene-conversion (GC) events, and the genealogy height (H) with coalescent simulations for the same parameters. As can be seen from Table 1, the expectations from the MCMC algorithm are in agreement with those obtained by straightforward Monte Carlo simulations under the coalescent.
Table 1 . Comparison between the MCMC program and coalescent simulations.
| ρa | γa | E(CO)b | E(CO)c | E(GC)b | E(GC)c | E(H)b | E(H)c | Mean tract | Sample size | Md | Le |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.000 | 0.300 | 0.0000 | 0.0000 | 4.4084 | 4.4233 | 2.4454 | 2.4572 | 5 | 8 | 4 | 4 |
| 0.000 | 0.100 | 0.0000 | 0.0000 | 1.2583 | 1.2614 | 1.9721 | 1.9781 | 2 | 8 | 4 | 4 |
| 0.100 | 0.100 | 0.3163 | 0.3162 | 0.5782 | 0.5777 | 1.2694 | 1.2682 | 5 | 2 | 4 | 4 |
| 0.300 | 0.300 | 2.4264 | 2.4281 | 2.7716 | 2.7760 | 4.4577 | 4.4683 | 5 | 8 | 4 | 4 |
| 0.100 | 0.300 | 0.8060 | 0.8061 | 2.5602 | 2.5605 | 4.4287 | 4.4282 | 5 | 8 | 4 | 4 |
| 0.001 | 0.001 | 29.884 | 30.646 | 3.3402 | 3.4341 | 5.7887 | 5.9864 | 100 | 8 | 8 | 8000 |
Population crossing-over and gene-conversion rates per base pair.
Average of crossing-over count, gene-conversion count, and height of genealogy from coalescent simulations.
Average of crossing-over count, gene-conversion count, and height of genealogy from MCMC algorithm.
Number of markers.
Sequence length in base pairs.
Test runs on simulated data:
Next, we tested our inference program on data sets simulated with gene conversion alone and compared the posterior distribution of γ as estimated by our program with the true value used in simulations. For this testing, we fixed the mean tract length and θ to their true values and ρ was fixed at 0. The results obtained are shown in supporting information, Figure S1. When gene-conversion rates are high, we expect that the power to detect gene conversion will be high and our posterior distribution will not include 0 most of the time (e.g., γ = 4000/Mb, Figure S1), which would tell us clearly that conversions have happened. On the other hand, when gene-conversion rates become lower it becomes difficult to infer whether a conversion event has happened in a particular sample’s history (because the posterior will often include 0). This is because not every gene-conversion event is necessarily detectable in a sample and with only a few conversion events in a sample history, by chance, we could have samples that are also consistent with one or more binary trees.
In Figure S1ii we show results when we run our joint estimation program on data sets simulated with gene conversion alone with the tract length fixed at its true value. We can see that estimates of ρ are low but nonzero and the credible intervals contain values close to 0. Note that estimates of γ under this joint estimation scenario can be considerably lower than the truth although the true value of ρ is 0 (which means that we interpret some of the gene conversions as crossing overs). In Figure S1iii we show results for the other boundary case, i.e., when data are simulated with crossing over alone with γ set at 0. For this scenario, we attempted to estimate both crossing-over and gene-conversion rates with the tract length fixed at different values. In addition, we also estimated the recombination parameters and mean tract lengths jointly. We can see that estimates of γ can be substantial although the true value is 0. When the mean conversion tract length becomes longer (e.g., 2000 bp, 8000 bp), we observe multiple peaks in the joint posterior distribution. This suggests that more than one combination of crossing-over and gene-conversion rates can explain the simulated data and there is difficulty in distinguishing between the effects of these two processes.
For Figure S2, we assumed that both gene-conversion rates and mean tract lengths are unknown and estimated them jointly. Subsequently, we performed simulations with both crossing over and gene conversion and attempted to jointly estimate both γ and ρ for these data sets with the mean tract length set to its true value. The corresponding results are shown in Figure S3. In all the figures (i.e., Figure S1, Figure S2, Figure S3), we can see that the modes of the posterior distribution are close to the true values of the recombination parameters. In Figure S3 too, we sometimes see more than one peak in the posterior distribution.
