Summary
The IL-17/IL-17 receptor family is the newest and least understood of the cytokine subclasses. Composed of ligands IL-17A-IL-17F and receptors IL-17RA-IL-17RE, these cytokines have many unique structural and functional features. Since the discovery of the “Th17” subset in 2005, particular attention has been paid to IL-17A and IL-17F and their cognate receptors. To date, far less is known about the rest of the family. This review discusses recent advances in the field, with an emphasis on IL-17A biology.
IL-17-producing cells: Th17 and beyond
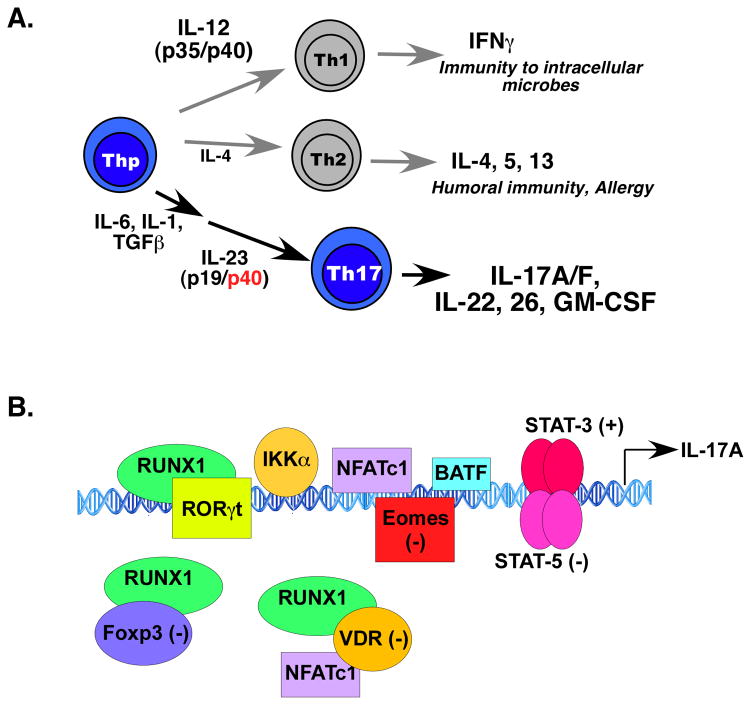
IL-17 (IL-17A, CTLA-8, Table 1) was cloned in 1993, but came into the limelight with the recognition of a third effector CD4+ T cell population distinct from classic Th1 and Th2 lineages, which produces IL-17 as a signature cytokine and responds potently to IL-23. Dubbed “Th17,” IL-17-producing CD4+ cells arise from multiple differentiation triggers, including TGFβ, IL-6, IL-1β, and IL-21 (Fig 1A). IL-23 is not required for Th17 differentiation per se, but is essential for maintenance and activity of these cells in vivo. The discovery of Th17 cells reconciled many of the long-standing discrepancies of the Th1/Th2 model, particularly the discordant roles of IFN γ and IL-12 (specifically, the IL-12p40 subunit shared with IL-23) [1]*. In addition, Th17 cells produce IL-17F, an IL-17A/F heterodimer, IL-22 and IL-26 (humans). Recent findings indicate that GM-CSF is another Th17-derived cytokine that contributes to the inflammatory pathology of Th17 cells, based on data from EAE models [2,3]. Understanding the integration and nuances of Th17-derived signals will doubtless continue to be an intensive area of research.
Table 1.
IL-17 receptor/ligand family
| Receptor complex | Ligand(s) |
|---|---|
| IL-17RA/RC | IL-17A, IL-17F, IL-17A/F, vIL-17* |
| IL-17RA/RB | IL-17E (a.k.a., IL-25) |
| IL-17RD (SEF) | unknown |
| IL-17RA/RD | unknown |
| IL-17RE | IL-17C |
| unknown | IL-17D |
viral IL-17
Figure 1. Th17 differentiation and IL-17A gene regulation.
A. Th17 cells arise from differentiative signals from IL-1, IL-6 and TGFβ, while stability and maintenance of Th17 cells is mediated by IL-23. In addition to IL-17A, Th17 cells produce IL-17F, IL-17A/F, IL-22, IL-26 (humans only) and GM-CSF. B. Summary of studies from the human and mouse il17a promoters indicates involvement of various positively (+) and negatively (−) acting transcription factors.
Although produced by T cells, IL-17 acts primarily on epithelial, endothelial and stromal cells. Genes induced by IL-17 encode antimicrobial proteins (AMPs; β defensins, cathelicidin, RegIII, lipocalin 2, salivary histatins), neutrophil-activating factors (G-CSF, CXC chemokines), and inducers of the acute phase response (IL-6) [4]. It was long evident that there must be innate sources of IL-17 that act rapidly during infection, prior to the onset of a bona fide adaptive T cell response [5]. Accordingly, a variety of studies have demonstrated IL-17 expression in γδ T cells, NKT cells, macrophages, LTi, TFh among others [6]**. These IL-17-producing cells play vital roles in mediating effects of IL-17, especially at mucosal surfaces [7].
