Abstract
The matrix protein VP40 of Marburg virus promotes the formation and release of virus-like particles (VLPs). Marburg virus VP40 interacts with cellular Tsg101 via its L domain motif; however, mutation of this motif does not affect VLP budding or the accumulation of VP40 in multivesicular bodies (MVBs), which are platforms for virus particle formation. To identify regions of Marburg virus VP40 that are important for VLP budding, we examined deletion mutants and alanine-scanning mutants at the N- and C-terminus of VP40 for their involvement in VLP budding. VLPs were not detected in the presence of alanine-replacement mutants at Ile39 and Thr40, and the level of VLP budding for the alanine mutant at Asn297 was decreased. Moreover, these mutants did not accumulate in MVBs. Our results suggest the involvement of a novel host factor(s) in VLP budding and VP40 transport to MVBs.
Marburg virus (MARV) is a member of the Filoviridae in the order Mononegavirales [1]. MARV and its close relative the Ebola virus cause severe fatal hemorrhagic fever in humans and nonhuman primates [2]. Enveloped MARV particles comprise 7 structural proteins and a nonsegmented negative-strand RNA genome of ∼19 000 base pairs in length [3]. The viral surface glycoprotein protrudes from the envelope and is assumed to be responsible for binding to cellular receptors and for fusion events [4–6]. VP40 is a major matrix protein that plays critical roles in viral assembly and budding [7, 8]. The expression of VP40 in mammalian cells leads to the budding of filamentous virus-like particles (VLPs) [9]. The accumulation of MARV VP40 in multivesicular bodies (MVBs) (as detected by colocalization with the MVB marker CD63 [10]), which are platforms for the formation of the viral envelope with glycoprotein, is followed by VP40-induced VLP budding [11].
MARV VP40 possesses an L domain motif, or 16PPPY19, near its N-terminus. L domains have been identified as regions critical for the late step of viral budding from retroviruses [12, 13]. A recent study indicates that MARV VP40 interacts with Tsg101, which is a component of the endosomal sorting complex required for transport I (ESCRT-I) and is involved in the MVB sorting pathway, which relies on the PPPY motif [14]. However, L domain motif mutants can bud into the supernatant as VLPs, albeit at levels ∼70% lower than that of wild-type VP40-induced VLPs [14]. Furthermore, L domain motif mutants are transported to the plasma membrane and colocalize with CD63, an MVB marker [11, 15]. These data suggest that MARV VP40 possesses other structural elements for VLP budding and for the accumulation of VP40 in MVBs. In fact, it has been reported that 53P and 96LPLGVA101 of Ebola virus VP40 and 84LPLGIM89 of MARV VP40 are critical for efficient VLP production [16, 17].
To obtain new insights into the role of other structural elements in MARV assembly, we searched for additional domains involved in MARV VP40-induced VLP budding and in the accumulation of VP40 in MVBs.
MATERIALS AND METHODS
Cells
Human embryonic kidney cells (293 and 293T cells) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and penicillin-streptomycin solution (SIGMA).
Plasmids
The VP40 coding region of the Musoke strain of MARV was inserted into pCAGGS/MCS [18, 19] with or without the FLAG or cMyc tag sequence at the N-terminus of the open reading frame [3]. Deletion mutants at the N-terminus or C-terminus of MARV VP40 were generated by polymerase chain reaction (PCR) and cloned into pCAGGS/MCS with a cMyc tag. L domain motif mutants, mutated from PPPY to PPPA and AAPA, and alanine-replacement mutants of single or double amino acids at positions 36–40 were generated in pcDNA™3.1/myc-HisA (Invitrogen) by using site-directed mutagenesis by PCR and were subcloned into pCAGGS/MCS with the cMyc tag. The double amino acid mutant at 39I and 40T was also fused with the FLAG tag and applied to immunoprecipitation assays. The single alanine-replacement mutants at positions 294–298 were generated in pcDNA™3.1/myc-HisA by using site-directed mutagenesis with PCR and were subcloned into pCAGGS/MCS with the cMyc tag.
