Abstract
In response to environmental stresses, cells activate stress-response genes and inhibit DNA replication. HBO1 histone acetylase is a coactivator both for AP-1 transcription factors responding to stress-activated JNK kinases and also for the Cdt1 licensing factor that ensures that DNA is replicated exactly once per cell cycle. In response to non-genotoxic stress, JNK phosphorylates Jun, an AP-1 transcription factor, leading to increased recruitment of HBO1 and increased transcription of target genes. In addition, JNK phosphorylates Cdt1 on threonine 29, leading to rapid dissociation of HBO1 from replication origins, thereby blocking initiation of DNA replication. Upon relief of stress, HBO1 re-associates with replication origins. Thus, regulated and reciprocal recruitment of the HBO1 co-activator to target genes and replication origins via JNK-mediated phosphorylation of the recruiting transcription and replication licensing factors coordinates the transcriptional and DNA replication response to cellular stress.
Keywords: HBO1, Cdt1, JNK signaling, DNA replication initiation, stress response, replication licensing
INTRODUCTION
Initiation of genome replication is the convergence point of extracellular and intracellular signals that control cell proliferation, quiescence, and differentiation. The initial step of DNA replication involves the assembly of the pre-replication complex (pre-RC) onto chromatin (Bell and Dutta, 2002). This complex is formed by the sequential loading of the origin recognition complex (ORC), licensing factors Cdt1 and Cdc6, and eventually the minichromosome maintenance complex (MCM), the presumptive replication helicase. This assembly process is tightly controlled to ensure that replication occurs during S phase and only once per cell cycle, with Cdt1 being the critical protein involved in this regulation. Cdt1 is rapidly degraded by the proteasome when cells enter S phase to prevent replication reinitiation (Arias and Walter, 2007). In addition, Cdt1 activity in S-phase is inhibited by Geminin via a direct interaction between these two proteins (Wohlschlegel et al., 2000; Tada et al., 2001; Miotto and Struhl, 2010). Cdt1 is also rapidly degraded when cells accumulate DNA damage, thereby preventing pre-RC assembly and origin activity under challenged conditions (Higa et al., 2003; Hu et al., 2004; Kondo et al., 2004).
HBO1, the major histone H4 acetylase in human cells (Doyon et al., 2006; Miotto and Struhl, 2010), is a co-activator of AP-1 and other transcription factors (Georgiakaki et al., 2006; Miotto and Struhl, 2006; Martinato et al., 2008) as well as a co-activator of the Cdt1 licensing factor (Miotto and Struhl, 2008; Wong et al., 2010). During G1, HBO1 is recruited to replication origins by a direct interaction with Cdt1 (Miotto and Struhl, 2008), and HBO1 is required for loading of the MCM complex (Iizuka et al., 2006; Miotto and Struhl, 2008). Replication licensing requires HBO1-mediated acetylation of H4 at the origin (Miotto and Struhl, 2010). The licensing inhibitor Geminin inhibits HBO1 histone acetylase activity in the context of the Cdt1-HBO1 complex, indicating that control of H4 acetylation at origins is critical for regulation of DNA replication (Miotto and Struhl, 2010). In addition, p53 has been suggested to interact directly with HBO1 and inhibit its catalytic activity and licensing activity (Iizuka et al., 2008).
Activation of JNK kinases is an important and universal response in eukaryotes to a variety of cellular and environmental stimuli (Karin and Gallagher, 2005; Bogoyevitch et al., 2010). Activation of JNK leads to phosphorylation of cytoplasmic and nuclear targets including transcription factors (e.g. the AP-1 factor Jun) and cell cycle regulators (e.g. Cdc25, Aurora B and Cdh1) to control cell cycle progression, cell proliferation and cell death (Bogoyevitch and Kobe, 2006; Gutierrez et al., 2010a; Gutierrez et al., 2010b). Phosphorylation of Jun leads to increased recruitment of HBO1 and increased transcription of AP-1 target genes (Miotto and Struhl, 2006). Furthermore, JNK activity has been linked to DNA replication (Rahmouni et al., 2006; Miyamoto-Yamasaki et al., 2007; Hu et al., 2008; Chen et al., 2010; Gutierrez et al., 2010a). Basal levels of JNK activities control cell proliferation and the G1-S transition time in various cell types (Tournier et al., 2000; Sabapathy et al., 2004). In serum-starved condition, high levels of AIB1 prevent activation of the JNK pathway, which correlates with DNA replication (Horiguchi et al., 2006), and osmotic stress inhibits loading of the MCM complex on chromatin (Iizuka et al., 2008). Nevertheless, the molecular events linking JNK activity and regulation of DNA replication remain enigmatic.
Here, we demonstrate that, in response to a variety of non-genotoxic stress conditions, JNK-mediated phosphorylation of Cdt1 inhibits its interaction with HBO1, thereby leading to rapid dissociation of HBO1 from replication origins. This JNK-mediated response is distinct from the response to DNA damage, in which Cdt1 is subject to proteolytic destruction. Thus, regulated and reciprocal recruitment of the HBO1 co-activator to target genes and replication origins via JNK-mediated phosphorylation of the recruiting transcription and replication licensing factors coordinates the transcriptional and DNA replication response to cellular stress.
RESULTS
HBO1 binding at replication origins is impaired under stress conditions
To examine the effect of stress on replication licensing, we investigated the effect of various chemical compounds on HBO1 association at origins. We used sorbitol and urea as models of hyper-osmotic shock, 1,4-dithiothreitol as a model of endoplasmic reticulum stress (or unfolded protein response), anisomicyn to inhibits protein synthesis, and actinomycin D as a genotoxic stress. As a control, we treated the cells with hydroxyurea for 12 hours to enrich the population in S-phase cells, as HBO1 loading is inhibited during S phase (Miotto and Struhl, 2008).
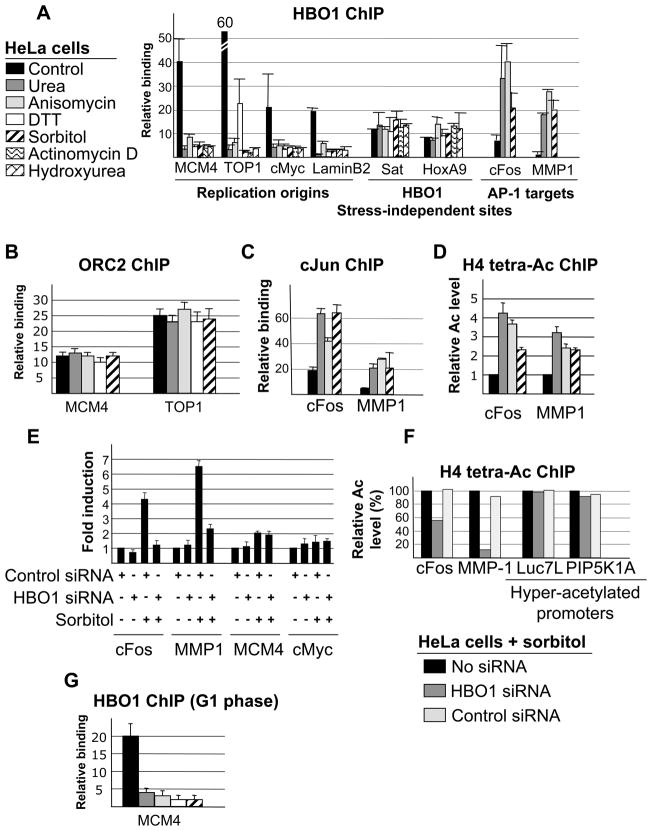
When HeLa cells are treated with any of these stress agents, HBO1 association at replication origins is not detected (Figures 1A and S1), whereas ORC2 association is not significantly affected (Figure 1B). As expected from the co-activator function of HBO1 at AP-1 target genes (Miotto and Struhl, 2006), these stress conditions result in increased association of HBO1 (Figure 1A), Jun (Figure 1C), and H4 acetylation (Figure 1D) at AP-1 target promoters, and depletion of HBO1 blocks transcriptional induction (Figure 1E) and H4 acetylation (Figure 1F). In contrast, expression of genes near the Mcm4 and cMyc replication origins is not significantly affected by osmotic stress or HBO1, indicating that HBO1 dissociation from origins is not related to transcription of nearby genes. Importantly, the decreased association of HBO1 with replication origins is not simply a redistribution of HBO1 towards AP-1 target genes. Overexpression of cJun, cFos, or an activated derivative of cJun (cJunAsp) does not affect HBO1 binding at origins (Figure S2A), and more importantly, stress does not affect HBO1 association with other HBO1 target loci (Figure 1A).