Interconnections among T cell subsets
Although Th17 cells are often depicted as a committed lineage, in fact there are many interrelationships with other subpopulations, including functional cooperation, cells that express signature cytokines from multiple subsets (e.g., IL-17+IFNγ+ cells) and lineage plasticity. For example, Th17 cells and Tregs both arise from TGFβ-dependent signals, although the importance of TGFβ in Th17 generation continues to be controversial [8–10]. Th17 cells can convert to Tregs and vice versa or Th1-type cells, a process known as “plasticity” [11,12]. This occurs in part by T-bet-mediated repression of RORγt [13], a transcription factor considered the “master regulator” of Th17 cells [14]. IL-2, which promotes Tregs, suppresses Th17 generation through STAT5 [15,16]*. Conversely, Tregs can promote Th17 cells by acting as a “sink” for IL-2 through the high affinity IL-2R, thereby relieving IL-2-mediated repression of Th17 differentiation [17,18]. Although Th1 cells can suppress Th17 generation via inhibitory signals from IFNγ [19,20], IL-17 can positively regulate Th1 cell differentiation in certain intracellular infections or vaccination settings, including M. tuberculosis, Francisella tularensis and chlamydial infections [21–23]*. In one instance this is mediated by direct IL-17 signaling on DCs, leading to upregulation of IL-12 [21]*. An elegant fate-mapping study recently showed that Th17 cells exhibit variable plasticity, dependent on setting. Whereas conversion of Th17 cells to other subsets was observed at a high frequency in chronic EAE, conversion occurred rarely during acute fungal infection [24]. At a molecular level, plasticity is related to epigenetic modifications of genes encoding the transcription factors that specify lineage choice, namely T-bet (Th1), GATA-3 (Th2), RORγt (Th17) and Foxp3 (Treg). Analysis of different CD4+ subsets by ChIP-Seq showed that “permissive” chromatin modifications are frequently found in the promoters of transcription factors different from the lineage analyzed, suggesting that these regulators are poised for rapid transcription [25–27]. Thus, the immune system exhibits considerable flexibility, presumably to allow appropriate fine-tuning of the immune response throughout the course of different immune challenges.
IL-17 gene regulation
The genes encoding IL-17A and IL-17F are closely linked, and a comprehensive picture of gene regulation is starting to emerge [28] (Fig 1B). Within the intergenic DNA between il17a and il17f lies a conserved noncoding sequence region. The chromatin in this locus is extensively remodelled upon receipt of signals from TGFβ and IL-6, helping to explain coordinate regulation of these cytokines [29]. Early studies demonstrated the importance of NFATc1 in regulating the human IL-17A promoter, consistent with the cyclosporin-A sensitivity of IL-17 expression [30]. RORγt is essential for regulating the murine il17a and il17f promoters, although RORα can also accomplish this [14,31,32]. An independent analysis of the mouse il17a promoter showed that RORγt and RUNX1 bind directly to conserved noncoding elements upstream of the il17a start site. Notably, interactions of RUNX1 with Foxp3 inhibit transcription, providing a molecular explanation for the reciprocal development of Th17 and Treg cells [33]. BATF, a member of the AP-1 transcription factor family, is implicated in regulation of il17a and Th17 differentiation, binding to both proximal and intergenic regulatory regions [34]. STAT3, activated by Th17-inductive cytokines such as IL-6, IL-21 and IL-23, also binds the il17a promoter [35], and consistently, humans with STAT3 defects show reduced IL-17 production and sensitivity to Th17-dependent infections (see below, Table 2) [36]. A new report indicates that the positive activities of STAT3 are offset by competitive binding of STAT5 at the same locus, accounting for the dose-dependent and competitive regulation of IL-17A production by IL-6 (which signals through STAT3) and IL-2 (which signals through STAT5) [16]*. The role of TGFβ in Th17 differentiation was recently shown to be mediated by SMAD-mediated suppression of eomesodermin, which regulates both the RORγt and il17a promoters [37]. IL-17-mediated inflammation can be tempered by signals from Vitamin D. Notably, 1,25-dihydroxyvitamin D3, the hormonally active form of Vitamin D, interacts with its receptor (VDR) to suppress mouse and human IL-17 transcription by blocking NFAT, sequestering Runx1 and enhancing Foxp3 [38]. In another study, it was shown that VDR in mice suppresses IL-17A expression in T cells mainly through a post-transcriptional signal, at least at physiological levels of Vitamin D [39]. The IKKα kinase was also shown to regulate mouse il17a (but interestingly not il17f), where it activates histone H3 phosphorylation [40]. A complete understanding of the factors involved in regulating IL-17A expression has important ramifications for rational drug design to target autoimmunity.