In general, estimating gene-conversion rates from population genomic data is a challenging problem. This is because the traces of this phenomenon in data are limited and subtle. Note that the ability to detect and quantify gene conversion in both experiments and population genetic analysis depends on the tract size. When tracts are short compared to the spacing between markers, a substantial fraction of gene-conversion events will not leave a trace in the sample, making it difficult to infer rates accurately. On the other hand, when tracts are long compared to the sequence of interest, some fraction of them will fall only partly inside the sequence and will be indistinguishable from crossing over. As Figure S1, ii and iii, and Figure S3 highlight, there can be substantial confounding when attempting to jointly estimate both these parameters and it is difficult to tease apart the effects of conversion and crossing over from one another.
Simulations with variable recombination rates:
We tested an implementation of InferRho that incorporates variation in crossing-over and gene-conversion rates on data sets simulated with recombination hotspots. We simulated 20 samples of 20-kb sequences with θ = 20.0, m = 500 bp, and a single hotspot between 9 kb and 11 kb. Both ρ and γ = 50/Mb outside hotspots, whereas within the hotspot these numbers were 5000/Mb. The ratio of gene conversion to crossing over (f) was assumed to be uniform along the sequence in these simulations with f = 1.0. Table 2 shows the results obtained from running our program on three independent replicates for these parameters.
Table 2 . Estimated locations of hotspots in three independent simulated data sets.
| Data set | Hotspot start, bp | Hotspot end, bp | Hotspot intensity estimate | |
|---|---|---|---|---|
| Replicate 1 | 9,486 (8,746–10,226) | 10,555 (8,911–12,199) | 4,030.71 | 2.375 (0.375–6.125) |
| Replicate 2 | 10,885 (5,240–16,530) | 11,879 (6,969–16,789) | 8,160.21 | 0.375 (0.125–7.375) |
| Replicate 3 | 9,822 (8,087–11,557) | 11,068 (9,240–12,896) | 3,968.12 | 3.625 (1.125–4.625) |
Each simulated data set consists of 20-kb sequences and 20 samples with a single hotspot of intensity 5000/Mb between 9 kb and 11 kb. The relative rate of gene conversion to crossing over (f) was uniform along the sequence and was equal to 1. The estimated locations for the start and end of hotspots and f are shown along with the 90% credible intervals in parentheses.
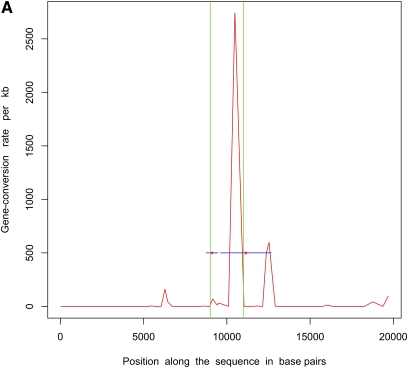
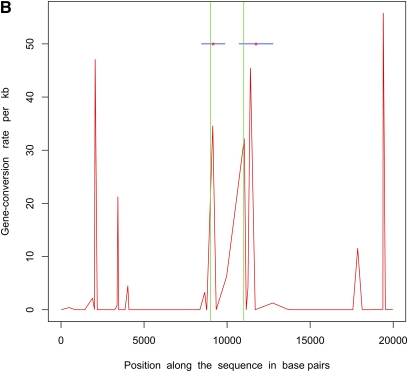
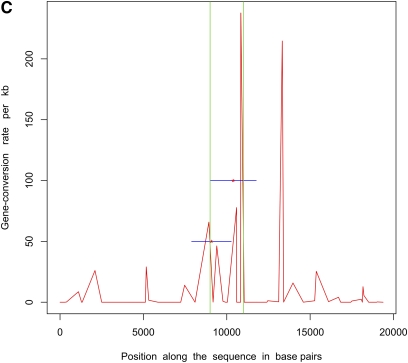
Next, we simulated 50 samples of 20-kb sequences with θ = 20.0, m = 500 bp, a single hotspot between 9 kb and 11 kb, and uniform f = 10.0 (parameters used in Gay et al. 2007). ρ = 50/Mb and γ = 500/Mb outside hotspots, whereas within the hotspot these numbers were 5000/Mb and 50,000/Mb, respectively. We estimated recombination rates for these data sets using the program of Gay et al. (2007), which implements a variable gene-conversion rate model. Note that this program does not try to identify hotspots but estimates a different crossing-over and gene-conversion rate between adjacent SNPs. We also inferred the locations of hotspots using InferRho. Figure 1 plots the results obtained using these two different methods on five independent replicates for these parameters. In both Table 2 and Figure 1, we can see that the estimated locations of the starts and ends of hotspots are close to the true locations.
