Abstract
Adenoviral (AdV) gene vectors offer efficient nucleic acid transfer into both dividing and non-dividing cells. However issues such as vector immunogenicity, toxicity and restricted transduction to receptor-expressing cells have prevented broad clinical translation of these constructs. To address this issue, engineered AdV have been prepared by both genetic and chemical manipulation. In this work, a polymer-coated Ad5 formulation is optimized by evaluating a series of N-(2-hydroxypropyl) methacrylamide (HPMA)-co-oligolysine copolymers synthesized by living polymerization techniques. This synthesis approach was used to generate highly controlled and well-defined polymers with varying peptide length (K5, K10 and K15), polymer molecular weight, and degradability to coat the viral capsid. The optimal formulation was not affected by the presence of serum during transduction and significantly increased Ad5 transduction of several cell types that lack the Coxsackie and Adenovirus Receptor (CAR) by up to 6-fold compared to unmodified AdV. Polymer-coated Ad5 also retained high transduction capability in the presence of Ad5 neutralizing antibodies. The critical role of heparan sulfate proteoglycans (HSPGs) in mediating cell binding and internalization of polymer-coated AdV was also demonstrated by evaluating transduction in HSPG-defective recombinant CHO cells. The formulations developed here are attractive vectors for ex vivo gene transfer in applications such as cell therapy. In addition, this platform for adenoviral modification allows for facile introduction of alternative targeting ligands.
1 Introduction
Adenovirus serotype 5 (Ad5) has been used extensively for gene delivery due to the ease with which it can be produced at high titer and its high transduction efficiency in many different cell types, both dividing and nondividing (1, 2). In humans, cell types that can be transduced by Ad5 include the heart, lung, liver, pancreas, central and peripheral nervous system, prostate, testis, and intestine (3–5). Efficient cell entry by Ad5 depends on high affinity interactions between the virus and the Coxsackievirus and Adenovirus Receptor (CAR) as well as with αV integrins, both of which are important for adenoviral transduction (6–9). Ad5 interacts with CAR through its globular homotrimeric fiber knob (4, 8–11). After this initial fiber knob-CAR interaction, the penton base pentamer binds to the αvβ3 and αvβ5 integrins through arginine-glycine-aspartic acid (RGD) sequences, resulting in integrin clustering and AdV cell entry (8, 11–13). However, certain cell types of interest for gene delivery, such as primary cancer cells and hematopoietic stem cells, have low expression levels of CAR and αV integrins on the surface, which can lead to poor transduction with Ad5 at lower multiplicity of infection (MOI) (14, 15). To address this challenge, alternative approaches to redirect Ad5 for CAR-independent cellular adhesion and internalization are important.
Synthetic materials such as cationic lipids and cationic polymers have been used as complexation reagents with AdV to improve cellular uptake (16–26). Most notably, Fasbender and colleagues complexed several commercially available cationic lipids and cationic polymers with Ad2 and showed increased cell transfection efficiencies for most formulations (16). Most approaches have used off-the-shelf materials such as PLL, polybrene, or PEI (16, 20, 22). To date, a detailed investigation to optimize cationic materials used to potentiate AdV infection of CAR-negative cells has, to our knowledge, not been reported.
We recently reported the synthesis of N-(2-hydroxypropyl) methacrylamide (HPMA)-peptide copolymers using a living polymerization technique, Reversible Addition-Fragmentation Chain Transfer (RAFT) polymerization (27). In RAFT polymerization, a chain transfer agent (CTA) is used in the presence of radical polymerization, which limits the growth of the polymer chains due to its higher reactivity with the free radical in comparison to the monomer (28). This characteristic of RAFT results in relatively monodisperse materials with controlled and facile incorporation of several different water-soluble peptides. Additional advantages include reproducibility of synthesis and the ability to produce well-characterized materials. Due to the hydrophilic HPMA backbone, these materials also impart colloidal stability when incorporated into nanoparticle formulations (27, 29). In this work, a series of HPMA-co-oligolysine materials with varying polymer molecular weight, oligolysine peptide length, and degradability were evaluated for application in Ad5 complexation and and interaction with heparan sulfate proteoglycans (HSPGs) was also investigated.
2 Materials and Methods
2.1 Virus
Adenovirus serotype 5 encoding for eGFP (Ad5-GFP) with known viral particle concentration and transducing unit concentration was purchased from UNC Vector Core (Gene Therapy Center UNC, Chapel Hill NC) or prepared as described previously (30, 31). Alexa Fluor 568-labeled Ad5-GFP was prepared by labeling 1 × 1012 vp of Ad5-GFP with five-fold excess of Alexa Fluor 568 succinimidyl ester (Invitrogen, Carlsbad, CA) with respect to the surface lysines present on the capsid (assumed to be 18,000 from (32)). The reaction was performed in sodium bicarbonate buffer (final concentration 50 mM, pH 8.0) at room temperature for one hour. Tris (tris(hydroxymethyl)aminomethane) buffer was added to a final concentration of 50 mM (pH 8.0) and allowed to react for one hour at room temperature to quench any unreacted amines. Viruses were purified using a PD-10 column (GE Biosciences, Piscataway, NJ). All viruses were stored at −80° C until use.
Viral particle concentration for Alexa Fluor 568-labeled Ad5-GFP was determined by PicoGreen fluorescence assay as reported previously with minor modification (33). Virus with known viral particle concentration was diluted 1:100 in Tris-EDTA (TE) buffer (10 mM Tris, 1 mM EDTA, pH 8.0). Sample virus was diluted 1:20, 1:50, and 1:100 with TE buffer. Viruses were denatured at 56°C for 30 minutes to release DNA from the viral capsid. After cooling to room temperature, 0.5% SDS in TE buffer was added to each virus sample. Virus with known viral particle concentration was diluted 1:2 to create a standard curve with known concentrations. 100 µL of 1:200 dilution of PicoGreen stock solution (Invitrogen, Carlsbad, CA) was added to virus samples in black 96-well plates and incubated for 2–3 minutes at room temperature before taking fluorescence measurements (Ex480 nm/Em520 nm) on a Safire2 plate reader (Tecan, Durham, NC).
2.2 Cell lines
HeLa cells (Human cervical carcinoma, ATCC CCL-2) were cultured in MEM medium supplemented with 10% fetal bovine serum (FBS), and 1% antibiotic/antimycotic solution. CHO-K1 cells (Chinese Hamster Ovary cells, ATCC CCL-61) were maintained in F12K medium supplemented with 10% FBS, and 1% antibiotic/antimycotic solution. CHO mutant cells CHO-pgs-A745 (34) (ATCC CRL-2242) and CHO-pgs-E606 (35) (ATCC CRL-2246) were cultured in CHO-K1 cell medium supplemented with 200 µM L-asparagine and 200 µM L-proline. BaF3 cells (mouse pro-B cells) were maintained in RPMI supplemented with 10% FBS and 1% antibiotic/antimycotic, with the addition of a final concentration of 5 ng/mL IL-3 for cell proliferation. RAW 264.7 cells (mouse macrophage cell line, ATCC TIB-71) were cultured in Dulbecco’s Minimum Essential Medium (DMEM) supplemented with 10% FBS, and 1% antibiotic/antimycotic solution.
2.3 Polymer Synthesis
2.3.1 Synthesis of peptide monomers
Oligolysine peptides with 5, 10 and 15 lysines (K5, K10, and K15 respectively) were synthesized with the stable linker, 6- aminohexanoic acid (Ahx). K10 was also synthesized with the reducible linker AEDP (3-[(2-aminoethyl)dithio]propionic acid·HCl). Cathepsin B-sensitive peptides used the cathepsin B substrate FKFL (phenylalanine-lysine-phenylalanine-leucine) flanked on both sides by an Ahx spacer as a linker, and peptides were synthesized using (d) and (l) amino acids and 6-aminohexanoic acid (Ahx): AhxFKFLAhxK10 (composed of only (l) amino acids); and AhxFKFLAhx(D)K10 (composed of (l) amino acid linker with oligo-D-lysine). Peptides were synthesized on a solid support with Rink amide resin following standard Fmoc/tBu chemistry on an automated PS3 peptide synthesizer (Protein Technologies, Phoenix, AZ). Prior to peptide cleavage from the resin, the amino terminus of the peptides were deprotected and coupled with N-succinimidyl methacrylate, providing methacrylamido functionality. These functionalized peptide monomers are respectively called MaAhxFKFLAhxK10 and MaAhxFKFLAhx(D)K10. Synthesized peptides were cleaved from resin by treatment of the solid support with a solution of TFA/triisopropylsilane (TIPS)/1,3-dimethoxybenzene (92.5:2.5:5, v/v/v) for 2.5 hours under gentle mixing. Cleaved peptide monomers were precipitated in cold ether, dissolved in methanol and re-precipitated twice in cold ether. Each peptide monomer was analyzed by RP-HPLC and MALDI-TOF MS and shown to have greater than 95% purity after cleavage.