Figure 1. HBO1 binding at replication origins is impaired under stress conditions.
(A) HBO1 association at the indicated replication origins, AP-1 promoters and stress-independent target sites in HeLa cells treated with 300 mM Sorbitol, 50 ng/ml anisomysin, 400 mM urea, 2 mM DTT and 0.2 μg/ml actinomycin D for 1 hour, or hydroxyurea for 12 hours. Data are presented as a fold-enrichment over a control genomic region.
(B) ORC2 association at the indicated replication origins in cells treated with the indicated drugs.
(C) Jun association at the cFos and MMP1 promoters in normal and stressed conditions.
(D) H4 acetylation at the cFos and MMP1 promoters in normal and stressed conditions. Data are normalized in each condition to nucleosome occupancy as measured by the level of bulk H3.
(E) RT-PCR analysis cFos and MMP-1 RNA levels in cells that were or were not treated with sorbitol (300 mM, 1 hr) and that were or were not depleted for HBO1.
(F) Effect of HBO1 depletion on histone H4 acetylation at AP-1 regulated (cFos and MMP-1) and hyper-acetylated (Luc7L and PIP5K1A) promoters in sorbitol-treated cells. Data are normalized in each condition to nucleosome occupancy as measured by the level of bulk H3.
(G) HBO1 association in HeLa cells staged in M phase by a nocodazole-block (14 hours incubation in 100nM nocodazole) and allowed to re-enter in G1 (8 hours after nocodazole released) were treated with the indicated drugs.
As HBO1 binding at origins is restricted to G1 phase, we performed several control to exclude stress-related effects on cell cycle progression (for all conditions but hydroxyurea). First, when cells were released from mitosis and allowed to enter G1 in a drug-free medium and then subjected to the various stress treatments, HBO1 association was not observed at origins (Figure 1G). Second, similar results were observed in three different cell lines (HCT116, 293T and K562; data not shown). Third, cell cycle profiles following drug treatments show similar distributions of cells at various stages as observed in non-treated cells (Figure S2B). Lastly, as drugs were applied to cells for an hour at most, the dissociation of HBO1 from origins is a direct effect of the stress condition rather than an indirect effect on cell cycle progression, cell proliferation, or cell death.
Recruitment of HBO1 is required for loading of the MCM complex but not other replication factors (Miotto and Struhl, 2008). As assayed by Western blot analysis and/or chromatin immunoprecipitation, exposure to sorbitol does not affect ORC2, Cdc6 and Cdt1 loading onto the chromatin, whereas association of the MCM complex is inhibited (Figure S3 and see below). Thus, under hyper-osmotic stress, HBO1 binding at origins is inhibited, resulting in a defect in DNA licensing as defined by the reduced loading of the MCM complex. In addition, as much of the chromatin-bound MCM complex is associated with DNA polymerase in the process of replication, this observation suggests that osmotic stress also inhibits the initiation of DNA replication.
Cdt1 is stable under stress conditions that do not affect the integrity of the DNA
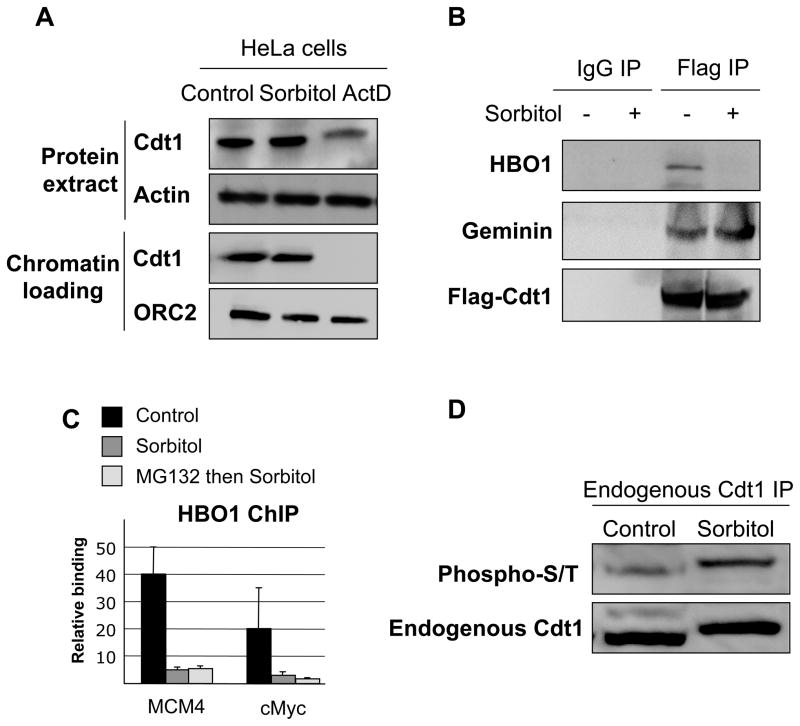
Cdt1 is degraded when cells enter S phase or in response to DNA damages (actinomycin D treatment), thereby preventing HBO1 binding at origins (Miotto and Struhl, 2008)(Figure 2A). In contrast, Cdt1 is not degraded following hyper-osmotic stress (Figure 2A), and the Geminin-Cdt1 interaction is not affected (Figure 2B). These observations indicate that the lack of HBO1 at origins is due to a defect in Cdt1-mediated recruitment, not a loss of Cdt1. Consistent with this observation, co-immunoprecipitation assays indicate that HBO1 does not interact with Flag-Cdt1 under hyper-osmotic conditions, whereas the two proteins co-purify under non-stressed conditions (Figure 2B). In addition, a proteasome inhibitor does not rescue HBO1 association at origins under conditions of osmotic shock (Figure 2C), whereas it does rescue Cdt1 destruction and HBO1 binding under conditions of DNA damage (Miotto and Struhl, 2008). As HBO1 and Cdt1 are not degraded under hyper-osmotic stress, a stress-induced post-translational modification of Cdt1 and/or HBO1 or regulation of a heretofore unknown partner interferes with the HBO1-Cdt1 interaction.
Figure 2. Cdt1 is stable under non-genotoxic stress conditions.
(A) Western blot analysis of Cdt1 expression level and chromatin binding in HeLa cells treated with sorbitol and actinomycin D. Actin and ORC2 were used as loading controls for total extract and chromatin fractions respectively.
(B) HeLa cells transfected with Flag-Cdt1 were or were not treated with sorbitol (300mM, 1 hour), and following immunoprecipitation of Flag-Cdt1, HBO1 and Geminin binding were analyzed by Western blotting.
(C) Effect of proteasome inhibition (MG132) on HBO1 association at the indicated replication origins.
(D) Western blot analysis of HA-Cdt1 immunoprecipitates in HeLa cells that were or were not treated with sorbitol. Myc-HBO1 binding, HA-Cdt1 acetylation (acetyl-K) and HA-Cdt1 phosphorylation (phospho-S/T) were investigated by Western blotting.