Figure 1 .
Estimated locations of hotspots in data sets simulated with a uniform ratio of gene conversion to crossing over (f) = 10. Simulations consisted of 20-kb sequences and 50 samples with a single hotspot between 9 kb and 11 kb. The red curve shows the gene-conversion rates as inferred by the program of Gay et al. 2007 under a variable rate model. The red asterisks mark the posterior means of the starts and ends of hotspots as estimated by InferRho, the blue lines represent 90% credible intervals, and the green lines show the true locations of hotspots. Most of the posterior samples obtained from InferRho contained a single hotspot.
Comparison with other methods:
We compared the performance of our method (implemented in the software package InferRho) with that of the methods described in Gay et al. (2007) and Yin et al. (2009). For this comparison, we simulated 100 data sets each for two different parameter combinations and estimated all three recombination parameters (i.e., the population crossing-over rate, the population gene-conversion rate, and the mean tract length) jointly for these data sets. Then, we calculated summaries of these estimates and compared them with those obtained from the other two methods. These comparisons are shown in Table 3. Note that the summaries for the methods of Gay et al. (2007) and Yin et al. (2009) are based on the same set of simulated data sets whereas those for our method are based on an independent sample of 100 data sets. When implementing all three methods, the starting values of the recombination parameters were set to the true value that was used in simulations.
Table 3 . Comparison between three different methods for recombination rate estimation.
| Method | ρa | γa | ma | MSE()b | MSE()b | MSE()b | #(:2)c | #(:2)c | #(:2)c |
|---|---|---|---|---|---|---|---|---|---|
| Gay et al. (2007) | 500 | 500 | 500 | 3.488 | 1,022,389 | 0.723 | 0.71 | 0.26 | 0.36 |
| Yin et al. (2009) | 500 | 500 | 500 | 0.079 | 0.661 | 0.105 | 0.71 | 0.55 | 0.88 |
| InferRho | 500 | 500 | 500 | 0.041 | 0.222 | 0.644 | 0.89 | 0.77 | 0.56 |
| Gay et al. (2007) | 500 | 1000 | 500 | 0.085 | 618,219 | 0.349 | 0.78 | 0.48 | 0.35 |
| Yin et al. (2009) | 500 | 1000 | 500 | 0.060 | 1.348 | 0.098 | 0.79 | 0.70 | 0.87 |
| InferRho | 500 | 1000 | 500 | 0.043 | 0.487 | 0.250 | 0.88 | 0.81 | 0.62 |
ρ denotes population crossing-over rate, γ denotes the population gene-conversion rate, and m denotes the mean conversion tract length for the 100 simulated data sets. Recombination rates and mean tract lengths are shown in units of per megabase and base pair, respectively.
MSE, the mean square error of the parameter estimates for the 100 simulated data sets × 10−6. For InferRho, we calculated the marginal estimate of each recombination parameter from the joint posterior distribution. Numbers for the other methods are taken from Yin (2010).
#(:2) represents the fraction of the 100 data sets for which estimates are within a factor of 2 of the true parameter value used in simulations (i.e., ρ). #(:2) and #(:2) are defined similarly.
For all the parameters tested, we find that the performance of the InferRho program and the method of Yin et al. (2009) are better than that of Gay et al. (2007). Presumably this is because the approach of Gay et al. (2007) ignores overlapping gene-conversion events while the other two methods do not assume this restriction. In theory, we would expect the Gay et al. (2007) method to perform worse if overlapping gene-conversion events are significantly frequent in the history of the sample. Compared to the method of Yin et al. (2009), InferRho appears to have higher mean squared error for tract length estimation but lower mean squared error for crossing-over and gene-conversion rate estimation. This may be due to confounding when attempting to estimate both gene-conversion rates and tract lengths jointly from population genetic data.
