Abstract
Double-stranded RNA (dsRNA) plays a centrally important role in antiviral innate immunity, both for the production of interferon (IFN) and also in the actions of IFN. Among the IFN inducible gene products are the protein kinase regulated by RNA (PKR) and the adenosine deaminase acting on RNA (ADAR1). PKR is an established key player in the antiviral actions of IFN, through dsRNA-dependent activation and subsequent phosphorylation of protein synthesis initiation factor eIF2α thereby altering the translational pattern in cells. In addition, PKR plays an important role as a positive effector that amplifies the production of IFN. ADAR1 catalyzes the deamination of adenosine in RNA with double-stranded character, leading to the destablization of RNA duplex structures and genetic recoding. By contrast to the antiviral and proapoptotic functions associated with PKR, the actions of ADAR1 in some instances are proviral and cell protective as ADAR1 functions as a suppressor of dsRNA-mediated antiviral responses including activation of PKR and interferon regulatory factor 3.
Introduction
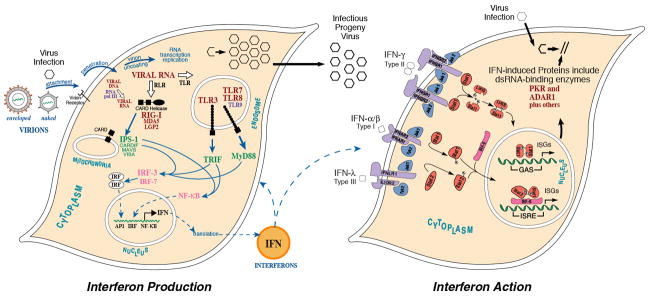
Interferon (IFN), the first cytokine discovered [1], derives its name from the robust biological activity for which it was discovered: the ability to interfere with virus growth. Interferon represents the founding cornerstone of antiviral innate immunity. Double-stranded RNA (dsRNA) has a long history in the interferon field. DsRNAs, both naturally occurring and synthetic, exemplified by reovirus genome RNA and poly rI: poly rC, respectively, were identified decades ago as potent inducers of IFN [2–4]. We now have significant understanding of the molecular mechanisms by which dsRNA produced during viral infections is detected as foreign by cellular nucleic acid sensors, thereby triggering signal transduction pathways that culminate in the transcriptional activation of IFN genes [5–8]. As shown schematically (Fig. 1, left), sensors of dsRNA in pathogen infected cells include the RIG-like family of receptors (RIG-I, MDA5) present in the cytosol [6,7] and the Toll-like receptor TLR3 that is endosomal membrane-associated [5]. RNA polymerase III also acts as a cytosolic sensor of DNA, leading to the production of dsRNA that is sensed by the RIG pathway [8–10].
Figure 1. Signaling pathways involved in the induction and action of interferon.
(left) Nucleic acid sensors that respond to viral infection detect viral nucleic acids as foreign, thereby leading to the production and action of IFN. Sensors include the RIG-I and MDA5 cytosolic helicases and their mitochondrial membrane-associated adaptor IPS-1; the Toll-like receptor TLR3 that acts through the TRIF adaptor; TLRs 7, 8 and 9 that act through the MyD88 adaptor; and, RNA polymerase III that acts through IPS-1. RIG-I, MDA5 and TLR3 sense dsRNA, and pol III senses cytosolic dsDNA to produce dsRNA. TLRs 7 and 8 sense ssRNA, and TLR9 CpG-rich DNA. These nucleic acid sensors trigger antiviral innate immunity through activation of factors that lead to transcriptional activation of IFN production. (right) Signaling by types I, II and III interferons through the canonical JAK-STAT pathway is illustrated, leading to the transcriptional activation of IFN-stimulated genes. Among the IFN-induced gene products are ADAR1 and PKR, both of which bind dsRNA and possess enzymatic activity, one (PKR) regulated by dsRNA and the other (ADAR1) utilizing dsRNA as a substrate. Adapted from Samuel [26].