Table 2.
Human Genetic diseases involving IL-17
| Infections, other disease manifestations | How IL-17 is affected | Gene affected | References | |
|---|---|---|---|---|
| Autoimmune polyendocrine syndrome-1 (APS-1) | CMCa | Neutralizing Abs to IL-17A, IL-17F (and IL-22) | Aire (LOF)b | [56,57] |
| Hyper-IgE Syndrome (Job’s Syndrome) | CMC, Staph, eczema, cold absesses, bone problems, etc. | Reduction in Th17 cells, low IL-17A levels | STAT-3 (DN)c | [49,50] |
| Dectin1-deficiency | CMC | Reduced Th17 numbers | Dectin-1 (LOF) | [55] |
| CARD9-deficiency | CMC | Reduced Th17 cell numbers | CARD9 (LOF) | [54] |
| IL-17RA-deficiency | CMC, Staph | No expression of IL-17RA, failure to signal to IL-17A, F | IL-17RA (LOF) | [58] |
| IL-17F deficiency | CMC, Staph | Inhibition of IL-17A/F and IL-17F signaling | IL-17F (DN) | [58] |
| IL-23R SNP (R381Q) | protection from various autoimmune diseases (GWAS)d | Decreased IL-23 signaling in T cells, leading to reduced IL-17/IL-22 production | IL-23 receptor | [63,64] |
Chronic mucocutaneous candidiasis
LOF, loss of function
DN, dominant negative
GWAS, genome wide association study
IL-17 in fungal infections
Numerous studies identified a role for IL-17 in immunity against extracellular pathogens [5]. In particular, several new reports implicate IL-17 as a particularly vital mediator of host defense against fungi. Candida albicans is an opportunistic yeast that colonizes skin and mucosal tissues, and can cause systemic infections that are a serious problem in hospitals. In mice, deficiency of IL-17A and IL-17RA but not IL-17F causes increased susceptibility to disseminated candidiasis [41,42]. Similarly, IL-17R in mice is essential for immunity to mucosal candidiasis in the oral cavity as well as skin [43]*[44,45]. A requirement for Th17 cells has been demonstrated in an experimental vaccine that targets both C. albicans and Staphylococcus aureus [46]. In contrast, IL-17 and IL-23 have been suggested to be pathogenic in a model of gastric candidiasis [47], and their roles in vaginal candidal infections are thus far unclear. In addition, IL-17-dependent immunity in vaccine responses to various other fungal pathogens has been demonstrated [48]**, indicating that antifungal immunity is a general feature of IL-17.
A number of “experiments of nature” have begun to reveal that IL-17 activity appears to be surprisingly limited to just a few infections, particularly those caused by Candida and Staphylococcus aureus (Table 2). Humans with hyper-IgE Syndrome (HIES, Job’s Syndrome) have mutations in STAT-3. Presumably because of defects in Th17-inducing cytokines such as IL-6, IL-21 and IL-23, these patients exhibit a selective paucity of Th17 cells [49–51]. Although they exhibit a wide spectrum of clinical manifestations, HIES patients are highly susceptible to mucosal candidiasis and staphylococcal abscesses, implicating Th17 cells in immunity to these organisms. Interestingly, this is associated with reduced salivary antifungal activity, implying a direct role for IL-17 on the salivary gland [52]. The major pattern recognition receptors for Candida are C-type lectin receptors including Dectin-1, -2 and Mincle. These recognize fungal cell wall carbohydrates, and stimulate potent Th17 responses [53]. Notably, patients with defects in Dectin-1 or its signaling effector CARD9 experience severe chronic mucocutaneous candidiasis (CMC) [54,55]. Further direct evidence for a role for IL-17 comes from APS-1, in which patients with mutations in Aire develop neutralizing autoantibodies against Th17 cytokines and experience recurrent CMC [53,56,57]**. Moreover, rare patients with mutations in IL-17RA or IL-17F exhibit similar susceptibility to CMC and Staphylococcus [58]. Thus, data in humans argues for an immunoprotective role of IL-17 signaling in controlling Candida and Staphylococcaal infections.