Estimates in real data
We applied our method to jointly estimate the crossing-over and gene-conversion rates in two genes, the su-w and su-s genes of D. melanogaster that were first studied by Langley et al. (2000). These genes are located close to the telomeric regions of the X chromosome and are ∼4.1 kb and 2.5 kb long, respectively. The su-s data set contains 50 haplotypes and 41 SNPs whereas the su-w data set that we used contains 50 haplotypes and 46 SNPs in an African-only sample. We did not try to estimate tract lengths in our initial analysis and fixed them to 352 bp (estimates from Hilliker et al. 1994). The value of θ was set to be equal to Watterson’s estimate for each data set. Table 4 shows the results obtained for this analysis (see also Figure S5).
Table 4 . Joint estimates of crossing over () and gene conversion () for m = 352 bp.
| Gene | Method | /Mb per Mb | /Mb per Mb | = / |
|---|---|---|---|---|
| su-s | Gay et al. (2007) | 1,700 | 12,000 | 7.10 |
| su-s | Yin et al. (2009) | 2,240 | 11,510 | 5.14 |
| su-s | InferRho | 1,500 (700–5,500) | 6,900 (3,500–10,300) | 4.60 (0.875–7.875) |
| su-w | Gay et al. (2007) | 570 | 28,000 | 48.0 |
| su-w | Yin et al. (2009) | 33 | 27,040 | 819.4 |
| su-w | InferRho | 4,300 (2,500–8,100) | 11,700 (5,300–16,100) | 2.72 (0.875–3.625) |
The 95% maximum posterior density credible intervals for InferRho are shown in parentheses. For InferRho, MCMC chains were run for 40 million iterations and the first 10 million iterations were discarded as burn-in. For other methods, the numbers are taken from Yin (2010).
Subsequently, we implemented a version of our inference program where we estimated both mean tract lengths and recombination rates jointly. These results are shown in Table 5. Figure S4 shows 95% credible intervals with maximum posterior density for the estimated parameter values. Note that the estimated rate of gene conversion depends on the mean tract length and for shorter tracts the inferred rates are expected to be higher (see also Figure S5).
Table 5 . Estimates of crossing over (), gene conversion (), and mean tract length ().
| Gene | Method | /Mb | /Mb | , bp |
|---|---|---|---|---|
| su-s | Gay et al. (2007) | 920 | 11,600 | 480 |
| su-s | Yin et al. (2009) | 1,290 | 9,860 | 560 |
| su-s | InferRho | 3,350 | 5,950 | 430 |
| su-w | Gay et al. (2007) | 8,520 | 251,130 | 5 |
| su-w | Yin et al. (2009) | 1,450 | 41,090 | 162 |
| su-w | InferRho | 5,550 | 28,950 | 70 |
For InferRho, MCMC chains were run for 41 million and 37 million iterations (for su-s and su-w, respectively) and the first 10 million iterations were discarded as burn-in. We show the marginal estimate of each parameter from the joint posterior distribution. For other methods, the numbers are taken from Yin (2010).
We also compared our results with those obtained by the methods in Gay et al. (2007) and Yin et al. (2009) in Tables 4 and 5. We observe that there is some difference between the point estimates provided by the three different programs. Presumably, this is because there is limited information in these data sets for joint estimation and there is difficulty in distinguishing between the effects of the different recombination mechanisms in the short sequences of the Drosophila genes.
Bayes factor in real data
We compared the posterior distribution of gene-conversion start points in the su-w and su-s genes with the distribution under an uninformative prior. The prior distributions were obtained by running the chain without any data (i.e., with likelihood ratio = 1) for the same sample size and marker positions as in the real data sets and assuming uniform recombination rates. Let p1 denote the probability that a conversion initiation point lies within a particular window for the prior sample of genealogies (This is the fraction of initiation points that lie within that window). Similarly, let p2 denote the corresponding probability for the posterior sample of genealogies. The odds for the prior and posterior for a window are given by p1/(1 − p1) and p2/(1 − p2), respectively. Formally, the Bayes factor (BF) is defined as the ratio of the posterior and prior odds; i.e.,
The log of the BF should be interpreted as the change in the evidence for gene-conversion initiation in a region due to the data. A large negative value indicates evidence against gene-conversion initiation and a large positive value indicates evidence for gene-conversion initiation. The BF for 10-bp windows is plotted along the genes, assuming the mean tract length = 352 bp (see Figure 2, A and B). Note that the Bayes factors for the windows were expected to indicate where gene-conversion events have occurred in the history of the sample but their values do not necessarily imply high or low rates. Thus, this analysis is unrelated to hotspot identification described in the Simulations with variable recombination rates section. For these Drosophila data sets, it seems that there is little information concerning the location of gene-conversion start points because we do not see very large or very small BFs (|log(BF)| < 2 generally).