2.3.2 RAFT copolymerization of HPMA and peptide monomers
HPMA-co-oligolysine polymers were synthesized as described previously (27, 36, 37). In brief, monomers (0.7 M in 1M acetate buffer, pH 5.1), RAFT chain transfer agent (CTA) ethyl cyanovaleric trithiocarbonate (ECT, molecular weight 263.4 g/mol) and VA-044 initiator (I) were dissolved together and polymerized at 44°C in air-free conditions for 48 hours. The molar ratios of total monomero:CTAo:Io at the start of polymerization were 190:1:0.1 for DP of 190, 150:1:0.1 for DP 150, 100:1:0.1 for DP 100, and 50:1:0.1 for DP 50. Polymers were dialyzed against distilled H2O to remove unreacted monomers and buffer salts before lyophilization.
2.3.3 Polymer characterization
Molecular weight analysis was carried out by size exclusion chromatography (SEC) in 150 mM acetate buffer, pH 4.4. Analysis was carried out on an OHpak SB-804 HQ column (Shodex) in line with a miniDAWN TREOS multi-angle light scattering detector (Wyatt, Santa Barbara, CA) and an OptiLab rEX refractive index detector (Wyatt). Absolute molecular weight averages (Mn, Mw) were calculated using ASTRA software (Wyatt). The actual incorporated amount of peptide and HPMA in the final copolymers was determined through modified amino acid analysis following the method of Bidlingmeyer and coworkers (38). Briefly, hydrolyzed lysine and HPMA (which results in 1-amino-2-propanol) were derivatized with o-phthalaldehyde/β-mercaptopropionic acid and run on a poroshell 120 EC-C18 (Agilent Technologies, Santa Clara, CA) HPLC column with pre-column derivatization to label hydrolyzed lysine and 1-amino-2-propanol. Calibration curves were generated using serial dilutions of (l)-lysine and reagent grade 1-amino-2-propanol.
2.3.4 Labeling of HPMA-co-MaAhxK10 with Alexa Fluor 488
Polymer was dissolved in phosphate buffered saline (PBS, pH 7.4), and 5 mole equivalents of TCEP (Tris (2-carboxyethyl)phosphine) was added to reduce the trithiocarbamate polymer termini and mixed with stirring. Two mole equivalents of Alexa Fluor 488 maleimide (Invitrogen, Carlsbad CA) were added, and the reaction was stirred at room temperature overnight in the dark. Unreacted dye was purified with a PD-10 column.
2.4 Preparation of polymer-coated virus and virus characterization
Polymer solutions (dissolved in dH2O) were added to virus solutions to achieve desired polymer to viral particle mole ratios in a total of 25 µL. After addition of polymer solution to virus, solution was incubated at room temperature for 30 minutes prior to further studies. The zeta potential of unmodified virus and polymer-coated virus was determined using dynamic light scattering (Brookhaven Instruments Zeta PALS analyzer, Holtsville, NY). Unmodified virus was diluted to a concentration of 1 × 1011 vp/mL, and polymer-coated virus was diluted to 6 × 1010 vp/mL. Samples were performed with 15 runs in triplicate.
2.5 Cell transduction
Cells were seeded in 24-well plates (40,000 cells/well) and allowed to incubate overnight. After incubation, adenovirus or freshly-prepared polymer-coated virus solutions were further diluted with complete culture medium (medium + 10% FBS + 1% Ab/Am) or culture medium without serum (medium + 1% Ab/Am) to a final volume of 300 µL per well. Multiplicity of infection (MOI) was calculated based on PFU/mL. Cells were washed one time with PBS before virus solutions were added. Virus solutions were incubated overnight and 1 mL of complete culture medium was added to each well the following day. 24 hours after additional culture medium was added, cells were washed one time with PBS before trypsinization. Cells were treated with 2 µg/mL propidium iodide for 5 minutes at 4°C or 7-AAD (7-amino-actinomycin D) as used according to manufacturer’s instructions for 10 min at room temperature before washing twice and resuspension in cell culture medium. Samples were performed in triplicate and were kept on ice prior to conducting flow cytometry. Flow cytometry was performed on a BD FACScan system in the UW Cell Analysis Facility.
2.6 Virus internalization studies
Cells were seeded at 80,000 per well in a 24-well plate and allowed to incubate overnight. Polymer-coated virus at a polymer to virus ratio of 5000 using Alexa Fluor 568-labeled Ad5-GFP were formulated as performed previously. Polymer-coated virus using Alexa Fluor 568-labeled Ad5-GFP were diluted in cell culture medium and added to cells at multiplicity of infection (MOI) of 50 (vp/cell) based on comparable viral particle numbers as unlabeled Ad5-GFP in a total volume of 300 µL. After a 30 minute incubation at 37°C, 5% CO2, virus solution was removed and cells were washed once with PBS. Cells were then trypsinized and washed one time with cell culture medium and kept on ice prior to conducting flow cytometry. Samples were performed in triplicate.
2.7 Transmission Electron Microscopy (TEM)
Unmodified and polymer-coated virus were diluted to 8.3 × 1011 vp/mL after formulation and deposited on silicon nitride support films (50 nm membrane thickness, 200 µm frame thickness, 0.25 × 0.25 mm window, Ted Pella, Redding CA). Samples were deposited on grids and incubated at room temperature for 15 minutes. Virus solutions were then wicked away, and grids were washed with PBS followed by double distilled water. 1% uranyl acetate (pHK10-Ad5) or 1% methylamine tungstate (Ad5) was then deposited on the grid and incubated for 1 minute, wicked away, and then deposited again for another 2 minute incubation. The final negative stain solution was wicked away, and the grids were allowed to air dry. TEM images were taken on an FEI Tecnai G2 F20 instrument at 200 kV, 17,000× and 39,000×.
2.8 Transduction of Ad5 or pHK10-Ad5 in the presence of neutralizing anti-Ad5 antibodies
HeLa cells were seeded at 20,000 cells/well in a 24-well plate 24 hours before transduction. CHO-K1 cells were seeded 40,000 cells/well in a 24-well plate 16 hours before transduction. pHK10-Ad5 were formulated as described previously at a polymer to virus ratio of 5000:1. Human serum containing AdV neutralizing antibodies (NAb) was diluted 1:20 in PBS and heated to 56°C for 20 minutes to inactivate complement. Diluted serum (100 µL) was then incubated with Ad5 or pHK10-Ad5 at 37°C for 20 minutes before culture medium was added to a final volume of 300 µL per sample. Cells were washed once with PBS before virus solutions were added. After overnight incubation, 1 mL of culture medium was added, and samples were assayed for transduction 24 hours later via flow cytometry.
Results
Optimization of polymer-coated adenovirus
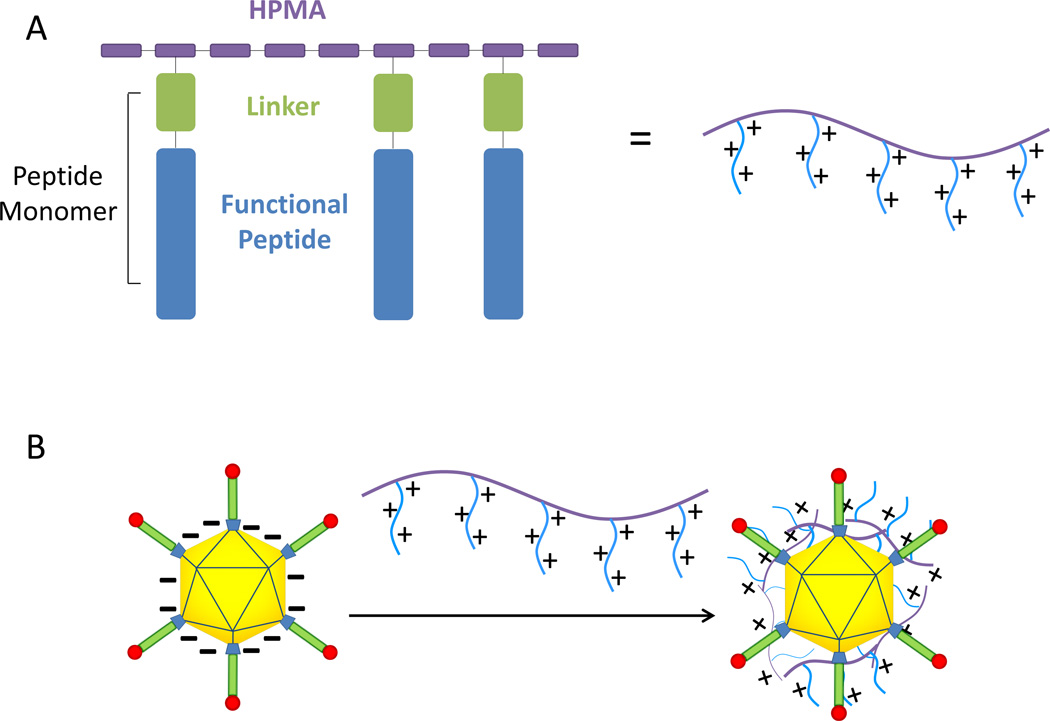
A series of HPMA-co-oligolysine polycations synthesized by RAFT polymerization (Table 1) were evaluated as surface-modification materials for Ad5. The polymer architecture (Scheme 1A) offers several parameters for optimization: pendant peptide length, overall polymer molecular weight, and polymer degradability at the linker site. Polymer-coated Ad5 was prepared by mixing polycation solutions with Ad5 solutions, resulting in polymer adsorption to viral capsid (Scheme 1B).