IL-17 in autoimmunity, therapies
Even before the discovery of Th17 cells, it was evident that IL-17 promotes autoimmunity, particularly in the context of rheumatoid arthritis (RA) [59]. Analyses of collagen-induced arthritis (CIA) confirmed that Th17 cells rather than Th1 cells promote disease [60], and blocking IL-17 or its receptor relieves experimental symptoms [61]. Studies in EAE similarly indicated a dominant role of Th17 cells over Th1 cells [62]. In humans, a SNP in the IL-23R (R381Q) is associated with reduced susceptibility to a variety of autoimmune disorders, and was recently linked to reduced IL-17 production in human T cells [63,64]. Accordingly, antibodies targeting IL-17A are in numerous clinical trials, and early reports are promising. Results, albeit from small numbers of patients, show efficacy in psoriasis, RA and autoimmune uveitis [65,66]*. Thus far side effects appear minimal, though much larger data sets will be needed to confirm this trend. However, findings from IL-17-deficient humans (Table 2) suggest that loss of IL-17 signaling may indeed predispose only to a limited number of infections.
IL-17 receptor structure and signaling
The receptor for IL-17A, IL-17F and the IL-17A/F heterodimer is composed of IL-17RA paired with IL-17RC [4]. The crystal structure of the human IL-17RA extracellular domain complexed to an IL-17F:F homodimer reveals an unusual fibronectin III domain conformation [67]**. Surface plasmon resonance studies showed stepwise association of receptor/ligand binding; namely, after either IL-17RA or IL-17RC bind their respective ligand, affinity for the reciprocal subunit is increased. This observation is consistent with studies showing ligand-inducible association of IL-17RA and IL-17RC on the cell surface [44,68]. There are notable differences between the human and mouse IL-17R systems, however. Human IL-17RA has much higher affinity for IL-17A than for IL-17F, and human IL-17RC binds both ligands with similar affinity. In contrast, murine IL-17RC alone binds primarily to IL-17F [67,69]. IL-17RA appears to be a component of at least one other receptor complex (IL-25R, Table 1), and may in fact be the common signaling subunit of the entire family [4,70].
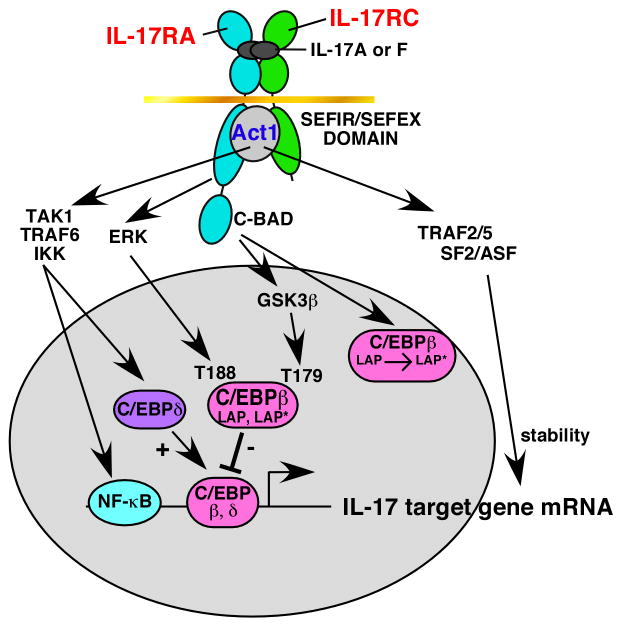
The signal transduction pathway mediated by IL-17 is still poorly defined (Fig 2). IL-17R family members contain a conserved “SEFIR” motif, which has homology to TIR domains found in Toll and IL-1 receptors [71]**. However, structure-function mapping studies in IL-17RA and IL-17RC indicate that the SEFIR domain does not encompass the entire requisite signaling motif. In addition, there are large nonconserved regions within these receptors also essential for signal transduction [44,72,73]. Consistently, IL-17 does not engage TIR-containing adaptors such as MyD88, TRIF or IRAKs [72]. Rather, IL-17 signals through Act1, an adaptor and U-box E3 ubiquitin ligase that contains both a SEFIR and TRAF-binding domain [74–76]. IL-17 induces rapid phosphorylation of Act1 [44], though the target sites and relevant kinases are not yet reported. IL-17 activates the classical NF-κB pathway, albeit modestly, an event requiring both Act1 and TRAF6 [74,77]. In addition to NF-κB, IL-17 activates IκBζ and C/EBP transcription factors (mainly C/EBPδ and C/EBPβ) [78], as well as MAPK and possibly PI3K [4].
Figure 2. IL-17R Signaling.
The IL-17R complex is composed of IL-17RA and IL-17RC. Major downstream signaling pathways mediated by this receptor are indicated.