VLP Budding Assay
We transfected 293T cells with each of the plasmids expressing the cMyc-tagged VP40 or VP40 mutants by using TransIT-293 (Mirus Bio) according to the manufacturer’s protocol. In some experiments, the cells were cotransfected with plasmids encoding FLAG-tagged VP40 and cMyc-tagged VP40 mutants or FLAG-tagged VP40 mutants and cMyc-tagged VP40. At 48 hours after transfection, the cell supernatants were harvested and cleared of cell debris by centrifugation at 1750 × g for 5 minutes at 4°C. The supernatants were then layered onto a 20% sucrose cushion. The VLPs were then pelleted by ultracentrifugation in an SW55Ti rotor (Beckman Coulter) at 303 800 × g for 2 hours at 4°C and suspended in sample buffer. The transfected cells were homogenized by means of supersonic treatment and lysed with sample buffer (Invitrogen). The VLPs and cell lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by Western blotting with a mouse anti-cMyc antibody (SIGMA) (dilution, 1:5000) or a mouse anti-FLAG antibody (SIGMA) (dilution, 1:5000).
Indirect Immunofluorescence Assay
At 24 hours after transfection with plasmids expressing cMyc-tagged VP40 or VP40 mutants, 293 cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The cells were then incubated with a rabbit anti-cMyc antibody and a mouse anti-CD63 antibody (Abcom) as primary antibodies for 30 minutes at room temperature. After washing 3 times with phosphate-buffered saline (PBS), the cells and nuclei were stained with Alexa Fluor 594 goat anti–rabbit IgG antibody, Alexa Fluor 488 goat anti–mouse IgG antibody, and Hoechst (Invitrogen) for 30 minutes at room temperature. Microscopic analysis was performed with an LSM510META (Carl Zeiss).
Immunoprecipitation
The 293T cells were cotransfected with plasmids encoding FLAG-tagged VP40 and cMyc-tagged VP40 mutants or FLAG-tagged VP40 mutants and cMyc-tagged VP40. At 48 hours after transfection, the cells were lysed in lysis buffer (50 mmol/L Tris-HCl [pH, 7.5], 150 mmol/L NaCl, 1 mmol/L EDTA, 0.5% Nonidet P-40, and protease inhibitor cocktail Complete Mini [Roche]) and incubated for 60 minutes at 4°C. After clarification by means of low-speed centrifugation, the supernatants were incubated with anti-FLAG M2 Affinity Gel (SIGMA) for 17 hours at 4°C. Then a fraction of supernatant was mixed with sample buffer and incubated for 5 minutes at 95°C. The FLAG beads were washed 3 times with lysis buffer, suspended in sample buffer, and then incubated for 5 minutes at 95°C. The samples were then subjected to SDS-PAGE, followed by Western blotting with a rabbit anti-FLAG antibody (SIGMA) or a rabbit anti-cMyc antibody (SIGMA).
RESULTS AND DISCUSSION
The L Domain Motif Is Not Essential for MARV VP40 Budding
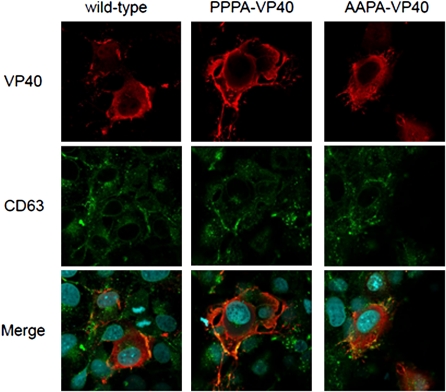
Single-alanine substitution mutants of the L domain motif of MARV VP40 (APPY, PAPY, and PPPA) were able to bud into supernatants as VLPs, albeit at a lower rate than that of wild-type VP40-induced VLPs [14]. To assess the intracellular localization of the L domain motif mutants PPPA and AAPA, 293 cells were transfected with wild-type VP40 or with each L domain motif mutant and detected with an anti-cMyc antibody 24 hours later. As shown in Figure 1, both the L domain motif mutants and wild-type VP40 were transported to the plasma membrane and colocalized with CD63, a marker for MVBs, where VLP formation occurs [11, 15]. We also constructed a deletion mutant that lacks 25 amino acids at the N-terminus of VP40 and examined its ability to promote VLP budding. We found that the level of VLP budding induced by this mutant was similar to that induced by wild-type VP40 (Figure 2B). Taken together, these data show that the L domain motif at positions 16–19 is not essential for VLP budding and is not required for VP40 accumulation in MVBs.
