Abstract
The Ebola virus (EBOV) protein VP24 inhibits type I and II interferon (IFN) signaling by binding to NPI-1 subfamily karyopherin α (KPNA) nuclear import proteins, preventing their interaction with tyrosine-phosphorylated STAT1 (phospho-STAT1). This inhibits phospho-STAT1 nuclear import. A biochemical screen now identifies heterogenous nuclear ribonuclear protein complex C1/C2 (hnRNP C1/C2) nuclear import as an additional target of VP24. Co-immunoprecipitation studies demonstrate that hnRNP C1/C2 interacts with multiple KPNA family members, including KPNA1. Interaction with hnRNP C1/C2 occurs through the same KPNA1 C-terminal region (amino acids 424–457) that binds VP24 and phospho-STAT1. The ability of hnRNP C1/C2 to bind KPNA1 is diminished in the presence of VP24, and cells transiently expressing VP24 redistribute hnRNP C1/C2 from the nucleus to the cytoplasm. These data further define the mechanism of hnRNP C1/C2 nuclear import and demonstrate that the impact of EBOV VP24 on nuclear import extends beyond STAT1.
Ebola virus (EBOV) is taxonomically classified in the genus Ebolavirus (family Filoviridae) and is a nonsegmented, negative-sense RNA virus that replicates in the cytoplasm of infected cells [1]. The EBOV genome encodes 8 proteins, one of which is the multifunctional VP24. VP24 has been described as a minor matrix protein and is present in viral particles [2, 3]. VP24 promotes the formation of filamentous nucleocapsids that are comprised mainly of 2 other viral proteins, NP and VP35 [4, 5]. VP24 also appears to be required for formation of fully infectious virus particles [3, 6, 7]. Other studies indicate that VP24 expression regulates a transition from viral transcription and replication to virion assembly [8].
In addition to these structural and assembly functions, VP24 also modulates host responses to infection. VP24 has been shown to acquire mutations during the experimental adaptation of EBOV in mice and guinea pigs [9–12]. These changes in VP24 appear to be critical for enhanced EBOV virulence in these species. EBOV VP24 also inhibits interferon (IFN) signaling by binding to karyopherin-α (KPNA) to block the nuclear accumulation of tyrosine-phosphorylated STAT1 (phospho-STAT1). This prevents the antiviral effects of IFN-α/β in VP24-expressing cells [13–15].
Nuclear import of many cytoplasmic proteins is mediated by KPNA (also called importin-α) [16–19]. There are 7 known members of the KPNA family in human cells. KPNA 1, 5, and 6 comprise the NPI-1 subfamily; KPNA 3 and 4 comprise the Qip-1 subfamily; and KPNA 2 and 7 belong to the Rch-1 subfamily [20, 21]. Each KPNA possesses an N-terminal importin-β binding domain, followed by a series of 10 armadillo (arm) repeats and a C-terminal nuclear export domain that is recognized and mediates export by the cellular apoptosis susceptibility protein [19, 22]. To facilitate nuclear import, the nuclear localization signal (NLS) of a cargo protein is recognized by arm repeats on the KPNA. These proteins interact with importin-β, which translocates the complex through the nuclear pore [19]. Most NLSs bind to KPNA arm repeats 1–8 [23]. Instead, VP24 and tyrosine-phosphorylated, dimerized STAT1 bind to the C-terminal arm repeats 8, 9, and 10 of the KPNA1 protein [15].
This study sought to identify additional KPNA-cargo protein interactions affected by the presence of VP24. We revealed that the protein complex heterogenous nuclear ribonuclear protein complex C1/C2 (hnRNP C1/C2) binds to several KPNA family members and that its binding and nuclear translocation are partially blocked by VP24. hnRNP C1/C2 is a nuclear protein complex involved in host mRNA transcript processing, but it does translocate to the cytoplasm during mitosis to facilitate translation, for example, by binding to the internal ribosome entry site (IRES) of c-myc [24]. Furthermore, poliovirus (PV), rhinovirus (RV), and vesicular stomatitis virus (VSV) infections disrupt the nuclear pore complex, relocalizing hnRNP C1/C2 to the cytoplasm, where it can interact with viral RNA and viral proteins [25–29] and enhance PV replication [30–32]. Our studies indicate that both VP24 and hnRNP C1/C2 bind to the same C-terminal amino acids of KPNA1, suggesting that VP24 may outcompete hnRNP C1/C2 for binding to KPNA1, resulting in its cytoplasmic retention. These data suggest a possible role for hnRNPC1/C2 in EBOV replication.