Table 1.
Physical properties of HPMA-co-oligolysine copolymers
| HPMA Oligolysine Copolymer |
Lysine Peptide Monomer |
Degree of Polymerization (DP) |
Mw/Mn | Degradability |
|---|---|---|---|---|
| pHK5 | MaAhxK5 | 190 | 1.17 | Exopeptidase |
| pHK10 | MaAhxK10 | 190 | 1.18 | Exopeptidase |
| pHK15 | MaAhxK15 | 190 | 1.27 | Exopeptidase |
| pHK10 DP 50 | MaAhxK10 | 50 | 1.09 | Exopeptidase |
| pHK10 DP 100 | MaAhxK10 | 100 | 1.10 | Exopeptidase |
| pHK10 DP 150 | MaAhxK10 | 150 | 1.14 | Exopeptidase, |
| pHSSK10 | MaAEDPK10 | 190 | 1.11 | Reducible exopeptidase |
| pH(d)K10 | MaAhx(d)K10 | 190 | 1.13 | None |
| pHCathK10 | MaAhxFKFLAhxK10 | 190 | 1.17 | Endopeptidase, Exopeptidase |
| pHCath(d)K10 | MaAhxFKFLAhx(d)K10 | 190 | 1.16 | Endopeptidase |
Schematic.
HPMA-peptide copolymers used for adenovirus polymer coating (A). Linkers can be nondegradable (Ahx), reducible (AEDP), or enzyme cleavable (AhxFKFLAhx). Negatively charged adenovirus interact with cationic HPMA-oligolysine polymers to form a polymer-coated virus (B).
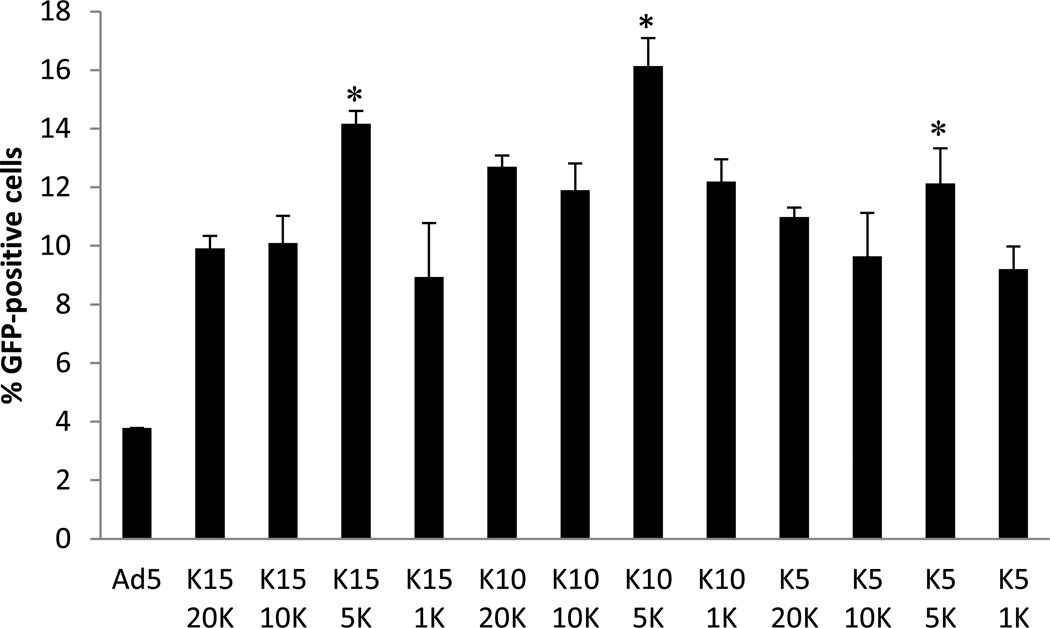
First, the effect of oligolysine length and polymer to virus ratio was determined by transfecting CAR-negative RAW 264.7 cells with coated Ad5. HPMA copolymers incorporating oligolysine of various lengths (K5, K10, K15) with a fixed degree of polymerization (DP=190) were tested at polymer to virus ratios 1k, 5k, 10k and 20k. For all three polymers, optimal transduction was obtained at a 5000 polymer: virus ratio. pHK10-Ad5 virus showed the highest transduction, resulting in 16% GFP positive cells, 4.3 -fold higher than unmodified virus, and less than 3% cell death, as assessed by propidium iodide (PI) staining (Figure 1).
Figure 1.
Transduction of RAW 264.7 cells with polymer-coated adenovirus. Adenovirus was coated with pHK15, pHK10, or pHK5 at different polymer to virus ratios (20K = 20,000:1, 10K = 10,000:1, 5K = 5000:1, 1K = 1000:1). * denotes p < 0.03 with respect to pHK10 5K. pHK5 5K showed statistically higher transduction than pHK510K, pHK51K and virus only (p<0.05). pHK15 5K showed statistically higher transduction than pHK15 20K, pHK15 1K and virus only (p<0.005). Student’s t-test was used to determine statistical significance.
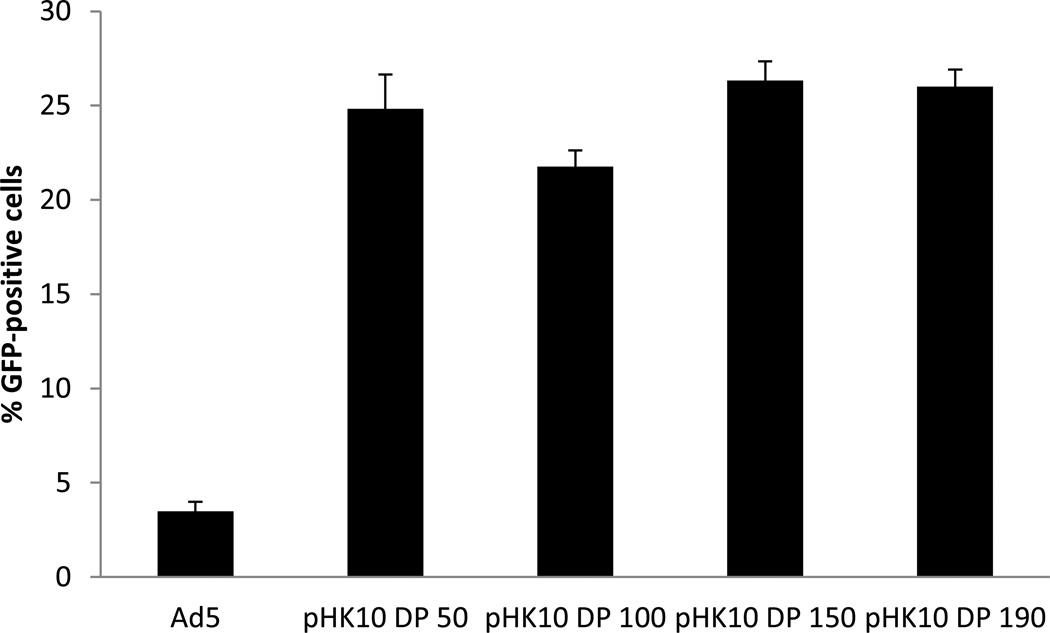
Next, the molecular weight of pHK10 was varied. Polymers were synthesized with target DP of 50, 100, 150 and 190 by adjusting the CTA to monomer ratio, resulting in polymers with molecular weights of 23 kDa, 44.91 kDa, 65.71 kDa, and 77.6 kDa, respectively. These polymers were used for Ad5 coating at the optimized 5000:1 polymer to virus ratio, and transduction efficiency to RAW 264.7 cells was compared (Figure 2). No significant difference was observed between pHK10 DP 50-, pHK10 DP 150-, and pHK10 DP190-Ad5 samples (transfection efficiency ~25%, 6-fold higher than unmodified virus, all polymers showing less than 6% PI positive cells).
Figure 2.
Transduction efficiency of pHK10-coated adenovirus at different degrees of polymerization. Polymer-coated viruses were formulated at MOI of 50, with transductions performed in RAW 264.7 cells. No statistical significance was found between pHK10 DP 190 and pHK10 DP 50 or pHK10 DP 150. pHK10 DP 190 showed statistically higher transduction than virus only and pHK10 100 (p<0.004). Statistical significance was based on student’s t-test.