Figure 1.
Intracellular localization of the L domain motif mutants. We transfected 293 cells with plasmids expressing cMyc-tagged VP40 and VP40 mutants. At 24 hours after transfection, cells were stained with an anti-cMyc antibody (red) and an anti-CD63 (a multivesicular body marker) antibody (green). Nuclei were stained with Hoechst (Invitrogen) (blue). L domain motif mutants were transported to the plasma membrane and colocalized with CD63 and with wild-type VP40.
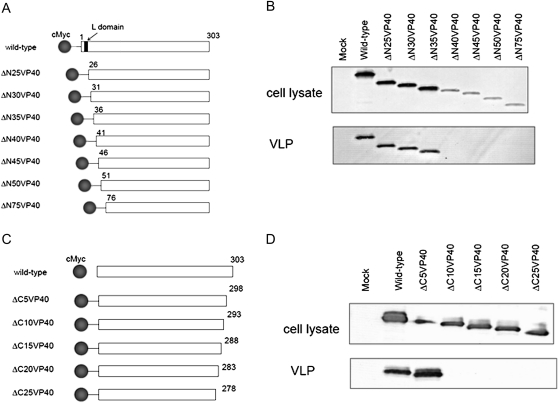
Figure 2.
Virus-like particle (VLP) budding induced by deletion mutants at the N- and C-terminus of VP40. Schematic diagrams of the deletion mutants at A, the N- terminus, and C, the C-terminus of VP40. B, D, Comparison of VLP budding induced by the deletion mutants. We transfected 293T cells with a plasmid expressing each of the deletion mutants. At 48 h after transfection, the VLP budding assay was performed. Thirty-five amino acids at the N-terminus and 5 amino acids at the C-terminus of VP40 were dispensable for VLP budding.
A series of cMyc-tagged deletion mutants was subjected to our VLP budding assay (Figure 2A). As shown in Figure 2B, the level of protein detected decreased as the extent of the deletion increased. VLP budding was detected with the mutant that lacked 35 amino acid residues but not with the mutants that lacked 40 or more amino acids (Figure 2B).
The Importance of Ile39 and Thr40 for VLP Budding and the Accumulation of VP40 in MVBs
The VLP budding assay with N-terminal deletion mutants showed that 35 residues are dispensable for VLP budding; therefore, we next evaluated the double-alanine mutants in amino acid residues 36–40 for their ability to produce VLPs (Figure 3A). VLPs were not detected with the V40 mutant possessing 3940A-VP40, whereas the levels of VLP production for the other mutants were similar to that for wild-type VP40 (Figure 3B).
Figure 3.
Double alanine-scanning of VP40 at amino acid residues 36–40. A, Schematic diagrams of alanine-scanning mutants. B, Comparison of virus-like particle (VLP) budding induced by the alanine-scanning mutants. We transfected 293T cells with a plasmid expressing each mutant. At 48 hours after transfection, the VLP budding assay was performed. We found that 3940A-VP40 did not produce VLPs in the supernatant. C, Intracellular localization of alanine-scanning mutants. We transfected 293 cells with a plasmid expressing a cMyc-tagged VP40 mutant. At 24 hours after transfection, cells were stained with an anti-cMyc antibody (red) and an anti-CD63 antibody (green). Nuclei were stained with Hoechst (Invitrogen) (blue). The 3940A-VP40 did not accumulate in multivesicular bodies and was not transported to the plasma membrane.