(E) Western blot analysis of endogenous Cdt1 immunoprecipitates for Cdt1 phosphorylation (phospho-S/T) in HeLa cells that were or were not treated with sorbitol
Cdt1 is phosphorylated under conditions of hyperosmotic shock
Cdt1 can be modified by acetylation (Glozak and Seto, 2009), phosphorylation (Sugimoto et al., 2004), and ubiquitylation (Arias and Walter, 2006). Although we observed no evidence for differential ubiquitylation or acetylation of Cdt1 (data not shown), we observed a marked increase in phosphorylation of endogenous Cdt1 in hyper-osmotic cells compared to control cells (Figure 2D). Thus, in conditions of osmotic stress, Cdt1 is hyper-phosphorylated, but its stability and modifications associated with its stabilization are not significantly altered. In accord with this observation, we do not observe an increase in Cdt1-associated proteins involved its destruction during S phase (PCNA) and in response to DNA damage (the E3 ubiquitin ligase SCF/Skp2) in Cdt1 immunoprecipitates in response to osmotic stress (Figure S4).
The HBO1 N-terminal domain has many serines, with several of them potentially phosphorylated (Beausoleil et al., 2004; Olsen et al., 2006; Matsuoka et al., 2007; Wu and Liu, 2008). We found that HBO1 is hyper-phosphorylated under normal conditions, but did not observe an increase in HBO1 phosphorylation following hyper-osmotic shock (data not shown). However, we cannot exclude that one or more residues in HBO1 might be phosphorylated upon osmotic shock.
JNK kinase activity is important for HBO1 binding at origins
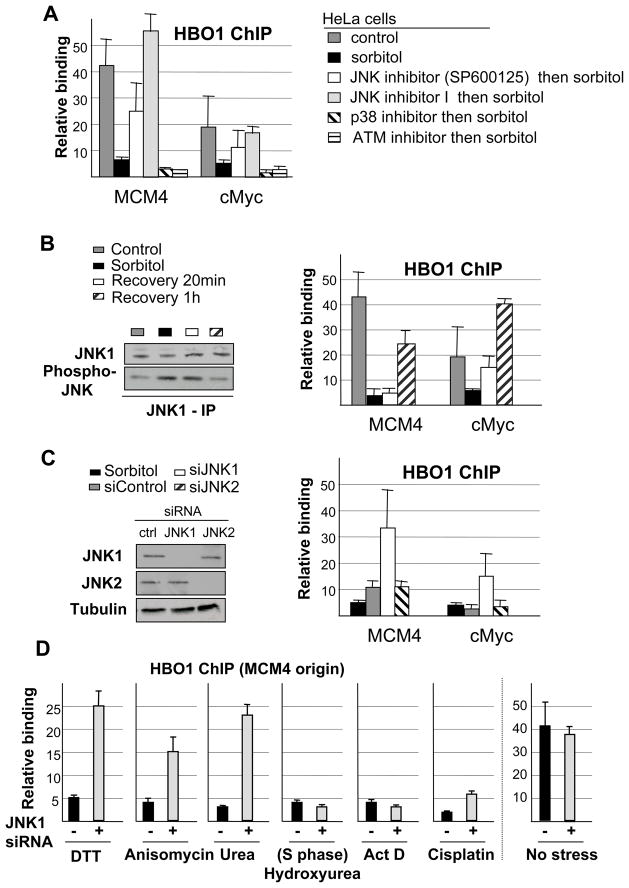
To identify the signal transduction pathway that mediates Cdt1 hyper-phosphorylation under hyper-osmotic condition, we examined the effect of chemical inhibitors that target kinases involved in the response to DNA damage (ATM), hyper-osmotic stress (p38 and JNK) and cell cycle regulation (Cdk2). Inactivation of the JNK, but not Cdk2, ATM or p38 kinases, prior to stress induction blocks HBO1 dissociation from origins in HeLa cells (Figure 3A, S5, S6). Inactivation of the JNK but not p38 kinase also blocks HBO1 dissociation from origins in a p53-positive cell line (Figure S6). JNK activity increases under hyper-osmotic stress and rapidly returns to its basal activity level when sorbitol is removed from the medium and the stressed state reversed (Figure 3B, left panel). In accord with changes in JNK activity, HBO1 association with origins is abolished in stressed cells, but it rapidly re-associates when sorbitol is removed (Figure 3B, right panel).
Figure 3. HBO1 binding at origins is controlled by JNK1 kinase activit.
(A) HBO1 association at the indicated origins in cells treated with sorbitol following a pre-treatment with JNK (2 different compounds), p38 or ATM kinase specific inhibitor (see Supplemental Figure S5 for level of kinase inhibition).
(B) HBO1 association at the indicated origins during the recovery phase following an hyper-osmotic shock (right panel). JNK activity (assayed by phosphorylation) in control, hyper-osmotic and recovery stages is illustrated in the left panel.
(C) HBO1 association at MCM4 origin in cells treated with siRNAs specific for JNK1 or JNK2 prior to hyper-osmotic shock (right panel). Western blot analysis of siRNA specificity and efficiency is shown in the left panel.
(D) HBO1 association at MCM4 origin in cells treated with a JNK1-specific siRNA, prior to exposure to DTT, anisomycin, urea, hydroxyurea, actinomycin D (ActD), cisplatin, or no treatment (asynchronously growing cells).
Using siRNA against JNK1 or JNK2, the ubiquitously expressed JNK proteins, we observe that depletion of JNK1 inhibits HBO1 dissociation while depletion of JNK2 has no effect (Figure 3C). Thus, under the hyper-osmotic condition, JNK1 activation inhibits the binding of HBO1 at replication origins, and this effect is reversed upon stress relief when JNK1 activity levels decrease. Depletion of JNK1 also permits HBO1 binding at origins in cells treated with DTT, urea and anisomycin (Figure 3D). In contrast JNK depletion does not allow HBO1 binding at origins in cells treated with actinomycin D, cisplatin, or hydroxyurea, consistent with Cdt1 being degraded under DNA-damaging conditions and during S phase. As expected from the low levels of JNK1 activity in non-stressed cells, HBO1 association in non-stressed cells is not affected by JNK1 depletion. Thus, JNK1 activity inhibits HBO1 binding at origins under various forms of stress that do not affect the integrity of the DNA.