The actions of IFNs are mediated by induced gene products and this too can involve dsRNA [11,12]. Transcriptional activation of IFN-stimulated gene (ISG) expression by the canonical JAK-STAT signaling pathway is illustrated in Figure 1 (right). Type I and III IFNs bind to their cognate receptors and activate a trimeric factor complex composed of STAT1 and 2 and IRF9 that then translocates to the nucleus and binds the DNA enhancer known as the interferon-stimulated response element (ISRE). Among the IFN inducible gene products are the PKR protein kinase [13–16] and the ADAR1 RNA adenosine deaminase [17–19]. Both PKR and ADAR1 are dsRNA binding proteins that possess multiple copies of a conserved dsRNA binding motif (Fig. 2A). In the case of PKR, dsRNA is a regulatory effector that either activates or antagonizes kinase activity; in the case of ADAR1, dsRNA is typically the substrate of the deaminase. Recent evidence suggests that PKR and ADAR1 function not only as mediators of the actions of IFN, but that they may also play roles in the production of IFN.
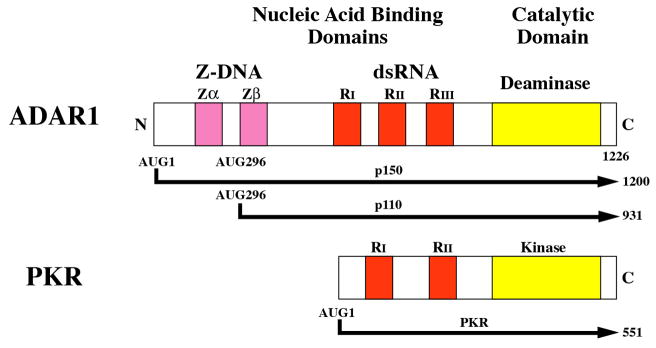
Figure 2. Domain organization of ADAR1 and PKR proteins from human cells and the enzymatic reactions catalyzed by them.
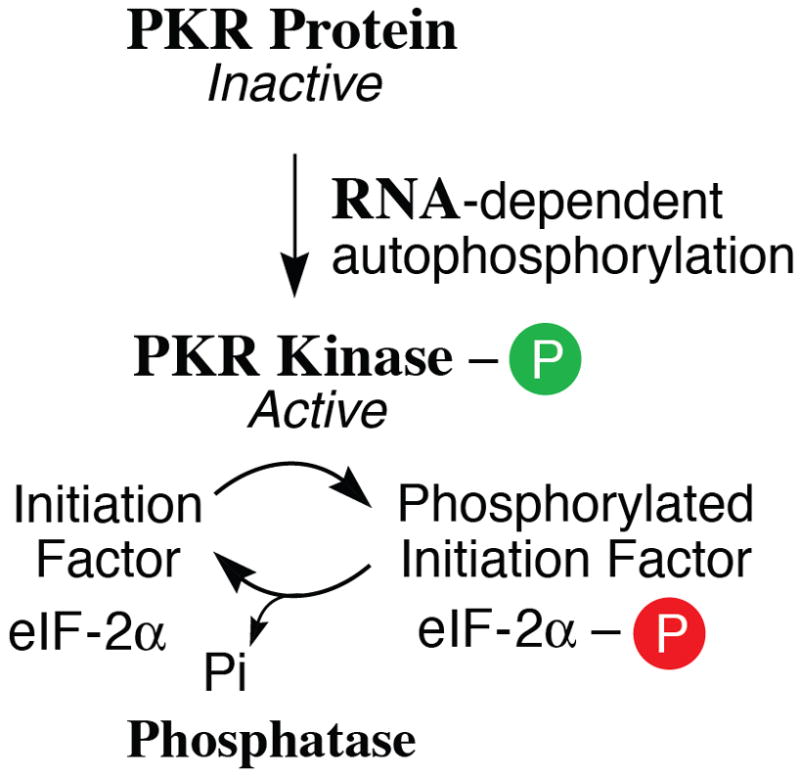
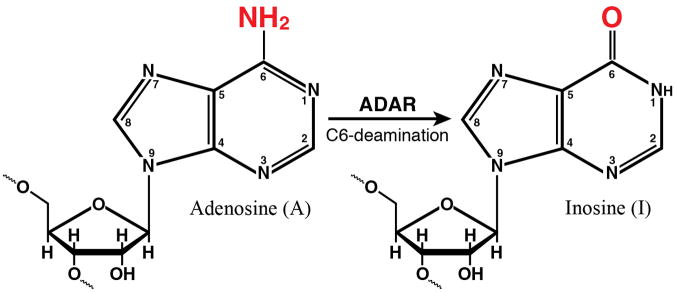
(A) Domains. Alternative promoters and alternative splicing give rise to two size isoforms of ADAR1, an IFN-inducible p150 protein and a constitutively expressed p110 protein. A single-sized form of PKR is known that is inducible by IFN. The N-terminal regions of ADAR1 and PKR include repeated nucleic acid binding domains and the C-terminal regions the catalytic domains responsible for their enzymatic activities. Multiple dsRNA binding domains (RI, RII, RIII), three present in both ADAR1 p110 and p150 and two in PKR, are shown in red. RNA adenosine deaminase and protein kinase catalytic domains are shown in yellow for ADAR1 and PKR, respectively. The N-terminal region of the p150 form of ADAR1 also possesses two Z-DNA binding domains (Zα and Zβ, and p110 the Zβ copy, as shown in pink. (B) dsRNA is the substrate of ADAR1. ADAR1 p110 and p150 catalyze the C-6 deamination of adenosine (A) to yield inosine (I) in RNA with double-stranded character. (C) dsRNA is an effector of PKR. RNA-dependent activation of eIF2α protein kinase activity is mediated by autophosphorylation (green P) of PKR; phosphorylation of serine 51 (red P) of eIF2α by PKR leads to an inhibition of protein synthesis.
