Abstract
Mice deficient for the gene encoding the RNA-binding protein CELF4 (CUGBP, ELAV-like family member 4) have a complex seizure phenotype that includes both convulsive and non-convulsive seizures, depending upon gene dosage and strain background, modeling genetically complex epilepsy. Invertebrate CELF is associated with translational control in fruit fly ovary epithelium and with neurogenesis and neuronal function in the nematode. Mammalian CELF4 is expressed widely during early development, but is restricted to the central nervous system in adult. To better understand the etiology of the seizure disorder of Celf4 deficient mice, we studied seizure incidence with spatial and temporal conditional knockout Celf4 alleles. For convulsive seizure phenotypes, it is sufficient to delete Celf4 in adulthood at seven weeks of age. This timing is in contrast to absence-like non-convulsive seizures, which require deletion before the end of the first postnatal week. Interestingly, selective deletion of Celf4 from cerebral cortex and hippocampus excitatory neurons, but not from inhibitory neurons, is sufficient to lower seizure threshold and to promote spontaneous convulsions. Correspondingly, Celf4 deficient mice have altered excitatory, but not inhibitory, neurotransmission as measured by patch-clamp recordings of cortical layer V pyramidal neurons. Finally, immunostaining in conjunction with an inhibitory neuron-specific reporter shows that CELF4 is expressed predominantly in excitatory neurons. Our results suggest that CELF4 plays a specific role in regulating excitatory neurotransmission. We posit that altered excitatory neurotransmission resulting from Celf4 deficiency underlies the complex seizure disorder in Celf4 mutant mice.
Keywords: epilepsy, genetics, RNA-binding proteins, neurotransmission, mice
Introduction
Common idiopathic epilepsy is thought to be largely genetic but with complex inheritance (Ottman et al. 1997; Ottman 2005). Rare, Mendelian idiopathic epilepsies are usually caused by mutations in ion channel or neurotransmitter receptor subunits (Gardiner 2005). Unfortunately, up to 30% of epilepsy patients do not achieve full seizure control with current drug therapies, which primarily modulate ion channel or gamma-aminobutyric acid (GABA) activity (Bialer & White 2010). These pharmacoresistant epilepsies could be due, in part, to more diverse underlying mechanisms.
Indeed, for known epilepsy genes, some exceptions to the “channelopathy” rule exist, including Lgi1 (Kalachikov et al. 2002) and Efhc1 (Suzuki et al. 2004). LGI1, a neuronal secreted protein, interacts with many synaptic proteins (Fukata et al. 2006; Schulte et al. 2006; Kunapuli et al. 2009) and regulates excitatory synaptic development and transmission (Fukata et al. 2006; Zhou et al. 2009; Fukata et al. 2010; Yu et al. 2010). EFHC1 is a microtubule-associated protein that regulates cell division and neuronal migration during development (de Nijs et al. 2009). These cases highlight roles of modulatory proteins that are one or more steps removed from neurotransmission itself.
One such mechanism for common epilepsy may involve dysregulation of RNA-binding proteins (RBPs) as implicated in humans and mice deficient for JRK/JH8 (Toth et al. 1995; Moore et al. 2001) and FMRP (Wisniewski et al. 1985; Musumeci et al. 2000). Ribonucleoprotein complexes (RNPs), comprised of proteins, mRNA, and non-coding RNA, perform essential roles in RNA metabolism (Glisovic et al. 2008). RBPs are key RNP components that recognize and bind specific subsets of RNAs. RBP dysregulation can affect the expression of many genes, often leading to disease. Significantly, RBPs have been implicated in many other complex neurological and neuropsychiatric disorders including amyotrophic lateral sclerosis and frontotemporal dementia (Lagier-Tourenne et al. 2010), mental retardation (Garber et al. 2008), autism (Kaufmann et al. 2004; Stein et al. 2006; Martin et al. 2007), and schizophrenia (Aberg et al. 2006).
Mice hypomorphic for CELF4 (CUGBP, ELAV-like family member 4), a brain-specific RBP, have a complex seizure disorder influenced by genetic background, making them an interesting model for common idiopathic epilepsy (Yang et al. 2007). Celf4 is expressed widely during development but is restricted to the adult CNS (Meins et al. 2002). Celf4 is a member of a RBP family in mammals, orthologous to an invertebrate RBP associated with translational control in Drosophila ovary epithelium and with neurogenesis and neuronal function in nematodes (Good et al. 2000; Ladd et al. 2001). Human CELF4 protein is 99.6% identical to mouse and 47% identical to the nematode ortholog, UNC-75 (Meins et al. 2002; Loria et al. 2003). UNC-75 mutations cause defects in neurotransmission that can be rescued by expression of human CELF4 (Loria et al. 2003), suggesting that they both regulate synaptic transmission.
To better understand the etiology of the seizure disorder in Celf4 mutant mice, here we study a gene-targeted null conditional allele, examining the effects of both temporal and spatial deletion on seizure type as well as on synaptic transmission.
Materials and Methods
Animals
All animals were fed standard National Institutes of Health diet containing 6% fat and acidified water ad libitum. All animal procedures followed Association for Assessment and Accreditation of Laboratory Animal Care guidelines and were approved by institutional Animal Care and Use Committee.
Generation of Celf4 gene-targeted null conditional allele
A 7 kb NheI/BsaA1 fragment of a 129S1/SvImJ BAC containing 4.2 kb 5’ and 2.2 kb 3’ to Celf4 exon 1 was subcloned into pBluescript. A loxP site was introduced 2.1 kb upstream and a loxP-frt-neo–frt fragment was inserted 444 bp downstream of exon 1 by recombineering (Celf4 reference sequence NM_001146292). R1 ES cells (129 strain background) were used for homologous recombination (Nagy et al. 1993), and ES cell clones were screened by PCR then confirmed by Southern blot before microinjecting two targeted ES cell clones into C57BL/6J blastocysts to make chimeras. The neomycin cassette was removed by mating to a mouse strain expressing flippase (FLPe). The mice were backcrossed at least 10 generations to C57BL/6J (B6J) or 129S1/SvImJ (129S1) to minimize variability in strain background that could affect phenotypic results. To distinguish the floxed and deleted alleles from wildtype, the following primers were used and gave the following products:
Primer 1: 5’AAGAGGAGATACTAGACACCTAGG 3’
Primer 2: 5’ AAGCATTTGCTACTACCAGAAGGG 3’
Primer 3: 5’ GATGCATGCTTTGCATACTTCTGC 3’
Primers 1 and 2 : 259bp (wildtype allele), 329bp (floxed allele with 5’ loxP insert)
Primers 1 and 3 : 459bp (deleted allele)
Generation of temporal and spatial conditional Celf4 mutants
Germline deletion of Celf4 on the B6J and 129S1 backgrounds was achieved by mating mice with the Celf4 null conditional allele to EIIA-cre (B6.FVB-Tg(EIIa-cre)C5379Lmgd/J) and Meox-cre (B6.129S4-Meox2tm1(cre)Sor/J) mice, respectively. Temporal and spatial conditional knockouts were made by mating mice with the Celf4 null conditional allele on the B6J background to ER-cre (B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J), Emx1-cre (B6.129S2-Emx1tm1(cre)Krj/J), CamK2a-cre (B6.Cg-Tg(Camk2a-cre)1Lfr/Mmcd), Pvalb-cre (B6;129P2-Pvalbtm1(cre)Arbr/J), or Viaat-cre (FVB-Tg(Slc32a1-cre)/Frk) mice. Pvalb-cre mice were incipient congenics backcrossed at least three generations to B6J when they were used. Mice to be tested for timing of absence seizure generation by Celf4 deletion at P7 and earlier were generated by crossing B6J mice with the Celf4 null conditional allele with Ubc-Cre mice (B6.Cg-Tg(UBC-cre/ERT2)1Ejb/J). These tamoxifen studies utilized Ubc-Cre instead of ER-Cre because Ubc-Cre has stronger, more uniform expression in the brain (our unpublished results).
Seizure threshold testing and observation of handling-associated seizures
Generalized seizure threshold was measured by ECT test as described previously (Boumil et al. 2010). Briefly, mice were restrained, a drop of anesthetic containing 0.5% tetracaine and 0.9% NaCl was placed onto each eye, and preset current was delivered via silver transcorneal electrodes using an electroconvulsive stimulator (Ugo Basile model 7801, Collegeville, PA, USA; rectangular wave pulses: 299 Hz, 0.2 s duration, 1.6 ms width, varying current). Individual mice were exposed once per day on sequential days until the first clear generalized seizure endpoint was observed - a minimal clonic forebrain seizure - and the average threshold for each group of mice was calculated. Mice were observed for handling-associated and spontaneous convulsive seizures during weekly cage change for at least 175 days. In order to minimize strain background effects and to maximize yield, with one exception ((FVB/NJ × B6J)F1-Viaat-cre)), all seizure threshold tests and comparison of handling-associated convulsions were done on a congenic B6J strain background.