A striking feature of IL-17A, IL-17F and IL-17A/F is their modest signaling activity, even in high concentrations of ligand. However, they exhibit potent synergy, particularly with TNFα but also LTα, IFNγ and IL-1β [4,73]. The mechanisms underlying synergy are not fully defined, but a major component is cooperative enhancement of mRNA stability of certain IL-17 target genes. For example, IL-6 and CXCL1 are both induced weakly at the transcriptional level by IL-17 alone, but their transcript half-life is enhanced dramatically by a combination of TNFα and IL-17. This signal is Act1-dependent, but occurs in the absence of TRAF6 [79]. Surprisingly, the well-characterized mRNA destabilization factor tristetraprolin, which reduces the stability of inflammatory transcripts induced by IL-1β, LPS and TNFα, is dispensable for IL-17-mediated enhancement of CXCL1 (KC) mRNA [80]*. New data indicate that a different mRNA destabilization factor, SF2/ASF, associates with TRAF5 and TRAF2 following IL-17 activation, leading to enhanced stabilization of CXCL1 (T. Hamilton, personal communication). Thus, despite many parallels to IL-1/TLR signaling, IL-17R activates distinct pathways mediated through unique receptor subdomains.
Given its pro-inflammatory propensities, it is not surprising that there are inhibitory signaling events mediated by IL-17 that modulate its activity. Mapping studies of IL-17RA revealed a distal cytoplasmic domain that mediates suppressive signaling through dual phosphorylation of C/EBPβ. These phosphorylation events are mediated by ERK and GSK3β, and many IL-17 target genes are enhanced upon exposure to pharmacological inhibitors of these kinases [81]. This IL-17RA subdomain also controls alternative translation of C/EBPβ, and hence is termed the “C/EBPβ activation domain” (C-BAD) [72]. Recently, the C-BAD was found to contain a TRAF3-binding element that mediates suppressive signaling, and TRAF3-transgenic mice indeed exhibit reduced IL-17-dependent pathology in EAE [82]. The full range of molecular signaling events mediated by IL-17 is still poorly understood, and is likely to continue to yield unexpected findings in the future.
Summary and Perspectives
The recognition of Th17 cells has stimulated a surge of interest in IL-17 cytokines. Although much remains to be learned about this enigmatic family, intensive studies in mice have provided much new information about this cytokine and its fascinating cellular and molecular regulation. The advent of clinical trials targeting IL-17 and its receptor will no doubt shed new light on its biology in humans as well.
Highlights.
IL-17, signature of the CD4+ “Th17” lineage, is also produced by innate cell types
il17a gene regulation and epigenetics helps explain Th cell plasticity
Data from animal models and humans reveals a vital role of IL-17 in fungal immunity
Anti-IL-17 therapeutics show early promise in the clinic
The IL-17 receptor activates noncanonical signal transduction pathways
Acknowledgments
SLG was supported by National Institutes of Health grant AR054389. We thank S. Christakos (UMDNJ, Newark NJ) for critical reading, and Dr. Tom Hamilton (Cleveland Clinic, Cleveland OH) for sharing unpublished information.
Abbreviations
- EAE
experimental autoimmune encephalomyelitis
- CIA
collagen-induced arthritis
- RA
rheumatoid arthritis
- CMC
chronic mucocutaneous candidiasis
- SF2/ASF
splicing factor 2/alternative splicing factor
- VDR
vitamin D receptor
- STAT
signal transducer and activator of transcription
- HIES
hyper-IgE Syndrome
- ChIP
chromatin immunoprecipitation
- SNP
single nucleotide polymorphism
- SEFIR
similar expression to FGF receptor/IL-17R
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1*.Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nature Med. 2007;13:139–145. doi: 10.1038/nm1551. This outstanding review describes the many discrepancies in the Th1/Th2 paradigm and how they were overlooked or ignored. [DOI] [PubMed] [Google Scholar]
- 2.El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, Rostami A. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12:568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 4.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peck A, Mellins E. Precarious Balance: Th17 cells in host defense. Infect Immun. 2010;78:32–38. doi: 10.1128/IAI.00929-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010;10:479–489. doi: 10.1038/nri2800. This comprehensive review article discusses the new literature describing innate cell sources of IL-17. [DOI] [PubMed] [Google Scholar]