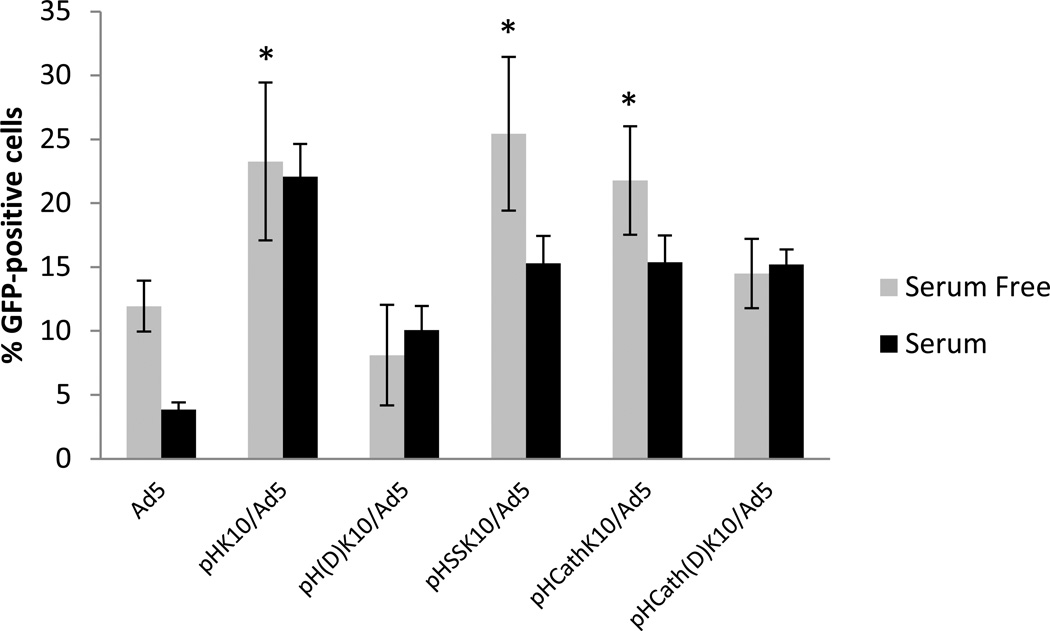
Finally, the effect of linker degradability on Ad5 transduction was assessed. Two degradable linkages were used: AEDP, an analog of the Ahx linker in pHK10 that contains a disulfide bond and a cathepsin B-cleavable linker, FKFL, flanked by Ahx spacers. These polymers are respectively called pHSSK10 and pHCathK10. Since (l)-amino acids can also be degraded by peptidases, (d)-amino acid analogues of pHK10 (pH(d)K10) and pHCathK10 (pHCath(d)K10) were also synthesized. Transductions were conducted in serum-free and 10% serum conditions under the optimal 5000:1 polymer: virus ratio (Figure 3). Serum significantly decreased transduction efficiency of unmodified Ad5 but not pHK10-, pH(d)K10-, nor pHCath(d)K10-coated Ad5. Transduction efficiency of pHSSK10- and pHCathK10-Ad5 was reduced by 40% and 30%, respectively, in serum conditions. pH(d)K10- and pHCath(d)K10-Ad5 did not impact Ad5 transduction efficiency in serum conditions. Transduction efficiency of pHK10-, pHSSK10-, and pHCathK10 -Ad5 were similar in serum-free conditions, increasing transduction by up to 113%.
Figure 3.
Transduction efficiency of pHK10-coated adenovirus with reducible linker (AEDP) and Cathepsin-B cleavable linker (AhxFKFLAhx) in serum free or serum-containing medium. Ad5, pHSSK10/Ad5, and pHCathK10/Ad5 showed statistically higher transduction in serum free conditions compared to serum conditions (p<0.008). *In serum free conditions, pHK10/Ad5, pH(d)K10/Ad5, and pHCathK10/Ad5 showed significantly higher transduction than Ad5, pH(d)K10/Ad5, and pHCath(d)K10/Ad5 (p<0.003), but no difference from each other. Ad5, pH(d)K10/Ad5, and pHCath(d)K10/Ad5 also showed no significant difference from each other in serum free conditions when p = 0.01.
Characterization of pHK10-coated adenovirus
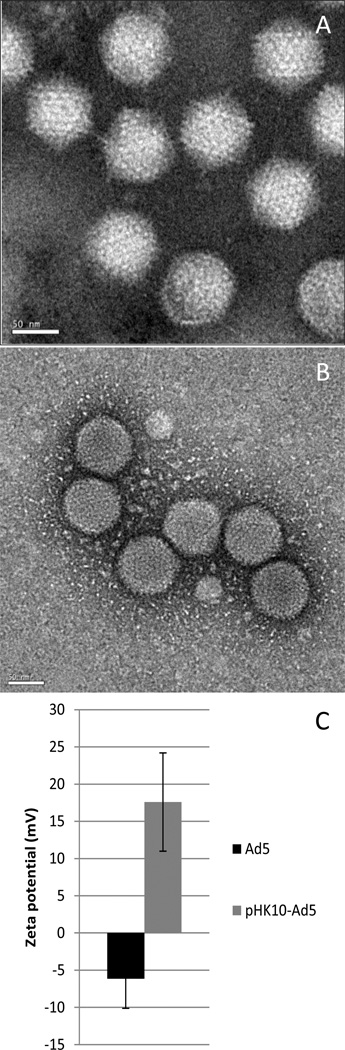
Ad5 and pHK10-Ad5 were characterized by transmission electron microscopy (TEM) to determine virus morphology and zeta potential measurements to determine surface charge after polymer coating (Figure 4). TEM imaging revealed that both Ad5 and pHK10-Ad5 had average diameters of 78.2 and 82.0 nm, respectively. Zeta potential measurements revealed that Ad5 had negative surface charge (−6 mV) while pHK10-Ad5 were positively charged (+17.6 mV).
Figure 4.
TEM (A, B) and zeta potential (C) of Ad5 and pHK10-Ad5. TEM images were conducted on silicon nitride membranes, with Ad5 stained with methylamine tungstate and pHK10-Ad5 stained with uranyl acetate. Images were taken at 39,000×. Scale bar represents 50 nm.
Ad5 and pHK10-Ad5 transduction in CAR-negative cell lines
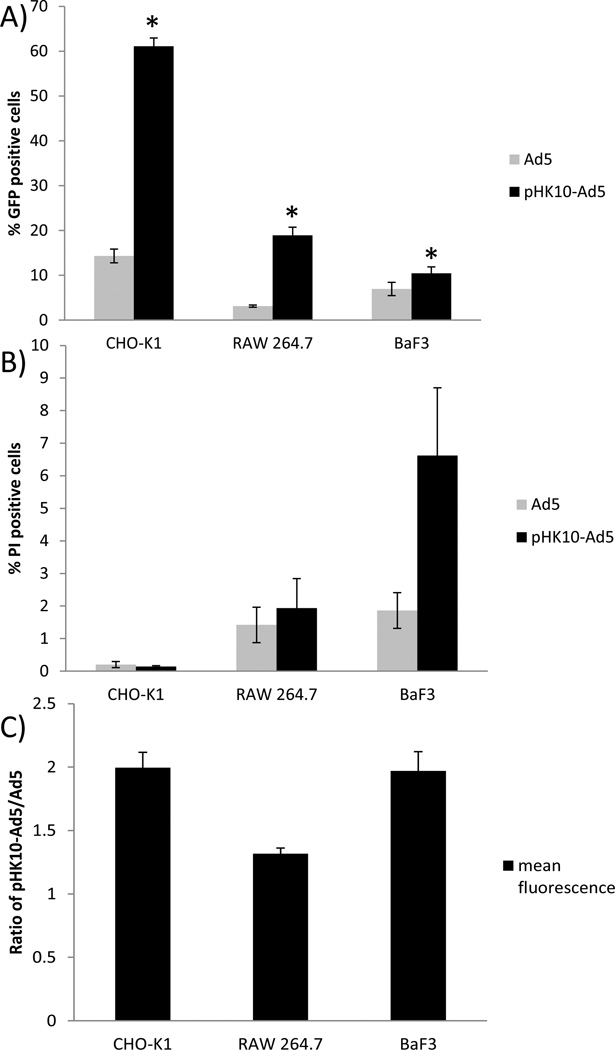
The transfection efficiency of the optimized pHK10-Ad5 and Ad5 was then compared in two additional CAR-negative cell lines, adherent CHO-K1 and suspension BaF3 cells (Figure 5A). Again, RAW cells showed over a 6-fold increase in transduction efficiency over naked virus using MOI 50 (18.9% vs 3.1% GFP-positive). A 4-fold increase in transduction efficiency was achieved with CHO-K1 cells using MOI 5 (61.1% vs 14.3% GFP-positive) and BaF3 cells showed a 1.5-fold increase at MOI 1000 (10.4% vs. 6.9% GFP-positive). Low cytotoxicity was observed for both polymer-coated and unmodified Ad5 in all three cell types (PI-positive <7%, Figure 5B). The mean GFP fluorescence levels in the gated transduced cell population was 2-fold, 1.3-fold, and 2-fold greater for CHO-K1, RAW 264.7, and BaF3 cells, respectively, for pHK10-Ad5 compared to naked Ad5 (Figure 5C).
Figure 5.
Transduction efficiency (A), cytotoxicity (B), and ratio of GFP positive mean fluorescence (C) of pHK10-Ad5 compared to Ad5 in three different CAR negative cell lines. Virus was used at MOI of 5, 50, and 1000 for CHO-K1, RAW, and BaF3 cells respectively. * pHK10-Ad5 showed statistically higher transduction compared to Ad5 in all cell types (p<0.05). There was no statistical difference in cell cytotoxicity between Ad5 and pHK10-Ad5 within each of the cell lines. pHK10-Ad5 showed significantly higher toxicity in BaF3 cells compared to RAW 264.7 and CHO-K1 cells (p<0.05).
