SUMMARY
Adenosine deaminases that act on RNAs (ADARs) interact with double-stranded RNAs, deaminating adenosines to inosines. Previous studies of Caenorhabditis elegans suggested an antagonistic interaction between ADAR and RNAi machineries, with ADAR defects suppressed upon additional knockout of RNAi. These results suggest a pool of common RNA substrates capable of engaging both pathways. To define and characterize such substrates, we examined small RNA and mRNA populations of ADAR mutants and identified a distinct set of loci from which RNAi-dependent short RNAs are dramatically upregulated. At these same loci, we observe populations of multiply edited transcripts, supporting a specific role for ADARs in preventing access to the RNAi pathway for an extensive population of dsRNAs. Characterization of these loci reveal an extensive overlap with non-coding and intergenic regions, suggesting that the landscape of ADAR targets may extend beyond previously annotated classes of transcripts.
INTRODUCTION
ADARs are a class of dsRNA binding proteins that catalyze the deamination of adenosines to inosines in double-stranded RNA, disrupting base-pairing at the editing site. A standard approach to characterize ADAR targets involves identifying A-to-G changes in cDNA sequences relative to the reference genome. Several well-characterized ADAR-editing events occur in coding regions and alter the amino acid sequence, including those in the glutamate ion channels, seratonin 2C receptor, and voltage-gated potassium channel (Kv1.1)1–5. Despite these prominent examples, genome-wide assays for A-to-I editing sites in mammals show that the majority of ADAR targets are disbursed in clusters amongst non-coding regions (e.g. 3′ UTRs) and genomic repeat structures (e.g. SINEs)6–16. Studies of ADAR activity in vitro show that ADARs edit perfect hairpins throughout the double-stranded structure17.
What is the functional consequence of promiscuous A-to-I editing for individual transcripts? Transcripts with inosine-containing 3′ UTRs have been suggested to undergo degradation, sequestration, additional processing, or enter other dsRNA-binding pathways18–24. However, several HeLa and C. elegans transcripts with inosine-containing 3′ UTRs showed no changes in protein levels, mRNA levels, or degree of ribosome association in animals lacking ADARs25, suggesting that some observed effects may not be direct consequences of loss of editing.
The ADAR class of enzymes is conserved across metazoans26, with loss of activity in mice and flies leading to a dramatic disruption: loss of ADAR1 in mice results in defective hematopoiesis and embryonic lethality27–30; Drosophila without dADAR exhibit adult-stage uncoordination and temperature-sensitive paralysis31,32. C. elegans adr mutant strains exhibit chemotaxis defects and reduced life-span, but remain viable and fertile33,34. As C. elegans adr mutants lack extreme phenotypes, they provide a valuable system for studying the molecular contributions of A-to-I editing in transcriptome regulation.
Transcript levels for C. elegans adr-1 and adr-2 reach peak expression in embryos and the developing vulva. Examination of editing targets in select 3′ UTRs in mutant backgrounds has implicated ADR-2 as the active A-to-I editing enzyme, while ADR-1 appears to control site specificity and editing frequency33.Physiological chemotaxis dependence has also been characterized, with defects evident in adr-1 mutant strains, more severe in adr-2, and most severe in adr-1;adr-2 double-mutants25.
In addition to the physiological phenotypes, C. elegans adr-1;adr-2 mutant animals also exhibit a modulated response to transgene expression. Reporter transgenes introduced into the animal exhibit reduced expression, independent of sequence composition of the transgene35. This phenomenon has been suggested to derive from unintended duplex structures produced from indiscriminant strand-nonspecific transcription of transgene arrays. The resulting duplex RNAs, which may serve as ADAR substrates in wild-type animals, would then trigger RNAi in the absence of active ADAR. Correspondingly, the modulated transgene effect is also rescued in adr-1;adr-2;rde-1 triple mutants, while transgene transcripts exhibit extensive A-to-I editing in the presence of ADAR35.
The competitive relationship between the ADAR and RNAi pathways in C. elegans appears to extend to regulation of endogenous genes. In particular, the chemotaxis defect of adr-1;adr-2 animals is rescued in strains additionally lacking critical components of the RNAi machinery (adr-1;adr-2;rde-4 and adr-1;adr-2;rde-1)36. These observations suggest that ADARs may act to unwind endogenous dsRNAs capable of entering the RNAi pathway36 (Fig. 1). Alternatively, it is possible that components of the RNAi pathway act upstream of the ADAR pathway, possibly modulating editing activity or the availability of editing targets. Additionally, ADAR activity may act in a specific or non-specific manner to antagonize or modulate small RNA processing and maturation37–39, and in such a role may act as a modulator of the miRNA pathway. These possibilities are certainly not mutually exclusive.
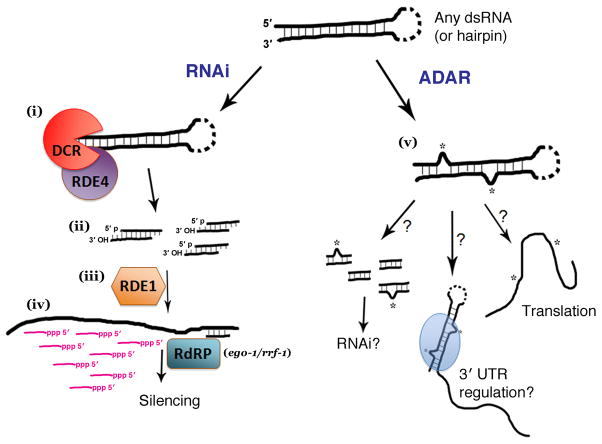
Figure 1. Diverse consequences of dsRNA formation.