Administration of tamoxifen
Tamoxifen (Sigma-Aldrich T5648) was dissolved at 20 mg/ml in corn oil by mixing overnight at room temperature and stored at 4°C for up to one week. The tamoxifen was administered to adult mice by oral gavage using a curved 22G animal feeding needle once a day for 5 consecutive days at the following doses, 6 mg/day (0.30 ml) for 20–24 g body weight and 7 mg/day (0.35 ml) for 25–30 g body weight. Mice were tested 12 days after the last treatment to ensure total loss of the gene product and avoid acute effects of tamoxifen treatment. Corn oil alone was administered as above as a sham treatment control. The tamoxifen was administered to P7 and younger mice by a single intraperitoneal injection of 0.05 ml of 10 mg/ml tamoxifen in corn oil (0.5 mg total tamoxifen).
EEG recording
EEG analysis was performed as previously described (Yang et al. 2007), all on a (B6J × 129S1) F2 hybrid background in order to both maximize survival of Celf4null homozygotes and yield of spike-wave discharges (SWD). Briefly, mice were anesthetized with tribromoethanol (400 mg/kg, intraperitoneal). Small burr holes were drilled on both sides of the skull, and the exposed tips of four Teflon-coated silver wires soldered onto a microconnector were placed between the dura and the brain, 1 mm on each side from midline, (two electrodes in front of bregma and two behind). A dental cap was then applied. The mice were given a post-operative analgesic of carprofen (5 mg/kg, subcutaneous) and were given a 48-hour recovery period before recordings were made. The general health of electrode-implanted mice was monitored daily. EEG activity was recorded for two 2-hour periods on consecutive days.
Immunohistochemistry
Mice were anesthetized with tribromoethanol and perfused with phosphate buffered saline (PBS pH 7.4), followed by paraformaldehyde (4%) in PBS. Brains were removed from the skull, post-fixed overnight in the same fixative (4°C), rinsed in PBS, and sectioned at 50 µM on a Leica VT1200 vibrating blade microtome (Leica Microsystems, Inc., Buffalo Grove, IL, USA). Sections were incubated in blocking buffer (0.3% TritonX-100, 1% BSA, 3% normal goat serum in PBS) for 2 hours at room temperature and transferred into primary antibody diluted in blocking buffer. After incubating for 44 hours at 4°C, sections were washed 3 times for 10 minutes each in PBST (PBS, 0.05% Tween 20) and incubated for 2 hours at room temperature in secondary antibody diluted in blocking buffer. The sections were washed as before, followed by a wash in PBS, and incubated in DAPI diluted in PBS for 5 minutes. The sections were mounted on lysine-coated glass slides in Fluorogel mounting media (Electron Microscopy Sciences, Hatfield, PA, USA). The following primary antibody was used: anti-CELF4, 1:400 (Santa Cruz sc84712, Santa Cruz, CA, USA). The secondary antibodies were: goat anti-rabbit Alexa Fluor 488, 1:1000 (Invitrogen A11070, Carlsbad, CA, USA) for CELF4- only IHC and goat anti-rabbit Alexa Fluor 647, 1:1000 (Invitrogen A21244, Carlsbad, CA, USA) for CELF4 IHC with tdTomato.
Western blot
Cortical or hippocampal tissue from wildtype and Celf4null homozygous (germline deletion with Meox-cre) mice was homogenized in RIPA lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors (Roche, complete mini; Indianapolis, IN, USA) and incubated for 30 minutes at 4°C. The lysate was centrifuged at 4,500g for 5 minutes at 4°C. Protein in the supernatant was quantified using the Bradford reagent (Bio-Rad; Hercules, CA, USA). Protein (300 µg from hippocampus, 500 µg from cortex) was subjected to SDS-PAGE, transferred to nitrocellulose, and probed with anti-CELF4 antibody, 1:200 (Santa Cruz sc84712, Santa Cruz, CA, USA) and goat anti-rabbit peroxidase-conjugated secondary antibody, 1:5000 (Bio-Rad; Hercules, CA, USA). Signal was detected with the ECL-plus kit (GE Healthcare). The blot was stripped with Restore stripping buffer (Thermo Scientific, Rockford, IL, USA) and reprobed with anti-β-tubulin antibody, 1:2000 (Sigma-Aldrich T4026) and goat anti-mouse peroxidase conjugated secondary, 1:5000 (Thermo Scientific 31430, Rockford, IL, USA).
Whole-cell patch-clamp recording and data analysis
Acute brain slices were prepared from Celf4null heterozygotes and homozygotes, and wildtype littermates between P14 and P21. To maximize the yield of homozygotes while minimizing strain background effects, all patch-clamp recording studies were done on the 129S1 strain background. Mice were anesthetized with tribromoethanol (250 mg/kg, i.p.) and decapitated. Brains were quickly removed and transferred into ice-cold solution containing: 210 mM sucrose, 3.0 mM KCl, 1.0 mM CaCl2, 3.0 mM MgSO4, 1.0 mM NaH2PO4, 26 mM NaHCO3, 10 mM glucose, saturated with 95% O2 and 5% CO2. Coronal slices were cut at 300 µm on a Leica VT 1200 vibrating blade microtome (Leica Microsystems, Inc., Buffalo Grove, IL, USA) and kept in artificial cerebral spinal fluid (ACSF) containing: 124 mM NaCl, 3.0 mM KCl, 1.5 mM CaCl2, 1.3 mM MgSO4, 1.0 mM NaH2PO4, 26 mM NaHCO3, and 20 mM glucose, saturated with 95% O2 and 5% CO2 at room temperature (21–23°C). Slices were allowed to recover for at least 1 hour before any recording. Each slice was transferred to a submerge-type chamber where it was continuously exposed to ACSF heated to 31–32°C, saturated with 95% O2 and 5% CO2, and flowing at a rate of ~2 ml/min. Whole-cell patch-clamp recordings were made at the soma of layer V pyramidal neurons of the visual cortex using a 40X water immersion objective (40X/0.80W, Zeiss). Patch pipettes were pulled from thick wall borosilicate glass (1.5/0.86 mm; Sutter Instruments, Novato, CA, USA) on a horizontal puller (P-97; Sutter Instruments, Novato, CA, USA). Resistance of electrodes was between 2 and 4 MΩ. The pipette solution contained: 100 mM CsCH3SO3, 4 mM ATP-Mg, 20 mM HEPES, 15 mM CsCl, 0.5 mM EGTA, pH to 7.2 with CsOH. Liquid junction potential was not corrected. Recordings were made with a Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA, USA). The series resistance (Rs), usually between 8 and 15 MΩ, was monitored throughout the recording, and data were not included in further analysis when Rs varied by 20% or more during recording. The quantal EPSCs and IPSCs were recorded in the presence of tetrodotoxin (TTX, 1–3 µM) on reverse potentials of GABA and glutamate receptors, respectively. Data were filtered at 2 kHz, sampled at 10 kHz and digitized by an ITC-18 interface. All analysis was performed using consumer routings in Matlab (MathWorks, Natick, MA, USA). Quantal EPSCs were detected and thresholded at 3.5 times the standard deviation of baseline noise. Quantal IPSCs were thresholded at 2 times of the standard deviation of baseline noise. The average of all isolated events in each cell was normalized and used as a template to fit each single event. Two hundred events in first 400 good fits were randomly selected and then averaged to give the mean response for each cell.
Results
Celf4null mice have a complex seizure phenotype
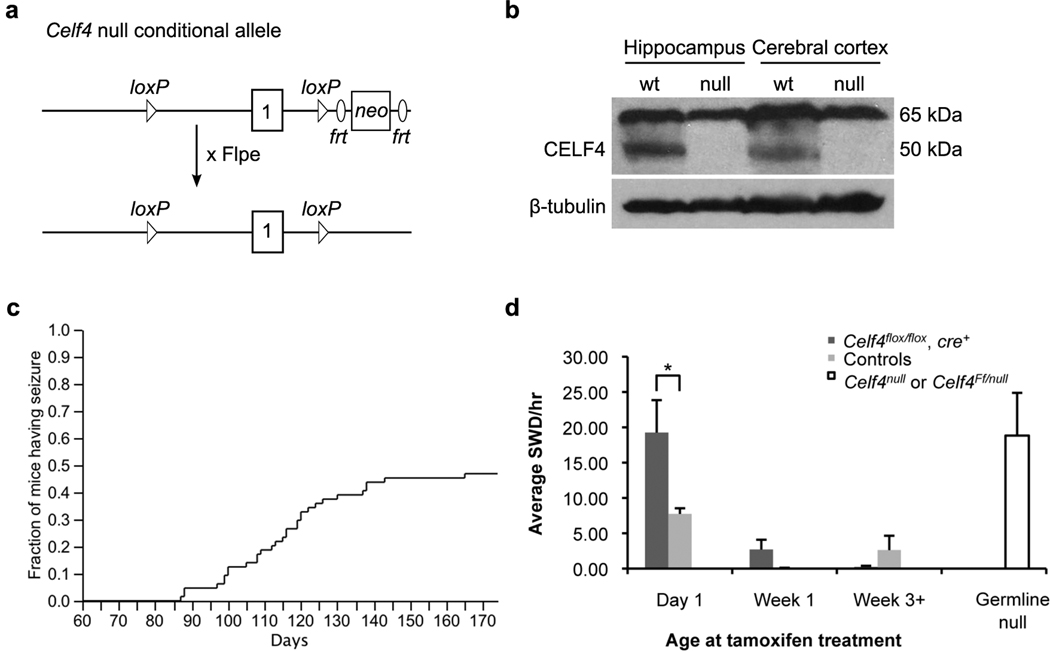
We created a gene-targeted null conditional allele of Celf4 (Figure 1a–b) using standard protocols as described in Materials and Methods. As expected based on the hypomorphic frequent-flyer allele of Celf4 (Yang et al. 2007), Celf4null mice have a genetically complex seizure disorder. Celf4null heterozygotes and homozygotes have recurrent limbic and tonic-clonic grand-mal like seizures that are often precipitated by routine handling, but can occasionally be observed spontaneously. Celf4null homozygotes experience tonic-clonic seizures starting as early as day 87 for mice on the 129S1/SvImJ (129S1) background, with 46% developing convulsive seizures by day 175 (Figure 1c). Celf4null homozygotes on the C57BL/6J (B6J) background have tonic-clonic seizures as early as day 21, with 5 out of 7 mice (71%) developing convulsive seizures thus far (observed up to 90 days, data not shown). Celf4null homozygotes also suffer from sporadic postnatal lethality, with B6J homozygotes showing a higher degree of penetrance than 129S1 homozygotes (Table 1). Interestingly, for Celf4 mutants, the B6J background permits a more severe convulsive phenotype than 129S1, despite the fact that B6J mice are generally resistant to experimentally-induced seizures (Frankel 2009), This may reflect the role of specific genetic effects of B6J alleles with reduced or absent Celf4; similar strain background effects were noted previously in Celf4Ff mice (Yang et al. 2007). Although Celf4null homozygotes show increased viability on the 129S1 genetic background, they are usually smaller than wildtype and heterozygote littermates. However, Celf4null mutants have an overtly normal brain morphology and normal lifespan (data not shown).