Protein Organization and Genetic Regulation of ADAR1
ADAR1, adenosine deaminase acting on RNA 1, catalyzes the C6 deamination of adenosine to produce inosine in RNA substrates with double-stranded (ds) character [14,18,20,21]. This reaction (Fig. 2B) is referred to as A-to-I RNA editing and is a form of nucleotide substitution editing. The generated I is decoded as G instead of A by ribosomes during translation and by viral polymerases during RNA-dependent RNA replication. A-to-I editing also alters the stability of RNA duplex structures because I:U mismatch base pairs are less stable than A:U base pairs [22–24]. Indeed, ADAR activity was first described as a dsRNA duplex unwinding activity, but instead of unwinding dsRNA, the deamination of adenosine in duplexes destabilized the dsRNA structure [18,25,26].
The ADAR1 deaminase is encoded by a single gene [14,18]. The major transcript seen in human cells is ~6.7-kb in size, is increased in steady-state amount by IFN treatment, and includes 17 exons and possesses an open reading frame (ORF) of 1226 amino acids [14,17,18]. Two differently sized ADAR1 proteins are expressed, an IFN inducible (p150) protein that is found in both the cytoplasm and nucleus and a constitutively expressed (p110) protein that is predominantly if not exclusively localized to the nucleus [17,18,27]. ADAR1 transcription is driven by multiple promoters: one is IFN inducible, and the others constitutively active [18,28,29]. The IFN inducible promoter possesses a consensus ISRE element; activation following type I IFN treatment is dependent upon the IFNAR receptor, JAK1 kinase and STAT2 factor, but surprisingly not on STAT1 [28–30]. ADAR1 transcripts undergo alternative splicing involving exons 1 and 7 to encode the inducible p150 protein (1200 amino acids) or the constitutively expressed p110 protein (931 amino acids). p110 is an N-terminally truncated version of p150 (Fig. 2A).