Phosphorylation of Cdt1-T29 by JNK1 upon hyperosmotic stress
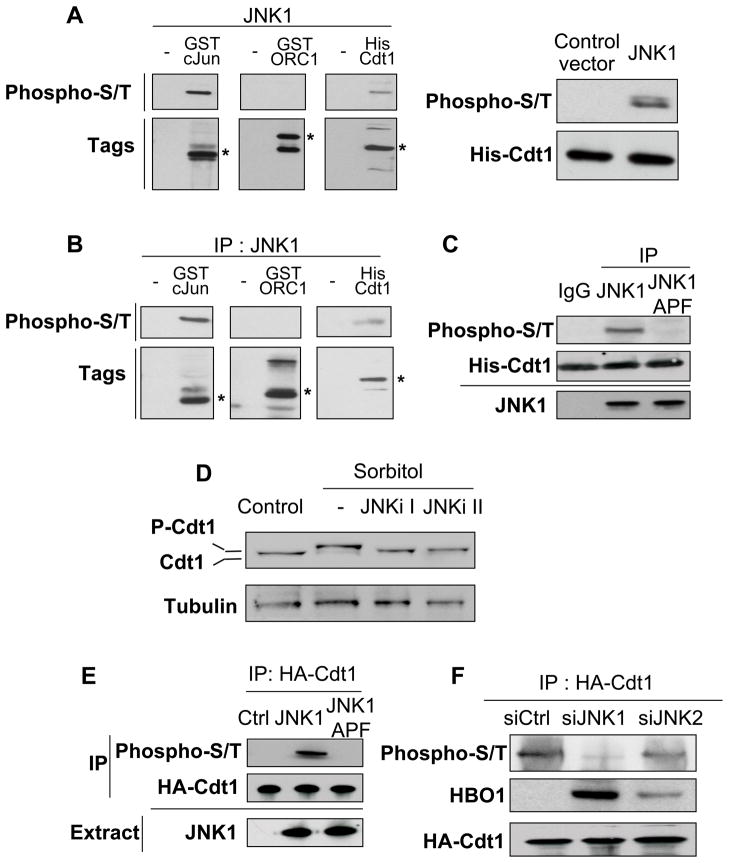
We next investigated whether JNK phosphorylates Cdt1 and ORC1, a subunit of the ORC complex that interacts with HBO1 (Iizuka and Stillman, 1999; Burke et al., 2001) and whose function is controlled by phosphorylation (Li et al., 2004; Saha et al., 2006). We first used an in vivo kinase assay in E. coli (Murata et al., 2008) in which human JNK1 was co-expressed with tagged versions of either full-length Cdt1, full-length ORC1, or the N-terminal JNK-interaction domain of cJun (Kallunki et al., 1994). We detected phosphorylation of Cdt1 and the cJun positive control, but not ORC1 (Figure 4A). We confirmed these results using an IP-kinase assay in which JNK1 was immuno-precipitated from sorbitol-treated HeLa cells and added to recombinant Cdt1, ORC1 or cJun (Figure 4B). When the experiment is repeated using a kinase-deficient JNK1 (JNK1-APF), phosphorylation of Cdt1 and cJun is not observed (Figure 4C). Thus, JNK1 phosphorylates Cdt1 in vitro.
Figure 4. Cdt1 is phosphorylated by JNK1 following stress.
(A) Left panel: Phosphorylation of purified Cdt1, ORC1 and cJun(Nter) by JNK1 upon co-transformation of E. coli was investigated by Western blot using a mix of phospho-serine and phospho-threonine antibodies (phospho-S/T). Asterisks indicate the expected size of fusion proteins. Right panel: Phosphorylation of purified His-Cdt1 by JNK1 upon co-transfection of E. coli.
(B) Phosphorylation status of recombinant Cdt1, ORC1, and cJun(Nter) phosphorylation status upon incubation with immunopurified JNK1 from HEK-293T cells treated with sorbitol.
(C) Phosphorylation of recombinant Cdt1 phosphorylation following incubation with immunopurified JNK1 or kinase-deficient JNK1 (JNK-APF) from HEK-293T cells treated with sorbitol. The amount of JNK1 and JNK1-APF is indicated on the bottom.
(D) Western blot analysis of endogenous Cdt1 in cells treated with sorbitol that had or had not been pre-treated with JNK inhibitors I or II.
(E) Western blot analysis of HA-Cdt1 phosphorylation after immunoprecipitation from HeLa cells expressing JNK1 or its kinase-deficient variant JNK1-APF. JNK1 and JNK1-APF expression in HeLa cells (proteins obtained from the crude extract prior to immunoprecipitation) is indicated.
(F) Western blot analysis of HA-Cdt1 phosphorylation after immunoprecipitation from HeLa cells treated with siRNA specific for JNK1 or JNK2 prior to sorbitol treatment.
Several observations indicate that JNK1 phosphorylates Cdt1 in vivo. First, phosphorylation of endogenous Cdt1 is blocked by two different JNK inhibitors (Figure 4D). Second, HA-Cdt1 is hyper-phosphorylated in cells transfected with JNK1 compared to control cells and cells transfected with JNK1-APF (Figure 4E). Third, HA-Cdt1 phosphorylation is abolished when JNK1 is depleted by siRNA, whereas it is only mildly reduced upon depletion of JNK2 (Figure 4F). Importantly, HBO1 interaction with Cdt1 is enhanced when JNK1 is removed (Figure 4F), consistent with HBO1 association at replication origins (Figure 3C). A slight increase in Cdt1-HBO1 interaction is also observed when JNK2 is removed (Figure 4F), even though rescue of HBO1 binding is not observed by ChIP (Figure 3C). Thus, Cdt1 is predominantly phosphorylated by JNK1 following hyper-osmotic shock, and JNK1-mediated phosphorylation of Cdt1 impairs the association of HBO1 with replication origins.
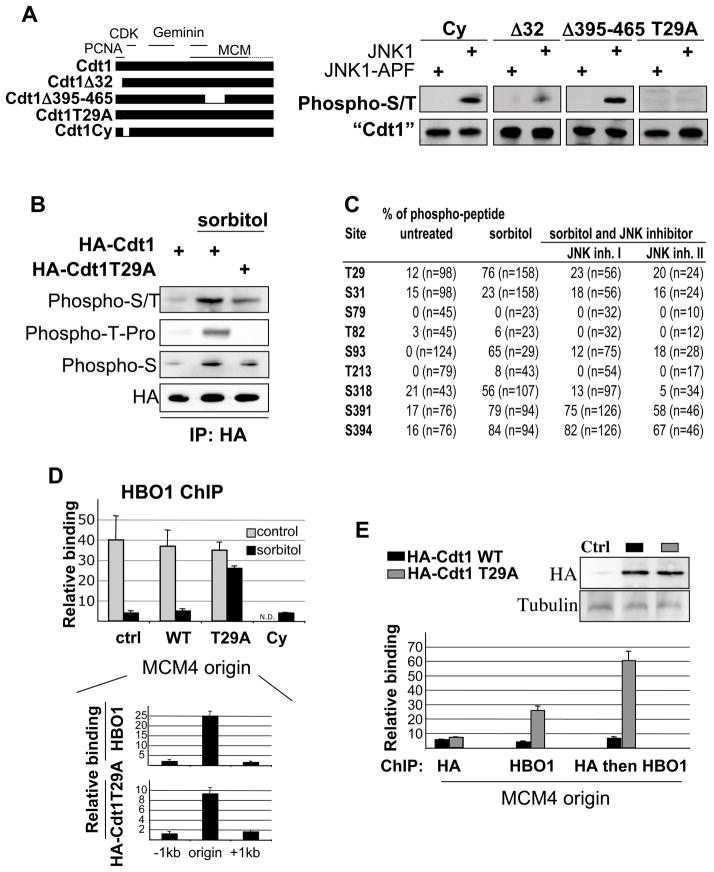
To identify Cdt1 residues phosphorylated by JNK1, we co-transfected JNK1 and JNK1-APF with various mutants of Cdt1: Δ32 (lacks the region necessary for replication-dependent destruction); Δ395–465 (lacks the MCM interaction domain); ΔCy (lacks the Cyclin-dependent kinase interaction domain); T29A (mutates a phosphorylation site essential for SCF/Skp2 interaction). As shown in Figure 5A, Cdt1 hyper-phosphorylation is reduced upon removal of the N-terminal amino acids, but not the MCM-interaction or CDK-interaction domains. More specifically, the T29A mutation abrogates Cdt1 hyper-phosphorylation by JNK1.
Figure 5. Cdt1 phosphorylation impairs HBO1 binding at origins.
(A) Phosphorylation of the indicated Cdt1 derivatives co-transfected with JNK1 or kinase-deficient JNK1 in HeLa cells. Description of the different Cdt1 variants is indicated on the left with interaction domains with PCNA, CDK, Geminin and MCM highlighted.
(B) Western blot analysis of Cdt1 and Cdt1 T29A phosphorylation under osmotic stress conditions with a context-specific phospho-T antibody.
(C) Cdt1 phosphorylation sites induced by JNK1 and sorbitol. Table represent the percentage of phospho-peptide for each serine and threonine residue recovered over the 3 samples. The total number of peptide for each site is indicated into bracket.