MATERIALS AND METHODS
Plasmids, Cells, and Antibodies
The mammalian expression plasmid pCAGGS was used for all expression studies, with the exception of VP24, which was cloned in pCDNA3. Both 293T and Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. The following antibodies were used in these studies: a monoclonal mouse M2 α-Flag and a rabbit polyclonal α-Flag (Sigma), mouse monoclonal anti-hnRNPC1/C2 clone 4F4 (Abcam), rabbit polyclonal α-VP24 (a generous gift from Dr. Viktor E. Volchkov; INSERM), a mouse monoclonal antibody to VP35 described elsewhere [33], and mouse anti–β-tubulin antibodies.
Coimmunoprecipitation Assays to Identify Karyopherin α-Interacting Proteins by Mass Spectrometry
HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, cells were harvested and washed in phosphate-buffered saline (PBS) and lysed in NP-40 lysis buffer (50 mmol/L Tris [pH, 7.5], 280 mmol/L NaCl, 0.5% NP-40, 0.2 mmol/L EDTA, 2 mmol/L EGTA, 10% glycerol, and protease inhibitors [Complete; Roche]). Lysates were incubated on ice for 30 min and centrifuged for 10 minutes at 4°C in a microcentrifuge, and the supernatant was collected. For immunoprecipitations, the α-Flag M2 monoclonal antibody cross-linked to agarose beads (Sigma) was added and incubated overnight at 4°C. The beads were then washed 3 times with lysis buffer, and immunoprecipitated proteins were eluted with 3 μL of 100 μg/mL stock of 3x Flag peptide (Sigma). The immunoprecipitated material was then analyzed by sodium dodecyl sulfide polyacrylamide electrophoresis (SDS-PAGE), followed by staining with Gel Code Blue (Pierce), a coomassie blue–based protein stain, to visualize protein bands. Protein bands were provided to the Northeast Biodefense Center (NBC) Proteomics Core at Yale University for mass spectrometric analysis and protein identification.
Protein Digestion
The excised gel bands were washed with 250 μL 50% acetonitrile and 50% water for 5 minutes, followed by 250 μL of 50 mmol/L ammonium bicarbonate, 50% acetonitrile, and 50% water for 30 minutes and a final 10 mmol/L ammonium bicarbonate, 50% acetonitrile, and 50% water wash for 30 minutes. After the removal of the washes, the gel was dried in a speedvac and rehydrated with 0.2 μg of modified porcine trypsin (Promega) in 26 μL of 10 mmol/L ammonium bicarbonate solution (enough volume to cover the gel pieces). Samples were digested at 37°C for 18 hours. After digestion, 10 μL of the digest supernatant was transferred to LC-MS/MS vials with 5 μL injected.
Liquid Chromatography and Mass Spectrometry (LC-MS/MS)
The analysis was performed on a Micromass Q-TOF Ultima equipped with a Waters nanoACQUITY UPLC system. A Waters Symmetry C18 180 μm × 20 mm trap column and a 1.7 μm, 75 μm × 250 mm nanoACQUITY UPLC column at 35°C were used for peptide separation. Trapping was done at 15 μL/min, 99% buffer A (100% water and 0.1% formic acid) for 1 minutes. Peptide separation was performed at 300 nL/min with buffer A (100% water and 0.1% formic acid) and buffer B (100% CH3CN and 0.075% formic acid). A linear gradient (51 min) was run with 5% buffer B at initial conditions, 50% B at 50 minutes, and 85% B at 51 minutes. Data-dependent acquisition was performed so that the mass spectrometer switched automatically from MS to 4 consecutive MS/MS modes when the total ion current increased to >1.5 counts/s threshold set point. To obtain good fragmentation, a collision energy ramp was set for the different mass sizes and charge states, giving preference to doubly and triply charged species for fragmentation.
Identification of Peptides
The LC-MS/MS data was searched using the Mascot Distiller and the Mascot search algorithms (Matrix Science). The Mascot Distiller program combines sequential MS/MS scans from profile data that have the same precursor ion. A charge state of +2 and +3 were preferentially located with a signal-to-noise ratio of ≥1.2, and a peak list was generated for database searching against the NCBInr database. The Mascot significance score match is based on a MOWSE score and relies on multiple matches to >1 peptide from the same protein. Parameters used for searching were a significance threshold of P < .5, a peptide tolerance of ±0.6 Da, MS/MS fragment tolerance of ±0.4 Da, 1 missed cleavage site, and peptide charges of +2 or +3. Variable modifications included propionamide for Cys and methionine oxidation.