Ad5 and pHK10-Ad5 transduction in the presence of neutralizing antibodies (NAb)
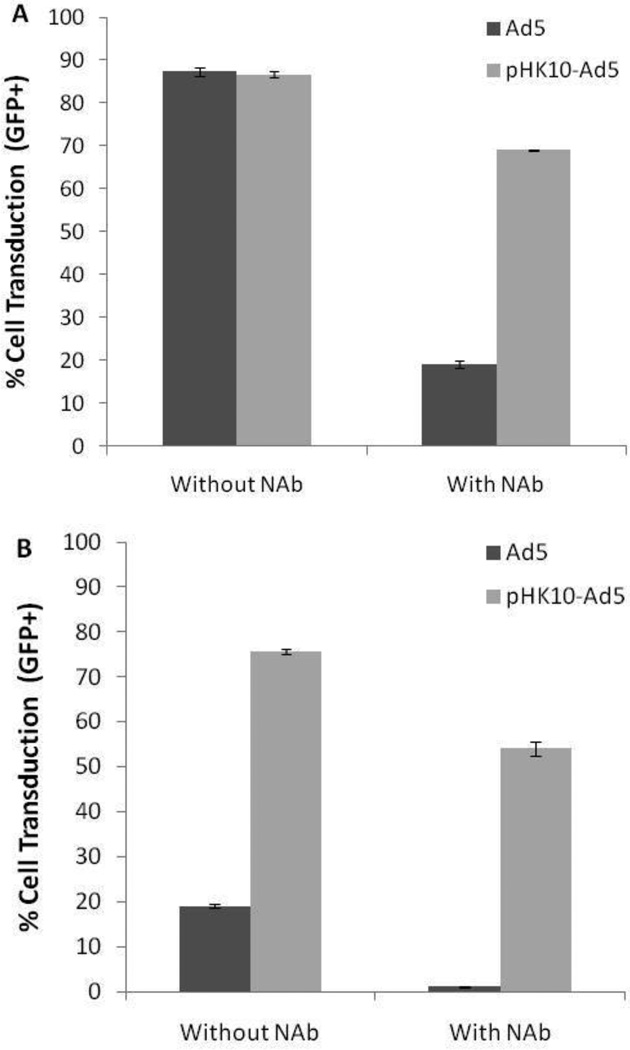
Human serum samples were first analyzed using a neutralization assay to determine the inhibition of adenoviral transduction by Ad5-specific NAb in serum diluted from 1:5 to 1:160. A corresponding decrease in Ad5 transduction was observed with decreasing serum dilution, thus confirming the presence of NAb (Supplementary Figure 1). Ad5 and pHK10-Ad5 pre-incubated with the NAb-containing human serum at 1:20 dilution were used to transduce CAR-positive HeLa and CAR-negative CHO-K1 cells at MOI=10 (Figure 6). Transduction to HeLa cells in the absence of NAb was efficient for both Ad5 and pHK10-Ad5 (87% GFP+). While NAb significantly inhibited Ad5 transduction (19% GFP+), polymer coating protected the virus against NAb binding (69% GFP+). Ad5 transduction of CHO-K1 cells decreased by ca. 95% in the presence of NAb (19% GFP+ to 1% GFP+) while NAb only decreased pHK10-Ad5 transduction in these cells by ca. 30% (76% GFP+ to 54% GFP+).
Figure 6.
Ad5 and pHK10-Ad5 transduction of CAR-positive HeLa (A) and CAR-negative CHO-K1 (B) cells in the presence and absence of neutralizing antibodies (NAb).
Virus uptake in HSPG-mutant cells
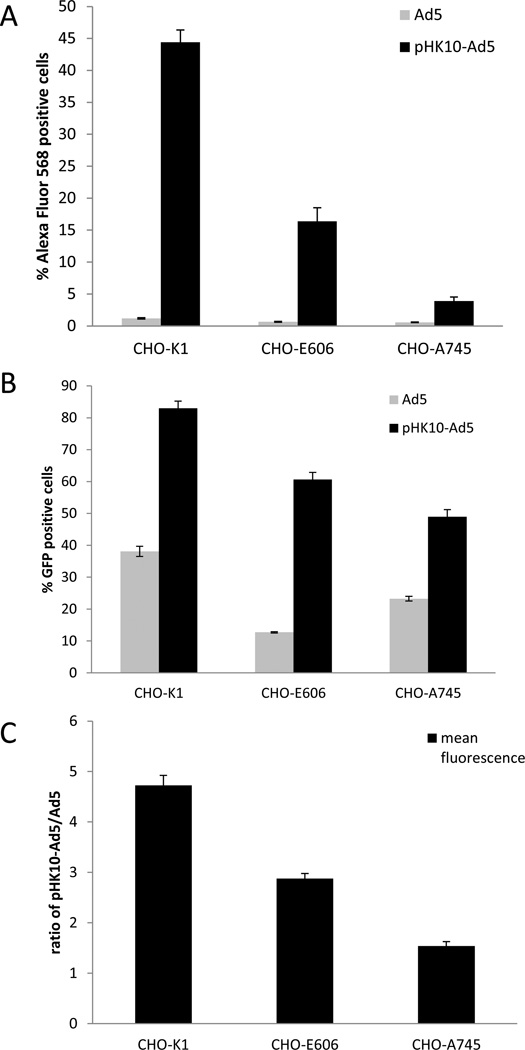
Virus internalization in engineered CHO-K1 cell lines with disrupted HSPG expression was assessed. CHO-pgs-E606 have undersulfated HSPGs, while CHO-pgs-A745 do not express HSPGs. Unmodified and pHK10-coated, Alexa Fluor 568-labeled Ad5 were exposed to cells for 30 minutes followed by flow cytometry analysis for cell association (Figure 7A). Uptake of unmodified Ad5 was low in all CHO cells (<1.5%). In contrast, significant cell association was observed with pHK10-coated Ad5 in CHO-K1 cells (44% of CHO-K1 cells associated with virus). The percent of cell-associated Ad5 decreased by 63% and 91% respectively in CHO-pgs-E606 (16.4%) and CHO-pgs-A745 cells (3.9%). All CHO cells showed slightly lower mean fluorescence in pHK10 coated Ad5 compared to unmodified Ad5 (Figure 7B).
Figure 7.
Cell uptake (A) and transduction (B, C) of pHK10-Ad5 and Ad5 in CHO-K1 and CHO mutant cell lines. Percent cells that are Alexa Fluor 568 or GFP positive (A, C) and the fluorescence ratio of pHK10-Ad5 to Ad5 (C) for the diffe rent cell types is shown. MOI 5 was used for all cells.
Transduction efficiency in these cells lines was assessed two days after Ad5 incubation and similar trends to cellular association were obtained. CHO-K1 cells were efficiently transduced by pHK10-Ad5 (83%) and lower transduction efficiencies were observed with CHO-pgs-E606 cells (60.6) and CHO-pgs-A745 cells (49.0%) (Figure 6C). CHO-K1 also showed the greatest fold increase in mean fluorescence with pHK10-Ad5 over Ad5 (4.7-fold), followed by CHO-pgs-E606 (2.9-fold), and the CHO-pgs-A745 cells (1.5-fold).
Discussion
Ad5 transduces cells mainly through interaction of the fiber protein with the Coxsackie and Adenovirus Receptor (CAR) and αV integrins present on cell surfaces (6, 13). Previous studies have shown that physical mixtures of AdV with cationic polymers or lipids improve AdV transduction to CAR-negative cells (19, 21, 23). The goal of this work was to systematically investigate the effects of polycation structure, molecular weight and degradation on Ad5 transduction. We previously demonstrated that these parameters could be independently controlled by RAFT polymerization of HPMA-co-oligolysine polymers (36). Here, ten HPMA-co-oligolysine polymers were evaluated for their relative ability to increase transduction efficiency of Ad5 by electrostatic coating of the virus.
First, the effect of oligolysine length was probed (Figure 1). All polycations (pHK5, pHK10, and pHK15) increased gene delivery efficiencies to CAR-negative RAW 264.7 cells over unmodified Ad5 when complexed with the vector. The optimal polymer to Ad5 ratio identified for all three polymers was 5000:1. Because the mol% lysine was kept relatively constant between the three polymers, the optimal average lysine per adenovirus (2 × 106) was also similar for all three polymers. This trend does not hold in CAR-positive cells or PC-12 cells; in these cells, the transduction efficiency of coated Ad5 decreases both with increasing peptide content in the polymer (mol% peptide) and with polymer to virus ratio (data not shown), possibly due to blocking of the CAR-binding domain in the fiber knob.
After selecting K10 as the optimal peptide length, the effect of polymer length was tested by synthesizing pHK10 polymers with molecular weights ranging from 23kDa-78kDa (Figure 2). There was no significant difference in transduction efficiency or toxicity among the four different molecular weights tested. Fasbender et al. also investigated coating Ad2 with poly(l)lysine of various molecular weight (9k, 25k, 55k and 233k), PEI, histone and spermine for transducing COS-1 cells and found that all agents except spermine similarly improved Ad2 transduction (16). The optimal PLL was 55k (~125k lysines per adenovirus) although the variation in luciferase activity only ranged between 1 to 5 million RLU for the tested range. These results show that a minimum polyvalency is likely required for high affinity virus binding, but that once this criterion is met, there is a broad range of polymer lengths that are effective for AdV coating for in vitro transductions. However, the effect of polymer MW may become critical in in vivo conditions where other proteins may displace weakly bound polymers. This phenomena has been reported in vivo for non-viral systems (39, 40). The lack of correlation between transduction efficiency and polymer MW within the tested MW range contrasts with our observation from non-viral transfections using these materials, where higher molecular weight cationic polymers have shown to be more cytotoxic than lower molecular weight polymers (41). One possible explanation is that cytotoxicity was correlated with polyplexes (cationic polymer/plasmid DNA complexes) formulated with higher molecular weight and higher concentrations of polycations while the amount of polymer used for Ad5 coating is approximately 5000-fold lower.