Figure 2 .
(A) Bayes factors along the su-w gene for 10-bp windows. The logarithm of the ratio of posterior to prior odds for gene-conversion initiation points is plotted along the gene. The su-w gene is 2.5 kb long. (B) Bayes factors along the su-s gene for 10-bp windows. The logarithm of the ratio of posterior to prior odds for gene-conversion initiation points is plotted along the gene. The su-s gene is 4.1 kb long.
Discussion
We have described a Bayesian MCMC method for jointly inferring the crossing-over and gene-conversion parameters from SNP data sets that extends the crossing-over estimation method originally proposed by Wang and Rannala (2008). In this method, we model the genealogy of a sample as a recombination graph and keep track of ancestral marker sites in the MA vectors of its nodes. Furthermore, we include only informative crossing-over and gene-conversion events that can potentially change the genealogy of the sample at the marker locations. These aspects of our algorithm make the full-likelihood estimation of recombination rates efficient in terms of both the running time and memory requirements. Assuming an uninformative prior for the parameters of interest, we can estimate their joint posterior distribution using a reversible-jump MCMC scheme. In the Metropolis–Hastings step, we propose changes to various features of the ancestral recombination graph such as the relative locations of the recombination or coalescence nodes, the number of crossing-over or gene-conversion nodes, the ancestral alleles, and the waiting times between consecutive events in the graph, etc. The method has been implemented in a new version of the software package InferRho (see Wang and Rannala 2008), which will become available online at http://rannala.org. Currently, we are checking the software’s performance for larger values of ρ and γ (≥50.0).
The MCMC method described here attempts to generate a posterior sample of genealogies for the data under the coalescent. It uses all the information contained in the haplotypes or genotypes for this purpose (because we propose and accept only genealogies consistent with the observed data) and theoretically, after a very large number of iterations, we expect samples from essentially the correct posterior distribution. In contrast, PAC-based models (e.g., Hellenthal 2006; Gay et al. 2007; Yin et al. 2009) evaluate likelihoods of the data assuming a heuristic model that is an approximation of the constant population size coalescent. The relationship between the PAC model and the coalescent process is not well understood theoretically although they appear to be analogous. Apart from the benefit of having a posterior sample of genealogies, the main advantage of our Bayesian approach compared to other alternatives is that we expect to obtain better confidence intervals for the estimated parameter values (if the coalescent is a closer approximation to reality). It is also worth mentioning that the PAC model developed by Yin et al. (2009) considers only a restricted class of gene-conversion events (with regard to overlaps between events) whereas our method is more general and models all the conversion events that can potentially affect the configuration of the sample. Finally, since the InferRho algorithm lies within the coalescent framework, it is flexible and in principle can be extended for estimation under demographic models other than constant sized ones (e.g., exponential growth, population bottlenecks, etc.).
We performed two different kinds of tests on simulated data to verify the correctness of our estimation program. First, we ran the program without any data using a likelihood ratio of 1 and compared the average of several genealogy-based summary statistics to those obtained by Monte Carlo simulations under the coalescent. Second, we simulated data sets for which the true parameter values are known and compared them with the mode of the posterior distributions as estimated by the program. From both these tests, it appears that the modified InferRho program is working correctly. In addition to these, we also compared the accuracy of our inference method with that of two other comparable methods (proposed in Gay et al. 2007 and Yin et al. 2009) for samples of 100 simulated data sets. For estimating crossing-over and gene-conversion rates, InferRho has smaller mean squared error compared to the methods of Gay et al. (2007) and Yin et al. (2009) for the parameters tested in this study. For estimation of tract length, the mean squared error of InferRho is lower than the program of Gay et al. (2007) but higher than that of Yin et al. (2009).
We used the new version of InferRho to jointly estimate crossing-over and gene-conversion rates in the su-w and su-s genes of D. melanogaster. Our analysis with a fixed mean tract length of 352 bp suggests that gene conversion occurs more frequently than crossing over in these regions while the local estimates of crossing-over rate are not low. Furthermore, we also attempted to jointly estimate recombination rates and gene-conversion tract lengths and from this analysis it appears that mean tract lengths are ∼70 bp and 430 bp for the su-w and su-s genes, respectively. Finally, we compared the posterior distribution of gene-conversion initiation points with an uninformative prior distribution, using the Bayes factor calculated for 10-bp windows along these genes. Plots of the logarithm of Bayes factors for gene-conversion start points do not indicate any strong deviations from a prior distribution expected under the uniform recombination rate model. This suggests that there is not sufficient information in the data to infer the locations of historical conversion breakpoints.