Co-immunoprecipitations and Western blot. HEK293T cells were transfected, lysed, and run on SDS-PAGE gels, as described above. The proteins were then transferred to a polyvinylidene difluoride membrane. The membrane was blocked in 5% nonfat dry milk and 0.1% Tween 20 in PBS and then probed with the aforementioned antibodies. Membranes were developed using a Western Lightning ECL kit (Perkin-Elmer) and Kodak BioMax film (Kodak).
Immunofluorescence. Immunofluorescence studies of hnRNPC1/C2 were based on a protocol described elsewhere [34]. Vero cells were transfected by using Lipofectamine 2000 with plasmids encoding proteins of interest and then plated on 12-mm–diameter glass coverslips. Twenty-four hours after transfection, the cells were fixed with 4% paraformaldehyde and were permeabilized with 0.1% triton-X for 10 minutes. The coverslips were then washed in PBS and blocked in PBS containing 4% normal goat serum and 2% BSA for 1 hour. The coverslips were incubated with mouse α-hnRNPC1/C2 and either a polyclonal rabbit α-Flag or VP24 for 1 hour and then washed 3 times. Next, the coverslips were incubated with a secondary rhodamine red-x-affinipure goat α-mouse IgG (Jackson) and an alexa 488 goat α-rabbit secondary (Invitrogen) for 30 minutes. To visualize nuclei, Hoechst 33342 (0.1 μg/mL; Molecular Probes) was added to one of the final washes. The coverslips were mounted with prolong gold antifade (Invitrogen) and imaged on a DM6000 microscope at the MSSM microscope facility.
RESULTS
hnRNP C1/C2 Interacts With Select Karyopherin α (KPNA) Proteins
Previous studies have revealed that the EBOV protein VP24 binds to all 3 members of the human NPI-1 KPNA (importin α) family (KPNA1, 5, 6) and that VP24 binds to a region of KPNA1 that is similar to that bound by phospho-STAT1 [13–15]. This interaction prevents STAT1 binding to KPNAs, its nuclear accumulation, and subsequent activation of IFN-responsive genes after IFN-β or IFN-γ treatment [15]. To build on these studies, we sought to identify additional cellular proteins that normally bind KPNA1 and to determine whether they were blocked in their ability to bind KPNA1 in VP24-expressing cells.
To identify cellular proteins that interact with KPNA1, previously described Flag-tagged KPNAs were overexpressed [14, 15]. Flag-KPNA 1, 2, and 3 (a member of each KPNA subfamily, although of these, VP24 only binds to KPNA 1) were transfected into 293T cells in parallel with Flag-GFP as a negative control. Twenty-four hours after transfection, cells were lysed, and the Flag-tagged proteins were immunoprecipitated. The eluted material was analyzed using SDS-PAGE and coomassie blue staining to visualize proteins that bound specifically to each Flag-KPNA. We were able to detect distinct and reproducible banding patterns for each of the KPNA-associated proteins (Figure 1). Next, we expressed either KPNA1 alone or KPNA1 in the presence of VP24, and compared the banding pattern of proteins. Of note, the intensity of one coprecipitating band in the middle of the gel was drastically reduced in the presence of VP24, compared with a coprecipitating protein band directly above this that was unaffected (Figure 1B). This band of interest was identified using mass spectrometry as hnRNP C1/C2 (Figure 1C). The interaction between Flag-KPNA1 and hnRNP C1/C2 was confirmed by Western blot using a monoclonal antibody specific for hnRNPC1/C2 (Figure 1D). Of interest, the banding pattern that corresponds to hnRNPC1/C2 on KPNA1 was also present on KPNA3, but not on KPNA2 (see Figure 1A). This indicated that hnRNPC1/C2 may be able to interact with multiple KPNA family members.
Figure 1.
hnRNP C1/C2 is a karyopherin α1 (KPNA1) interacting protein that may be displaced by VP24. A, Coprecipitation of host cell proteins with Flag-tagged KPNA1, 2, and 3. The bands corresponding to the KPNAs and GFP are labeled. The single asterisks beside the 2 bands in figure 1A indicate bands that corresponded to hnRNPC by Mass Spectrometry analysis. B, Identification of a protein band that interacts with KPNA1 in the absence but not the presence of VP24 (band labeled with a single asterisk). The band directly above this band (labeled with 2 asterisks) is unaffected by the presence of VP24 (top). Western blot of the same lysates as above showing the interaction of VP24 interacting with KPNA1 (bottom). C, Mass spectrometry results for bands labeled with single asterisks were identified as either hnRNP C1 or C2. The complete sequence of hnRNPC1 is shown with the underlined bold text indicating the peptides that were identified by mass spectroscopy. D, FLAG-KPNA1 co-immunoprecipitates hnRNPC1/C2. Marburg virus nucleoprotein (MARV NP) was used as a negative control in this study.