We then assessed the intracellular localization of the double-alanine mutants by using an immunofluorescence assay (IFA) (Figure 3C). We found that 3940A-VP40 did not accumulate in MVBs and was not transported to the plasma membrane. The localization of 3839A-VP40 was different from that of wild-type VP40. The other mutants colocalized with CD63, as did the wild type. These results indicate that Ile39 and Thr40 are important for VLP budding and play a role in the accumulation of VP40 in MVBs.
To further map the residues important for VLP budding, the single-alanine mutants were assessed for their ability to produce VLPs and for their intracellular localization (Figure 4A). As shown in Figure 4B, none of the mutations affected VLP budding. The intracellular localizations of I39A-VP40 and T40A-VP40 were different from that of the wild type. We found that I39A-VP40 did not accumulate in MVBs (Figure 4C).
Figure 4.
Single alanine-scanning of VP40 at amino acid residues 36–40. A, Schematic diagrams of alanine-scanning mutants. B, Comparison of virus-like particle (VLP) budding induced by the alanine-scanning mutants. We transfected 293T cells with a plasmid expressing each mutant. At 48 hours after transfection, the VLP budding assay was performed. All mutants produced VLPs in the supernatant as effectively as wild-type VP40. C, Intracellular localization of single-alanine mutant 293 cells were transfected with a plasmid expressing a cMyc-tagged VP40 mutant. At 24 hours after transfection, cells were stained with an anti-cMyc antibody (red) and an anti-CD63 antibody (green). Nuclei were stained with Hoechst (Invitrogen) (blue). The intracellular localizations of I39A-VP40 and T40A-VP40 were different from that of the wild type.
A-VP40 Forms Oligomer and Has No Effect on VLP Budding Induced by the Wild Type
MARV and Ebola virus VP40 form hexameric or octameric structures; the N-terminal domain motif of VP40 is thought to be responsible for this oligomerization [20, 21]. To evaluate the involvement of Ile39 and Thr40 in the oligomerization of VP40, we analyzed the interaction between 3940A-VP40 and wild type or 3940A-VP40 and 3940A-VP40 by using an immunoprecipitation assay. The 293T cells were cotransfected with a cMyc-tagged plasmid and a FLAG-tagged plasmid. At 48 h after transfection, cells were harvested and subjected to immunoprecipitation with anti-FLAG M2 affinity gel (SIGMA). The 3940A-VP40 coprecipitated with the wild-type and 3940A-VP40, and the wild-type VP40 coprecipitated with 3940A-VP40 and the wild type (Figure 5A). This result indicates that Ile39 and Thr40 are not involved in the oligomerization of VP40.
Figure 5.
Oligomerization of VP40 mutants. We cotransfected 293T cells with plasmids encoding the FLAG-tagged VP40 and a cMyc-tagged VP40 mutant or a FLAG-tagged VP40 mutant and the cMyc-tagged VP40. At 48 hours after transfection, the immunoprecipitation assay (A) and virus-like particle (VLP) budding assay (B) were performed. The 3940A-VP40 had no effect on oligomer formation or VLP budding induced by the wild type.
We next examined whether the expression of 3940A-VP40 inhibits VLP budding induced by wild-type VP40. We cotransfected 293T cells with a cMyc-tagged plasmid and a FLAG-tagged plasmid. At 48 hours after transfection, cells and supernatants were harvested and subjected to the VLP budding assay. As shown in Figure 5B, the expression of 3940A-VP40 had no effect on VLP budding induced by wild-type VP40.
Asn297 Is Important for VLP Budding and the Accumulation of VP40 in MVBs
To identify the region of VP40 important for VLP budding at the C-terminus, C-terminal deletion mutants of VP40 were constructed and subjected to the VLP budding assay (Figure 2C). Although deleting 5 amino acid residues at the C-terminus of the protein had no effect on VLP budding, the deletion of an additional 5 amino acids or more at the C-terminus eliminated VLP budding (Figure 2D). This result suggests that amino acid residues 299–303 are dispensable for VLP budding.