The domain structure of the ADAR1 proteins includes within the C-terminal region the deaminase catalytic domain, and within the N-terminal region two kinds of nucleic acid binding domains, for dsRNA and for Z-DNA (Fig. 2A). Three copies of the dsRNA binding domain (RI, RII, RIII) are found in both p150 and p110; they are similar to the prototypical dsRNA binding R-domain (dsRBM) first identified in PKR [31]. Two Z-DNA binding domains (Zα, Zβ) are found in p150, but only Zβ is present in p110 [14,17,18,32]. p150 also includes a nuclear export signal [33]. Mutational analyses have established the importance of key residues in the ADAR1 functional domains, including the H910Q, E912A double mutant that inactivates deaminase catalytic activity; mutations of a conserved lysine at positions K554, K665 and K777 that affect RNA binding activity; the K418R mutation that abolishes sumoylation; and, mutations including Y177F that affect Z-DNA binding activity [14,34–36].
A-to-I editing is of two general types [19,20,24,25]. Editing can be highly site-selective with deamination occurring at one or very few specific A’s in the RNA substrate such as is seen with the glutamate and serotonin receptor pre-mRNAs and the hepatitis delta virus (HDV) antigenome RNA. These selective edits alter the genetic decoding of mRNA during translation, as the I pairs with C, whereas A pairs with U. A-to-I editing can also occur at multiple A’s in RNA substrates with near perfect duplex structure, such as observed when ADAR activity was discovered [18,25,26] or with synthetic dsRNA or viral dsRNA [14,18]. Hence, RNA substrate selectivity for adenosine deamination ranges from highly site-selective to non-selective, and is dependent in part on the duplex structure of the substrate RNA and possibly also interacting protein partners [18]. Little information is available regarding RNA as an effector rather than substrate, although adenovirus VAI RNA does antagonize ADAR1 editing activity [18,26]. A-to-I editing can affect how viruses interact with their hosts, either directly through genetic recoding, or indirectly through perturbations of RNA structures qualitatively if not quantitatively [26]. The importance of ADAR1 proteins to mammals furthermore is revealed by the embryonic lethality seen following genetic disruption of Adar1 expression. Independent Adar1 disruptions that knockout both p150 and p110 expression [37–40] or only p150 expression [41] all display embryonic lethality.
Protein Organization and Genetic Regulation of PKR
The IFN inducible, dsRNA-activated protein kinase (PKR, also known as eukaryotic translation initiation factor 2-alpha kinase 2 [EIF2AK2], P1 kinase, or p68 kinase) is a member of the eIF2α family of protein kinases that also include the PKR-like endoplasmic reticulum kinase (PERK, or EIF2AK3); the general control non-derepressible 2 kinase (GCN2, or EIF2AK4); and the hemin-regulated inhibitor (HRI, or EIF2AK1) of translation [42,43]. These eIF2α kinases are activated under different conditions of cellular stress, which is viral infection in the case of PKR, and catalyze the phosphorylation of serine 51 of eIF2α [42]. PKR expression is inducible by IFN [13]; the Pkr gene promoter, like the inducible ADAR1 promoter, possesses a consensus ISRE element [14,16]. The major PKR gene transcript seen in human cells includes 17 exons and specifies an ORF of 551 amino acids [14–16]. In contrast to the embryonic lethality observed by genetic disruption of mouse Adar1, neither of the two differently targeted Pkr homozygous null disruptions is lethal [44,45].
Two RNA binding motifs (RI, RII) are present within the N-terminal portion of PKR; the C-terminal region includes the kinase catalytic domain [14,16,46] as shown by Figure 2A. Mutational analyses established the importance of the PKR functional domains, including the K296R mutation in the catalytic subdomain II that impairs kinase activity; mutations of the highly conserved lysine of the RNA-binding domain (K64 in RI) that impair RNA binding activity; and the T446A phosphorylation site mutant that impairs in vivo kinase activity [12,14,15]. Binding of dsRNA leads to a PKR conformational change —which is believed to dissociate the catalytic domain from the autoinhibitory N-terminal domain— permitting dimerization and autophosphorylation [14,15,47]. RNA-mediated activation of PKR involves phosphorylation of both serine and threonine sites [14], including the T446 residue within the catalytic region that is commonly used as a measure of activation. Activation of PKR also has been described to include phosphorylation of tyrosine residues [48]. In addition to synthetic and natural duplex RNAs that activate PKR, naturally occurring viral RNAs with double-stranded character function either as PKR activators or antagonists as illustrated by reovirus s1 mRNA and adenovirus VAI RNA, respectively [12,14,49]. RNA binding is believed to occur in a non-sequence specific but RNA structure-dependent manner [11,12,50].