- 7.Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2:403–411. doi: 10.1038/mi.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatton R. TGF-beta in Th17 cell development: The truth is out there. Immunity. 2011;34:288–290. doi: 10.1016/j.immuni.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutcher I, Donkor M, Ma Q, Rudensky A, Flavell R, Li M. Autocrine transforming growth factor-β1 promotes in vivo Th17 cell differentiation. Immunity. 2011;34:396–408. doi: 10.1016/j.immuni.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov I, McKenzie B, Zhou L, Tadokoro C, Lepelley A, Lafaille J, Cua D, Littman D. The orphan nuclear receptor RORgammaT directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 15.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 16*.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. This study shows for the first time that STAT3 and STAT5 exert reciprocal positive and negative activation of the il17a gene locus in response to activating or repressive cytokine signals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandiyan P, Conti H, Zheng L, Peterson A, Mathern D, Hernandez-Santos N, Edgerton M, Gaffen S, Lenardo M. CD4+CD25+Foxp3+ regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 infection model. Immunity. 2011;34:422–434. doi: 10.1016/j.immuni.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Haines CJ, Gutcher I, Hochweller K, Blumenschein WM, McClanahan T, Hammerling G, Li MO, Cua DJ, McGeachy MJ. Foxp3(+) regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity. 2011;34:409–421. doi: 10.1016/j.immuni.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 21*.Lin Y, Ritchea S, Logar A, Slight S, Messmer M, Rangel-Moreno J, Guglani L, Strawbridge H, Park S, Nguyen N, et al. IL-17 is required for Th1 immunity and host resistance to the intracellular pathogen Francisella tularensis LVS. Immunity. 2009;183:799–810. doi: 10.1016/j.immuni.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4(+) T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol. 2007;8:369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- 23*.Bai H, Cheng J, Gao X, Joyee AG, Fan Y, Wang S, Jiao L, Yao Z, Yang X. IL-17/Th17 promotes type 1 T cell immunity against pulmonary intracellular bacterial infection through modulating dendritic cell function. J Immunol. 2009;183:5886–5895. doi: 10.4049/jimmunol.0901584. These 3 articles show that Th17/IL-17 responses can induce Th1/IFNγ responses in the context of intracellular infections. [DOI] [PubMed] [Google Scholar]
- 24.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowell E, Wilson C. Programming perpetual T helper cell plasticity. Immunity. 2009;30:7–8. doi: 10.1016/j.immuni.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Hirahara K, Ghoreschi K, Laurence A, Yang XP, Kanno Y, O’Shea JJ. Signal transduction pathways and transcriptional regulation in Th17 cell differentiation. Cytokine Growth Factor Rev. 2010;21:425–434. doi: 10.1016/j.cytogfr.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling at IL17-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007 doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Lin X, Gaffen SL. Crucial role for nuclear factor of activated T cells (NFAT) in T cell receptor-mediated regulation of the human interleukin-17 gene. J Biol Chem. 2004;279:52762–52771. doi: 10.1074/jbc.M405764200. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov, Zhou L, Littman DR. Transcriptional regulation of Th17 cell differentiation. Semin Immunol. 2007;19:409–417. doi: 10.1016/j.smim.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F, Meng G, Strober W. Interactions among the transcriptoin factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schraml B, Hildner K, Ise W, Lee W-L, Smith W, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, et al. The AP-1 transcription factor Batf controls Th17 differentiation. Nature. 2009;460:405–410. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, Tato C, Yoshimura A, Hennighausen L, O’Shea JJ. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci U S A. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minegishi Y, Karasuyama H. Genetic origins of hyper-IgE syndrome. Curr Allergy Asthma Rep. 2008;8:386–391. doi: 10.1007/s11882-008-0075-x. [DOI] [PubMed] [Google Scholar]
- 37.Ichiyama K, Sekiya T, Inoue N, Tamiya T, Kashiwagi I, Kimura A, Morita R, Muto G, Shichita T, Takahashi R, et al. Transcription factor Smad-independent T helper 17 cell induction by transforming growth factor-beta is mediated by suppression of eomesodermin. Immunity. 2011 doi: 10.1016/j.immuni.2011.02.021. in press. [DOI] [PubMed] [Google Scholar]
- 38.Joshi S, Pantalena L-C, Liu XK, Gaffen SL, Ono M, Sakaguchi S, Liu H, Rohowsky-Kochan C, Ichiyama K, Yoshimura A, et al. 1,25-Dihydroxyvitamin D3 Ameliorates Th17 Autoimmunity Via Transcriptional Modulation of IL-17A. 2011 doi: 10.1128/MCB.05020-11. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang S, Chung Y, Dong C. Vitamin D suppresses Th17 cytokine production by inducing C/EBP homologous protein (CHOP) expression. J Biol Chem. 2010;285:38751–38755. doi: 10.1074/jbc.C110.185777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Ruan Q, Hilliard B, DeVirgiliis J, Karin M, Chen Y. Transcriptional regulation of the Th17 immune response by IKKalpha. J Exp Med. 2011;208:787–796. doi: 10.1084/jem.20091346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis. 2004;190:624–631. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 43*.Conti H, Shen F, Nayyar N, Stocum E, JN S, Lindemann M, Ho A, Hai J, Yu J, Jung J, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299–311. doi: 10.1084/jem.20081463. This paper shows for the first time that IL-23 and IL-17 mediate immunity to oral mucosal Candida albicans infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho A, Shen F, Conti H, Patel N, Childs E, Peterson A, Hernandez-Santos N, Kolls J, Kane L, Ouyang W, et al. IL-17RC is required for immune signaling via an extended SEFIR domain in the cytoplasmic tail. J Immunol. 2010;185:1063–1070. doi: 10.4049/jimmunol.0903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Kagami S, Rizzo HL, Kurtz SE, Miller LS, Blauvelt A. IL-23 and IL-17A, but not IL-12 and IL-22, are required for optimal skin host defense against Candida albicans. J Immunol. 2010;185:5453–5462. doi: 10.4049/jimmunol.1001153. This paper shows for the first time that IL-23 and IL-17 mediate immunity to mucosal Candida albicans infections of skin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin L, Ibrahim AS, Xu X, Farber JM, Avanesian V, Baquir B, Fu Y, French SW, Edwards JE, Jr, Spellberg B. Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PLoS Pathog. 2009;5:e1000703. doi: 10.1371/journal.ppat.1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zelante T, De Luca A, Bonifazi P, Montagnoli C, Bozza S, Moretti S, Belladonna ML, Vacca C, Conte C, Mosci P, et al. IL-23 and the Th17 pathway promote inflammation and impair antifungal immune resistance. Eur J Immunol. 2007;37:2695–2706. doi: 10.1002/eji.200737409. [DOI] [PubMed] [Google Scholar]
- 48**.Wuthrich M, Gern B, Hung CY, Ersland K, Rocco N, Pick-Jacobs J, Galles K, Filutowicz H, Warner T, Evans M, et al. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J Clin Invest. 2011;121:554–568. doi: 10.1172/JCI43984. This thorough study demonstrates that Th17 responses are required for effective responses to experimental vaccines against human fungal infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, Fulcher DA, Tangye SG, Cook MC. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205:1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 52.Conti H, Baker O, Freeman A, Jang W, Li R, Holland S, Edgerton M, Gaffen S. New mechanism of oral immunity to mucosal candidiasis in hyper-IgE syndrome. Mucosal Immunol. 2011 doi: 10.1038/mi.2011.5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puel A, Picard C, Cypowyj S, Lilic D, Abel L, Casanova JL. Inborn errors of mucocutaneous immunity to Candida albicans in humans: a role for IL-17 cytokines? Curr Opin Immunol. 2010;22:467–474. doi: 10.1016/j.coi.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glocker EO, Hennigs A, Nabavi M, Schaffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, et al. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med. 2009;361:1760–1767. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56**.Kisand K, Boe Wolff AS, Podkrajsek KT, Tserel L, Link M, Kisand KV, Ersvaer E, Perheentupa J, Erichsen MM, Bratanic N, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57**.Puel A, Doffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, Cobat A, Ouachee-Chardin M, Toulon A, Bustamante J, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207:291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Puel A, Cypowji S, Bustamante J, Wright J, Liu L, Lim H, Migaud M, Israel L, Chrabieh M, Audry M, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011 doi: 10.1126/science.1200439. in press. These experiments of nature demonstrate that loss of IL-17 signaling via neutralizing Abs (in Aire deficiency) or via naturally occurring mutations cause CMC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lubberts E. IL-17/Th17 targeting: on the road to prevent chronic destructive arthritis? Cytokine. 2008;41:84–91. doi: 10.1016/j.cyto.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 60.Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lubberts E, Koenders MI, Oppers-Walgreen B, van den Bersselaar L, Coenen-de Roo CJ, Joosten LA, van den Berg WB. Treatment with a neutralizing anti-murine interleukin-17 antibody after the onset of collagen-induced arthritis reduces joint inflammation, cartilage destruction, and bone erosion. Arthritis Rheum. 2004;50:650–659. doi: 10.1002/art.20001. [DOI] [PubMed] [Google Scholar]
- 62.McGeachy MJ, Cua DJ. The link between IL-23 and Th17 cell-mediated immune pathologies. Semin Immunol. 2007;19:372–376. doi: 10.1016/j.smim.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 63.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A Genome-Wide Association Study Identifies IL23R as an Inflammatory Bowel Disease Gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarin R, Wu X, Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1017854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Hueber W, Patel DD, Dryja T, Wright AM, Koroleva I, Bruin G, Antoni C, Draelos Z, Gold MH, Durez P, et al. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci Transl Med. 2010;2:52ra72. doi: 10.1126/scitranslmed.3001107. [DOI] [PubMed] [Google Scholar]