EBOV VP24 Disrupts hnRNP C1/C2 Binding to KPNA1
We next confirmed that hnRNPC1/C2 binding to KPNA1 was blocked by VP24. KPNA1 was transfected with either empty plasmid or with increasing amounts of a VP24-expressing plasmid. Consistent with our initial pull-down studies, increasing amounts of VP24 reduced the ability of endogenous hnRNP C1/C2 to bind to Flag-KPNA1 in 293T cells (Figure 2). The total levels of hnRNPC1/C2 were unaffected, as reflected by the amount of hnRNPC1/C2 in the total cell lysates (Figure 2). These data indicate that, as is the case with phospho-STAT1, VP24 blocks the ability of hnRNPC1/C2 to bind to KPNA1.
Figure 2.
hnRNP C1/C2 binding to KPNA1 is inhibited by VP24. Cells were cotransfected with KPNA1 and either empty plasmid or increasing amounts of EBOV VP24 plasmid. Endogenous hnRNPC1/C2 binding to KPNA1 is partially blocked in the presence of VP24 as assessed by Western blot, as indicated (top). Whole cell extracts from A were analyzed by Western blot, as indicated; VP24 does not affect total levels of hnRNPC1/C2 (bottom).
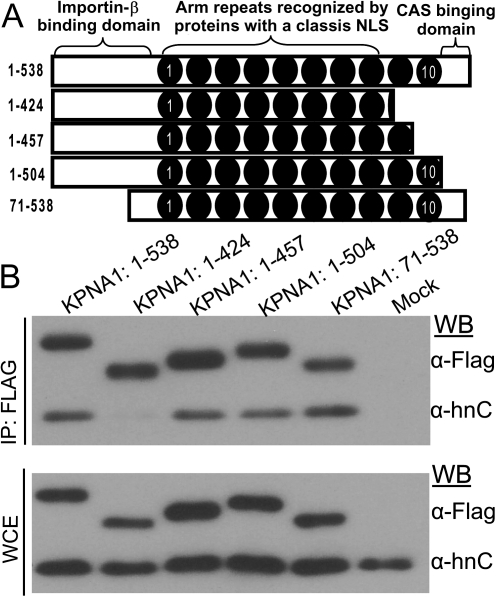
Mapping the KPNA1 Domains Required for hnRNPC1/C2 Binding.
Previous coprecipitation experiments that used deletion mutants of KPNA1 showed that VP24 binds to regions of KPNA1 that are similar to those of phospho-STAT1, thereby blocking STAT1 nuclear import [14, 15]. This panel of KPNA1 mutants has C-terminal deletions,which either eliminate Arm repeat 10 or Arm repeat 10 plus the majority of Arm repeat 9 (Figure 3A and [21]). To test whether VP24 blocks hnRNP C1/C2 binding to KPNA1 by a similar mechanism, we used the same constructs to test their ability to bind hnRNPC1/C2. Either full-length KPNA1 or individual mutants truncated from the C-terminus were expressed in 293T cells, and co-immunoprecipitations were performed to test whether any of these constructs lost the ability to interact with hnRNPC1/C2. Of interest, the Flag-KPNA1 deletion mutant lacking amino acids 425–457 was unable to bind to hnRNPC1/C2, whereas the full-length 538 amino acid protein and all other truncations that either retained the C-terminal region or that had smaller C-terminal truncations were able to interact with hnRNPC1/C2 (Figure 3B). Reid et al [15] also demonstrated that KPNA1 constructs 1–424 and 1–457 are unable to interact with VP24, whereas deletion mutants 1–424, 1–457, and 1–504 do not bind STAT1. A summary of the proteins that interact with each KPNA1 mutant is provided in Table 1. These data indicate that VP24 binds the same region of KPNA1 as phospho-STAT1 and hnRNPC1/C2, suggesting that when VP24 is present, it may outcompete hnRNPC1/C2 for binding to KPNA1.