Figure shows an arbitrary dsRNA (or hairpin) with a number of possible downstream processes. Left: RNA interference (RNAi). In the current model of RNAi, RDE-4 is involved in the recruitment of DCR-1 following recognition of a double-stranded RNAi trigger, resulting in its cleavage (i). (ii) siRNA duplexes produced by Dicer have characteristic structure with 5′ monophosphate, 3′ hydroxyl RNA termini for each strand, and a 2 nt 3′ overhang in the duplex. (iii) These cleaved duplexes subsequently program the Argonaute factor RDE-1 to recognize cognate mRNAs. (iv) Following RDE-1 interaction, target mRNAs serve as templates for the transcription of a pool of “secondary” siRNAs (short antisense transcripts templated from targeted mRNAs that carry a 5′ triphosphate terminus). Secondary siRNAs (shown in magenta) are produced by one of two cellular RNA-directed RNA polymerase (RdRP) enzymes, rrf-1 (somatic tissue) and ego-1 (germline). The RNAi process results in efficient and rapid loss of the pool of cognate mRNAs. (v) ADARs also target double-stranded RNA in vivo, converting a subset of adenines to inosine by deamination and resulting in the unwinding of the dsRNA or in other potential consequences including alterations in mRNA stability, localization, translation, and engagement in other RNA-based machineries such as RNAi43,42. Genetic evidence in C. elegans suggests that ADAR and RNAi pathways compete for a population of substrates36.
Assignment of ADARs to distinct cellular roles has been constrained by the lack of an identifiable sequence motif at editing sites40 and by challenges in definitive identification of functional ADAR targets41. Nevertheless, conserved characteristics of binding and activity appear to implicate ADARs in a broad regulation of dsRNA populations42,43. In mice, for example, the liver failure associated with defective hematopoiesis in ADAR mutants is suggested to arise from dsRNA-triggered interferon response27. Mutant phenotypes in organisms lacking ADAR could thus arise as a secondary consequence of the deregulation of a much broader class of transcripts. Although the salient phenotype of C. elegans ADAR mutants is neurological (resulting in defective chemotaxis), the broad role of ADAR editing may or may not be directly related to regulation of transcripts involved neurological development.
Taken in this view, elucidation of ADAR function will entail general characterization of broad classes of ADAR targets. Antagonistic interactions between the ADAR and RNAi pathways in C. elegans suggest that such a population of endogenous ADAR-targets could be evident by examining endogenous small RNA populations in editing-deficient animals. We introduce a genome-wide approach to identify highly edited loci that trigger a dramatic RNAi response in C. elegans in the absence of ADAR, characterizing a population of transcripts that naturally engage the ADAR mechanism.
RESULTS
A class of 5′ p 23–24 nt RNAs are enriched in adr mutants
We sequenced small RNAs from C. elegans wild type populations and from animals carrying deletions in one or both adr genes (Supplementary Table 1). Since the chemotaxis defect in editing-deficient mutants is suppressed in adr-1;adr-2;rde-1 and adr-1;adr-2;rde-4 strains36, we additionally sequenced small RNA populations from these animals, reasoning that a similar rescue of molecular phenotypes might also be observed. Based on expression patterns of the adr transcripts, we queried populations at the embryonic and L4 stages. Small RNAs (18–30 nt) were extracted and captured by procedures44 that required substrates to have 5′ monophosphate and 3′ hydroxyl termini. These samples were sequenced on the Illumina platform, yielding an average of 5.8 million high-quality reads per sample that align to the C. elegans genome.
Initial examination revealed genomic regions for which small RNA counts were dramatically increased in the absence of ADAR activity (e.g. Fig. 2). These increases were most prominent among 23–24 nt small RNAs. To identify additional regions with these characteristics, we divided the genome into regular segments and determined the number of reads aligning to each segment (Supplementary Methods online). We normalized for repetitive content by dividing the contribution of each read alignment by the total number of genomic positions aligned by the corresponding read. ADAR-deficient strains exhibited a size-specific enrichment for 23–24 nt RNAs over many loci throughout the genome (Supplementary Fig. 1).
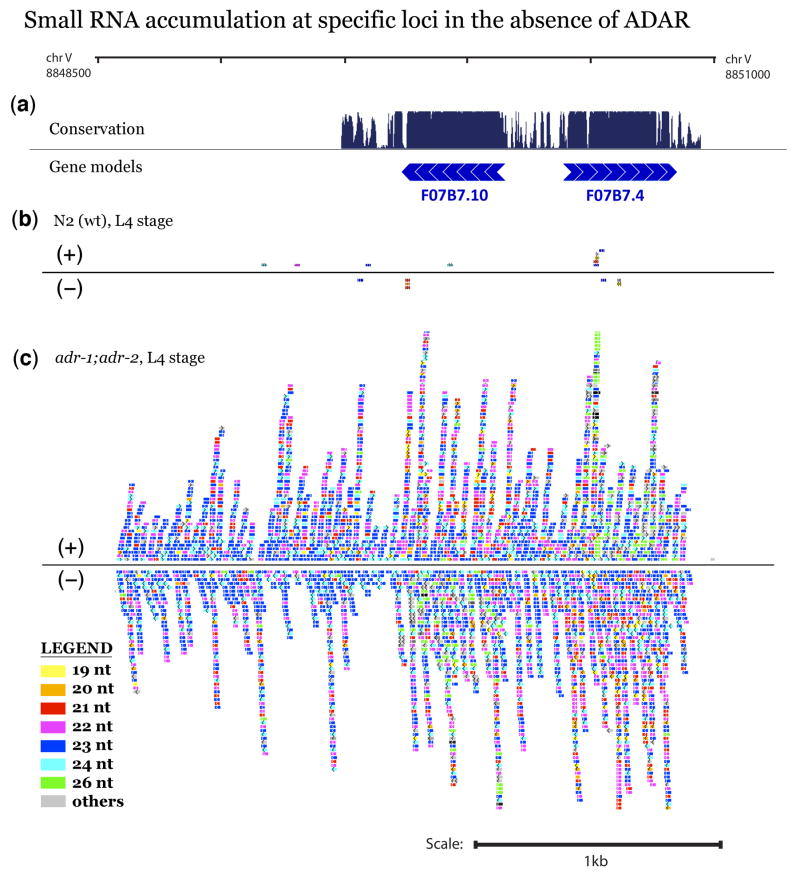
Figure 2. Small RNA accumulation at the F07B7 histone locus in the absence of ADAR activity.