(D) HBO1 association at the MCM4 origin in HeLa cells transfected with the indicated Cdt1 derivatives and subjected or not to sorbitol treatment (ND, not determined). Bottom panel: Association of HBO1 and HA-Cdt1-T29A at the MCM4 origin and surrounding regions in cells subject to sorbitol treatment.
(E) Sequential ChIP analysis of HBO1+HA-Cdt1 (black) or HBO1+HA-Cdt1-T29A (grey) binding at the MCM4 replication origin in HeLa cells treated with sorbitol. Top panel: Western blot analysis of HA-fusions expression.
Phosphorylation of T29 by JNK1 in vitro was confirmed by phospho-peptide mapping (Figure S7) and western blotting using an antibody specific for phospho-Thr-Pro (Figure 5B). Furthermore, phospho-peptide analysis of Flag-Cdt1 obtained from control and sorbitol-treated cells identified several phosphorylation sites including T29 (Figure 5C). Upon treatment with two different JNK inhibitors prior to stress induction, only T29, S93, and S318 are phosphorylated in a JNK1-dependent manner under stress (Figure 5C). Two additional serines, S391 and S394, adjacent to the MCM interacting domain are phosphorylated under stress conditions, but insensitive to both JNK inhibitors (Figure 5C and Figure S7). These results are consistent with Western analysis showing a total loss of Thr-Pro phosphorylation and a significant decrease of phospho-serine and global phosphorylation by removing T29 (Figure 5B, C). These observations demonstrate that T29 is a direct JNK1 target under conditions of osmotic stress.
Phosphorylation of Cdt1-T29 impairs HBO1 binding at origins
To investigate whether Cdt1 phosphorylation is important for HBO1 regulation, we expressed JNK-insensitive (Cdt1-T29A) or JNK-sensitive (Cdt1-ΔCy) Cdt1 mutants in cells, and monitored HBO1 binding at origins in response to hyperosmotic shock. Expression of wild-type Cdt1 does not rescue HBO1 binding at replication origins (Figure 5D), indicating that its amount in the cell is not limiting. In contrast, expression of the Cdt1-T29A, but not the Cdt1-ΔCy, derivative rescues HBO1 binding at origins under conditions of stress (Figure 5D). As expected, expression of Cdt1 or Cdt1-T29A does not affect the level of HBO1 association at origins under non-stressed condition. Thus T29 is a JNK-phosphorylated site that is critical to control HBO1 binding at origins under stress conditions.
To further demonstrate that this effect is direct, we performed sequential ChIP analysis in which DNA fragments bound by HA-Cdt1 or HA-Cdt1-T29A were immunoprecipitated with an HBO1 antibody (Figure 5E). We observed that HA-Cdt1 and HA-Cdt1-T29A associate with replication origin and are expressed at similar levels in the cell (Figure 5E, top panel). Importantly, HBO1 and HA-Cdt1-T29A co-occupy the same origin; i.e. the fold-enrichments of the sequential ChIP samples are significantly higher than the fold-enrichment for HBO1 alone. In contrast, HA-Cdt1 and HBO1 do not co-occupy the origin, as the fold-enrichment is roughly equal to HA-Cdt1 alone. Thus, HA-Cdt1-T29A directly rescues HBO1 binding by recruiting HBO1 at origins under condition of stress.
DISCUSSION
JNK1 phosphorylation of Cdt1-T29 inhibits recruitment of the HBO1 co-activator to replication origins in response to non-genotoxic stress
DNA replication is tightly regulated by licensing factors that ensure that the genome is replicated exactly once per cell cycle. Replication licensing is restricted to a specific time of the cell cycle, the G1-S transition, and this restriction is mediated by regulation of the Cdt1 licensing factor by proteolysis and by Geminin, which inhibits Cdt1 function via a direct interaction. In addition to such cell-cycle regulation, the initiation of DNA replication is delayed or prevented by environmental stress. In response to UV light and other forms of DNA damage, the Cdt1 and Cdc6 licensing factors are degraded, thereby inhibiting DNA replication (Higa et al., 2003; Yim et al., 2003; Hu et al., 2004; Kondo et al., 2004; Duursma and Agami, 2005; Higa et al., 2006; Hu and Xiong, 2006).
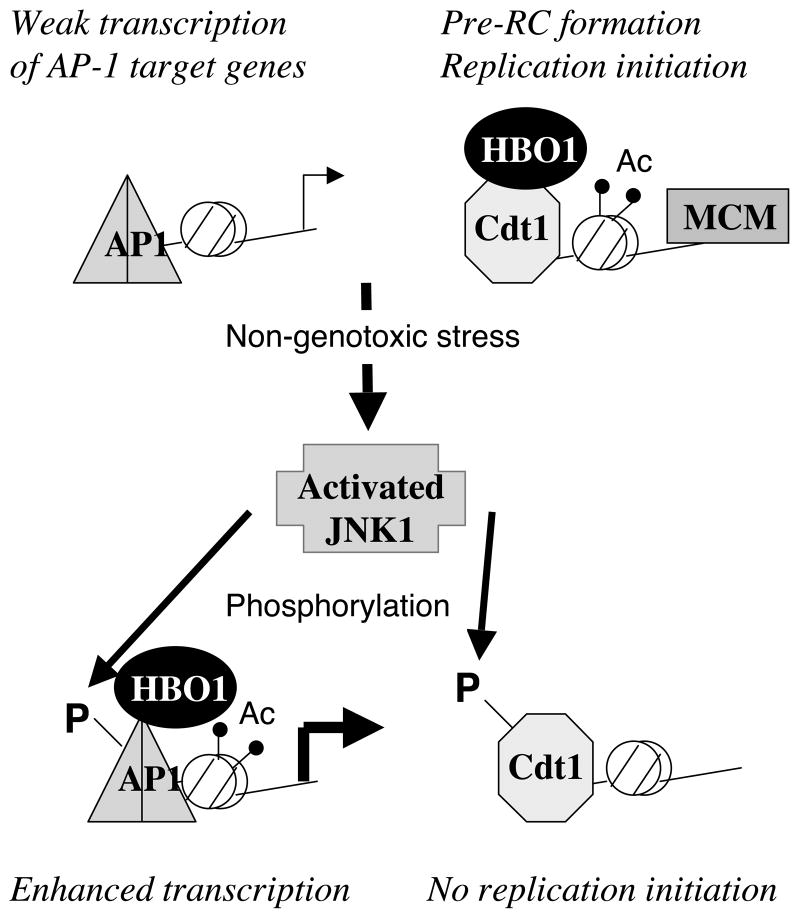
Here, we describe a distinct molecular pathway for regulating replication licensing in response to a variety of non-genotoxic stresses that are activated by events at the cell membrane or cytoplasm (Figure 6). In this pathway, stress activates JNK1, which phosphorylates Cdt1 on T29, thereby inhibiting the interaction with HBO1, a co-activator of Cdt1 necessary for licensing. As expected, the rapid dissociation of HBO1 from replication origins leads to decreased association of the MCM complex from replication origins and bulk chromatin, indicating a block in replication licensing and initiation of DNA replication. Importantly, and in contrast to cell-cycle regulation and the response to DNA damage, JNK1-mediated phosphorylation does not affect the level of Cdt1. Furthermore, the inhibition of HBO1 association with origins is rapid and reversible upon removal of the stress, indicating that stress-activated JNK1 directly affects the activity of Cdt1 via phosphorylation. Lastly, the T29A derivative of Cdt1, unlike the wild-type protein, can recruit HBO1 to replication origins under stress conditions. A key feature of this regulatory mechanism is that, because Cdt1 protein levels are unaffected, it permits both the rapid disassociation and reassociation of HBO1 in changes in stress conditions that affect phosphorylation of T29.