Figure 1. Summary of Celf4null allele construction and seizure incidence.
a) Schematic of targeting construct for generation of the Celf4 null conditional allele. A loxP site was introduced upstream of Celf4 exon 1, and a loxP-frt-neo-frt site was introduced downstream, creating the Celf4flox allele. The neo cassette was removed by mating to an FLPe mouse strain. Celf4flox mice were backcrossed at least ten generations to either C57BL/6J or 129S1. b) CELF4 protein expression is abolished in a Celf4null homozygote (129S1 background strain shown). Cortices from wildtype or Celf4null homozygous mice were homogenized in lysis buffer, and total protein was extracted and processed for western blot. The blot was probed with CELF4 antibody and signal was detected using a peroxidase-conjugated secondary antibody. CELF4 protein is observed as an approximately 50 kDa band in the wildtype lanes only. A nonspecific band of 65 kDa is observed in all lanes. The blot was stripped and probed with β-tubulin as a loading control. c) Celf4null homozygotes exhibit handling-associated spontaneous seizures. Mice were observed for spontaneous convulsive seizures after weekly cage change for at least 175 days. A curve showing the fraction of mice (129S1) with at least one observed seizure at each timepoint is shown (64 mice total). d) Celf4 must be deleted early to generate the non-convulsive (absence) seizure phenotype. EEG recordings were made from mice receiving tamoxifen by intraperitoneal injection (Day 1, Week 1) or oral gavage (Week 3+). Experimental mice were Celf4flox/flox and cre+, and control mice were either Celf4+/+ with or without cre expression or Celf4flox/flox without cre expression. The average number of spike-and-wave discharges (SWD) per hour ± SEM is shown (total number of mice recorded: Day 1, n = 5; Week 1, n = 16; Week 3+, n = 12). The average number of SWD/hr for a germline Celf4null homozygote or compound heterozygote (Celf4Ff/null) is shown for comparison (total number of mice recorded: 6). In Celf4null mutants, SWD are not observed on strain backgrounds that are predominantly B6J; all mice shown here are on the (B6J × 129S1)F2 hybrid background.
Table 1.
Survival of Celf4null homozygotesa
| Celf4 genotype | 129S1 | C57BL/6J |
|---|---|---|
| +/+ | 112 | 30 |
| +/− | 224 | 56 |
| −/− | 74 | 7 |
| (%) | (18) | (7.5) |
The Celf4 gene-targeted null conditional allele was generated on the 129S1 background, and mice were backcrossed 10 generations to 129S1 or C57BL/6J. For deletion, 129S1 and C57BL/6J null conditional mice were crossed with Meox-cre or EIIA-Cre mice, respectively. The number of animals with a specific genotype (examined at weaning age) is shown. The percent that were homozygous null is shown in the bottom row in parentheses (expect 25% from Mendelian law).
Just like the Celf4Ff allele (Yang et al. 2007), Celf4null homozygotes also experience non-convulsive, or absence, seizures on a mixed B6J ×129S1 F2 hybrid strain background and in inbred 129S1 mice, as indicated by spike-and-wave discharges (SWD) in the electroencephalogram (EEG; Supplemental Figure 1). Here, by using temporal conditional mice, we find that Celf4 must be deleted very early in postnatal development to generate an appreciable incidence of SWD (Figure 1d). EEG recordings from mice with tamoxifen-inducible cre expression (Ruzankina et al. 2007) revealed that deletion of Celf4 by postnatal day one is required for emergence of a high incidence of SWD (Figure 1d). Deletion at P5-7 (Week 1, Figure 1d) or later does not increase the SWD to a rate appreciably higher than what is seen after tamoxifen treatment alone (Figure 1d).
Celf4 deletion in adulthood or from excitatory neurons is sufficient to promote convulsive seizures
Seizures associated with epilepsy are thought to occur because of primary defects in excitability or synaptic transmission, or developmental miscues that create an imbalance between specific excitatory and inhibitory circuits (Noebels 2003). We studied both temporal and spatial conditional knockout mice to explore the etiology of the seizure disorder in Celf4null mice. We deleted Celf4 in adult mice using tamoxifen-inducible ER-Cre (Hayashi & McMahon 2002) and assessed their seizure susceptibility by measuring electroconvulsive threshold (ECT) - often a component of epilepsy susceptibility (Frankel 2009) - and by observing grand-mal like convulsions after routine handling. Control mice for these experiments consisted of a combination of wildtype mice with or without cre expression, mice expressing floxed Celf4 but not cre, or mice that were sham treated (N.b. Supplemental Table 1 specifies the numbers and genotypes of the controls for each group undergoing ECT testing). We also tested mice with a germline deletion of Celf4 generated with EIIA-Cre (Lakso et al. 1996), as well as mice with excitatory or inhibitory neuron-specific somatic deletions.
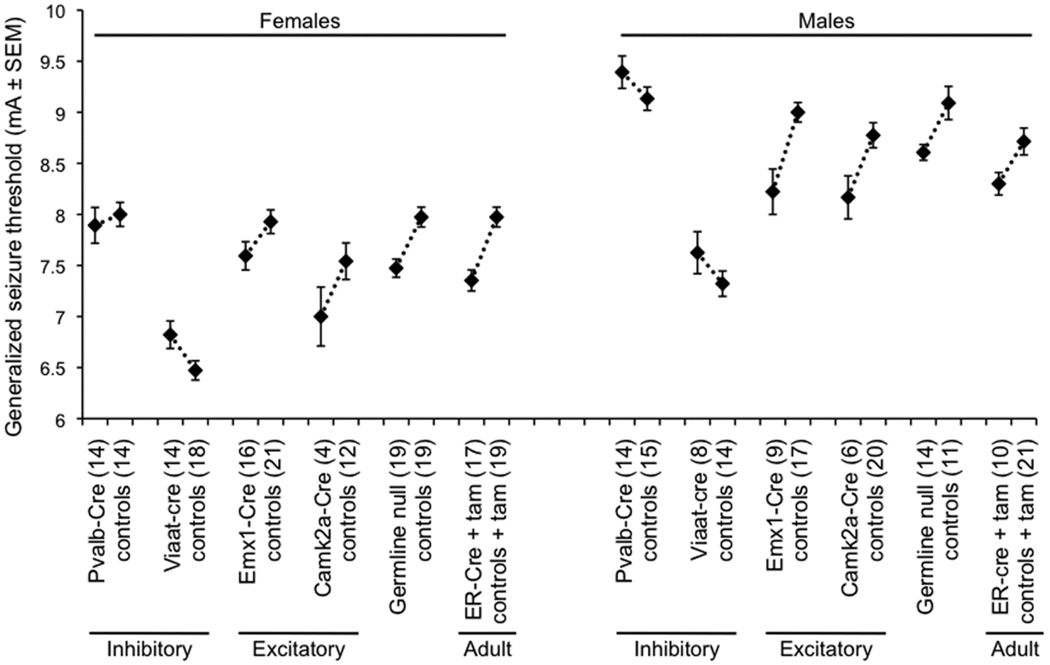
In ER-cre positive, Celf4null heterozygotes or homozygotes treated with tamoxifen at seven weeks, specific deletion of Celf4 conferred the same low ECT as that of germline mutants (Figure 2), and it also induced convulsions after routine handling (Table 2). Mice were tested twelve days after completing treatment to avoid complication of test results due to acute effects of tamoxifen. Mice with specific Celf4 deletion from excitatory neurons, conferred by Camk2a-cre (Xu et al. 2000) or Emx1-cre (Gorski et al. 2002), showed a very low ECT (Figure 2), and many mice of the same genotype also developed convulsions after routine handling (Table 2). In contrast, mice with inhibitory neuron-specific deletion, conferred by Pvalb-cre (Hippenmeyer et al. 2005) or Viaat-cre (Chao et al. 2010), showed no ECT decrease (Figure 2) nor have any handling-associated convulsions been observed to date (Table 2).