Finally, the effect of polymer degradability was assessed. A polymer with reducible linkers, pHSSK10, and two polymers with enzymatically-cleavable linkers, pHCathK10 and pHCath(d)K10 were synthesized and tested as viral coatings. Ideally, the adsorbed polymer would not affect Ad5 trafficking after cellular internalization, and intracellular polymer degradation would facilitate displacement of materials from the adenoviral capsid. Disulfide bond reduction has been reported to occur in the endosome for some systems (42) and cathepsin B, a cysteine protease, functions primarily in the endo/lysosomal compartments (43, 44). In addition, the pendant peptides synthesized with (l)-amino acids are susceptible to exopeptidase digestion by serum proteases (45). In serum-free conditions, all polymers synthesized with only HPMA and (l)-amino acids (pHK10, pHSSK10 and pHCathK10) increased Ad5 transfection efficiency when used as a coating, regardless of the presence of a degradable linker (Figure 3).
The reducible pHSSK10 polymer, when used directly as a non-viral gene delivery material, transfects poorly compared to the pHK10 polymer. This effect was attributed in part to the instability of the disulfide bond possibly due to metal-catalyzed oxidation and crosslinking of reduced polymer (37). When used as a virus coating, the concentration of polymers is ~5000-fold lower so that polymer crosslinking is less likely to occur. The pHCathK10, pHCath(d)K10, and pH(d)K10 polymers transfect cells with similar efficiency to pHK10 when used directly in polyplex formulation (unpublished data). When applied as an Ad5 coating, pHCathK10 has a similar effect on Ad5 serum free transfection efficiency as pHK10. In contrast, the polymers containing (d)-amino acids had no significant effect on Ad5 transfection in serum-free conditions. We previously showed that HPMA-co-oligolysine copolymers with (d)-lysine are stable against enzymatic degradation whereas copolymers with (l)-lysine undergo exopeptidase degradation. Therefore, polymer degradation is critical for efficient Ad5 transfection but the mechanism (disulfide reduction or enzymatic hydrolysis) and degradation site (linker region or peptide carboxy terminus) is not as important. Degradation may facilitate polymer uncoating of virus after cellular internalization, thereby minimizing further disruption to the adenovirus infection pathway. For example, defensins are antimicrobial peptides that also exert antiviral activity against adenovirus by interacting with a negatively charged region at the interface of the penton base and fiber, blocking viral uncoating (46). Adsorbed polymer may similarly interfere with viral capsid disassembly that is critical for efficient intracellular and gene transfer by the virus (47).
The inclusion of 10% serum during transduction decreased delivery efficiency of Ad5 by 65%. The negative effect of serum on in vitro adenovirus transduction has also been reported by others in the literature (16, 19). In the absence of CAR receptor, Ad5 interacts with HSPG through a KKTK motif on the fiber shaft, resulting in low efficiency transduction (48, 49). Serum proteins may therefore interfere with this low affinity interaction. pHK10-Ad5, on the other hand, transduces cells with similar efficiencies both in the presence and absence of serum. This is in contrast to two other polymer-coated virus systems that report decreased transduction efficiency in serum conditions (16, 19) as well as to pHK10-based polyplexes that have an order of magnitude decrease in transfection efficiency in the presence of serum (37). The retention of high transfection efficiency using pHK10-Ad5 in serum conditions may be attributed to the HPMA backbone of the pHK10. This hydrophilic polymer has been used to modify both non-viral particles and AdV to improve particle stability (50, 51). Indeed, the size of pHK10-Ad5 increases only ~25% in the presence of serum as measured by dynamic light scattering (data not shown). The difference between the effect of serum on pHK10 polyplex delivery compared to pHK10-Ad5 may be due to the importance of uncomplexed polymer on non-viral gene delivery (52); free polymer has been shown to enhance gene transfer efficiency by affecting intracellular trafficking of polyplexes but may be bound up by serum proteins. However, free polymer is not expected to impact adenoviral trafficking.
Transduction in the presence of serum decreases Ad5 efficiency by ~40% and 30% for pHSSK10- and pHCathK10-coated Ad5, respectively, although these polymer-coated viruses are still more effective than Ad5 in the presence of serum. The polyvalency of these materials is expected to provide a higher affinity to HSPGs than the Ad5 KKTK motif. The decreased effectiveness of pHSSK10- Ad5 in serum may be due to premature polymer degradation by extracellular thiols in the serum (53), which could affect the polymer-coating of the virus and thus decrease transfection gains afforded by the polymer-coating. Similarly, the pHCathK10 polymer has been shown to undergo both exopeptidase and slow non-specific endopeptidase degradation at the FKFL linker site in the presence of serum (45). While intracellular degradation facilitates polymer release from Ad5, degradation before the polymer can facilitate cellular uptake is undesirable.
The optimized polymer-coated Ad5 formulation was characterized by zeta potential measurements and TEM imaging (Figure 4). Unmodified adenovirus is negatively charged (−6.14 mV zeta potential) while pHK10-coated Ad5 is positively charged (+17.6 mV zeta potential). TEM imaging show that both Ad5 and pHK10-Ad5 were similar in size with diameters around 80nm. Therefore, virus structure remains intact after polymer coating.
The optimized pHK10-Ad5 was tested in two other cell lines: CHO-K1 cells (Chinese Hamster Ovary epithelial cells), and BaF3 cells (mouse pro-B cell line). pHK10-Ad5 virus was able to improve transduction efficiency into these cell types over Ad5 (Figure 5). An MOI that resulted in low but reproducibly detected transduction was first selected for each cell line. CHO-K1 cells were relatively easy to transduce. At MOI of 5, pHK10-Ad5 transduction was increased 4.3-fold over Ad5 (61% compared to 14.3% GFP-positive). Of the GFP-expressing cells, more transgene expression was also achieved; the mean fluorescence per cell was two-fold higher using pHK10-Ad5 as the vector. RAW 264.7 macrophage cells required MOI of 50 to achieve just 3% cell transfection, although pHK10-Ad5 improved transduction efficiency 6.1-fold to 19%. Transgene expression in macrophage cells have been reported to be 100–1000 fold lower than in CAR-positive cells; this effect is likely due to impaired endosomal escape in these types of scavenging cells (54). Kaner et al. previously showed that introduction of CAR to alveolar macrophage increases subsequent Ad5 transfection but only by around 5-fold (55), similar to the increase achieved by pHK10-Ad5. Pro-B BaF3 cells were the most resistant to transfection. At MOI of 1000, only 6.9% of cells were transfected by unmodified Ad5 and pHK10-Ad5 transfected 10.4%, a 1.5-fold increase. Unlike macrophages, ectopic expression of CAR in BaF3 cells and other lymphocyte cell lines rescues adenovirus transfection, indicating that adenoviral uptake is the limiting barrier in transfection of these cells (56). However, BaF3 cells also do not express HSPGs (57), which have been shown to be a major partner that mediates cellular entry of cationic polymer-based particles (58). Therefore, HSPG may also be required for transduction using polymer-coated AdV.
Neutralizing antibodies generated after adenoviral infection inhibit subsequent transduction efficiency, thus either limiting the efficacy of the initial adenoviral gene therapy or preventing vector readministration. This is particularly an issue for Ad5 serotypes because of the prevalence of anti-Ad5 NAb. In the United States for example, about 33–60% of the population already have anti-Ad5 NAb due to prior adenoviral infection (59). Direct polymer conjugation to adenovirus has been shown to decrease antibody induction as well antibody binding;(60) however, this is often achieved at a significant cost to transduction efficiency.(61, 62) For example, Subr and colleagues conjugated an HPMA copolymer with quaternary ammoniums (to increase association via electrostatic interaction) to AdV. The polymer-conjugated polymer exhibited over three orders of magnitude decrease in transduction efficiency but also reduced binding of vector to erythrocytes, indicating protection against antibody and complement binding.(62) The vectors developed in their work are therefore promising formulations on which retargeting functionality can be introduced. In this work, coating of Ad5 with pHK10 significantly protected the virus from inactivation due to NAb without compromising transduction efficiency (Figure 6).
To test the hypothesis that HSPG may be required for pHK10-Ad5 transduction, pHK10-Ad5 and unmodified Ad5 were used to transduce CHO-K1 and CHO cells with deficiencies in HSPG synthesis. CHO-pgs-E606 is deficient in heparan sulfate N-sulfotransferase activity (35), which sulfates 40–50% of N-glucosamine residues in HSPGs (63). Without heparan sulfate N-sulfotransferase, only ~20% of the N-glucosamine residues are sulfated resulting in under-sulfation. CHO-pgs-A745 is defective in xylosyltransferase activity, which catalyzes the first sugar transfer reaction necessary in the formation of glycosaminoglycans (34). Because of the loss of this enzyme, these mutants lack both heparan sulfate and chondroitin sulfate proteoglycans (CSPGs) (48).