Figure 6.
Model for the coordination between transcriptional and replication responses to non-genotoxic stress mediated by HBO1.
Although T29 phosphorylation prevents the interaction of Cdt1 with HBO1, T29 does not appear essential for the Cdt1-HBO1 interaction because a truncated Cdt1 protein lacking the first 32 amino-acids can recruit HBO1 to replication origins (Miotto and Struhl, 2008). One possible explanation for these observations is that T29 phosphorylation induces a conformation change in Cdt1 that blocks interaction with HBO1. In this view, the surface of Cdt1 that directly interacts with HBO1 does not include T29 and indeed may be located in a distinct part of the protein. Alternatively, T29 might be important for JNK-mediated phosphorylation of other sites within Cdt1 (e.g. S93 and S318). Whatever the precise mechanism, our results indicate that Cdt1 phosphorylation at T29 by JNK1 is critical for dissociation of HBO1 from replication origins, and hence reduction of Cdt1 licensing function, in response to non-genotoxic stress.
The role of HBO1 in regulating the initiation of DNA replication
HBO1 is a co-activator of Cdt1, and it plays a direct role at origins for replication licensing (Miotto and Struhl, 2008). As HBO1 is recruited to origins by Cdt1, its association with origins is determined by the level of Cdt1, which is subject to cell-cycle regulated proteolysis. Here, we show that the Cdt1-HBO1 interaction itself is regulated by a variety of non-genotoxic stresses, specifically via JNK1-mediated phosphorylation of T29. In this regard, Cdt1 resembles transcriptional activator and repressor proteins that are modified by physiological conditions in a manner that alters recruitment of co-activator or co-repressor proteins. Among chromatin-modifying co-activators, HBO1 is also subject to a unique form of regulation involving binding to an inhibitor (Geminin) that reduces its histone acetylase activity (Miotto and Struhl, 2010). Thus, HBO1 does not function solely as a co-activator to allow the initiation of DNA replication to occur efficiently in the context of chromatin, but its recruitment and enzyme activity are regulated by physiological effectors that are linked to the cell cycle and the response to environmental conditions.
After this work was completed, it was reported that mice and primary mouse embryo fibroblasts lacking HBO1 had severe defects in acetylation of H3-K14 but not H4, and did not show defects in DNA replication or cell proliferation (Kueh et al., 2011). This extremely unexpected observation is in apparent contradiction to results in numerous publications from multiple laboratories involving cultured cells and it also appears inconsistent with the strong preference of HBO1 for acetylation of H4 vs. H3 in vitro. The bases of these apparent discrepancies are unknown, and it seems likely that the preferential defect in H3-K14 acetylation is an indirect consequence of a role in transcriptional activation. Interestingly, HBO1 levels are significantly higher in tumors and established cancer cell lines than in non-transformed cells (Iizuka et al., 2009), suggesting the possibility that the role of HBO1 in replication licensing is more important in cancer cells than in normal cells.
HBO1 coordinates the transcriptional and replication response to stress
In response to stress, cells activate transcriptional programs to relieve the stress, and they also inhibit DNA replication to prevent cell proliferation under non-optimal conditions. It is important that the transcription and replication responses are coordinated so that replication is inhibited during stress but permitted upon the relief of the stress. As a co-activator of both stress-regulated transcriptional activators and the Cdt1 licensing factor, HBO1 serves to coordinate the transcriptional and replication responses (Figure 6). In both cases, stress-activated JNK1 phosphorylates proteins that recruit HBO1 to their target sites. However, JNK1-mediated phosphorylation of Jun (and perhaps other transcription factors) increases HBO1 association with target promoters, whereas phosphorylation of Cdt1 inhibits HBO1 association with origins.
Two aspects of this coordination are noteworthy. First, as is often true for stress responses, JNK1-mediated transcriptional responses are typically transient, because relief of stress lowers the activity of JNK1. Such reduction of JNK1 activity upon the transcriptional response will kinetically link the resumption of DNA licensing with stress relief. In mutant cell types or environmental situations in which JNK activation is persistent, loss of HBO1 from origins would likely lead to DNA damage checkpoint activation and eventually apoptosis. Second, down-regulation of replication licensing may provide cells with additional time to mount a sufficient transcriptional response to adapt to the stress.
As JNK1 activity can be affected by a variety of physiological and pathological stimuli, JNK1-dependent regulation of Cdt1-HBO1 interaction may be important during cellular differentiation, cell propagation, viral infection and oncogenic transformation. For instance, JNK activity is down-regulated when cells enter in G1 (Bakiri et al., 2000; Gutierrez et al., 2010a), which may favor the interaction between HBO1 and Cdt1 and perhaps pre-RC assembly. In cells infected by Helicobacter pylori, JNK inhibition allows cells arrested in G1 to progress through S phase and G2 (Ding et al., 2007), leading to the speculation that this might be coupled with a DNA licensing defect. In these other biological contexts, the coordination between transcriptional and replication responses may involve JNK1-dependent phosphorylation of non-AP-1 transcription factors and increased recruitment of HBO1 to the relevant target promoters.
EXPERIMENTAL PROCEDURES
Plasmids and siRNA reagents
DNA constructs BK28-cFos (Morlon and Sassone-Corsi, 2003); pRSV-cJun, pRSV-cJunAsp (Musti et al., 1997); pGEX-cJun residues 1–253 (Ferguson and Goodrich, 2001); pcDNA3-Flag-Cdt1, pcDNA3-HA-Cdt1, pET28a-Cdt1 (Xouri et al., 2007); pGEX-ORC1 (Takayama et al., 2000); pAHT-Cdt1T29 and pAHT-Cdt1Δ32 (Takeda et al., 2005); pcDEBdelta-T7-Cdt1Cy (Sugimoto et al., 2004); pEFF-Flag-hsCdt1delta395–465 (Teer and Dutta, 2008) have been described elsewhere. JNK1 and kinase-deficient JNK1-APF were purchased from Add-gene. HBO1, JNK1, JNK2 and control siRNA duplexes were purchased from Ambion. The JNK1 vector for expression in E. coli and its use in the kinase assays was obtained from RIKEN DNA bank. Other plasmids and siRNA reagents were previously described (Miotto and Struhl, 2006).
Antibodies
Antibodies were obtained from the following suppliers: Santa Cruz biotechnology for anti-cJun, -cFos, -JNK1, -JNK2, -GAPDH; Cell Signaling for anti-phospho-JNK (Thr-183/Tyr185), anti-phospho-p38, -actin, -IgG, -ATM, -phospho-ATM, -phospho-CREB; Upstate for anti-H4 tetra-acetylated, -Serine/Threonine phosphorylation; Abcam for anti-acetyl-lysine; Sigma-Aldrich for anti-ubiquitin; Cell signaling for anti-phospho-ATM and phospho-Thr-Pro. Antibodies directed against pre-RC subunits (Cdt1, Cdc6, Geminin, HBO1, ORC2 and MCM2–7) were previously described (Miotto and Struhl, 2008). Protein A and protein G sepharose beads as well as control IgG-sepharose beads were purchased from Amersham Biosciences.
Cell culture, cell cycle synchronization, stress induction and transfection assays
HeLa, HCT116 and HEK293 cells were grown in Dulbecco’s modified eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics. Lymphoblastoid K562 cells were grown in RPMI-1640 medium supplemented with 10% serum, 2mM glutamine and antibiotics. HeLa cells were staged in mitosis by a nocodazole block (14 hours incubation in 100 nM nocodazole) and releases in normal culture media. To collect cells in G1 cells were harvested 8 hours after release from nocodazole block (Miotto and Struhl, 2008).