Figure 2. Reduced seizure threshold in excitatory neuron-specific or adult conditional Celf4null mice.
Generalized seizure threshold was measured by ECT test in conditional knockout mice. The average seizure threshold in milliamps (mA) ± SEM for individual mice in each group is shown; threshold values for female (left) and male (right) mice are shown separately because female mice typically have a lower standard threshold (Frankel et al. 2001). The number of mice in each test group is indicated in parentheses beside the group description on the x-axis. Experimental mice were cre+ and either Celf4flox/flox homozygotes or Celf4flox/+ heterozygotes. Experimental ER-Cre mice and littermate controls were treated with tamoxifen (tam) by oral gavage at seven weeks of age for five sequential days and tested twelve days after completion of treatment. Control mice for each experiment were either Celf4flox/flox homozygotes or Celf4flox/+ heterozygotes that were cre−, Celf4+/+ with or without cre expression, or a combination of these groups (see Supplemental Table 1). All mice were on a C57BL/6J strain background, except for the Viaat-Cre mice which were tested on the (C57BL/6J × FVB/NJ)F1 hybrid background, which has a significantly lower seizure threshold than C57BL/6J itself as seen in the figure. A regression fit model was used to assess statistical significance incorporating both sex and treatment (experimental versus control) as independent covariates. The reduced ECTs of the germline, the adult, and the excitatory neuron-specific deletions were statistically significant. P-values were: germline (EIIa-Cre, p < 0.0001); adult (ER-Cre, p < 0.0001); excitatory (Camk2a-Cre, p < 0.01; Emx1-Cre, p < 0.001); inhibitory (Pvalb-Cre, p > 0.5; Viaat-Cre, p < 0.01).
Table 2.
Seizure incidence in Celf4 spatial and temporal conditional mutantsa
| Condition | Cre driver |
% mice ≥1 seizure |
Total no. of mice observed |
Strain backgroundb |
p-value |
|---|---|---|---|---|---|
| Spatial deletion | |||||
| flox/flox or flox/+, cre+ | Pvalb | 0 | 25 | C57BL/6J | ns |
| controls | Pvalb | 0 | 21 | ” | |
| flox/flox or flox/+, cre+ | Emx1 | 12.9 | 31 | ” | 0.06 |
| controls | Emx1 | 0 | 29 | ” | |
| flox/flox or flox/+, cre+ | Camk2a | 26.4 | 53 | ” | 0.00004 |
| controls | Camk2a | 3.3 | 61 | ” | |
| (F1 hybrid background) | |||||
| flox/+, cre+ (males) | Viaat | 0 | 8 | (C57BL/6J × FVB)F1 | 0.21 |
| Ff/+ (males) | N/A | 20 | 30 | “ | |
| Temporal deletion | |||||
| flox/+, cre+ | EIIa | 55.6 | 36 | C57BL/6J | 0.00008 |
| controls | EIIa | 4.8 | 21 | ” | |
| flox/flox or flox/+, cre+ (+tam) | ER | 20 | 30 | ” | 0.02 |
| controls | ER | 0 | 25 | ” | |
Spatial Cre Celf4 null conditional deletions were driven by inhibitory neuron-specific promoters (Pvalb-cre and Viaat-cre) or excitatory neuron-specific promoters (Emx1-cre and Camk2a-cre). Temporal deletion included germline (EIIa-cre) and adult (ER-cre expression induced at seven weeks). Incidence of handling-associated spontaneous seizures during weekly cage change was observed for each condition until mice were 25 weeks old. Experimental ER-cre mice were treated with tamoxifen (tam) by oral gavage at seven weeks of age for five sequential days. Control mice were either cre−, Celf4+/+, sham treated, or a combination of these groups. P-values were determined using the Fisher Exact test, ns = not significant.
Because the Viaat-Cre mouse was available on an FVB/NJ background, the experimental Viaat-Cre mice have a different strain background than the others: (C57BL/6J × FVB)F1.
Celf4 is expressed predominantly in excitatory neurons
We hypothesized that deletion of CELF4 from excitatory neurons in cerebral cortex and hippocampus was primarily responsible for the low seizure threshold and routine handling-associated seizures seen in the CamK2a-cre and Emx1- cre mice since the Viaat- cre mice were unaffected. One possibility for this excitatory neuron-specific effect is that Celf4 may be expressed only in excitatory neurons, at least in cerebral cortex and hippocampus, and thus may only exert a function in that neuron type. Alternatively, Celf4 may be expressed in several types of neurons, including inhibitory neurons, reflecting a selective function in each neuron type. Immunohistochemistry was performed to discriminate between these possibilities.
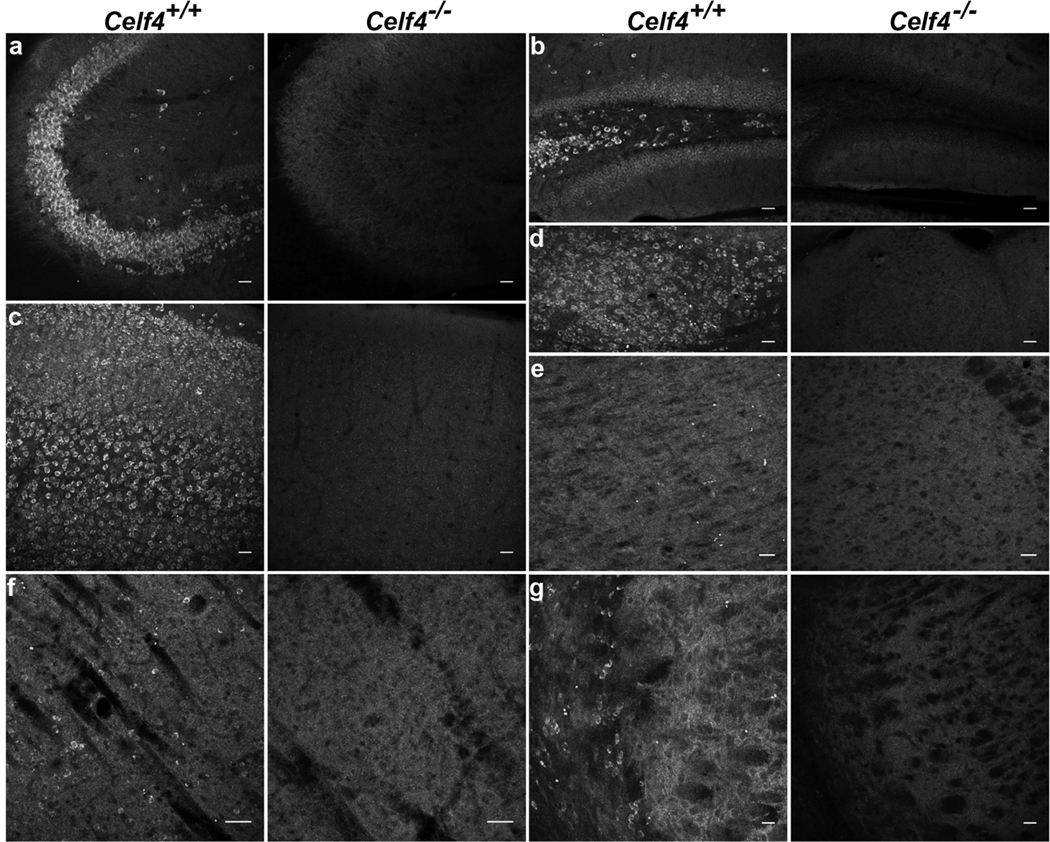
Immunostaining of CELF4 protein in 129S1 adult mouse brain showed that CELF4 is expressed highly in key regions involved in epilepsy circuits, namely cerebral cortex and hippocampus, and is also significantly expressed in hypothalamus but not in thalamus (Figure 3a–e). Notably, CELF4 expression is highest in pyramidal neurons in the CA3 region of the hippocampus and in layers II/III, V, and VI of the cerebral cortex, which are all highly glutamatergic areas of the brain. In contrast, CELF4 is expressed sparsely in highly GABAergic regions, such as striatum and reticular thalamus (Figure 3f–g).
Figure 3. CELF4 protein expression in adult mouse brain.
a–g: Sagittal sections from 129S1 adult wildtype (Celf4+/+) and homozygous null (Celf4−/−) mouse brains were examined for CELF4 protein expression by immunohistochemistry with α-CELF4 antibody. Confocal maximum projections of CELF4 staining showed that CELF4 is strongly expressed in pyramidal neurons in the CA3 region of the hippocampus (a), in the dentate gyrus (b), in the cerebral cortex (c), and in the hypothalamus (d). No specific staining is shown in thalamus (e), and CELF4 is expressed sparsely in striatum (f) and reticular thalamus (g). Scale bars: 50 µm.