An exemplary region spanning 2.6 kb (overlapping the F07B7.10 and F07B7.4 regions of C. elegans chromosome V) is shown. (a) Identified coding regions and conservation are diagrammed as UCSC Genome Browser tracks (C. elegans genome version WS190)59. F07B7.10 encodes an H2A histone; F07B7.4 encodes an H2B histone. Direction of transcription is depicted by arrows. (b–c) Small RNAs mapping to this region from (b) N2 and (c) adr-1;adr-2 animals. Each colored rectangle represents up to 10 instances of a distinct small RNA sequence per five million sequenced sRNAs. Small RNAs aligning to the (+) strand are drawn above the line and those aligning to the (−) strand are drawn below the line. Overall numbers of aligned reads for the wild type and adr mutant datasets in this example were 9.2 million and 10.1 million, respectively, (Supplementary Table 1) with comparable representation of miRNAs, 21-U RNAs, and endo siRNAs in the two samples. Additional examples of small RNA coverage are shown in Supplementary Figure 4. Colors indicate sizes (as on figure legend): yellow=19 nt, orange=20 nt, red=21 nt, magenta=22 nt, blue=23 nt, cyan=24 nt, green=26 nt, grey=all other lengths.
To generate a provisional list of affected loci, we identified regions that exhibited extensive coverage by 23–24 nt RNAs in the adr-1;adr-2 datasets and for which 23–24 nt RNAs were enriched over RNAs of other lengths that map to the same loci (Supplementary Fig. 2a and Supplementary Methods online). Of the regions discovered from adr-1;adr-2 datasets (786 regions totaling 505 kb), over 80% showed enrichment of 23–24 nt small RNA signal relative to wild-type levels. As a control, the same initial discovery procedure was applied to wild-type datasets, yielding few equivalent regions (49 regions of at most 400 bp each, totaling 5 kb, of which none were differentially expressed between datasets).
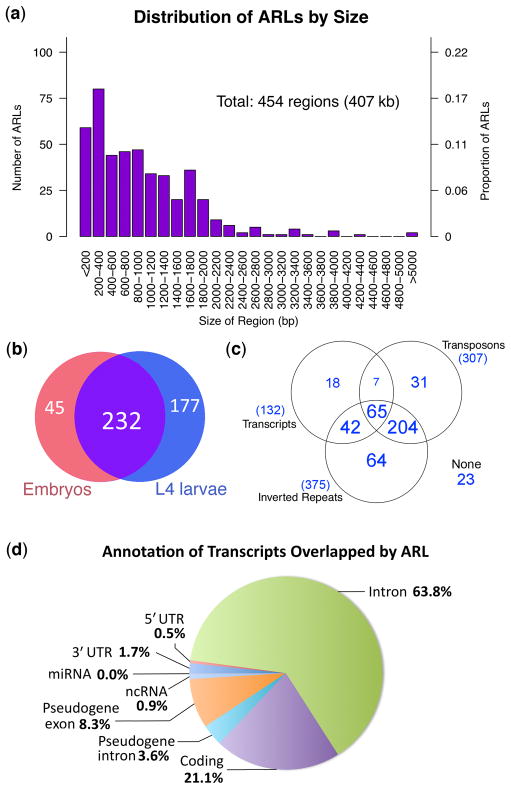
We provisionally term these (23–24 nt) sRNA-enriched regions "ADAR-modulated RNA loci” (ARLs). The 454 identified ARLs range from 0.1 to 9 kb (Fig. 3a, Supplementary Table 2), spanning a total of 407 kb (0.4% of the genome), distributed across all C. elegans chromosomes. The majority of defined ARLs were readily detected in both embryonic and L4 larval tissue (Fig. 3b).
Figure 3. Characteristics of ADAR-modulated-RNA-Loci (ARLs).
(a) Histogram showing the distribution of sizes of ARLs. ARLs ranging from 100 bp to 9 kb were detected. (b) Venn diagram showing overlap between ARLs detected at embryo and L4 stages, based on sRNA enrichment (p<0.05) of adr-1;adr-2 animals over wild type levels at embryo and L4 larval stages, respectively. Most ARLs are represented at both stages. (c–d) Overlap between ARLs and genomic repeats and features. (c) 82% of detected ARLs overlap annotated inverted repeats, while 67% overlap transposons. Fewer ARLs (18) overlap transcripts alone. A few ARLs (23) do not overlap any annotation assayed. (d) Partitioning of ARL annotations among annotated transcripts. Each ARL is divided into 100 bp segments, which are then indexed to the annotated genome59. Overlaps with 5′ UTR, coding, 3′ UTR, introns, miRNA, and pseudogenes are then tallied, with segments overlapping two or more different annotation categories being split between the relevant classes.
ARLs overlap numerous inverted repeats and transposons
Genomic annotations of regions overlapped by ARLs were significantly enriched for inverted repeats and transposons (Fig. 3c). A total of 375 ARLs (82%) overlapped annotated inverted repeats, while 307 ARLs (67%) overlapped annotated transposons. By contrast, a significantly smaller fraction of random fragments of equivalent length to ARLs overlap annotated inverted repeats (31.2%) or annotated transposons (18.5%) (p-value< 1e-40, Monte Carlo simulation; Supplementary Table 3 and Supplementary Methods online). ARLs were further depleted in annotated transcribed regions (actual: 29.7%; expected: 60.7%; p< 1e-40), with over 60% of transcript-associated ARLs falling into introns (Fig. 3d).
Small RNAs aligning within ARLs represented a substantial fraction of the small RNA pool in adr-deficient animals (up to 11.7% of aligned distinct sequences in each sample, in contrast to <1.1% from wild-type samples). Although repetitive, copy numbers for ARL regions were not extreme, with over 80% of ARL-associated RNAs aligning to four locations or less within ARLs (Supplementary Fig. 2b). Most ARLs could thus be classified as “low-copy” repetitive.