Figure 3.
hnRNP C1/C2 binding to KPNA1 requires KPNA1 amino acids 424–457. A, Diagram of the KPNA1 truncation mutants used. B, Co-immunoprecipitation of KPNA1 truncation mutants with hnRNPC1/C2. The presence of Flag-KPNA1 proteins and hnRNP C1/C2 was assessed by Western blot. The full length and the mutants are able to interact with hnRNPC1/C2, with the exception of the KPNA1 mutant possessing only amino acids 1–424. C, Western blot analysis of whole cell extracts from the experiment in panel B.
Table 1.
Synopsis of the Ability of KPNA1 C-Terminal Truncation Mutants to Bind to VP24, Phospho-STAT-1, or hnRNPC1/C2 From This and Previous Studies
| KPNA1 Construct | VP24a | Phospho-STAT-1a | hnRNP C1/C2b |
| KPNA1 1-538 (FL) | + | + | + |
| KPNA1 1-424 | – | – | – |
| KPNA1 1-457 | – | – | + |
| KPNA1 1-504 | + | – | + |
| KPNA1 71-538 | + | + | + |
NOTE. a Binding data performed previously [15].
Binding data performed in this study.
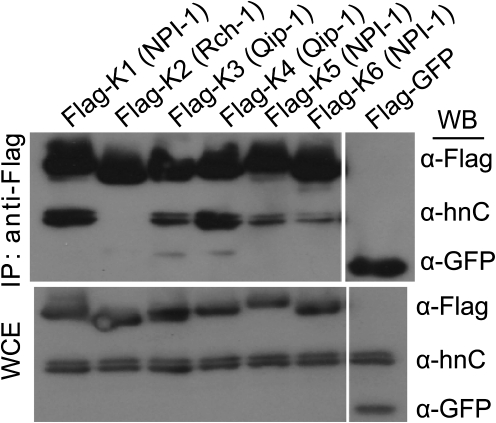
hnRNP C1/C2 Binds to Multiple KPNA Family Members
hnRNP C1/C2 function and localization has been extensively studied, but how it translocates into the nucleus is incompletely defined [35]. Because VP24 has been shown to bind to all members of the NPI-1 subfamily of KPNAs (KPNA 1, 5, and 6), we decided to test which NPI-1 subfamily members bind to hnRNPC1/C2. We therefore transfected 6 of the 7 known KPNA subfamily members into 293T cells and tested for their ability to interact with hnRNPC1/C2 by co-immunoprecipitation studies. hnRNP C1/C2 bound to KPNA 1, 3, 4, 5, and 6 with varying efficiencies (members of the NPI-1 and Qip-1 subfamilies) (Figure 4). However, we did not detect efficient binding of hnRNPC1/C2 to KPNA2, a member of the Rch-1 subfamily (Figures 1 and 4). Because hnRNPC1/C2 can bind to multiple KPNA family members, VP24 may not be able to completely block hnRNPC1/C2 trafficking in cells that express members of both the NPI-I and Qip-I subfamilies. However, it is still possible that VP24 can partially block hnRNPC1/C2 nuclear import.
Figure 4.
hnRNP C1/C2 binds to multiple KPNA subfamily members. The top is a co-immunoprecipitation of 6 of the 7 known human KPNA members and hnRNPC1/C2. The bottom is the whole cell extract corresponding to immunoprecipitated sample. The presence of Flag-KPNA proteins and hnRNP C1/C2 was assessed by Western blotting. All KPNA1 subfamily members interact with hnRNPC1/C2 with the exception of KPNA2, the sole member of the Rch-1 subfamily.