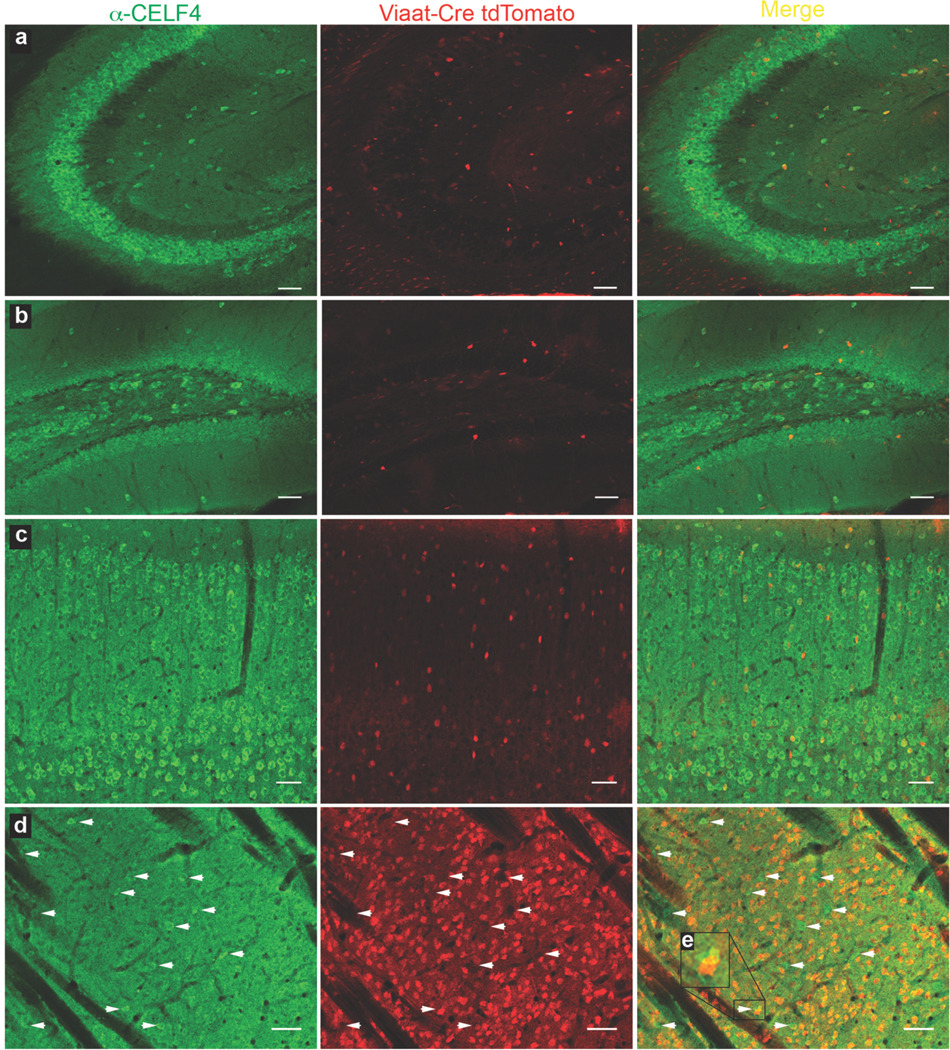
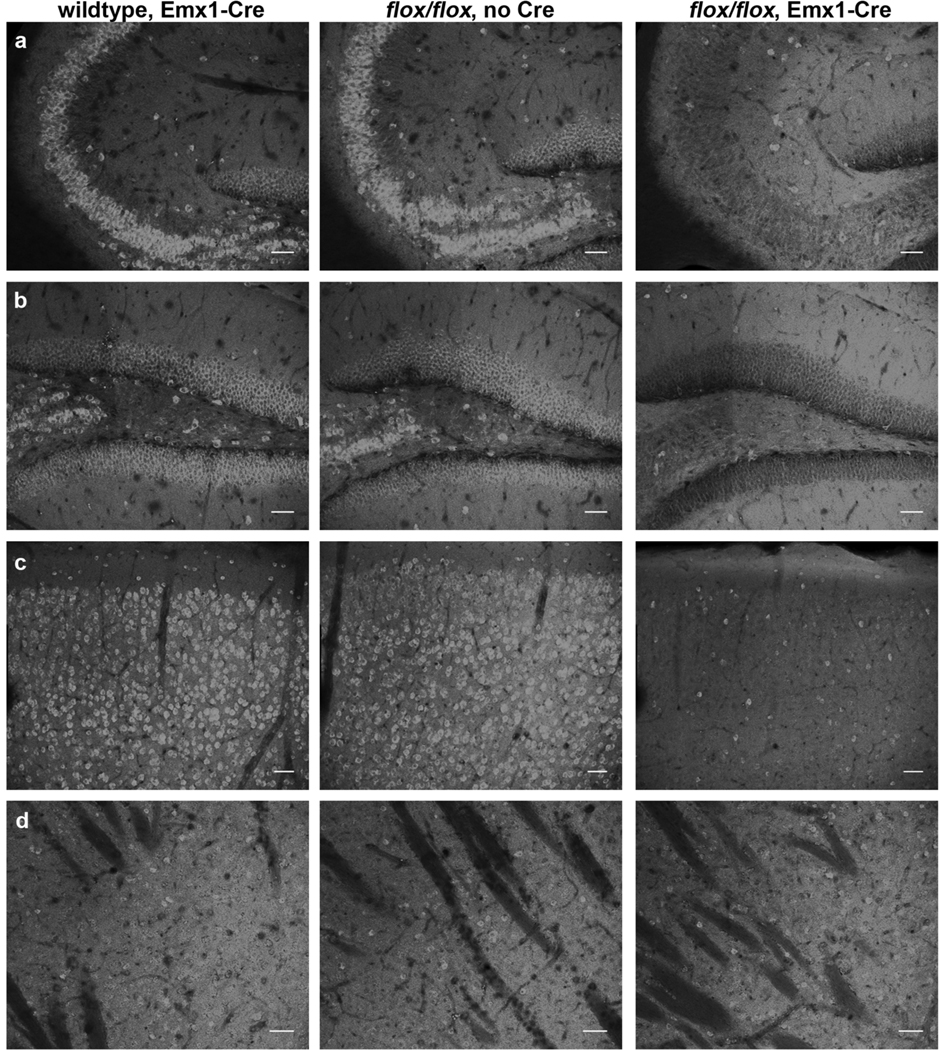
To determine the extent to which, if at all, CELF4 is present in inhibitory neurons in adult mice, we utilized a Cre-responsive fluorescent reporter strain. In this mouse line, CAG promoter-driven expression of the red fluorescent protein variant tdTomato is inhibited by a floxed STOP cassette; Cre-mediated recombination results in tdTomato fluorescence in the cre-expressing tissue (Madisen et al. 2010). We crossed Viaat-cre mice with the Cre-responsive reporter strain to mark inhibitory neurons and immunostained for CELF4. In cerebral cortex, hippocampus, striatum, and reticular thalamus, CELF4 is expressed predominantly in excitatory neurons while only expressed marginally in inhibitory neurons (Figure 4a–d). Additionally, in Celf4flox/flox mice with Emx1-cre expression, the number of CELF4-staining excitatory neurons in cerebral cortex and hippocampus is greatly reduced while the number of inhibitory CELF4-staining neurons remains unchanged (Figure 5a–d). Neither Celf4flox/flox nor Emx1-cre expression alone alters the expression pattern of CELF4 (Figure 5a–d).
Figure 4. CELF4 protein is expressed predominantly in excitatory neurons.
a–d: Sagittal sections from B6J adult wildtype mice with Viaat-cre recombinase-induced tdTomato expression were examined by immunohistochemistry with α-CELF4 antibody for colocalization between CELF4 (green) and inhibitory neurons (red). Confocal imaging showed that in the CA3 region of the hippocampus (a), the dentate gyrus (b), and the cerebral cortex (c), the majority of cells are CELF4-positive excitatory neurons. Only a small subset of the inhibitory neurons in these regions is CELF4-positive. In the striatum (d), the majority of neurons are CELF4-negative inhibitory neurons. The small population of CELF4-positive neurons (designated by white arrows) in the striatum is distinct from the tdTomato expressing population, indicating that CELF4-positive neurons in the striatum are excitatory. In some cases, overlap of a green pseudo-colored CELF4-positive neuron with a red pseudo-colored tdTomato-positive inhibitory neuron resulted in yellow signal when merged, however upon close inspection the two signals clearly come from different cells (inset e). Scale bars: 50 µm.
Figure 5. The expression of CELF4 protein is greatly reduced in Celf4flox/flox mice with Emx1-cre.
a–d: Sagittal sections from B6J adult mice with either wildtype Celf4 and Emx1-cre expression, floxed Celf4 expression (flox/flox) without Cre, or floxed Celf4 and Emx1-cre expression were examined for CELF4 protein expression by immunohistochemistry with α-CELF4 antibody. Confocal imaging showed that expression of either Emx1-Cre or floxed Celf4 alone did not affect the high levels of CELF4 expression observed in the highly glutamatergic CA3 region of hippocampus (a), dentate gyrus (b), or cerebral cortex (c) nor the sparse expression of CELF4 observed in the highly GABAergic striatum (d). However, expression of floxed Celf4 in conjunction with Emx1-cre greatly reduces CELF4 protein expression specifically in excitatory neurons in hippocampus and cerebral cortex (a–c), but does not affect CELF4 expression in inhibitory neurons, such as those seen in abundance in the striatum. This result was predicted from the known expression pattern of the excitatory-neuron specific Emx1 promoter, and shows that the seizure phenotype of Celf4flox/flox, Emx1-cre+ mice is indeed correlated with specific deletion of CELF4 from excitatory neurons. Scale bars: 50 µm.