PKR Acts to Amplify Induction of Interferon following Virus Infection
PKR has long been known as a key player in the actions of IFNs [11,12]. Evidence is accumulating that PKR also plays an important role in the induction of type I IFNs, particularly IFNβ during viral infections [16,44,51–53]. The cytoplasmic RIG-I like receptors (RLRs) are principal sensors of foreign or non-self viral RNAs (Fig. 1, left). A complex forms involving the mitochondrial adaptor IPS-1 (also known as VISA, CARDIF, or MAVS) and activated RLRs, and together with TRAFs and IKK-related kinases, signal the activation of IRF3 and NFκB [5–7] to drive IFNβ gene expression [54].
Recent studies show that activation of PKR accompanies IRF3 activation, and that knock-down of PKR reduces activated IRF3 levels and IFNβ induction by transfected dsRNA [55]. Furthermore, the effect of PKR on activation of IRF3 is dependent on the adaptor IPS-1; siRNA mediated knock-down of either IPS-1 or PKR reduces IFNβ gene expression in response to measles virus (MV) infection to similar levels [51]. It is not yet known whether PKR directly phosphorylates IRF3, or indirectly contributes to the assembly or activation of the RIG-I/IPS-1 dependent signaling complex. For several positive-stranded RNA viruses including West Nile virus (WNV), encephalomyocarditis virus (EMCV), Theiler’s murine encephalomyelitis virus and Semliki Forest virus, PKR also is required for production of IFNβ [52, 53]. But for JFH1 hepatitis C virus (HCV), which likewise is a positive-stranded RNA virus, phosphorylation of PKR and eIF2α leads to an inhibition of IFNβ induction at the level of translation [56]. HCV, whose IRES-mediated protein synthesis initiation is independent of eIF2α phosphorylation, thus utilizes PKR to inhibit both the production [56] and action [57] of IFN. For bovine rotavirus UK, a double-stranded RNA virus, optimal induction of IFNβ production also is dependent upon PKR as well as RLRs, IPS-1, and IRF3; PKR deficiency leads to a defect in UK-infected cell secretion of IFNβ [58]. The mechanisms are not yet resolved, but one possibility in EMCV-infected cells involves PKR-mediated regulation of IFN mRNA stability [52].
The PKR dependency for optimal IFNβ induction by MV correlates with enhanced activation of NFκB and ATF2, and with WNV the PKR effect also involves enhanced NFκB activation [53]. The possibility that the PKR amplification of IFNβ expression is a translational control effect through eIF2α phosphorylation cannot be excluded, for example affecting the level of an activated transcription factor such as NFκB through control of synthesis of a rapidly degraded inhibitor like IκBα. To the extent that RNA structural features overlap for recognition by the RLRs and PKR also is not yet clear, but length of dsRNA and presence of a 5′-triphosphate on ssRNA are important for RLR sensing [5,59]. However, studies with reovirus ssRNA transcripts and genome dsRNA indicate that a 5′-triphospate is neither necessary nor sufficient for PKR activation [12,60], although 5′-triphosphate dependent activation of PKR by synthetic RNAs with short stem loops has been reported [61]. Interaction of PKR with members of the TRAF protein family has been described, with TRAF2 and TRAF3 interacting sites located around RII [62]. These interactions possibly stabilize the dsRNA-induced active (open) conformation of PKR by preventing the autoinhibitory function of RII. Furthermore, TRAF5 and 6 have been shown to interact with PKR, presumably indirectly and mediated by TRAF3 and 2, respectively [63]. The assembly of a TRAF-platform might enable the formation of an alternative signaling complex leading to phosphorylation of IRF3 or conceivably mediate an interaction with the RIG-I/IPS-1 signaling complex, thereby enhancing NFκB activation. Activation of NFκB by recruitment of the IKK complex [64], consisting of the inhibitor of kappaB kinases IKKα, IKKβ and IKKγ (also known as NEMO), could also be mediated by TRAFs. The IFNβ enhanceosome, besides NFκB and IRF3, also consists of ATF2/c-Jun activated by the mitogen-activated protein kinase (MAPK) pathways, including p38 and Erk1/2. PKR has been shown to be involved in the activation of p38 and Erk1/2 [51,65], but the biochemical mechanisms underlying this activation are not yet fully resolved.