To pinpoint the VP40 C-terminal amino acid residues that are important for VLP budding, we constructed single-alanine or glycine mutants at amino acid residues 294–298 and performed the VLP budding assay (Figure 6A). The level of VLP budding for N297A-VP40 was reduced to ∼20% of that of the wild type, whereas the other mutations had no effect on VLP budding (Figure 6B).
Figure 6.
Virus-like particle (VLP) budding induced by the single-alanine mutants. A, Schematic diagram of the single-alanine mutants. B, Comparison of VLP budding induced by the alanine mutants. We transfected 293T cells with a plasmid expressing each mutant. At 48 hours after transfection, the VLP budding assay was performed. The intensity of the VP40 bands was quantified in relation to the amount of VP40 in lane 1, which was set to 100%. The result presented is the mean of 3 independent experiments. The level of VLP budding for N297A-VP40 was decreased to ∼20% of that of the wild type. C, Intracellular localization of single-alanine mutants. We transfected 293 cells with a plasmid expressing a cMyc-tagged VP40 mutant. At 24 hours after transfection, cells were stained with an anti-cMyc antibody (red) and an anti-CD63 antibody (green). Nuclei were stained with Hoechst (Invitrogen) (blue). The intracellular localization of N297A-VP40 was different from that of the wild type.
The intracellular localization of these mutants was assessed with an IFA. As shown in Figure 6C, VP40 was not transported to the plasma membrane in the cells expressing N297A-VP40, although the localization patterns of the other mutants were similar to that of the wild type. These results indicate that Asn297 is important for VLP budding and the transportation of VP40 to the plasma membrane.
The L domain motif was originally defined as an amino acid sequence whose mutation inhibits virus particle release [12]. MARV VP40 contains an L domain motif, or 16PPXY19, that interacts with the host protein Tsg101 [14]. Tsg101 forms part of a high molecular weight complex, termed ESCRT-I, that is involved in the sorting pathway of MVBs [13]. In this study, Ile39, Thr40, and Asn297 were identified as components of MARV VP40 that are important for VLP budding induced by VP40. In 3940A-VP40–expressing cells, VLP was not detected in the supernatant and VP40 did not accumulate in MVBs (Figure 3B and 3C). The alanine replacement of Asn297 reduced VLP budding to ∼20% and affected the subcellular localization of VP40 (Figure 6B and 6C). On the other hand, the levels of VLPs for the L domain motif deletion mutants were similar to that for the wild-type, and the alanine-replacement mutants accumulated in MVBs (Figures 1 and 2B). These results suggest that MARV VP40 possesses amino acid sequences other than the L domain motif that are important for VLP budding and play a role in the intracellular transport of VP40. Ile39, Thr40, and Asn297 of VP40 are conserved among all strains of MARV. Given the previous report that the nucleocapsid protein of MARV interacts with Tsg101 via its L domain motif to enhance VP40-induced VLP production [22], it is likely that the L domain motif of MARV VP40 is dispensable for VLP budding.
Because MARV glycoprotein and VP40 colocalize in MVBs and the budding of progeny virions into MVBs has been detected in MARV-infected cells, it is suggested that MVBs are platforms for the formation of viral particles of MARV [10]. However, the single-alanine mutants at Ile39, Thr40, and Asn297 produced VLPs without the accumulation of VP40 in MVBs. This result suggests that the accumulation in MVBs may not be necessary for VP40-induced VLP budding per se.
In this study, we identified regions of MARV VP40 that are important for VLP budding and the subcellular transport of VP40. These domains may be involved in interactions with a host factor or factors that play roles in the transport of VP40 to MVBs and the plasma membrane. The identification of amino acids that are critical for VP40-induced VLP budding contributes to our understanding of the mechanism of MARV budding.
Funding
This work was supported by the National Institutes of Health (NIH; R01 AI055519) and the Region V ‘‘Great Lakes’’ Regional Center of Excellence (NIH grant 1-U54-AI-057153).
Acknowledgments
We thank Susan Watson for editing the manuscript. We are grateful to Jiro Yasuda for providing us with a plasmid encoding Marburg virus VP40.