Genetic requirements in accumulation of ADAR-modulated sRNA
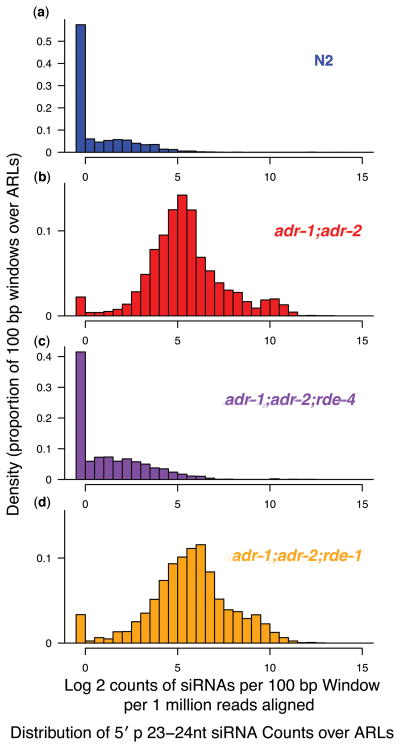
To elucidate key players in the biogenesis of these small RNAs, we examined their distribution in populations carrying loss-of-function mutations for rde-1 or rde-4 in addition to the adr-1;adr-2 double mutation. Mutants for rde-1 and rde-4 were chosen for this analysis based on the availability of viable null mutants that lack a functional RNAi response45,46. We found that 23–24 nt sRNAs levels from ARLs were restored to wild-type in adr-1;adr-2;rde-4 but not adr-1;adr-2;rde-1 animals (Fig. 4, Supplementary Fig. 2c, and Supplementary Methods online). These results suggested that the 5′ p ARL-associated sRNAs may be analogous to primary siRNAs generated during RNAi responses to exogenous dsRNA triggers47. Interestingly, ARL-associated small RNAs from adr-1;adr-2;rde-1 animals were at almost twice the level observed in adr-1;adr-2 animals. It is conceivable that in the absence of RDE-1, the passenger strand of the siRNA duplex may be retained48, resulting in double the available small RNAs over any region.
Figure 4.
Dependence of ARL sRNAs on the RNAi machinery. Distribution of small RNA abundances (23–24 nt only) for all 100 bp windows contained in ARLs. Small RNA values are shown as counts per million total reads aligned. Counts for each small RNA alignment was normalized to the total number of distinct genomic alignments of the associated sequence read. Distribution of small RNA counts over ARLs were calculated for each of (a) N2 (blue), (b) adr-1;adr-2 (red), (c) adr-1;adr-2;rde-4 (purple), and (d) adr-1;adr-2; rde-1 (orange). Graphs in this figure aggregate L4 and embryo data (individual distributions for L4 and embryo comparisons show a comparable difference; data not shown). Regions with high sRNA counts in wild-type also have comparable levels in adr-1;adr-2 animals, as is evident when small RNA abundances for each 100 bp region are normalized to wild-type levels (Supplementary Fig. 2c).
Since DICER has the property of cleaving dsRNA into small 21–25 bp duplexes with characteristic 2nt 3′ overhangs49,50, we examined alignment offsets for reads mapping to ARLs. Characterization of alignment offsets for reads from opposite strands equates to calculating 5′-to-3′ distance over a hypothetical siRNA duplex (Supplementary Methods). For 23–24 nt small RNAs, this analysis revealed a set of prominent +2nt alignment offsets, consistent with a 2nt 3′ overhang (Supplementary Fig. 3).
Secondary siRNA generation from transcripts overlapping ARLs
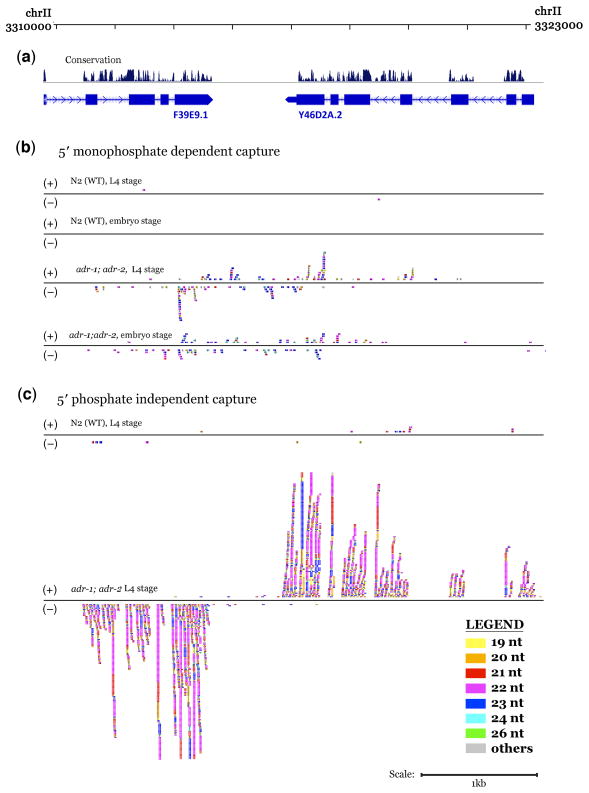
Although most ARLs overlap transposons or inverted repeat structures, a proportion also overlap annotated transcripts. In such cases, ADAR-modulated sRNAs in C. elegans might serve as triggers for RdRP-dependent secondary siRNA generation in C. elegans, templating off transcripts that overlap these ARLs (e.g. Fig. 5). Since secondary siRNAs generally contain 5′ triphosphates and 3′ hydroxyl51, a modified small RNA cloning protocol was used to capture these substrates in a manner independent of the presence of 5′ phosphates, 44,51. Using this 5′ phosphate independent cloning technique, small RNA libraries were constructed for each sample listed in Supplementary Table 1 and sequenced to generate an average 3.7 million aligned reads per library.
Figure 5.
A substantial class of additional ARL-associated siRNAs are evident in 5′ phosphate independent capture and sequencing. (a) UCSC Genome Browser map (C. elegans genome version WS190) displaying an ARL in the intergenic region between genes F39E9.1 (split into F39E9.1 and F39E9.22 in WS215) and Y46D2A.2 (split into Y46D2A.2 and Y46D2A.5 in WS215), overlapping the last exon of both genes. Direction of transcription is depicted by arrows. (b) Populations of 5′ phosphorylated small RNAs. A substantial increase in small RNA accumulation in adr-1;adr-2 animals over wild-type levels can be seen at both the embryo and L4 stages. (c) Populations of small RNAs that have been exposed at the 5′ end by sequential treatment with alkaline phosphatase followed by polynucleotide kinase. Small RNAs accumulate (with size preference for 21–22 nt and a distinct strand preference) in adr-1;adr-2 animals over wild-type levels at both transcribed loci overlapping the ARL.