ADAR1 Acts as a Suppressor of PKR and Displays Proviral Properties
Although ADAR1 is an IFN-inducible gene product, a growing body of evidence shows that ADAR1 functions in a proviral manner during acute infection in cell culture, notably with some RNA viruses [19,66]. These include HDV [67], MV [68], vesicular stomatitis virus (VSV) [27,69] and human immunodeficiency virus (HIV-1) [70–73]. In the case of HDV, site-selective A-to-I editing changes an amber UAG termination codon to a tryptophan UIG codon that permits synthesis of large delta antigen [67]. In the case of MV acute infection, depletion of ADAR1 in MEFs by genetic knockout of p150 and in human cells by knockdown of both p110 and p150 results in enhanced apoptosis and virus-induced cytotoxicity, and in the human ADAR1-deficient cells reduced MV growth [41,68]. In the case of HIV-1, overexpression of either ADAR1 [70,71,73] or ADAR2 [72] increases viral replication by both editing-dependent and-independent mechanisms.
While multiple mechanisms may be responsible for the proviral activity of ADAR1, one relates to the inhibition of PKR [74]. ADAR1 interferes with the activation of PKR and reduces the phosphorylation of eIF2α [27,68,69,75]. Overexpression of ADAR1, either the full-length p150 protein or the region with the RNA-binding and Z-DNA binding domains alone, impairs both PKR autophosphorylation and eIF2α phosphorylation [69,73,75]. Perhaps more physiologically relevant than overexpression are loss of function studies, where stable knockdown of ADAR1 leads to enhanced PKR autophosphorylation and eIF2α phosphorylation following infection with Cko mutant MV or VSV [27,68]. That is, in ADAR1 sufficient cells, PKR autophosphorylation is suppressed following infection, but in ADAR1 deficient cells is enhanced possibly because of the lack of editing-mediated destablization of dsRNA, lack of sequestration of dsRNA by ADAR1, or lack of formation of inactive heterodimeric ADAR:PKR complexes.
By virtue of the ability to impair PKR activation, ADAR1 would be expected to suppress PKR-mediated biological activities including antiviral, proapoptotic, and IFN induction amplification activities. Some of these predictions have been tested, either by ADAR1 loss of function or overexpression strategies. Depletion of ADAR1 by knockdown in human cells or by genetic knockout in mouse MEFs leads to enhanced apoptosis and cell cytotoxicity following infection with a number of different viruses of the Paramyxoviridae families and the DNA virus, polyoma [41,68,76]. In the case of MV, growth of both wild-type and Vko mutant virus are reduced in ADAR1 deficient cells compared to ADAR1 sufficient cells [68]. Furthermore, the inhibition of VSV growth by IFN is about 1 log10 further reduced in ADAR1 deficient compared to sufficient cells, and the reduced VSV yield correlates with enhanced PKR activation [27]. Using an overexpression screening strategy in which more than 380 human ISGs were tested for their antiviral activity against a number of medically important viruses, different categories of ISGs were identified: some acted broadly with an inhibitory effect, and a few enhanced viral replication [77]. Among the most potent proviral ISGs was ADAR1 that significantly enhanced the replication of all viruses tested including HIV-1, WNV, chikungunya virus, Venezuelan equine encephalitis virus and yellow fever virus [77].