Cell association of pHK10-Ad5 and Ad5 was determined by exposing cells to Alexa Fluor 568-labeled vectors for 30 minutes, followed by flow cyometry analysis (Figure 7A). Interaction of pHK10-Ad5 with CHO-K1 was efficient and rapid, whereas low Ad5 association was observed for all 3 cell lines. Cell association of pHK10 correlated with the levels on HSPG sulfation in the cell lines, confirming that the primary mechanism of cell binding is likely through electrostatic interactions with surface proteoglycans. Binding is significantly reduced but not eliminated in HSPG- and CSPG-negative CHO-pgs-A745 cells, indicating that pHK10-Ad5 do also have other binding partners in addition to these proteoglycans.
Transduction efficiency to the three CHO cell lines follows similar trends as cell association (Figure 7B). For these studies, virus was incubated with cells for 36 hours. Transduction studies were performed at subconfluent conditions (50% confluency) because the extent of HSPG-mediated AdV internalization depends on cell confluency (48). As reported previously, CHO cells with defective HSPGs are more resistant to AdV infection (48). In addition, CSPG does not play a significant role in Ad5 infection since CSPG-negative CHO-pgs-A745 cells do not show impaired Ad5 transduction compared to CHO-pgs-E606. In contrast, pHK10-Ad5 infection is facilitated by both HSPG and CSPG, with 24% and 41% decrease in transduction efficiency to CHO-pgs-E606 and CHO-pgs-A745 cells, respectively, compared to CHO-K1. Both the percent of transduced cells and the average transgene expression per cell is reduced in the mutant CHO cell lines. Mislick and Baldeschwieler also previously showed that both HSPGs and CSPGs play a role in uptake of polyplexes (58). Therefore sulfated proteoglycans are the primary binding partners for pHK10-Ad5, although other binding interactions can still mediate pHK10-Ad5 transduction.
Conclusion
In summary, we have synthesized and evaluated ten cationic HPMA-oligolysine polymers for polymer-coating of adenovirus to transduce CAR-negative cells. The pHK10-Ad5 formulation provides substantial increase in transduction efficiency over Ad5 in the tested CAR negative cell lines. Notably, transduction efficiency of pHK10-Ad5 is not affected by the presence of 10% serum. The primary internalization pathway of polymer-coated Ad5 was shown to be redirected from CAR to sulfated proteoglycans. This facile virus modification approach is therefore a potential solution for improving Ad5 transduction to a variety of CAR-negative cell types.
Supplementary Material
Acknowledgments
This work was supported by NSF DMR 0706647 and NIH 1R01NS064404. Julie Shi is supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-0718124. We thank Profs. Anthony Convertine and Patrick Stayton for providing ECT chain transfer agent and Roma Yumal for Ad5 preparation. Flow cytometry was conducted at the Cell Analysis Facility and TEM at the UW Nanotechnology User Facility (NTUF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Benihoud K, Yeh P, Perricaudet M. Adenovirus vectors for gene delivery. Curr Opin Biotechnol. 1999;10(5):440–447. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- 2.Campos SK, Barry MA. Current advances and future challenges in adenoviral vector biology and targeting. Curr Gene Ther. 2007;7(3):189–204. doi: 10.2174/156652307780859062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McConnell MJ, Imperiale MJ. Biology of adenovirus and its use as a vector for gene therapy. Hum Gene Ther. 2004;15(11):1022–1033. doi: 10.1089/hum.2004.15.1022. [DOI] [PubMed] [Google Scholar]
- 4.Howitt J, Anderson CW, Freimuth P. Adenoviruses: Model and Vectors in Virus-Host Interactions. Berlin: Springer-Verlag Berlin; 2003. Adenovirus interaction with its cellular receptor CAR; pp. 331–364. [DOI] [PubMed] [Google Scholar]
- 5.Meier O, Greber UF. Adenovirus endocytosis. J Gene Med. 2003;5(6):451–462. doi: 10.1002/jgm.409. [DOI] [PubMed] [Google Scholar]
- 6.Bergelson JM, Cunningham JA, Droguett G, KurtJones EA, Krithivas A, Hong JS, et al. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275(5304):1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 7.Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286(5444):1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 8.Nemerow GR, Stewart PL. Role of alpha v Integrins in Adenovirus Cell Entry and Gene Delivery. Microbiol Mol Biol Rev. 1999;63(3):725–734. doi: 10.1128/mmbr.63.3.725-734.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomko RP, Xu RL, Philipson L. HCAR and MCAR: The human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci U S A. 1997;94(7):3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law LK, Davidson BL. What does it take to bind CAR? Mol Ther. 2005;12(4):599–609. doi: 10.1016/j.ymthe.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Medina-Kauwe LK. Endocytosis of adenovirus and adenovirus capsid proteins. Adv Drug Deliv Rev. 2003;55(11):1485–1496. doi: 10.1016/j.addr.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrin-alpha-V-beta-3 and integrin-alpha-V-beta-5 promote adenovirus internalization but not virus attachment. Cell. 1993;73(2):309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 13.Nemerow GR, Stewart PL. Role of alpha(v) integrins in adenovirus cell entry and gene delivery. Microbiol Mol Biol Rev. 1999;63(3):725–734. doi: 10.1128/mmbr.63.3.725-734.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M, Zinn KR, Barnett BG, Sumerel LA, Krasnykh V, Curiel DT, et al. The therapeutic efficacy of adenoviral vectors for cancer gene therapy is limited by a low level of primary adenovirus receptors on tumour cells. Eur J Cancer. 2002;38(14):1917–1926. doi: 10.1016/s0959-8049(02)00131-4. [DOI] [PubMed] [Google Scholar]
- 15.Rebel VI, Hartnett S, Denham J, Chan M, Finberg R, Sieff CA. Maturation and lineage-specific expression of the coxsackie and adenovirus receptor in hematopoietic cells. Stem Cells. 2000;18(3):176–182. doi: 10.1634/stemcells.18-3-176. [DOI] [PubMed] [Google Scholar]
- 16.Fasbender A, Zabner J, Chillon M, Moninger TO, Puga AP, Davidson BL, et al. Complexes of adenovirus with polycationic polymers and cationic lipids increase the efficiency of gene transfer in vitro and in vivo. J Biol Chem. 1997;272(10):6479–6489. doi: 10.1074/jbc.272.10.6479. [DOI] [PubMed] [Google Scholar]
- 17.Toyoda K, Ooboshi H, Chu Y, Fasbender A, Davidson BL, Welsh MJ, et al. Cationic polymer and lipids enhance adenovirus-mediated gene transfer to rabbit carotid artery. Stroke. 1998;29(10):2181–2187. doi: 10.1161/01.str.29.10.2181. [DOI] [PubMed] [Google Scholar]
- 18.Toyoda K, Nakane H, Heistad DD. Cationic polymer and lipids augment adenovirus-mediated gene transfer to cerebral arteries in vivo. J Cereb Blood Flow Metab. 2001;21(9):1125–1131. doi: 10.1097/00004647-200109000-00010. [DOI] [PubMed] [Google Scholar]
- 19.Kim PH, Kim TI, Yockman JW, Kim SW, Yun CO. The effect of surface modification of adenovirus with an arginine-grafted bioreducible polymer on transduction efficiency and immunogenicity in cancer gene therapy. Biomaterials. 2010;31(7):1865–1874. doi: 10.1016/j.biomaterials.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 20.Arcasoy SM, Latoche JD, Gondor M, Pitt BR, Pilewski JM. Polycations increase the efficiency of adenovirus-mediated gene transfer to epithelial and endothelial cells in vitro. Gene Ther. 1997;4(1):32–38. doi: 10.1038/sj.gt.3300349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasman LM, Barua S, Lu P, Rege K, Voelkel-Johnson C. Polymer-Enhanced Adenoviral Transduction of CAR-Negative Bladder Cancer Cells. Mol Pharm. 2009;6(5):1612–1619. doi: 10.1021/mp9000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodds E, Piper TA, Murphy SJ, Dickson G. Cationic lipids and polymers are able to enhance adenoviral infection of cultured mouse myotubes. J Neurochem. 1999;72(5):2105–2112. doi: 10.1046/j.1471-4159.1999.0722105.x. [DOI] [PubMed] [Google Scholar]
- 23.Han J et al. Combination of adenovirus and cross-linked low molecular weight PEI improves efficiency of gene transduction. Nanotech. 2010;21(10):105–106. doi: 10.1088/0957-4484/21/10/105106. [DOI] [PubMed] [Google Scholar]
- 24.Park JW, Mok H, Park TG. Physical adsorption of PEG grafted and blocked poly-L-lysine copolymers on adenovirus surface for enhanced gene transduction. J Control Rel. 2010;142(2):238–244. doi: 10.1016/j.jconrel.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Singh R, Tian BW, Kostarelos K. Artificial envelopment of nonenveloped viruses: enhancing adenovirus tumor targeting in vivo. Faseb J. 2008;22(9):3389–3402. doi: 10.1096/fj.08-103275. [DOI] [PubMed] [Google Scholar]