From the 5′ phosphate independent capture, transcripts overlapping ARLs exhibited significant upregulation of antisense small RNAs in adr-1;adr-2 animals (Fig. 6a). This enrichment was particularly prominent for transcripts that overlap an ARL by a minimum of three 100 bp windows. We used this threshold to define an exemplary set of such ARL-affected loci, yielding 63 transcripts (Supplementary Table 4). These transcripts include a large class of histone genes (e.g. Fig. 2) as well as instances of paralogous genes with intergenic inverted duplications (e.g. Fig. 5; Supplementary Fig. 4). The small RNA populations generated from these transcripts were mostly 21–22 nt in size and exhibited restriction to the antisense strand, extending along the transcript beyond the region of overlap with the ARL. This contrasts with the ADAR-modulated sRNAs seen using the 5′ monophosphate dependent capture, which were enriched in the 23–24 nt size range and did not exhibit strand specificity. The absence of these antisense 21–22 nt ARL-associated RNAs from 5′ monophosphate dependent libraries suggested that their capture is facilitated by 5′ phosphatase-kinase treatment, consistent with the signature 5′ triphosphate termini expected of direct RdRP (RNA-directed RNA Polymerase) products.
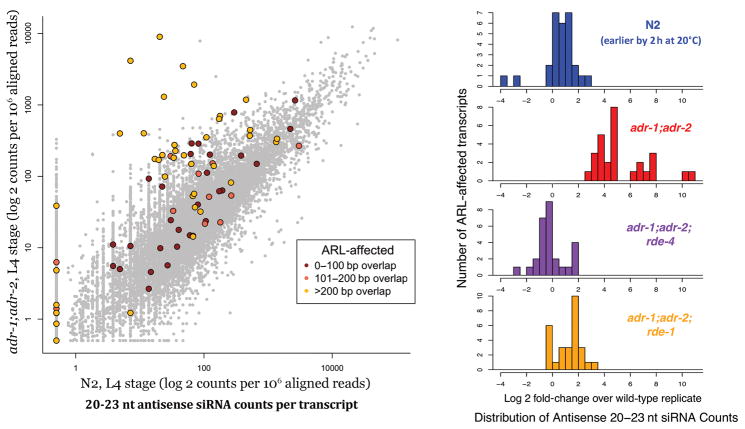
Figure 6. Accumulation of a second population of siRNAs in cis to ARLs.
(a) Genome-wide characterization of antisense siRNAs isolated using a 5′ phosphate independent capture protocol. Secondary siRNAs (antisense, 20–23nt) were tallied over each transcript. Transcripts overlapping ARLs are colored in dark red, orange, and yellow, based on the size of the total overlap (one, two, or three or more 100 bp segments, respectively). Genes that overlapped ARLs by at least three 100 bp segments and that exhibited a minimum 6-fold increase of secondary siRNAs in adr-1;adr-2 samples were deemed “ARL-affected transcripts”, and were considered as potential beneficiaries of ADAR-RNAi competition (without ADAR, they would be subject to populations of siRNAs produced by the RNAi machinery). (b) Effects of RNAi mutants on the secondary siRNA levels of ARL-affected transcripts. Antisense siRNA counts for ARL-affected transcripts were normalized to their respective wild-type levels. The top row (blue) denotes a separate biological preparation of wild-type animals at an earlier L4 stage (2 h earlier at 20°C). Secondary siRNA levels return to wild-type levels in adr-1;adr-2;rde-4 and adr-1;adr-2;rde-1 triple mutants. Gene-by-gene comparisons of expression-changes between different mutant backgrounds are depicted in Supplementary Figure 5.
To determine whether accumulation of this second class of ARL-associated siRNAs required RDE-1 and RDE-4, we carried out 5′ phosphate independent capture on RNA from adr-1;adr-2;rde-4 and adr-1;adr-2;rde-1 strains at equivalent developmental stages. We focused our analysis on transcripts that exhibited upregulation of antisense siRNAs in adr-1;adr-2 animals to at least 6 fold of wild-type. In both adr-1;adr-2;rde-1 and adr-1;adr-2;rde-4 genetic backgrounds, antisense siRNAs from these transcripts displayed a dramatic reversion to near wild-type levels (Fig. 6b, Supplementary Fig. 5). Hence, it appears that generation of these secondary siRNAs is dependent not only on the presence of siRNA duplexes, but also on the cleavage of these siRNA duplexes from a longer dsRNA—possibly transcribed from the overlapping ARLs.
Examining siRNA counts from 5′ phosphate independent capture more broadly (for all transcripts from transcriptome version WS215), we found that many of the genes with most drastic upregulation of secondary siRNAs in editing-deficient animals also overlap ARLs (Supplementary Table 4). This suggests a situation in which the most extreme differential siRNA responses in editing-deficient animals are cis effects due to secondary siRNA production from trigger siRNAs generated by Dicer cleavage of RNA duplexes.
Do ARL-associated siRNAs induce downregulation of targets?
The observation that some transcripts overlapping ARLs appear to serve as templates for RdRP activity (triggered by ADAR-modulated sRNAs) led us to ask whether this had an effect on steady-state mRNA levels of the corresponding transcripts. We examined RNA-Seq data from adr-1;adr-2 mutant C. elegans animals at the L4 stage and compared the expression level of our candidate transcripts in these samples with that from wild-type animals. Two transcripts overlapping ARLs that showed the greatest upregulation of antisense secondary siRNAs in adr-1;adr-2 mutant animals also exhibited a significant decrease in captured mRNA counts (Y46D2A.2 and Y46D2A.5, with mRNA decreased on average by 70% and 91% of wild type levels, respectively; Supplementary Table 4). Transcripts that displayed less dramatic increases of secondary siRNA levels (<5 fold relative to wild type levels) showed no evident downregulation at the mRNA level. Downregulation of Y46D2A.2 mRNA levels (accompanied by upregulation of antisense secondary siRNAs) was also observed in an adr-2 mutant background (data not shown).