Finally, for some RNA viruses including human respiratory syncytial virus and lymphocytic choriomeningitis virus, isolates have been obtained with A-to-G and U-to-C nucleotide substitutions in viral glycoproteins, sequence changes consistent with editing by ADAR1 [19]. Conceivably, limited low level editing by ADAR1 might be proviral in an infected animal if changes in a surface antigen occur in a manner that alters an epitope structure sufficient to allow escape from immune surveillance and neutralization. By contrast, when extensive editing occurs leading to hypermutations and inhibition of protein production, whether the editing is by ADAR1, or APOBEC3G in the case of retroviruses, then the effect could be antiviral.
Inosine-containing RNA as an Effector of the Innate Immune Response: Activator or Suppressor?
Poly rI:poly rC was one of the first dsRNA inducers of IFN discovered [3], and remains one of the most potent and efficient inducers [5–7]. Poly rI: poly rC also is bound by PKR, and dependent upon the concentration, either activates or inhibits of PKR autophosphorylation as well as activation of the 2′,5′-oligoadenylate synthetases [11,12,42]. ADAR1 action on dsRNA produces inosine (I) from adenosine (A) (Fig. 2B). Thus, A-to-I editing of dsRNA would be expected to produce inosine-containing dsRNA with I:U pairs in place of A:U pairs. Synthetic dsRNA with I:U base pairs unexpectedly did not induce, but rather suppressed the IFN response [78]. Induction of IFN-stimulated gene expression and apoptosis by poly rI: poly rC was suppressed by IU-dsRNA that contained multiple I:U pairs [78]. Furthermore, the IRF3 activation was inhibited by IU-dsRNA, possibly by inhibition of RIG-like receptor signaling, although the precise mechanism remains unresolved [78]. The observation that synthetic IU-dsRNA inhibits IRF3 activation is consistent with prior studies with virus-infected cells. Infection of ADAR1-deficient cells with MV results in an enhanced activation of IRF3 compared to the activation seen in ADAR1-sufficent cells [68].
The p150 isoform of ADAR1 has emerged as an important component in the host response to infection by a number of RNA viruses that replicate either in the cytoplasm or nucleus [27,41,68], whereas the p110 isoform plays a role for some DNA viruses that replicate in the nucleus [76]. A common theme is that the presence of ADAR1 correlates with a cell protective response, and in some cases even enhances virus replication. By contrast, under conditions of ADAR1 deficiency either in genetically null MEFs or knockdown cells, PKR activation is increased and the replication of VSV [27,69] and MV [68] are reduced. These results further establish that ADAR1 can display antiapoptotic and even proviral behavior in cell culture. Additionally, foreign DNA present in the cytosol, like foreign RNA, can trigger an innate immune response [8,79]. Among the DNA sensors in addition to RNA polymerase III [8] is DAI, DNA-dependent activator of IFN [79]. ADAR1 strongly suppresses the activity of DAI and reduces IFNβ induction by herpes simplex virus infection, effects that might enhance virus replication.
ADAR1 and Development of the Hematopoietic system
ADAR1 plays an important role in the development of the immune system. The presence of ADAR1 protein is an obligate requirement in mice for development of the liver and bone marrow hematopoietic system as established by knock-out studies. In mice homozygous for the Adar1 null mutation, embryonic lethality occurs at day 11.5–12.5 with liver disintegration and widespread apoptosis in many tissues [37]. MEFs from Adar1−/− embryos deficient in both p110 and p150 are prone to apoptosis due to stress induced by serum deprivation [39], and MEFs deficient in only p150 show enhanced cytotoxicity following viral infection [41]. Furthermore, the absence of ADAR1 p110 and p150 proteins results in a global upregulation of type I and II IFN-inducible transcripts and apoptosis [38], consistent with analyses that show ADAR1 suppresses the IFN response [27,68,69]. Studies of inducible Adar1 deficient mouse lines indicate that ADAR1 suppresses the deleterious effects of a robust activation of the IFN response [38]. ADAR1 either protects hematopoietic stem cells from apoptosis [38] or is necessary for differentiation of hematopoietic progenitor cells [40]. The selective knockout of p150 [41], like the knockouts that disrupt both p110 and p150 [37–40], is embryonic lethal, suggesting that the IFN inducible p150 isoform of ADAR1 is the form that regulates IFN production and protects against stress-induced cytotoxicity, thereby facilitating cell survival and maintenance of the hematopoietic stem cells.