References
- 1.Sanchez A, Geisbert TW, Feldmann H. Filoviridae. In: Knipe DM, Howley PM, Griffin DM, editors. Fields virology. 5th ed. Philadelphia: Lippincott, Williams & Wilkins; 2007:. pp. 1409–48. [Google Scholar]
- 2.Towner JS, Khristova ML, Sealy TK, et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J Virol. 2006;80:6497–516. doi: 10.1128/JVI.00069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldmann H, Mühlberger E, Randolf A, et al. Marburg virus, a filovirus: messenger RNAs, gene order, and regulatory elements of the replication cycle. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- 4.Becker S, Spiess M, Klenk HD. The asialoglycoprotein receptor is a potential liver-specific receptor for Marburg virus. J Gen Virol. 1995;76:393–9. doi: 10.1099/0022-1317-76-2-393. [DOI] [PubMed] [Google Scholar]
- 5.Becker S, Klenk HD, Mühlberger E. Intracellular transport and processing of the Marburg virus surface protein in vertebrate and insect cells. Virology. 1996;225:145–55. doi: 10.1006/viro.1996.0582. [DOI] [PubMed] [Google Scholar]
- 6.Feldmann H, Will C, Schikore M, Slenczka W, Klenk HD. Glycosylation and oligomerization of the spike protein of Marburg virus. Virology. 1991;182:353–6. doi: 10.1016/0042-6822(91)90680-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolesnikova L, Bugany H, Klenk HD, Becker S. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J Virol. 2002;76:1825–38. doi: 10.1128/JVI.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolesnikova L, Bamberg S, Berghöfer B, Becker S. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J Virol. 2004;78:2382–93. doi: 10.1128/JVI.78.5.2382-2393.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swenson DL, Warfield KL, Kuehl K, et al. Generation of Marburg virus-like particles by co-expression of glycoprotein and matrix protein. FEMS Immunol Med Microbiol. 2004;40:27–31. doi: 10.1016/S0928-8244(03)00273-6. [DOI] [PubMed] [Google Scholar]
- 10.Kolesnikova L, Bohil AB, Cheney RE, Becker S. Budding of Marburgvirus is associated with filopodia. Cell Microbiol. 2007;9:939–51. doi: 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 11.Kolesnikova L, Berghöfer B, Bamberg S, Becker S. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J Virol. 2004;78:12277–87. doi: 10.1128/JVI.78.22.12277-12287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A. 1991;88:3195–9. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed EO. Viral late domain. J Virol. 2002;76:4679–87. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J Virol. 2007;81:4895–9. doi: 10.1128/JVI.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramer B, Pelchen-Matthews A, Deneka M, Garcia E, Piguet V, Marsh M. HIV interaction with endosomes in macrophages and dendritic cells. Blood Cells Mol Dis. 2005;35:136–42. doi: 10.1016/j.bcmd.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Yamayoshi S, Kawaoka Y. Mapping of a region of Ebola virus VP40 that is important in the production of virus-like particles. J Infect Dis. 2007;196:S291–5. doi: 10.1086/520595. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Cocka L, Okumura A, Zhang YA, Sunyer JO, Harty RN. Conserved motifs within Ebola and Marburg virus VP40 proteins are important for stability, localization, and subsequent budding of virus-like particles. J Virol. 2010;84:2294–303. doi: 10.1128/JVI.02034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobasa D, Rodgers ME, Wells K, Kawaoka Y. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J Virol. 1997;71:6706–13. doi: 10.1128/jvi.71.9.6706-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–9. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 20.Timmins J, Schoehn G, Kohlhaas C, Klenk HD, Ruigrok RW, Weissenhorn W. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology. 2003;312:359–68. doi: 10.1016/s0042-6822(03)00260-5. [DOI] [PubMed] [Google Scholar]
- 21.Hoenen T, Biedenkopf N, Zielecki F, et al. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J Virol. 2010;84:7053–63. doi: 10.1128/JVI.00737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolnik O, Kolesnikova L, Stevermann L, Becker S. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J Virol. 2010;84:7847–56. doi: 10.1128/JVI.00476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]