Transcripts generated from ARLs exhibit A-to-I editing
Based on our evidence for engagement of ARLs with the ADAR machinery, it follows that transcripts originating from ARLs might be edited in a wild type background (allowing potential capture in RNA-Seq libraries). In a separate study (unpublished), we performed a comprehensive survey of A-to-I editing sites by looking for high-frequency A-to-G changes in the transcriptome that were recurrent across multiple biological replicates, using aligned 33-base RNA-Seq reads. We did not find a significant enrichment of individual specific editing sites for ARLs (data not shown). However, given evidence from in vitro assays which demonstrate that ADARs multiply edit double-stranded duplexes with high efficiency and low site specificity, we suspected that conventional alignment algorithms might fail to properly assign RNA-Seq reads from a class of heavily edited ADAR targets to the genome. With the aim of capturing this subset of ADAR targets, we took an alternative approach of in silico “pre-editing” or “collapsing” all adenosines to guanosines in both the RNA-Seq dataset and the genome sequence prior to alignment, in a manner similar to alignment techniques employed for bisulfite sequencing studies (Methods52,53). To evaluate all possible editing modalities, we carried out such collapsed comparisons using all possible nucleotide pairs: A+G, A+T, A+C, T+G, T+C, and G+C. To enrich for multiply edited sequences while accounting for sequencing error, we applied this approach using only RNA-Seq reads that failed to align to the genome with one mismatch or less. Reverting each read to the original 4-base sequence after alignment to the collapsed genome generates a set of candidate editing sites for each modality. To increase the signal for reads that represent frequently edited sequences (rather than spurious alignments), we restricted our analysis to reads which (i) contain only one class of mismatch throughout the sequence (e.g. A-to-G), (ii) contain at most 6 mismatches in total, (iii) aligned to less than twenty genomic locations, and (iv) for which at least one of the mismatches along the read was supported by an additional distinct sequence (Supplementary Fig. 6a and Methods for a complete description of criteria). Using these filtered alignments to generate a list of putative editing sites, we saw a significant enrichment of A-to-G edits over all other possible edits in the wild-type sample, and over A-to-G edits in the adr-1;adr-2 sample (Supplementary Fig. 6b). In total, we found ~15,000 putative A-to-G editing sites that were present in the wild-type and not the adr-1;adr-2 sample (numbers were equivalent for both stages assessed). Of these, ~7000 (~50%) putative editing sites from each stage fall into a total of ~130 ARLs (Supplementary Table 5). These edited ARLs include those that overlap the F07B7 histone locus and the downregulated Y46D2A.2 gene locus, from Figures 2 and 5, respectively (Supplementary Fig. 6c and 6d). Validation of multiple editing was carried out for the F07B7 histone locus using RT-PCR followed by Sanger sequencing (Supplementary Fig. 7 and Supplementary Table 6), which also provided a clearer picture of the editing signature over single transcripts. The global distribution of A-to-G putative editing sites over genomic annotations resembles the distribution exhibited by ARLs, with the majority of edits overlapping either a transposon or an inverted repeat (Fig. 7). This enrichment is not seen for any other class of nucleotide change, suggesting that the observations from this technique likely uncover a genome-wide population of specific A-to-I editing activity, rather than arising merely from spurious alignments. The striking similarity in genomic distributions of putative A-to-G editing loci (detected using mRNA sequence) and the ARLs (detected using small RNA sequence), in addition to the significant overlap between the two datasets, suggest that these two sets of loci may be common targets of ADAR as part of the same pathway.
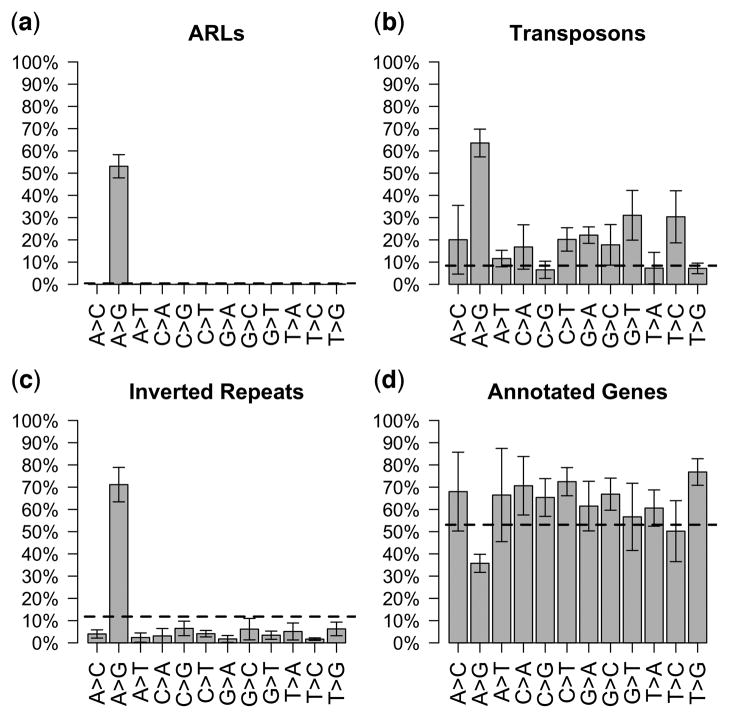
Figure 7. A-to-G changes in mRNA exhibit unique and significant enrichment for ARL, inverted repeat, and transposon regions.
Nucleotide changes were assayed using RNA-Seq data from both wild-type and adr-1;adr-2 animals, using a stringent set of criteria for candidate editing sites. Sites that matched these criteria and that were absent in the adr-1;adr-2 sample were reported as putative ADAR editing events. In addition to the RNA-Seq analysis of embryo and L4 animals described herein, a wild-type RNA-Seq dataset from L4 larvae prepared in an independent study56 was used as a replicate. Distribution of genomic annotations for each class of editing event reveal that A-to-G edits are enriched for (a) ADAR-modulated RNA loci as defined from small RNA sequences, (b) Transposon regions, and (c) regions with inverted repeat structure, but not for annotated transcribed regions (d). Dotted lines indicate the fraction of editing events expected to overlap each annotation based on a random distribution across the genome (calculated using a Monte Carlo simulation as described in Methods). Error bars denote one s.d. No other class of editing events detected show similar enrichment patterns. Within ARLs, the distribution of annotations overlapped by A-to-G editing sites is consistent: 89% of edits fall in transposon regions and 78% fall in inverted repeats (data not shown). Note that the distributions over separate annotations are not mutually exclusive (i.e. an editing site may overlap both a transposon and an inverted repeat).