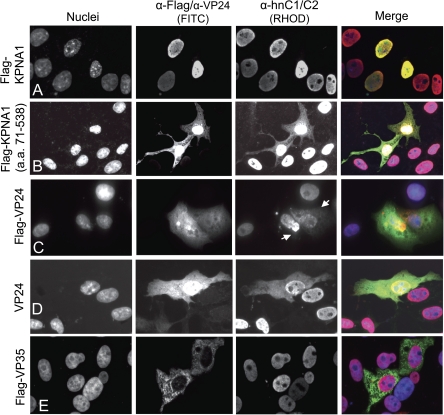
VP24 Expression Partially Relocalizes hnRNP C1/C2 From the Nucleus to the Cytoplasm
We next tested whether hnRNPC1/C2 is redistributed from the nucleus to the cytoplasm in the presence of VP24. Vero cells were transfected with full-length Flag-tagged KPNA1, a KPNA1 deletion mutant missing the importin-β binding domain (71–538), and VP24. Twenty-four to 48 hours after transfection, cells were fixed and stained with either α-Flag, α-hnRNP C1/C2, or α-VP24 antibodies (Figure 5). Because we had shown that KPNA1 interacts with hnRNPC1/C2, we also transfected Flag-tagged KPNA1 and the KPNA1(71–538) as controls for hnRNPC1/C2 redistribution. Overexpressed KPNA1 predominately localized in the nucleus of transfected cells along with hnRNP C1/C2 (Figure 5). In addition, the KPNA1(71–538) mutant, which lacks the importin-β–binding domain and is therefore impaired in its ability to translocate from the cytoplasm to the nucleus, was transfected. This mutant partially redistributed hnRNP C1/C2 to the cytoplasm (Figure 5). Finally, in VP24-expressing cells, hnRNP C1/C2 is partially but reproducibly redistributed (Figure 5). We observed hnRNPC1/C2 redistribution in cells transfected with both untagged and Flag-tagged VP24. Cytoplasmic hnRNPC1/C2 in VP24-transfected cells was observed to be diffusely distributed and in punctuate dots that did not colocalize with the nuclear staining. The significance of these patterns remains to be determined.
Figure 5.
Redistribution of hnRNPC1/C2 from the nucleus to the cytoplasm by VP24 and a KPNA mutant in Vero cells. Indirect immunoflourescense in Vero cells to characterize hnRNPC1/C2 subcellular distribution. A, Overexpressed full length KPNA1 localizes to the nucleus. In these cells hnRNPC also localizes to the nucleus. B, A KPNA mutant that is unable to traffic to the nucleus (encoding amino acids 71–538) redistributes hnRNP C1/C2 to the cytoplasm. C, Flag-VP24 expression partially relocalizes hnRNPC1/C2 from the nucleus to the cytoplasm. D, Untagged VP24 also redistributes hnRNPC1/C2 to the cytoplasm. E. Flag-VP35 does not alter hnRNPC1/C2 nuclear localization to the same extent as either the KPNA1 mutant or VP24.
DISCUSSION
Both previous data [13–15] and our current studies indicate that VP24 binds to KPNA1 and prevents the association of KPNA1 with other cellular factors and impairing their nuclear accumulation. Although disrupting phospho-STAT1 nuclear translocation by VP24 results in an inhibited type I IFN response [13], the significance of redistributing hnRNP C1/C2 from the nucleus to the cytoplasm is not known. We currently hypothesize that EBOV VP24 blocks binding and nuclear import of hnRNP C1/C2 and that this may facilitate EBOV replication in primate cells. Consistent with this hypothesis, hnRNP C1/C2 plays a role in the replication cycle of several other RNA viruses [30–32,36–38].
In the present study, we revealed that hnRNP C1/C2 interacts with KPNA1. Furthermore, VP24 and hnRNP C1/C2 bind to overlapping regions of KPNA1, and VP24 expression blocks hnRNP C1/C2 binding to KPNA1. VP24 and hnRNP C1/C2 (and STAT1) bind to KPNA1 at an atypical site on the far C-terminus, whereas most proteins with a basic NLS enter the nucleus by binding KPNA arm repeats 1–8 [15, 23]. Therefore, our study extends the list of cellular proteins that bind in this atypical fashion to KPNA1. This extends our knowledge of the mechanism by which hnRNP C1/C2 nuclear import occurs.
hnRNP C mRNA is translated into 2 isoforms, referred to as C1 and C2, which form a heterotetramer (3 copies of C1 and 1 copy of C2) [39, 40]. During normal cellular processes, hnRNP C1/C2 localizes in the nucleus and binds unprocessed mRNA both to aid in its stability and to facilitate splicing before the mRNA is exported into the cytoplasm for translation [41]. During mitosis, hnRNP C1/C2 is relocalized to the cytoplasm, where it binds to IRES elements to facilitate cap-independent, IRES-dependent translation [24, 36]. In both situations, hnRNP C1/C2 binding to RNA is sequence specific, such that the protein complex binds to at least 4 consecutive uridine (U) nucleotides [42]. Both EBOV genomic RNA and mRNAs encode multiple poly U tracts, which are potential targets for hnRNPC1/C2 [43, 44].