One area of concern with using full-likelihood methods is their high computational expense when data sets become large. Note that the execution time of the InferRho algorithm is much higher (orders of magnitude) than those of Gay et al. (2007) and Yin et al. (2009). Therefore, it would be desirable to make such inference methods as optimal as possible in terms of run-time efficiency. To improve speed, we implemented several optimizations in the current version of our program. In our MA vectors, we keep track of marker locations that are ancestral to the sample and exclude uninformative gene-conversion and crossing-over events in the ARGs. We use arrays of size equal to the number of markers to represent the MA vectors. Operations on MA vectors (e.g., merging two MA vector arrays during coalescence, finding which marker positions have coalesced in a node, etc.) take linear time in terms of the total number of markers. Second, we use lookup tables to keep track of the coalescent prior likelihood of a graph and the likelihood of observed data given a graph once they have been calculated. Since a majority of newly proposed genealogies do not get accepted, these values are reused multiple times when calculating the acceptance probabilities in the algorithm. Finally, modifying some part of the graph (e.g., adding, deleting, or moving a node) during a proposal entails that we update the MA vectors of the subsequent nodes in the graph. While implementing this step, we first mark all the nodes that are ancestral to the modified node and then update only the MA vectors of the marked nodes. This previous step as well as parallelization of the code when running multiple MCMC chains can lead to further improvements in run-time efficiency.
Acknowledgments
This research project was supported by National Human Genome Research Institute grant HG-01988 to B.R.
Literature Cited
- Allers T., Lichten M., 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 [DOI] [PubMed] [Google Scholar]
- Andolfatto P., Wall J. D., 2003. Linkage disequilibrium patterns across a recombination gradient in African Drosophila melanogaster. Genetics 165: 1289–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C. S., Eberle M. A., Kruglyak L., Nickerson D. A., 2004. Mapping complex disease loci in whole-genome association studies. Nature 429: 446–452 [DOI] [PubMed] [Google Scholar]
- Coop G. C., Wen X., Ober C., Pritchard J. K., Przeworski M., 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319: 1395–1398 [DOI] [PubMed] [Google Scholar]
- Crawford D. C., Bhangale T., Li N., Hellenthal G., Rieder M. J., et al. , 2004. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36: 700–706 [DOI] [PubMed] [Google Scholar]
- Dunham I., Shimizu N., Roe B. A., Chissoe S., Hunt A. R., et al. , 1999. The DNA sequence of human chromosome 22. Nature 402: 489–495 [DOI] [PubMed] [Google Scholar]
- Fearnhead P., Smith N. G., 2005. A novel method with improved power to detect recombination hotspots from polymorphism data reveals multiple hotspots in human genes. Am. J. Hum. Genet. 77: 781–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S., Mortimer R. K., Lusnak K., 1983. Meiotic gene conversion in yeast: molecular and experimental perspectives, pp. 67–107 Yeast Genetics, edited by Spencer J. F. T., Spencer D. M., Smith A. R. W. Springer-Verlag, New York [Google Scholar]
- Frisse L., Hudson R. R., Bartoszewicz A., Wall J. D., Donfack J., et al. , 2001. Gene-conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69: 831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton S. M., Harding R. M., Boyce A. J., Clegg J. B., 1994. Molecular and population genetic analysis of allelic sequence diversity at the human-globin locus. Proc. Natl. Acad. Sci. USA 91: 1805–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay J., Myers S., McVean G., 2007. Estimating meiotic gene conversion rates from population genetic data. Genetics 177: 881–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. C., Marjoram P., 1996. Ancestral inference from samples of DNA sequences with recombination. J. Comput. Biol. 3: 479–502 [DOI] [PubMed] [Google Scholar]
- Hellenthal G., 2006. Exploring rates and patterns of variability in gene conversion and crossover in the human genome. Ph.D. Thesis, University of Washington, Seattle [Google Scholar]