Single gene effects of adr isoforms on ADAR-modulated sRNAs
To assess the relative contributions of the individual adr isoforms to the observed molecular phenotype, we repeated our analysis of ARL-associated small RNAs on samples from animals with single mutations in adr-1 and adr-2 (Supplementary Fig. 2d). We note that the adr-2 mutants exhibit slightly greater upregulation of regions with ARL-associated RNAs than adr-1 mutants, with the adr-1;adr-2 double mutant showing the most extreme molecular phenotype from ARLs defined either from this set (Supplementary Methods) or from the adr-1 or adr-2 data alone (data not shown). Secondary siRNAs originating from transcripts overlapping ARLs also exhibit greater accumulation in adr-2 mutants than in adr-1 mutants (Supplementary Table 4). The observation that adr-2 leads to stronger mutant effects than adr-1 corroborates with previous phenotypic characterizations of the C. elegans adr mutants, with the chemotaxis defect being most extreme in the double mutant, but being more evident in adr-2 single mutants than adr-1 single mutants33.
DISCUSSION
In this study, we describe a set of loci throughout the C. elegans genome from which RNA transcripts are extensively edited in wild type genetic backgrounds, and which serve as origins of abundant small RNAs in the absence of ADAR activity. ADAR-modulated “primary” siRNAs are distinctive, with (a) a 23–24 nt length enrichment, (b) 5′ monophosphate and 3′ hydroxyl termini, (c) a dependence on RNAi factor RDE-4 but not on the RNAi argonaute RDE-1, (d) an alignment offset indicative of a 2 nt 3′ overhang, (e) a distributed coverage over ARLs, and (f) an extensive overlap with a diverse set of low-to-moderate copy number inverted repeats. These molecular characteristics suggest that these small RNAs are likely generated by the RNAi machinery from double-stranded RNAs formed from large inverted repeat structures in the genome.
In C. elegans, the robustness of RNAi is attributed to the production of “secondary” siRNAs, which are templated from the target transcript and generated by a set of enzymes called RdRPs51,54,55. We observed such a population of putative secondary siRNAs with the expected characteristics for (1) 5′ triphosphate structure, (2) position relative to ARLs, (3) antisense orientation to transcripts, (4) length enrichment for 21–22 nt RNAs, and (5) dependence on both RDE-1 and RDE-4. Transcribed regions overlapping ADAR-modulated loci exhibit significant upregulation of secondary siRNAs, suggesting an effective engagement of the RNAi machinery that arises in consequence to the production of a population of primary siRNAs in the absence of ADAR activity. Furthermore, the transcripts exhibiting the most extreme upregulation of secondary siRNAs in adr-1;adr-2 animals were also significantly downregulated at the steady-state mRNA level. Other transcripts showing more moderate upregulation of siRNA populations exhibit only modest differences in levels of steady-state mRNA. The limited effects could be a reflection of a steep dose response between siRNA levels and mRNA downregulation; alternatively, homeostatic mechanisms (e.g. feedback regulation for histone mRNA levels) may act to compensate for any siRNA-induced downregulation.
Among the potential targets defined in this study, it is conceivable that one or more transcripts participate in determining the chemotactic behavior of animals, which is deviant in adr mutant animals and restored upon loss of RNAi activity36. Although such effects could conceivably reflect a single definitive misregulated target, no clear candidate emerges from a comparison of our lists with previous genetic analyses. As one possibility, the chemotactic behaviors in adr mutants could involve partial (or local) perturbation-of-function for one or more genes with broader roles in the organism; alternatively, other spheres of RNA metabolism in which RNAi and ADAR might compete could contribute.
The striking changes in the small RNA pool in ADAR-deficient animals provide an explicit view of the role of ADAR and the molecular consequences of A-to-I editing on a set of targets in vivo. Previously, assignments of functional ADAR targets have largely been based on detection of single nucleotide A-to-G changes in the cDNA. Extrapolations of functional and phenotypic implications from single nucleotide changes in non-coding regions have been difficult. In particular, since ADARs often lack sequence specificity and edit a dsRNA duplex in a largely promiscuous fashion, the levels of editing over many detected target sites may not recur across samples or result in downstream functional consequences. Analysis of small RNA pools appearing in ADAR mutant backgrounds provide an alternative approach to genome-wide identification of a set of ADAR targets, complementing traditional methods of looking for A-to-I changes on stable transcripts. By identifying loci that serve as sources of siRNA production in the absence of ADAR, we have uncovered a large set of ADAR targets at regions that are not necessarily highly represented in the mRNA pool. This signal is striking in C. elegans (though may occur to different extents in other organisms), with ARL-derived RNAs from <0.5% of the genome accounting for a substantial fraction (>10%) of all captured small RNAs in ADAR mutants.
In a complementary approach for detection of genome-wide ADAR targets using high-throughput mRNA-sequencing data, we applied a modified alignment technique that definitively identifies highly edited RNA segments. This analysis provided evidence for frequent A-to-I editing in mRNA originating from a large fraction of ARLs, suggesting that the two approaches we utilize here reveal a set of loci that are part of the same class of ADAR targets. Enrichment of these edits for intergenic regions with low mRNA coverage further suggests that ADARs act upon a population of targets that may be elusive to traditional methods of A-to-I editing target discovery.
Interestingly, the boundaries for our list of ARLs do not seem to correlate with annotated boundaries of coding regions. Although many ARLs are located near annotated genes, RNA-Seq results also do not provide convincing evidence of co-transcription of ARLs with adjacent transcripts. It is possible that transcripts originating from ARLs may be transcribed independently of neighboring transcripts. This would be consistent with a set of ADAR-engaging transcripts with a high rate of turnover or low levels of transcription—an attribute that would allow potential regulatory roles and perhaps preclude their detection using traditional methods of capture. Collectively, the results presented here support a model in which an extensive family of endogenous double-stranded RNA duplexes formed from transcription over a series of low-to-moderate copy inverted repeat regions are efficiently edited by ADAR and unwound, sequestered, specifically degraded, or directed down another salvage pathway before they can trigger RNAi. In consideration of emerging evidence in C. elegans and in mammals, we favor the idea that one main contribution of ADARs to evolutionary fitness may be to regulate accumulation of dsRNA from basally-transcribed (and possibly non-functional) regions throughout the genome in order to limit engagement by other dsRNA pathways in the cell.