Stress conditions were induced in presence of 10% serum, which requires higher concentration of stress agents but avoids complications due to stress due to serum deprivation. The final concentrations of stress agents (all obtained from Sigma-Aldrich) in the medium were 300 mM Sorbitol, 50 ng/ml anisomysin, 2 mM DTT, 400 mM urea, 100 μM cisplatin, and 0.2 μg/ml actinomycin D. MG132 (Sigma-Aldrich), JNK II inhibitor (SP600125, 20 μM; Calbiochem), JNK I inhibitor (20 μM; Calbiochem), p38 inhibitor (SP203580, 40 μM; Calbiochem), Cdk2 inhibitor (Santa Cruz, sc-221409), and ATM inhibitor (caffeine, 15 mM) were diluted prior to use and added 30 minutes before stress induction to allow optimal inhibition of proteasome, JNKs, p38 and ATM kinases respectively. For transfection cells were plated at a density of 2×106 per well in 10 cm tissue culture plates and transfected using lipofectamine using a standard protocol provided by the manufacturer.
Co-immunoprecipitation assays
In vivo immunoprecipitation assays in HeLa cells were performed as previously described (Miotto and Struhl, 2006). Whole cellular extract were pre-cleared at 4°C using protein G sepharose beads in binding buffer 150 mM KCl, 20 mM Tris-HCl (pH 7.5), 20% glycerol, 5 mM DTT complemented with anti-protease inhibitors. Specific antibodies were coupled to protein G sepharose beads at 4°C and then incubated for 2 hours at 4°C with pre-cleared cellular lysate. Washes (5 times) were carried out with the same buffer, bound material eluted by boiling in electrophoresis sample buffer and proteins of interest were by Western blotting.
Chromatin loading
Cells (2×107) were scraped in ice-cold PBS (Sibani et al., 2005). After centrifugation, they were re-suspended in lysis buffer (10mM HEPES pH 7.9, 100mM NaCl, 300mM Sucrose, 0.1% triton X-100) containing protease inhibitors (Roche) and incubated on ice for 10 minutes. After centrifugation at 1000 rpm for 5 minutes, cells were washed in lysis buffer and re-suspended in extraction buffer (100mM HEPES pH 7.9, 200mM NaCl, 300mM sucrose, 0.1% triton X-100, 5mM MgCl2 containing 100U of DNAse I (Promega). Following incubation at 25°C for 30 minutes, the chromatin-enriched fraction was isolated by centrifugation at 2500 rpm for 5 minutes.
Chromatin immunoprecipitation
ChIP assays were performed on sonicated chromatin (average length 300 bp) from 5×107 cells using primary antibodies or IgG as previously described (Cawley et al., 2004). After immunoprecipitation overnight, DNA was purified with the QIAquick PCR purification kit (Qiagen). For sequential analysis (Geisberg and Struhl, 2004), chromatin-bound material after the first immunoprecipitation was heat-eluted for 20 minutes at 65°C, and the resulting material diluted and then immunoprecipitated with a second antibody overnight. Quantitative real-time PCR was performed using SYBR Green I. Enrichment for a specific DNA sequence was calculated using the comparative Ct method as previously described (Cawley et al., 2004). Data are normalized to a background region (GAPDH coding region) and expressed as relative binding value. All ChIP experiments involving chromatin from 3 independent preparations of cells, and hence involve 3 independent replicates. PCR primer pairs are available on request.
Phosphorylation assays
The E. coli phosphorylation assay (Murata et al., 2008) involved cells inducibly expressing JNK1 and constitutively expressing His- or GST-tagged substrates. JNK1 production was initiated by adding IPTG to the medium for 30 minutes, after which His-Cdt1, GST-cJun(1–133) and GST-ORC1 were purified with appropriate resins. The phosphorylation levels were analyzed by Western blotting. The IP/Kinase assay in mammalian cells was conducted using a protocol developed by manufacturers for cJun phosphorylation by JNKs. HEK 293T cells were treated with sorbitol for 1 hour, and JNK1 (or JNK1-APF) purified by affinity immunoprecipitation as described above. GST-ORC1, His-Cdt1 and GST-cterJun fusion proteins were immobilized on glutathione/nickel agarose beads at 4°C overnight. After washing, the bound proteins were incubated in a kinase buffer containing ATP (200 μM) for 30 min at 30°C. JNK activity was determined by western blot using a serine/threonine-phosphorylation antibody.
Mapping Cdt1 phosphorylation sites
For phosphorylation in vitro, His-Cdt1 was purified from E. coli and incubated for 30 min with active-JNK1 purified from HEK293 cells treated with sorbitol. After the reaction, His-Cdt1 was purified on Ni-NTA agarose and washed 3 times in buffer (300mM NaCl, 20mM imidazole) prior to elution. For phosphorylation in vivo, Flag-Cdt1 was immunoprecipitated from cells treated with sorbitol, sorbitol + JNK I or II inhibitor, or left untreated. Immunoprecipitation and washes were performed in RIPA buffer (0.1% SDS) supplemented with a cocktail of phosphatase inhibitors (20 mM sodium fluoride, 1 mM dosium orthovanadate, 50 μM sodium molybdate). All samples were run on a SDS-Page gel and stained with Coomassie Blue. The band corresponding to Cdt1 was excised from the gel and subjected to an in-gel tryptic digest followed by tandem mass spectrometry (in house; Université de Paris 7).
FACS analysis
In accord with standard procedures, 1×105 cells stained with propidium iodide (Sigma) were analyzed for each condition using a FACS-Calibur machine. Data were processed using the CellQuest software (Becton Dickinson).
Statistical analyses
To determine statistical significance a student t-test for comparison of two different groups was performed. P<0.05, *, was considered significant.
Supplementary Material
Highlights.