Celf4 deletion boosts excitatory synaptic transmission
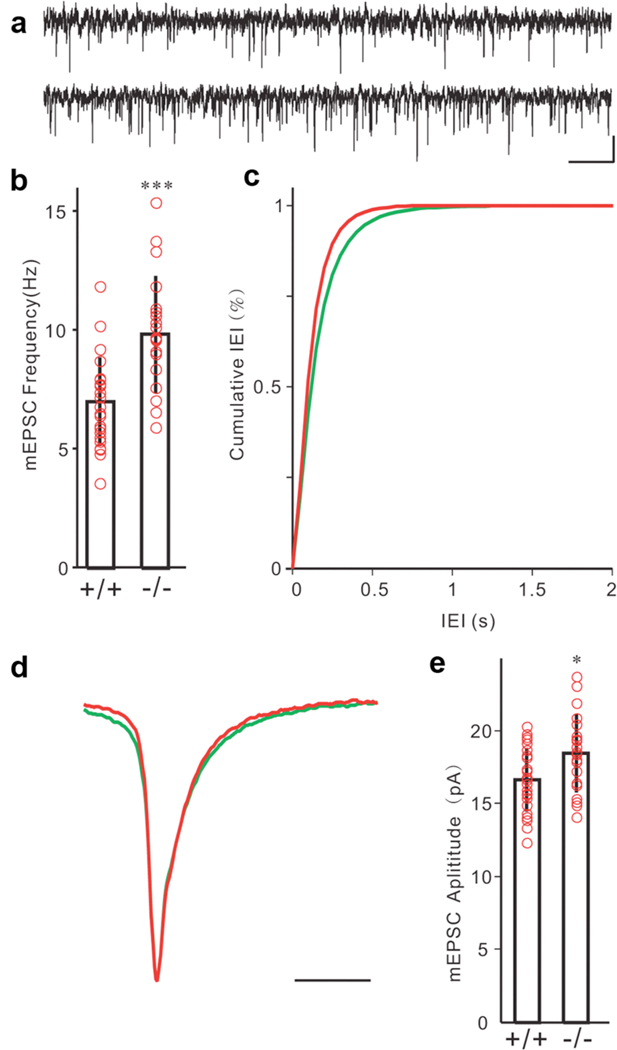
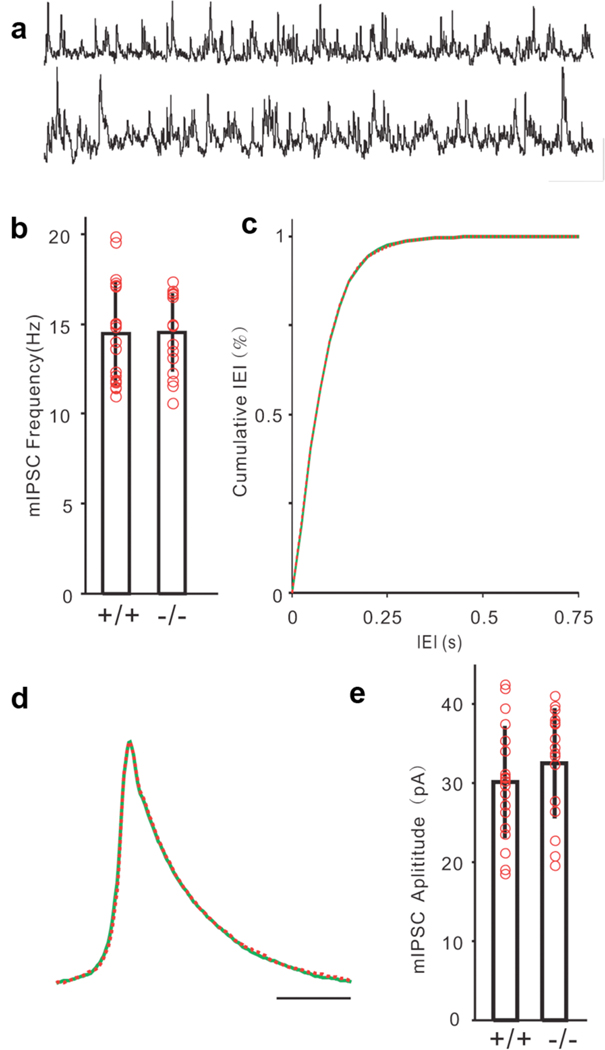
Since CELF4 was expressed mainly in excitatory neurons, we hypothesized that Celf4 deficiency would only directly alter the function of excitatory synapses, potentially resulting in neural network hyperexcitability that could induce seizures. To test this hypothesis, we recorded quantal synaptic activities to examine the possible change of synaptic inputs to cortical layer V pyramidal neurons. Miniature excitatory post-synaptic currents (mEPSCs) were recorded at the reverse potential of GABAergic channels (−70 mV) in the presence of tetrodotoxin (TTX; Figure 6a, upper: Celf4+/+, lower: Celf4null). Quantal analysis of mEPSCs showed that the frequency of mEPSCs was dramatically increased in the Celf4null homozygotes (n= 20) compared to the wildtype animals (n = 24, Figure 6b–c), which indicated increased glutamate release or increased glutamatergic synapse number. The amplitude of mEPSCs was also larger in Celf4 deficient mice (Figure 6e), suggesting alterations in the response of post-synaptic components. However, we did not detect any changes in mEPSC kinetics (Figure 6d). Miniature inhibitory post-synaptic currents (mIPSCs) were recorded at the reverse potential of glutamate receptors with TTX (Figure 7a, upper: Celf4+/+, lower: Celf4null). We detected no difference between wildtype and mutant mice in release frequency (Figure 7b–c), kinetics (Figure 7d) or amplitude (Figure 7e).
Figure 6. Altered excitatory synaptic transmission in Celf4 mutants.
a) Samples of mEPSCs recorded from a wildtype (+/+, upper trace) and a Celf4null homozygote (−/−, lower trace) layer V pyramidal neuron in the presence of TTX (scale bars: 10 pA, 500 ms). b) Frequency of mEPSCs was significantly higher in Celf4null homozygotes compared with the wildtype mouse (*** p = 0.00014, Mann-Whitney-Wilcoxon test). c) Cumulative distribution of mEPSC frequency also showed significant difference between mutant and wildtype control mice (bin = 50 ms). d) Averaged mEPSCs from 24 wildtype cells (green) and 20 Celf4null cells (red) aligned with normalized peaks. e) Amplitudes of mEPSCs were larger in the mutant mice than in wildtype control (* p = 0.0207, Mann-Whitney-Wilcoxon test). Total number of mice: 8 +/+, 7 −/−. Total number of cells: 24 +/+, 20 −/−. Histograms represent mean ± standard deviation. Red circles represent individual cells.
Figure 7. Normal inhibitory synaptic transmission in Celf4 mutants.
a) Samples of mIPSCs recorded from a wildtype (+/+, upper trace) and a Celf4null homozygote (−/−, lower trace) layer V pyramidal neuron in the presence of TTX (Scale bars: 30 pA, 250 ms). b) Frequency of mIPSCs was similar in Celf4null compared with the wildtype mice (p = 0.467, Mann-Whitney-Wilcoxon test). c) Cumulative distribution of mIPSC frequency failed to show significant difference between mutant and wildtype control mice (bin = 25 ms, p = 0.999, Kolmogorov-Smirnov test). d) Averaged mIPSCs from 19 wildtype cells (green) and 16 Celf4null cells (red) aligned with normalized peaks. e) Amplitudes of mIPSCs were similar in the mutant mice to wildtype control (p = 0.1726, Mann-Whitney-Wilcoxon test). Total number of mice: 7 +/+, 6 −/−. Total number of cells: 19 +/+, 16 −/−. Histograms represent mean ± standard deviation. Red circles represent individual cells.
Discussion
Loss of the brain-specific RNA-binding protein CELF4 causes a complex seizure disorder in mice, and this disorder is associated with dysregulation of excitatory synaptic transmission. Germline deletion of Celf4 lowers generalized seizure threshold and promotes handling-associated convulsive seizures as well as non-convulsive seizures, subject to gene dosage and genetic background, suggesting that CELF4 functions in various types of seizure circuits. CELF4 is expressed broadly during development but is brain-specific in adults. Strikingly, deletion of Celf4 at seven weeks of age is sufficient to significantly lower seizure threshold and to cause handling-associated convulsive seizures. This result, together with the observation that Celf4 mutant mice show no obvious morphological brain defects, strongly suggests that the convulsive seizure disorder in Celf4 mutants is caused by a functional deficit rather than abnormal development.