Supplementary Material
Acknowledgments
We thank Jonathan Gent, S. Gu, M. Stadler, H. Zhang, K. Artiles, J. Pak, L. Gracey, A. Sidow, Z. Weng, P. Lacroute, and P. Parameswaran for help and suggestions; the Caenorhabditis Genetics Center, National Bioresource Project, and Brenda Bass (University of Utah) for strains; and Stanford Graduate Fellowship (D.W.), Stanford Dean’s Fellowship and Machiah Foundation (A. L.), and NIH (R01GM37706) for support.
Footnotes
CONTRIBUTIONS
D.W. and A.Z.F. designed experiments and wrote the paper. D.W. prepared samples, created small RNA libraries, and performed data analysis. A.T.L. created mRNA libraries and participated in intellectual discussions.
References
- 1.Werry TD, Loiacono R, Sexton PM, Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol Ther. 2008;119:7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Nicholas A, et al. Age-related gene-specific changes of A-to-I mRNA editing in the human brain. Mech Ageing Dev. 2010;131:445–7. doi: 10.1016/j.mad.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 4.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 5.Rula EY, Emeson RB. Mouse models to elucidate the functional roles of adenosine-to-inosine editing. Meth Enzymol. 2007;424:333–367. doi: 10.1016/S0076-6879(07)24016-9. [DOI] [PubMed] [Google Scholar]
- 6.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osenberg S, et al. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS ONE. 2010;5:e11173. doi: 10.1371/journal.pone.0011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–3. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 9.Levanon K, Eisenberg E, Rechavi G, Levanon EY. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep. 2005;6:831–5. doi: 10.1038/sj.embor.7400507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–5. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 11.Barak M, et al. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res. 2009;37:6905–15. doi: 10.1093/nar/gkp729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Research. 2004;14:2379–87. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DDY, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Research. 2004;14:1719–25. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paz-Yaacov N, et al. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci USA. 2010;107:12174–9. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurent GS, Savva YA, Reenan R. Enhancing non-coding RNA information content with ADAR editing. Neurosci Lett. 2009;466:89–98. doi: 10.1016/j.neulet.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Kleinberger Y, Eisenberg E. Large-scale analysis of structural, sequence and thermodynamic characteristics of A-to-I RNA editing sites in human Alu repeats. BMC Genomics. 2010;11:453. doi: 10.1186/1471-2164-11-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–84. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 18.Zinshteyn B, Nishikura K. Adenosine-to-inosine RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2009;1:202–9. doi: 10.1002/wsbm.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 20.DeCerbo J, Carmichael GG. Retention and repression: fates of hyperedited RNAs in the nucleus. Curr Opin Cell Biol. 2005;17:302–8. doi: 10.1016/j.ceb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Desterro JMP, et al. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–18. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 22.Scadden D. A NEAT way of regulating nuclear export of mRNAs. Mol Cell. 2009;35:395–6. doi: 10.1016/j.molcel.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Scadden ADJ. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 2005;12:489–96. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 24.Scadden ADJ. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol Cell. 2007;28:491–500. doi: 10.1016/j.molcel.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hundley HA, Krauchuk AA, Bass BL. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA. 2008;14:2050–60. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y, Zhang W, Li Q. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life. 2009;61:572–8. doi: 10.1002/iub.207. [DOI] [PubMed] [Google Scholar]
- 27.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA Editing Deaminase ADAR1 Gene for Embryonic Erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, et al. Stress-induced Apoptosis Associated with Null Mutation of ADAR1 RNA Editing Deaminase Gene. Journal of Biological Chemistry. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 30.Hartner JC, et al. Liver Disintegration in the Mouse Embryo Caused by Deficiency in the RNA-editing Enzyme ADAR1. Journal of Biological Chemistry. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 31.Jepson JE, Reenan RA. Unraveling pleiotropic functions of A-To-I RNA editing in Drosophila. Fly (Austin) 2010;4:154–8. doi: 10.4161/fly.4.2.11232. [DOI] [PubMed] [Google Scholar]
- 32.Jepson JEC, Reenan RA. Adenosine-to-inosine genetic recoding is required in the adult stage nervous system for coordinated behavior in Drosophila. J Biol Chem. 2009;284:31391–400. doi: 10.1074/jbc.M109.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tonkin LA, et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–35. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebastiani P, et al. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans. PLoS ONE. 2009;4:e8210. doi: 10.1371/journal.pone.0008210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol Cell. 2002;10:809–17. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- 36.Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heale BS, Keegan LP, O’Connell MA. ADARs have effects beyond RNA editing. cc. 2009;8:4011–4012. doi: 10.4161/cc.8.24.10214. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, et al. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280:3946–53. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heale BSE, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–56. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ensterö M, Daniel C, Wahlstedt H, Major FC, Ohman M. Recognition and coupling of A-to-I edited sites are determined by the tertiary structure of the RNA. Nucleic Acids Res. 2009;37:6916–26. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wulff B, Sakurai M, Nishikura K. Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat Rev Genet. 2011;12:81–85. doi: 10.1038/nrg2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci. 2010;35:377–83. doi: 10.1016/j.tibs.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–49. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gent JI, et al. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–89. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 46.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 47.Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–18. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:207–11. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- 49.Industrial Enzymes. Springer Netherlands: Dordrecht; 2007. < http://www.springerlink.com/content/h665788873j73902/>. [Google Scholar]
- 50.Nicholson AW. Structure, reactivity, and biology of double-stranded RNA. Prog Nucleic Acid Res Mol Biol. 1996;52:1–65. doi: 10.1016/s0079-6603(08)60963-0. [DOI] [PubMed] [Google Scholar]
- 51.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–4. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 52.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Briefings in Bioinformatics. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 55.Alder MN, Dames S, Gaudet J, Mango SE. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA. 2003;9:25–32. doi: 10.1261/rna.2650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamm AT, Stadler MR, Zhang H, Gent JI, Fire AZ. Multimodal RNA-seq using single-strand, double-strand, and CircLigase-based capture yields a refined and extended description of the C. elegans transcriptome. Genome Research. 2011;21:265–275. doi: 10.1101/gr.108845.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.