Upon non-genotoxic stress, JNK1 phosphorylates Cdt1 on threonine 29
Cdt1-T29 phosphorylation causes rapid dissociation of HBO1 from origins
Regulated and reciprocal recruitment of HBO1 to enhancers and origins
JNK1, via HBO1, coordinates the transcriptional and replication response to stress
Acknowledgments
We thank Dirk Bohmann, Roger Davis, Anyndia Dutta, Hideo Nishitani, Hiroyoshi Ariga, Masatoshi Fujita and Take Murata for reagents. Johannes Walter, Jonathan Weitzman and Moshe Yaniv are acknowledged for insightful comments during the course of this work and manuscript redaction. This work was supported by grants to K.S. from the National Institutes of Health (GM30186).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 2000;19:2056–2068. doi: 10.1093/emboj/19.9.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bogoyevitch MA, Kobe B. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiol Mol Biol Rev. 2006;70:1061–1095. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch MA, Ngoei KR, Zhao TT, Yeap YY, Ng DC. c-Jun N-terminal kinase (JNK) signaling: recent advances and challenges. Biochim Biophys Acta. 2010;1804:463–475. doi: 10.1016/j.bbapap.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Burke TW, Cook JG, Asano M, Nevins JR. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem. 2001;276:15397–15408. doi: 10.1074/jbc.M011556200. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Smentchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of non-coding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chen P, O'Neal JF, Ebelt ND, Cantrell MA, Mitra S, Nasrazadani A, Vandenbroek TL, Heasley LE, Van Den Berg CL. Jnk2 effects on tumor development, genetic instability and replicative stress in an oncogene-driven mouse mammary tumor model. PloS one. 2010;5:e10443. doi: 10.1371/journal.pone.0010443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Côté V, Selleck W, Lane WS, Tan S, Yang XJ, Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Duursma AM, Agami R. CDK-dependent stabilization of Cdc6: linking growth and stress signals to activation of DNA replication. Cell cycle (Georgetown, Tex. 2005;4:1725–1728. doi: 10.4161/cc.4.12.2193. [DOI] [PubMed] [Google Scholar]
- Ferguson HA, Goodrich JA. Expression and purification of recombinant human c-Fos/c-Jun that is highly active in DNA binding and transcriptional activation in vitro. Nucl Acids Res. 2001;29:E98. doi: 10.1093/nar/29.20.e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucl Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiakaki M, Chabbert-Buffet N, Dasen B, Meduri G, Wenk S, Rajhi L, Amazit L, Chauchereau A, Burger CW, Blok LJ, et al. Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol Endocrinol. 2006;20:2122–2140. doi: 10.1210/me.2005-0149. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Seto E. Acetylation/deacetylation modulates the stability of DNA replication licensing factor Cdt1. J Biol Chem. 2009;284:11446–11453. doi: 10.1074/jbc.M809394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez GJ, Tsuji T, Chen M, Jiang W, Ronai ZA. Interplay between Cdh1 and JNK activity during the cell cycle. Nat Cell Biol. 2010a;12:686–695. doi: 10.1038/ncb2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez GJ, Tsuji T, Cross JV, Davis RJ, Templeton DJ, Jiang W, Ronai ZA. JNK-mediated phosphorylation of Cdc25C regulates cell cycle entry and G(2)/M DNA damage checkpoint. J Biol Chem. 2010b;285:14217–14228. doi: 10.1074/jbc.M110.121848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell cycle (Georgetown, Tex. 2006;5:1675–1680. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat Cell Biol. 2003;5:1008–1015. doi: 10.1038/ncb1061. [DOI] [PubMed] [Google Scholar]
- Horiguchi K, Arai S, Nishihara T, Nishikawa J. AIB1 promotes DNA replication by JNK repression and AKT activation during cellular stress. J Biochem. 2006;140:409–419. doi: 10.1093/jb/mvj167. [DOI] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Hu J, Xiong Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem. 2006;281:3753–3756. doi: 10.1074/jbc.C500464200. [DOI] [PubMed] [Google Scholar]
- Hu W, Hofstetter W, Guo W, Li H, Pataer A, Peng HH, Guo ZS, Bartlett DL, Lin A, Swisher SG, et al. JNK-deficiency enhanced oncolytic vaccinia virus replication and blocked activation of double-stranded RNA-dependent protein kinase. Cancer Gene Ther. 2008;15:616–624. doi: 10.1038/cgt.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol. 2006;26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Sarmento OF, Sekiya T, Scrable H, Allis CD, Smith MM. Hbo1 links p53-dependent stress signaling to DNA replication licensing. Mol Cell Biol. 2008;28:140–153. doi: 10.1128/MCB.00662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- Iizuka M, Takahashi Y, Mizzen CA, Cook RG, Fujita M, Allis CD, Frierson HF, Jr, Fukusato T, Smith MM. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene. 2009;436:108–114. doi: 10.1016/j.gene.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T, Su B, Tsigelny I, Sluss HK, Derijard B, Moore G, Davis R, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- Karin M, Gallagher E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB life. 2005;57:283–295. doi: 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kobayashi M, Tanaka J, Yokoyama A, Suzuki S, Kato N, Onozawa M, Chiba K, Hashino S, Imamura M, et al. Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J Biol Chem. 2004;279:27315–27319. doi: 10.1074/jbc.M314023200. [DOI] [PubMed] [Google Scholar]
- Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol. 2011;31:845–860. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex's largest subunit (Orc1) from binding to chromatin during mitosis. Mol Cell Biol. 2004;24:5875–5886. doi: 10.1128/MCB.24.13.5875-5886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PloS one. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Miotto B, Struhl K. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol Cell Biol. 2006:5969–5982. doi: 10.1128/MCB.00696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K. HBO1 histone acetylase is a co-activator of the replication licensing factor Cdt1. Genes & Dev. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto-Yamasaki Y, Yamasaki M, Tachibana H, Yamada K. Induction of endoreduplication by a JNK inhibitor SP600125 in human lung carcinoma A 549 cells. Cell biology international. 2007;31:1501–1506. doi: 10.1016/j.cellbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Morlon A, Sassone-Corsi P. The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc Natl Acad Sci USA. 2003;100:3977–3982. doi: 10.1073/pnas.0735923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Shinozuka Y, Obata Y, Yokoyama KK. Phosphorylation of two eukaryotic transcription factors, Jun dimerization protein 2 and activation transcription factor 2, in Escherichia coli by Jun N-terminal kinase 1. Anal Biochem. 2008;376:115–121. doi: 10.1016/j.ab.2008.01.038. [DOI] [PubMed] [Google Scholar]
- Musti AM, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Rahmouni S, Cerignoli F, Alonso A, Tsutji T, Henkens R, Zhu C, Louis-dit-Sully C, Moutschen M, Jiang W, Mustelin T. Loss of the VHR dual-specific phosphatase causes cell-cycle arrest and senescence. Nat Cell Biol. 2006;8:524–531. doi: 10.1038/ncb1398. [DOI] [PubMed] [Google Scholar]
- Sabapathy K, Hochedlinger K, Nam SY, Bauer A, Karin M, Wagner EF. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol Cell. 2004;15:713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Saha T, Ghosh S, Vassilev A, DePamphilis ML. Ubiquitylation, phosphorylation and Orc2 modulate the subcellular location of Orc1 and prevent it from inducing apoptosis. J Cell Sci. 2006;119:1371–1382. doi: 10.1242/jcs.02851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibani S, Price GB, Zannis-Hadjopoulos M. Decreased origin usage and initiation of DNA replication in haploinsufficient HCT116 Ku80+/- cells. Journal of cell science. 2005;118:3247–3261. doi: 10.1242/jcs.02427. [DOI] [PubMed] [Google Scholar]
- Sugimoto N, Tatsumi Y, Tsurumi T, Matsukage A, Kiyono T, Nishitani H, Fujita M. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J Biol Chem. 2004;279:19691–19697. doi: 10.1074/jbc.M313175200. [DOI] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama MA, Taira T, Tamai K, Iguchi-Ariga SM, Ariga H. ORC1 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc-SNF5/INI1 interaction. Genes Cells. 2000;5:481–490. doi: 10.1046/j.1365-2443.2000.00338.x. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Parvin JD, Dutta A. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J Biol Chem. 2005;280:23416–23423. doi: 10.1074/jbc.M501208200. [DOI] [PubMed] [Google Scholar]
- Teer JK, Dutta A. Human Cdt1 lacking the evolutionarily conserved region that interacts with MCM2–7 is capable of inducing re-replication. J Biol Chem. 2008;283:6817–6825. doi: 10.1074/jbc.M708767200. [DOI] [PubMed] [Google Scholar]
- Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wong PG, Glozak MA, Cao TV, Vaziri C, Seto E, Alexandrow M. Chromatin unfolding by Cdt1 regulates MCM loading via opposing functions of HBO1 and HDAC11-geminin. Cell cycle (Georgetown, Tex. 2010;9:4351–4363. doi: 10.4161/cc.9.21.13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZQ, Liu X. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc Natl Acad Sci USA. 2008;105:1919–1924. doi: 10.1073/pnas.0712063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xouri G, Squire A, Dimaki M, Geverts B, Verveer PJ, Taraviras S, Nishitani H, Houtsmuller AB, Bastiaens PI, Lygerou Z. Cdt1 associates dynamically with chromatin throughout G1 and recruits Geminin onto chromatin. EMBO J. 2007;26:1303–1314. doi: 10.1038/sj.emboj.7601597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H, Jin YH, Park BD, Choi HJ, Lee SK. Caspase-3-mediated cleavage of Cdc6 induces nuclear localization of p49-truncated Cdc6 and apoptosis. Mol Biol Cell. 2003;14:4250–4259. doi: 10.1091/mbc.E03-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.