Redistribution of hnRNP C1/C2 after virus infection has been documented with PV, RV, and VSV [25, 26, 28]. Both PV and RV infection degrade the nuclear pore complex resulting in several hnRNP proteins normally localized in the nucleus to diffuse into the cytoplasm [25, 26]. In contrast, the VSV matrix protein prevents nuclear export of mRNA, a process that results in hnRNP K, A1, and C1/C2 to redistribute from the nucleus into the cytoplasm [28]. Alternatively, EBOV VP24 may prevent entry of newly synthesized hnRNP C1/C2 to the nucleus by blocking its binding to KPNA1. During virus infection, hnRNP C1/C2 binds to RNA of several viruses, including VSV, hepatitis C virus, human papilloma virus, and PV, to facilitate either viral RNA replication or IRES-dependent translation of viral proteins [29, 32, 36, 37]. In addition, hnRNP C1/C2 binds to the dengue virus NS1 protein, but the function of this interaction is not defined [27]. To date, there are no reports describing associations between EBOV viral RNA or EBOV proteins with hnRNP C1/C2. On the basis of our data, it will be interesting to test whether VP24 redistributes hnRNPC1/C2 during filovirus infection and whether altering cellular levels of hnRNPC1/C2 modulate EBOV replication.
Funding
This work was supported by National Institutes of Health (AI059536 and U54 AI 057158, Northeast Biodefense Center-Lipkin to CFB, and 5F32AI084453 to RSS) and the Northeast Center of Excellence for Biodefense and the Emerging Infectious Diseases Research Proteomics Core (U54 AI 057158; PI: Ian W. Lipkin; NBC Proteomics Core PI: EEG).
Acknowledgments
We thank Dr Lawrence Leung (Mount Sinai School of Medicine) for his assistance with the immunoflourescence studies and Dr Viktor E. Volchkov (Claude Bernard University and INSERM) for the α-VP24 antibody.
References
- 1.Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1410–48. [Google Scholar]
- 2.Elliott LH, Kiley MP, McCormick JB. Descriptive analysis of Ebola virus proteins. Virology. 1985;147:169–76. doi: 10.1016/0042-6822(85)90236-3. [DOI] [PubMed] [Google Scholar]
- 3.Licata JM, Johnson RF, Han Z, Harty RN. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J Virol. 2004;78:7344–51. doi: 10.1128/JVI.78.14.7344-7351.2004. 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell. 2002;10:307–16. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- 5.Noda T, Halfmann P, Sagara H, Kawaoka Y. Regions in Ebola virus VP24 that are important for nucleocapsid formation. J Infect Dis. 2007;196:S247–S250. doi: 10.1086/520596. doi:10.1086/520596. [DOI] [PubMed] [Google Scholar]
- 6.Hoenen T, Groseth A, Kolesnikova L, et al. Infection of naive target cells with virus-like particles: implications for the function of Ebola virus VP24. J Virol. 2006;80:7260–4. doi: 10.1128/JVI.00051-06. 10.1128/JVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoenen T, Jung S, Herwig A, Groseth A, Becker S. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology. 2010;403:56–66. doi: 10.1016/j.virol.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J Infect Dis. 2007;196 doi: 10.1086/520582. doi:10.1086/520582. [DOI] [PubMed] [Google Scholar]
- 9.Prins KC, Delpeut S, Leung DW, et al. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–15. doi: 10.1128/JVI.02459-09. 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebihara H, Takada A, Kobasa D, et al. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog. 2006;2:e73. doi: 10.1371/journal.ppat.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volchkov VE, Chepurnov AA, Volchkova VA, Ternovoj VA, Klenk H-D. Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology. 2000;277:147–55. doi: 10.1006/viro.2000.0572. [DOI] [PubMed] [Google Scholar]
- 12.Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis. 1999;179:S248–S258. doi: 10.1086/514292. doi:10.1086/514292. [DOI] [PubMed] [Google Scholar]
- 13.Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol. 2010;84:1169–75. doi: 10.1128/JVI.01372-09. 10.1128/JVI.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid SP, Leung LW, Hartman AL, et al. Ebola virus VP24 binds karyopherin {alpha}1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–67. doi: 10.1128/JVI.02349-05. 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin {alpha} proteins with activated STAT1. J Virol. 2007;81:13469–77. doi: 10.1128/JVI.01097-07. 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chook Y, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–15. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 17.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin [alpha]: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–14. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin {alpha} J Biol Chem. 2007;282:5101–5. doi: 10.1074/jbc.R600026200. 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 20.Miyamoto Y, Imamoto N, Sekimoto T, et al. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–81. doi: 10.1074/jbc.272.42.26375. 