Interestingly, the absence, or non-convulsive, seizure phenotype of Celf4 deficient mice requires deletion prior to the end of the first postnatal week. The SWDs associated with absence seizures result from oscillations produced in the corticothalamic neural network, which includes projections from the cerebral cortex to specific thalamic and reticular thalamic nuclei, and these oscillations have been shown to initiate in the cortex in the GAERS (Genetic Absence Epilepsy Rat from Strasbourg) rat (Polack et al. 2007). Corticothalamic projections form early in development, therefore early deletion of CELF4 may negatively impact development of the corticothalamic circuitry, leading to absence seizures. Conversely, deletion of CELF4 after corticothalamic circuit development is complete does not generate absence seizures, suggesting that CELF4 may be involved in the regulation of corticothalamic circuit development but not maintenance. However, both germline and adult deletion of CELF4 are sufficient for generation of convulsive seizures, suggesting that CELF4 may be involved in maintaining proper neurotransmission in neural networks involved in convulsive seizures.
Our studies of spatial, neuron-specific conditional knockout mice show that specific deletion of Celf4 from primarily excitatory neurons, driven by either Emx1-cre or Camk2a-cre is also sufficient to induce a very low seizure threshold and handling-associated convulsions. These Cre drivers are expressed in some other cell types in various brain regions; Camk2a-cre is expressed in GABAergic neurons in the striatum and Emx1-cre is also expressed in glia, for example. However, both Cre drivers are selective for excitatory neurons in the cortex and hippocampus, two areas of the brain that are critically important in epilepsy. Furthermore, we find that CELF4 expression is neuron-specific and highest in highly glutamatergic regions, such as cortex and hippocampus but very sparse in highly GABAergic regions, such as striatum, suggesting that excitatory neurons would be most affected by Celf4 deletion.
In contrast, deletion of Celf4 from inhibitory neurons is not sufficient to lower seizure threshold or induce handling-associated convulsions. Deletion of Celf4 driven by Pvalb-cre, which is present in ~40% of inhibitory neurons (Gonchar et al. 2007), or Viaat-cre, which is expressed at the majority of GABAergic synapses (Chaudhry et al. 1998), does not lower seizure threshold and does not induce convulsive seizures. Interestingly, deletion of Celf4 by Viaat-cre actually increases seizure threshold, implying that Celf4 may also function in the few inhibitory neurons in which it is expressed – a possibility that requires further inquiry. We do note that the (C57BL/6J × FVB/NJ)F1 hybrid strain background of the Viaat-cre mice is less permissive for convulsive seizures than C57BL/6J, based on previous observations in Celf4Ff mutants (seizure incidence of 20% for Celf4Ff F1 hybrid males versus 68% on B6J). Still, even using the Celf4null allele we have not observed any convulsive seizures in the experimental Viaat-cre mice to date. Together, the data point strongly to the conclusion that Celf4 deletion primarily from excitatory neurons promotes convulsive seizure phenotypes. This is also a most plausible suggestion, as effective neurotransmission in the brain requires a balance between excitatory and inhibitory signals.
Consistent with the clinical results in live mice, patch-clamp slice recordings show that deletion of Celf4 has no effect on synaptic inhibition of cortical layer V pyramidal neurons, but instead dramatically boosts excitatory synaptic function by increasing both response frequency and amplitude, suggesting that CELF4 specifically regulates excitatory neurotransmission. The increased frequency of excitatory synaptic events could be due to an increased number of excitatory synapses in Celf4 deficient mice or increased quantal release from excitatory synaptic vesicles. Since inhibition is not altered, the dysregulation of excitatory neurotransmission may enhance cortical excitability and lead to seizure development in Celf4 deficient mice.
In donning Celf4 mutant mice as modeling a “complex seizure disorder,” it is useful to be explicit about the multiple ways in which the term “complex” applies to this particular animal model. First, Celf4 loss mutants model two different kinds of epilepsy – convulsive (grand-mal like), and non-convulsive (absence, or petit-mal like); clearly an example of phenomenological complexity. Second, although we have not focused on pursuing strain differences in the present study, Celf4 mutants appear to model genetically complex disease (i.e. variants in modifier alleles), because the penetrance of seizure and survival phenotypes is highly dependent on strain background (i.e. homozygosity for B6J alleles worsens the convulsive seizure and lethality phenotypes, but resolves the absence seizures). Last, the sequence of behaviors during the convulsive seizures appear to model ”complex partial seizures with secondary generalizations,” the most common type of grand-mal seizure.
Such a “trifecta” of complexity is very much an outcome that one might expect for an RNA-binding protein whose loss may lead to the dysregulation of multiple gene targets and consequently a diversity of pathological effects, including epileptogenesis. For example, the neuronal RBP fragile X mental retardation protein (FMRP) regulates activity-dependent translation in neuronal dendrites by stabilizing mRNAs at dendritic synapses for local, on-demand synthesis (De Rubeis & Bagni 2010). Loss of FMRP results in aberrant translation of FMRP targets and reduced synaptic plasticity (Bassell & Warren 2008) and leads to fragile X syndrome, a disorder that includes a spectrum of symptoms including mental retardation, features of autism, and altered neuronal excitability that can lead to epilepsy (Wisniewski et al. 1985). Recently, FMRP was shown to associate with potassium channel Kv4.2 mRNA and regulate its translation, resulting in reduced levels of Kv4.2 protein in hippocampus and cerebral cortex of mice deficient for FMRP (Gross et al. 2011). A Kv4.2 loss-of-function truncation has been identified in a human case of temporal lobe epilepsy (TLE) (Singh et al. 2006), and decreased Kv4.2 expression is associated with increased neuronal excitability in animal models of TLE (Bernard et al. 2004).
Our finding that Celf4 deficiency leads to enhanced excitatory neurotransmission underscores the importance of mRNA transcript regulation in the brain, including the contribution of RBPs to neurological function. Microarray analysis using Celf4Ff homozygotes showed that mRNA transcripts of several factors involved in synaptic transmission were down-regulated compared to wildtype mice, including α-synuclein (Snca), serotonin receptor 2c (Htr2c), N-ethylmaleimide-sensitive factor (Nsf), and synapsin II (Syn2) (Yang et al. 2007). We are currently characterizing the subset of mRNAs bound to and regulated by CELF4 in more detail to elucidate a mechanism for altered excitatory neurotransmission and seizure generation in Celf4 deficient mice. Because numerous genes can be modulated by a defect in a single RBP, dysregulation of RBPs seems likely to be an important upstream player in many genetically complex neurological disorders, such as idiopathic generalized epilepsy.
Supplementary Material
Shown are example SWD episodes from mutant, cre-positive Celf4 mice with SWD in the day 1 tamoxifen-treated group, described as part of Figure 1d. On the left is a SWD that occurred during active wake phase and is associated with a brief behavioral arrest, and is flanked by typical movement-artifacts in the EEG upon cessation and resumption of normal locomotor activity. On the right is a SWD that occurred during quiet wake – no movement artifact but still fast, low-voltage interictal EEG. Key: 1s, one second; differential electrode montage abbreviations are: FL, front-left; FR, front-right; BL, back-left; BR, back-right, as described in Materials and Methods.
Acknowledgements
Viaat-cre mice were generated in the laboratory of Huda Y. Zoghbi, to whom we are grateful for sharing prior to publication. We thank Barbara Beyer, Verity Letts, Satoko Tokuda, Carolyne Dunbar and Nathalie Bérubé for assistance with mouse studies and EEG recordings. We also thank Rebecca Boumil, Gregory Cox, Verity Letts, and Zhong-wei Zhang for helpful comments and advice. The Jackson Laboratory’s Cell Biology and Microinjection services, Gene Expression and Sequencing services, and Imaging Sciences service were subsidized by an NCI core grant [5 P30 CA034196-27]. This work was supported by grants from the National Institutes of Health [NS061971 and NS061971-2Z to WNF].