- Hilliker A. J., Clark S. H., Chovnick A., 1991. The effect of DNA sequence polymorphisms on intragenic recombination in the rosy locus of Drosophila melanogaster. Genetics 129: 779–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A. J., Harauz G., Reaume A. G., Gray M., Clark S. H., et al. , 1994. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics 137: 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 1983. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23: 183–201 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 2001. Two-locus sampling distributions and their application. Genetics 159: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Padhukasahasram B., Nordborg M., 2003. The pattern of polymorphism on human chromosome 21. Genome Res. 13: 1158–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, 2005. A haplotype map of the human genome. Nature 437: 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., May C. A., 2004. Intense and highly localized gene-conversion activity in human meiotic crossover hotspots. Nat. Genet. 36: 151–156 [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Kauppi L., Neumann R., 2001. Intensely punctuate meiotic recombination in the class II region of the major histocompatibilty complex. Nat. Genet. 29: 217–222 [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Neumann R., Panayi M., Myers S., Donnelly P., 2005. Human recombination hotspots hidden in regions of strong marker associations. Nat. Genet. 37: 601–606 [DOI] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982. The coalescent. Stoch. Proc. Appl. 13: 235–248 [Google Scholar]
- Langley C. H., Lazzaro B. P., Phillips W., Heikkinen E., Braverman J. M., 2000. Linkage disequilibrium and the site frequency spectra in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Genetics 156: 1837–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Stephens M., 2003. Modeling linkage disequilibrium, and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165: 2213–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. S., Sabatti C., Teng J., Keats B. J., Risch N., 2001. Bayesian analysis of haplotypes for linkage disequilibrium mapping. Genome Res. 11: 1716–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A. T., Myers S. R., Hunt S., Deloukas P., Bentley D. R., et al. , 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584 [DOI] [PubMed] [Google Scholar]
- Morris A. P., Whittaker J. C., Balding D. J., 2002. Fine-scale mapping of disease loci via shattered coalescent modeling of genealogies. Am. J. Hum. Genet. 70: 686–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G. A. T., Donnelly P., 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324 [DOI] [PubMed] [Google Scholar]
- Niu T., Qin Z. S., Xu X., Liu J. S., 2002. Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am. J. Hum. Genet. 70: 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhukasahasram B., Marjoram P., Nordborg M., 2004. Estimating the rate of gene-conversion on human chromosome 21. Am. J. Hum. Genet. 75: 386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhukasahasram B., Wall J. D., Marjoram P., Nordborg M., 2006. Estimating recombination rates from single-nucleotide polymorphisms using summary statistics. Genetics 174: 1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak S. E., Voelpel K., Przeworski M., 2004. Insights into recombination from patterns of linkage disequilibrium in humans. Genetics 167: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. W., 1994. The Holliday junction on its thirtieth anniversary. Genetics 138: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W., 1983. The double-strand-break repair model for recombination. Cell 33: 25–35 [DOI] [PubMed] [Google Scholar]
- Tiemann-Boege I., Calabrese P., Cochran D., Sokol R., Arnheim N., 2006. High resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PLoS Genet. 2: e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. D., 2004. Estimating recombination rates using three-site likelihoods. Genetics 167: 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. D., Pritchard J. K., 2003. Haplotype blocks and linkage disequilibrium in the human genome. Nat. Rev. Genet. 4: 587–597 [DOI] [PubMed] [Google Scholar]
- Wang Y., Rannala B., 2008. Bayesian inference of fine-scale recombination rates using population genomic data. Philos. Trans. R. Soc. B 363: 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Rannala B., 2009. Population genomic inference of recombination rates and hotspots. Proc. Natl. Acad. Sci. USA 15: 6215–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf C., Hein J., 2000. The coalescent with gene-conversion. Genetics 155: 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., 2010. Computational methods for meiotic recombination inference. Ph.D. Dissertation, University of California, Berkeley [Google Scholar]
- Yin J., Jordan M. I., Song Y. S., 2009. Joint estimation of gene conversion rates and mean conversion tract lengths from population SNP data. Bioinformatics 25: i231–i239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangenberg G., Huang M. M., Arnheim N., Erlich H., 1995. New HLA-DPB1 alleles generated by interallelic gene conversion detected by analysis of sperm. Nat. Genet. 10: 407–444 [DOI] [PubMed] [Google Scholar]