- 26.Zhong Z, Shi S, Han J, Zhang Z, Sun X. Anionic Liposomes Increase the Efficiency of Adenovirus-Mediated Gene Transfer to Coxsackie-Adenovirus Receptor Deficient Cells. Mol Pharm. 2009;7(1):105–115. doi: 10.1021/mp900151k. [DOI] [PubMed] [Google Scholar]
- 27.Johnson RN, Burke RS, Convertine AJ, Hoffman AS, Stayton PS, Pun SH. Synthesis of Statistical Copolymers Containing Multiple Functional Peptides for Nucleic Acid Delivery. Biomacromol. 2010;11(11):3007–3013. doi: 10.1021/bm100806h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick CL, Lowe AB. Aqueous RAFT polymerization: Recent developments in synthesis of functional water-soluble (Co)polymers with controlled structures. Accounts Chem Res. 2004;37(5):312–325. doi: 10.1021/ar0302484. [DOI] [PubMed] [Google Scholar]
- 29.Burke RS, Pun SH. Synthesis and Characterization of Biodegradable HPMA-Oligolysine Copolymers for Improved Gene Delivery. Bioconjugate Chem. 2010;21(1):140–150. doi: 10.1021/bc9003662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shayakhmetov DM, Papayannopoulou T, Stamatoyannopoulos G, Lieber A. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J Virol. 2000;74(6):2567–2583. doi: 10.1128/jvi.74.6.2567-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shayakhmetov DM, Li ZY, Ternovoi V, Gaggar A, Gharwan H, Lieber A. The interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of adenoviruses. J Virol. 2003;77(6):3712–3723. doi: 10.1128/JVI.77.6.3712-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espenlaub S, Corjon S, Engler T, Fella C, Ogris M, Wagner E, et al. Capsomer-Specific Fluorescent Labeling of Adenoviral Vector Particles Allows for Detailed Analysis of Intracellular Particle Trafficking and the Performance of Bioresponsive Bonds for Vector Capsid Modifications. Hum Gene Ther. 2010;21(9):1155–1167. doi: 10.1089/hum.2009.171. [DOI] [PubMed] [Google Scholar]
- 33.Murakami P, McCaman MT. Quantitation of adenovirus DNA and virus particles with the PicoGreen fluorescent dye. Anal Biochem. 1999;274(2):283–288. doi: 10.1006/abio.1999.4282. [DOI] [PubMed] [Google Scholar]
- 34.Esko JD, Stewart TE, Taylor WH. Animal-cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A. 1985;82(10):3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bame KJ, Esko JD. Undersulfated heparan-sulfate in a chinese-hamster ovary cell mutant defective in heparan-sulfate N-sulfotransferase. J Biol Chem. 1989;264(14):8059–8065. [PubMed] [Google Scholar]
- 36.Johnson RN, Chu DSH, Shi J, Schellinger JG, Carlson PM, Pun SH. HPMA-oligolysine copolymers for gene delivery: Optimization of peptide length and polymer molecular weight. J Control Rel. 2011 doi: 10.1016/j.jconrel.2011.07.009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, Johnson RN, Schellinger JG, Carlson PM, Pun SH. Reducible HPMA-co-oligolysine copolymers for nucleic acid delivery. Int J Pharm. 2011 doi: 10.1016/j.ijpharm.2011.08.015. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino-acids using pre-column derivatization. J Chromatography. 1984;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- 39.Burke RS, Pun SH. Extracellular barriers to in Vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjugate Chemistry. 2008 Mar;19(3):693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- 40.Buyens K, Meyer M, Wagner E, Demeester J, De Smedt SC, Sanders NN. Monitoring the disassembly of siRNA polyplexes in serum is crucial for predicting their biological efficacy. J Control Rel. 2010 Jan;141(1):38–41. doi: 10.1016/j.jconrel.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 41.Fischer D, Li YX, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 42.Yang J, Chen H, Vlahov IR, Cheng JX, Low PS. Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proc Natl Acad Sci. 2006 Sep;103(37):13872–13877. doi: 10.1073/pnas.0601455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Authier F, Metioui M, Bell AW, Mort JS. Negative regulation of epidermal growth factor signaling by selective proteolytic mechanisms in the endosome mediated by cathepsin B. J Biol Chem. 1999;274(47):33723–33731. doi: 10.1074/jbc.274.47.33723. [DOI] [PubMed] [Google Scholar]
- 44.Blum JS, Fiani ML, Stahl PD. Proteolytic cleavage of ricin A chain in endosomal vesicles-evidence for the action of endsomal proteases at both neutral and acidic pH. J Biol Chem. 1991;266(33):22091–22095. [PubMed] [Google Scholar]
- 45.Chu DSH, Johnson RN, Pun SH. Cathepsin B-sensitive polymers for compartment-specific degradation and nucleic acid release. 2011. doi: 10.1016/j.jconrel.2011.10.016. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith JG, Silvestry M, Lindert S, Lu WY, Nemerow GR, Stewart PL. Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization. PLoS Pathog. 2010;6(6) doi: 10.1371/journal.ppat.1000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol. 2005;79(4):1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dechecchi MC, Melotti P, Bonizzato A, Santacatterina M, Chilosi M, Cabrini G. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J Virol. 2001;75(18):8772–8780. doi: 10.1128/JVI.75.18.8772-8780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith TAG, Idamakanti N, Rollence ML, Marshall-Neff J, Kim J, Mulgrew K, et al. Adenovirus serotype 5 fiber shaft influences in vivo gene transfer in mice. Hum Gene Ther. 2003;14(8):777–787. doi: 10.1089/104303403765255165. [DOI] [PubMed] [Google Scholar]
- 50.Fisher KD, Stallwood Y, Green NK, Ulbrich K, Mautner V, Seymour LW. Polymer-coated adenovirus permits efficient retargeting and evades neutralising antibodies. Gene Ther. 2001;8(5):341–348. doi: 10.1038/sj.gt.3301389. [DOI] [PubMed] [Google Scholar]
- 51.Green NK, Herbert CW, Hale SJ, Hale AB, Mautner V, Harkins R, et al. Extended plasma circulation time and decreased toxicity of polymer-coated adenovirus. Gene Ther. 2004;11(16):1256–1263. doi: 10.1038/sj.gt.3302295. [DOI] [PubMed] [Google Scholar]
- 52.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004 Oct;6(10):1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 53.Sun W, Davis PB. Reducible DNA nanoparticles enhance in vitro gene transfer via an extracellular mechanism. J Control Rel. 2010;146(1):118–127. doi: 10.1016/j.jconrel.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leopold PL, Crystal RG. Intracellular trafficking of adenovirus: Many means to many ends. Adv Drug Deliv Rev. 2007;59(8):810–821. doi: 10.1016/j.addr.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 55.Kaner RJ, Worgall S, Leopold PL, Stolze E, Milano E, Hidaka C, et al. Modification of the genetic program of human alveolar macrophages by adenovirus vectors in vitro is feasible but inefficient, limited in part by the low level of expression of the coxsackie/adenovirus receptor. Am J Respir Cell Mol Biol. 1999;20(3):361–370. doi: 10.1165/ajrcmb.20.3.3398. [DOI] [PubMed] [Google Scholar]
- 56.Leon RP, Hedlund T, Meech SJ, Li SB, Schaack J, Hunger SP, et al. Adenoviral-mediated gene transfer in lymphocytes. Proc Natl Acad Sci. 1998;95(22):13159–13164. doi: 10.1073/pnas.95.22.13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoneda A, Asada M, Oda Y, Suzuki M, Imamura T. Engineering of an FGF-proteoglycan fusion protein with heparin-independent, mitogenic activity. Nat Biotech. 2000;18(6):641–644. doi: 10.1038/76487. [DOI] [PubMed] [Google Scholar]
- 58.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci U S A. 1996;93(22):12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nwanegbo E, Vardas E, Gao WT, Whittle H, Sun HJ, Rowe D, et al. Prevalence of neutralizing antibodies to adenoviral serotypes 5 and 35 in the adult populations of The Gambia, South Africa, and the United States. Clin Diagn Lab Immunol. 2004;11(2):351–357. doi: 10.1128/CDLI.11.2.351-357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Croyle MA, Chirmule N, Zhang Y, Wilson JM. PEGylation of E1-deleted adenovirus vectors allows significant gene expression on readministration to liver. Hum Gene Ther. 2002;13(15):1887–1900. doi: 10.1089/104303402760372972. [DOI] [PubMed] [Google Scholar]
- 61.Kim PH, Sohn JH, Choi JW, Jung Y, Kim SW, Haam S, et al. Active targeting and safety profile of PEG-modified adenovirus conjugated with herceptin. Biomaterials. 2011;32(9):2314–2326. doi: 10.1016/j.biomaterials.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 62.Subr V, Kostka L, Selby-Milic T, Fisher K, Ulbrich K, Seymour LW, et al. Coating of adenovirus type 5 with polymers containing quaternary amines prevents binding to blood components. J Control Rel. 2009;1352:152–158. doi: 10.1016/j.jconrel.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 63.Gorsi B, Stringer SE. Tinkering with heparan sulfate sulfation to steer development. Trends Cell Biol. 2007;17(4):173–177. doi: 10.1016/j.tcb.2007.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.