Abbreviations
- CELF
CUGBP, ELAV-like family
- ECT
electroconvulsive threshold
- EEG
electroencephalogram
- GABA
gamma-aminobutyric acid
- mEPSC
miniature excitatory post-synaptic current
- mIPSC
miniature inhibitory post-synaptic current
- mRNA
messenger RNA
- RBP
RNA-binding protein
- RNA
ribonucleic acid
- RNP
ribonucleoprotein complex
- SWD
spike-and-wave discharge
- TLE
temporal lobe epilepsy
- TTX
tetrodotoxin
References
- Aberg K, Saetre P, Lindholm E, Ekholm B, Pettersson U, Adolfsson R, Jazin E. Human QKI, a new candidate gene for schizophrenia involved in myelination. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:84–90. doi: 10.1002/ajmg.b.30243. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C, Anderson A, Becker A, Poolos NP, Beck H, Johnston D. Acquired dendritic channelopathy in temporal lobe epilepsy. Science. 2004;305:532–535. doi: 10.1126/science.1097065. [DOI] [PubMed] [Google Scholar]
- Bialer M, White HS. Key factors in the discovery and development of new antiepileptic drugs. Nat Rev Drug Discov. 2010;9:68–82. doi: 10.1038/nrd2997. [DOI] [PubMed] [Google Scholar]
- Boumil RM, Letts VA, Roberts MC, Lenz C, Mahaffey CL, Zhang ZW, Moser T, Frankel WN. A missense mutation in a highly conserved alternate exon of dynamin-1 causes epilepsy in fitful mice. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry FA, Reimer RJ, Bellocchio EE, Danbolt NC, Osen KK, Edwards RH, Storm-Mathisen J. The vesicular GABA transporter, VGAT, localizes to synaptic vesicles in sets of glycinergic as well as GABAergic neurons. J Neurosci. 1998;18:9733–9750. doi: 10.1523/JNEUROSCI.18-23-09733.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nijs L, Leon C, Nguyen L, Loturco JJ, Delgado-Escueta AV, Grisar T, Lakaye B. EFHC1 interacts with microtubules to regulate cell division and cortical development. Nat Neurosci. 2009;12:1266–1274. doi: 10.1038/nn.2390. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, Bagni C. Fragile X mental retardation protein control of neuronal mRNA metabolism: Insights into mRNA stability. Mol Cell Neurosci. 2010;43:43–50. doi: 10.1016/j.mcn.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Frankel WN. Genetics of complex neurological disease: challenges and opportunities for modeling epilepsy in mice and rats. Trends Genet. 2009;25:361–367. doi: 10.1016/j.tig.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel WN, Taylor L, Beyer B, Tempel BL, White HS. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science. 2006;313:1792–1795. doi: 10.1126/science.1129947. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Lovero KL, Iwanaga T, Watanabe A, Yokoi N, Tabuchi K, Shigemoto R, Nicoll RA, Fukata M. Disruption of LGI1-linked synaptic complex causes abnormal synaptic transmission and epilepsy. Proc Natl Acad Sci U S A. 2010;107:3799–3804. doi: 10.1073/pnas.0914537107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber KB, Visootsak J, Warren ST. Fragile X syndrome. Eur J Hum Genet. 2008;16:666–672. doi: 10.1038/ejhg.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner M. Genetics of idiopathic generalized epilepsies. Epilepsia. 2005;46 Suppl 9:15–20. doi: 10.1111/j.1528-1167.2005.00310.x. [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonchar Y, Wang Q, Burkhalter A. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat. 2007;1:3. doi: 10.3389/neuro.05.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good PJ, Chen Q, Warner SJ, Herring DC. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J Biol Chem. 2000;275:28583–28592. doi: 10.1074/jbc.M003083200. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Yao X, Pong DL, Jeromin A, Bassell GJ. Fragile X Mental Retardation Protein Regulates Protein Expression and mRNA Translation of the Potassium Channel Kv4.2. J Neurosci. 2011;31:5693–5698. doi: 10.1523/JNEUROSCI.6661-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalachikov S, Evgrafov O, Ross B, Winawer M, Barker-Cummings C, Martinelli Boneschi F, Choi C, Morozov P, Das K, Teplitskaya E, Yu A, Cayanis E, Penchaszadeh G, Kottmann AH, Pedley TA, Hauser WA, Ottman R, Gilliam TC. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet. 2002;30:335–341. doi: 10.1038/ng832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Cortell R, Kau AS, Bukelis I, Tierney E, Gray RM, Cox C, Capone GT, Stanard P. Autism spectrum disorder in fragile X syndrome: communication, social interaction, and specific behaviors. Am J Med Genet A. 2004;129A:225–234. doi: 10.1002/ajmg.a.30229. [DOI] [PubMed] [Google Scholar]
- Kunapuli P, Jang GF, Kazim L, Cowell JK. Mass spectrometry identifies LGI1-interacting proteins that are involved in synaptic vesicle function in the human brain. J Mol Neurosci. 2009;39:137–143. doi: 10.1007/s12031-009-9202-y. [DOI] [PubMed] [Google Scholar]
- Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria PM, Duke A, Rand JB, Hobert O. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr Biol. 2003;13:1317–1323. doi: 10.1016/s0960-9822(03)00532-3. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM, Rao K, Cook E, Geschwind DH. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Meins M, Schlickum S, Wilhelm C, Missbach J, Yadav S, Glaser B, Grzmil M, Burfeind P, Laccone F. Identification and characterization of murine Brunol4, a new member of the elav/bruno family. Cytogenet Genome Res. 2002;97:254–260. doi: 10.1159/000066619. [DOI] [PubMed] [Google Scholar]
- Moore T, Hecquet S, McLellann A, Ville D, Grid D, Picard F, Moulard B, Asherson P, Makoff AJ, McCormick D, Nashef L, Froguel P, Arzimanoglou A, LeGuern E, Bailleul B. Polymorphism analysis of JRK/JH8, the human homologue of mouse jerky, and description of a rare mutation in a case of CAE evolving to JME. Epilepsy Res. 2001;46:157–167. doi: 10.1016/s0920-1211(01)00275-3. [DOI] [PubMed] [Google Scholar]
- Musumeci SA, Bosco P, Calabrese G, Bakker C, De Sarro GB, Elia M, Ferri R, Oostra BA. Audiogenic seizures susceptibility in transgenic mice with fragile X syndrome. Epilepsia. 2000;41:19–23. doi: 10.1111/j.1528-1157.2000.tb01499.x. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noebels JL. The biology of epilepsy genes. Annu Rev Neurosci. 2003;26:599–625. doi: 10.1146/annurev.neuro.26.010302.081210. [DOI] [PubMed] [Google Scholar]
- Ottman R. Analysis of genetically complex epilepsies. Epilepsia. 2005;46 Suppl 10:7–14. doi: 10.1111/j.1528-1167.2005.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottman R, Hauser WA, Barker-Cummings C, Lee JH, Risch N. Segregation analysis of cryptogenic epilepsy and an empirical test of the validity of the results. Am J Hum Genet. 1997;60:667–675. [PMC free article] [PubMed] [Google Scholar]
- Polack PO, Guillemain I, Hu E, Deransart C, Depaulis A, Charpier S. Deep layer somatosensory cortical neurons initiate spike-and-wave discharges in a genetic model of absence seizures. J Neurosci. 2007;27:6590–6599. doi: 10.1523/JNEUROSCI.0753-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte U, Thumfart JO, Klocker N, Sailer CA, Bildl W, Biniossek M, Dehn D, Deller T, Eble S, Abbass K, Wangler T, Knaus HG, Fakler B. The epilepsy-linked Lgi1 protein assembles into presynaptic Kv1 channels and inhibits inactivation by Kvbeta1. Neuron. 2006;49:697–706. doi: 10.1016/j.neuron.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Singh B, Ogiwara I, Kaneda M, Tokonami N, Mazaki E, Baba K, Matsuda K, Inoue Y, Yamakawa K. A Kv4.2 truncation mutation in a patient with temporal lobe epilepsy. Neurobiol Dis. 2006;24:245–253. doi: 10.1016/j.nbd.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Stein JM, Bergman W, Fang Y, Davison L, Brensinger C, Robinson MB, Hecht NB, Abel T. Behavioral and neurochemical alterations in mice lacking the RNA-binding protein translin. J Neurosci. 2006;26:2184–2196. doi: 10.1523/JNEUROSCI.4437-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Delgado-Escueta AV, Aguan K, Alonso ME, Shi J, Hara Y, Nishida M, Numata T, Medina MT, Takeuchi T, Morita R, Bai D, Ganesh S, Sugimoto Y, Inazawa J, Bailey JN, Ochoa A, Jara-Prado A, Rasmussen A, et al. Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat Genet. 2004;36:842–849. doi: 10.1038/ng1393. [DOI] [PubMed] [Google Scholar]
- Toth M, Grimsby J, Buzsaki G, Donovan GP. Epileptic seizures caused by inactivation of a novel gene, jerky, related to centromere binding protein-B in transgenic mice. Nat Genet. 1995;11:71–75. doi: 10.1038/ng0995-71. [DOI] [PubMed] [Google Scholar]
- Wisniewski KE, French JH, Fernando S, Brown WT, Jenkins EC, Friedman E, Hill AL, Miezejeski CM. Fragile X syndrome: associated neurological abnormalities and developmental disabilities. Ann Neurol. 1985;18:665–669. doi: 10.1002/ana.410180607. [DOI] [PubMed] [Google Scholar]
- Xu B, Zang K, Ruff NL, Zhang YA, McConnell SK, Stryker MP, Reichardt LF. Cortical degeneration in the absence of neurotrophin signaling: dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron. 2000;26:233–245. doi: 10.1016/s0896-6273(00)81153-8. [DOI] [PubMed] [Google Scholar]
- Yang Y, Mahaffey CL, Berube N, Maddatu TP, Cox GA, Frankel WN. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YE, Wen L, Silva J, Li Z, Head K, Sossey-Alaoui K, Pao A, Mei L, Cowell JK. Lgi1 null mutant mice exhibit myoclonic seizures and CA1 neuronal hyperexcitability. Hum Mol Genet. 2010;19:1702–1711. doi: 10.1093/hmg/ddq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YD, Lee S, Jin Z, Wright M, Smith SE, Anderson MP. Arrested maturation of excitatory synapses in autosomal dominant lateral temporal lobe epilepsy. Nat Med. 2009;15:1208–1214. doi: 10.1038/nm.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown are example SWD episodes from mutant, cre-positive Celf4 mice with SWD in the day 1 tamoxifen-treated group, described as part of Figure 1d. On the left is a SWD that occurred during active wake phase and is associated with a brief behavioral arrest, and is flanked by typical movement-artifacts in the EEG upon cessation and resumption of normal locomotor activity. On the right is a SWD that occurred during quiet wake – no movement artifact but still fast, low-voltage interictal EEG. Key: 1s, one second; differential electrode montage abbreviations are: FL, front-left; FR, front-right; BL, back-left; BR, back-right, as described in Materials and Methods.