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- 21.Kelley J, Talley A, Spencer A, Gioeli D, Paschal B. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010;11:63. doi: 10.1186/1471-2121-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin [alpha] from the nucleus is mediated by a specific nuclear transport Factor. Cell. 1997;90:1061–71. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 23.Fontes MRM, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-{alpha} J Biol Chem. 2003;278:27981–7. doi: 10.1074/jbc.M303275200. 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- 24.Kim JH, Paek KY, Choi K, et al. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol Cell Biol. 2003;23:708–20. doi: 10.1128/MCB.23.2.708-720.2003. 10.1128/MCB.23.2.708-720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustin KE, Sarnow P Inhibition of nuclear import and Alteration of nuclear pore complex composition by rhinovirus. J Virol. 2002;76:8787–96. doi: 10.1128/JVI.76.17.8787-8796.2002. 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gustin KE, Sarnow P. Effects. of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001;20:240–9. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noisakran S, Sengsai S, Thongboonkerd V, et al. Identification of human hnRNP C1/C2 as a dengue virus NS1-interacting protein. Biochem Biophys Res Commun. 2008;372:67–72. doi: 10.1016/j.bbrc.2008.04.165. [DOI] [PubMed] [Google Scholar]
- 28.Pettit Kneller EL, Connor JH, Lyles DS. hnRNPs relocalize to the cytoplasm following infection with vesicular stomatitis virus. J Virol. 2009;83:770–80. doi: 10.1128/JVI.01279-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeifer I, Elsby R, Fernandez M, et al. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc Natl Acad Sci. 2008;105:4173–8. doi: 10.1073/pnas.0711222105. 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ertel KJ, Brunner JE, Semler BL. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J Virol. 2010;84:4229–42. doi: 10.1128/JVI.02198-09. 10.1128/JVI.02198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunner JE, Ertel KJ, Rozovics JM, Semler BL. Delayed kinetics of poliovirus RNA synthesis in a human cell line with reduced levels of hnRNP C proteins. Virology. 2010;400:240–7. doi: 10.1016/j.virol.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunner JE, Nguyen JHC, Roehl HH, Ho TV, Swiderek KM, Semler BL. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J Virol. 2005;79:3254–66. doi: 10.1128/JVI.79.6.3254-3266.2005. 10.1128/JVI.79.6.3254-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid SP, Cárdenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–89. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hossain M, Fuji M, Miki K, Endoh M, Ayusawa D. Downregulation of hnRNP C1/C2 by siRNA sensitizes HeLa cells to various stresses. Mol Cell Biochem. 2007;296:151–7. doi: 10.1007/s11010-006-9308-2. [DOI] [PubMed] [Google Scholar]
- 35.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–73. doi: 10.1083/jcb.134.6.1365. 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gontarek R, Gutshall L, Herold K, et al. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3'NTR of the HCV RNA genome. Nucl Acids Res. 1999;27:1457–63. doi: 10.1093/nar/27.6.1457. 10.1093/nar/27.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokolowski M, Schwartz S. Heterogeneous nuclear ribonucleoprotein C binds exclusively to the functionally important UUUUU-motifs in the human papillomavirus type-1 AU-rich inhibitory element. Virus Res. 2001;73:163–175. doi: 10.1016/s0168-1702(00)00238-0. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Oberg D, Rush M, Fay J, Lambkin H, Schwartz SA. 57-Nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein. J Virol. 2005;79:4270–88. doi: 10.1128/JVI.79.7.4270-4288.2005. 10.1128/JVI.79.7.4270-4288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lahiri DK, Thomas JO. A cDNA clone of the hnRNP C proteins and its homology with the single-stranded DNA binding protein UP2. Nucl Acids Res. 1986;14:4077–94. doi: 10.1093/nar/14.10.4077. 10.1093/nar/14.10.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagawa TY, Swanson MS, Wold BJ, Dreyfuss G. Molecular cloning of cDNA for the nuclear ribonucleoprotein particle C proteins: a conserved gene family. Proc Natl Acad Sci U S A. 1986;83:2007–11. doi: 10.1073/pnas.83.7.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 42.Wilusz J, Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol Cell Biol. 1990;10:6397–407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mühlberger E, Trommer S, Funke C, Volchkov V, Klenk H-D, Becker S. Termini of all mRNA species of Marburg virus: sequence and secondary Structure. Virology. 1996;223:376–80. doi: 10.1006/viro.1996.0490. [DOI] [PubMed] [Google Scholar]
- 44.Feldmann H ME, Randolf A, Will C, Kiley MP, Sanchez A, Klenk HD. Marburg virus, a filovirus: messenger RNAs, gene order, and regulatory elements of the replication cycle. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]