- 66*.Genovese M, Van den Bosch F, Roberson S, Bojin S, Biagini I, Ryan P, Sloan-Lancaster J. LY2439821, a humanized anti-interleukin-17 monoclonal antibody, in the treatment of patients with rheumatoid arthritis. Arth Rheum. 2010;62:929–939. doi: 10.1002/art.27334. These papers show evidence for the efficacy of anti-IL-17A Abs in treating autoimmunity. [DOI] [PubMed] [Google Scholar]
- 67**.Ely LK, Fischer S, Garcia KC. Structural basis of receptor sharing by interleukin 17 cytokines. Nat Immunol. 2009;10:1245–1251. doi: 10.1038/ni.1813. The first reported crystal structure of an IL-17R family receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu Y, Ota N, Peng I, Refino CJ, Danilenko DM, Caplazi P, Ouyang W. IL-17RC is required for IL-17A- and IL-17F-dependent signaling and the pathogenesis of experimental autoimmune encephalomyelitis. J Immunol. 2010;184:4307–4316. doi: 10.4049/jimmunol.0903614. [DOI] [PubMed] [Google Scholar]
- 69.Kuestner R, Taft D, Haran A, Brandt C, Brender T, Lum K, Harder B, Okada S, Osatrander C, Kreindler J, et al. Identification of the IL-17 receptor related molecule, IL-17RC, as the receptor for IL-17F. J Immunol. 2007;179:5462–5473. doi: 10.4049/jimmunol.179.8.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rickel EA, Siegel LA, Yoon BR, Rottman JB, Kugler DG, Swart DA, Anders PM, Tocker JE, Comeau MR, Budelsky AL. Identification of functional roles for both IL-17RB and IL-17RA in mediating IL-25-induced activities. J Immunol. 2008;181:4299–4310. doi: 10.4049/jimmunol.181.6.4299. [DOI] [PubMed] [Google Scholar]
- 71**.Novatchkova M, Leibbrandt A, Werzowa J, Neubuser A, Eisenhaber F. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem Sci. 2003;28:226–229. doi: 10.1016/S0968-0004(03)00067-7. This foresightful paper identified the SEFIR in the IL-17R family, and predicted the role of Act1 as a key adaptor for the IL-17R family. [DOI] [PubMed] [Google Scholar]
- 72.Maitra A, Shen F, Hanel W, Mossman K, Tocker J, Swart D, Gaffen SL. Distinct functional motifs within the IL-17 receptor regulate signal transduction and target gene expression. Proc Natl Acad Sci, USA. 2007;104:7506–7511. doi: 10.1073/pnas.0611589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Onishi R, Park S, Hanel W, Maitra A, Gaffen S. The SEFIR is not enough: An extended region downstream of the Interleukin-17RA SEFIR domain is required for IL-17-dependent signal transduction. J Biol Chem. 2010;285:32751–32759. doi: 10.1074/jbc.M110.121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol. 2007;8:247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 75.Chang SH, Park H, Dong C. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem. 2006;281:35603–35607. doi: 10.1074/jbc.C600256200. [DOI] [PubMed] [Google Scholar]
- 76.Liu C, Qian W, Qian Y, Giltiay NV, Lu Y, Swaidani S, Misra S, Deng L, Chen ZJ, Li X. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci Signal. 2009;2:ra63. doi: 10.1126/scisignal.2000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwandner R, Yamaguchi K, Cao Z. Requirement of tumor necrosis factor-associated factor (TRAF)6 in interleukin 17 signal transduction. J Exp Med. 2000;191:1233–1239. doi: 10.1084/jem.191.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruddy MJ, Wong GC, Liu XK, Yamamoto H, Kasayama S, Kirkwood KL, Gaffen SL. Functional cooperation between interleukin-17 and tumor necrosis factor-α is mediated by CCAAT/enhancer binding protein family members. J Biol Chem. 2004;279:2559–2567. doi: 10.1074/jbc.M308809200. [DOI] [PubMed] [Google Scholar]
- 79.Hartupee J, Liu C, Novotny M, Sun D, Li X, Hamilton TA. IL-17 signaling for mRNA stabilization does not require TNF receptor-associated factor 6. J Immunol. 2009;182:1660–1666. doi: 10.4049/jimmunol.182.3.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80*.Datta S, Novotny M, Pavicic PG, Jr, Zhao C, Herjan T, Hartupee J, Hamilton T. IL-17 regulates CXCL1 mRNA stability via an AUUUA/tristetraprolin-independent sequence. J Immunol. 2010;184:1484–1491. doi: 10.4049/jimmunol.0902423. This report shows that induction of IL-17-dependent mRNA stability in the target gene CXCL1 occurs independently of TTP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen F, Li N, Gade P, Kalvakolanu DV, Weibley T, Doble B, Woodgett JR, Wood TD, Gaffen SL. IL-17 Receptor Signaling Inhibits C/EBP{beta} by Sequential Phosphorylation of the Regulatory 2 Domain. Sci Signal. 2009;2:ra8. doi: 10.1126/scisignal.2000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu S, Pan W, Shi P, Gao H, Zhao F, Song X, Liu Y, Zhao L, Li X, Shi Y, et al. Modulation of experimental autoimmune encephalomyelitis through TRAF3-mediated suppression of interleukin 17 receptor signaling. J Exp Med. 2010;207:2647–2662. doi: 10.1084/jem.20100703. [DOI] [PMC free article] [PubMed] [Google Scholar]