Stress, RNA Granules, PKR and ADAR
Stress granules (SG), a form of RNA granules found in the cytoplasm of cells, can be both a cause and a consequence of stress-induced alteration in translation [80]. SG form within cells during conditions of stress, including viral infection, and among the consequences are the global downregulation of translation and the production of proteins necessary for cell survival [81,82]. Multiple mechanisms may be involved in the SG-associated modulation of translation, and among them is the phosphorylation of eIF2α. Formation of SG during viral infection often, but not always, is associated with activation of PKR and phosphorylation of eIF2 [80–82]. While some viruses mediate PKR activation and eIF2α phosphorylation, other viruses encode gene products that antagonize these processes [12,83].
Vaccinia virus E3L protein antagonizes PKR activation and facilitates virus growth, but mutant virus lacking E3L grows poorly, activates PKR and eIF2α phosphorylation and induces the formation of cytoplasmic SG-like structures [84,85]. Effective PKR-mediated restriction of E3L mutant virus growth requires SG-like complex formation subsequent to eIF2α phosphorylation [84]. Mammalian reovirus also induces the formation of SG in an eIF2α phosphorylation dependent manner early during infection, but at later times the SG structures become disrupted which correlates with the release of viral, but not cellular, mRNA from translation inhibition [86]. PKR activation and eIF2α phosphorylation also are associated with induction of SG formation by respiratory syncytial virus, but in a manner that facilitates virus replication [87]. In the case of WNV and dengue virus, the NS3 protein and viral dsRNA co-localize with the SG components TIA-1 and TIAR, and interaction with TIAR facilitates genome RNA synthesis and inhibits SG formation [88].
Among the SG-associated proteins is ADAR1 [80]. Furthermore, synthetic IU-dsRNA associates with a SG-like complex and downregulates gene expression in cultured cells [89]. Over-expression of ADAR1 increases gene expression at the translational level by decreasing PKR-dependent eIF2α phosphorylation [75]. What is not yet clear is whether the overexpression of ADAR1 simply impairs PKR activation, or alternatively alters SG-formation and function, to affect gene expression. It also is not yet known whether naturally occurring I-containing RNAs, similar to the synthetic IU-dsRNAs, or cytosolic RNA sensor components of the RIG-like receptor-IPS signaling complex, also associate with SG-like complexes to modulate gene expression and the innate immune response.
Conclusions
PKR, an IFN-inducible protein, is firmly established as a regulator of translation in virus-infected cells through phosphorylation of protein synthesis initiation factor eIF2α. Increasing evidence further positions PKR as a positive effector of IFN production triggered by infection and elicited via the RIG-like receptor pathway. PKR-mediated amplification of IFNβ expression is described for several RNA viruses, although there is the counter exception illustrated by HCV that displays a PKR-dependent impairment of IFNβ production. For PKR-dependent IFNβ expression, increased phosphorylation of PKR correlates with increased activation of IRF3, NFκB and ATF2, and enhanced IFN induction.
ADAR1, likewise is an IFN-inducible protein, is best known for its A-to-I RNA editing activity, whereby adenosine in dsRNA structures is deaminated to produce inosine. Increasing evidence implicates ADAR1 as an important modulator of the innate antiviral response, down-regulating the IFN response. ADAR1 suppresses activation of both PKR and IRF3, for example. In addition to the effects of A-to-I editing on RNA structure and function, ADAR1 also may conceivably affect the innate immune response via mechanisms that are dependent upon the RNA-binding or protein-interaction properties of ADAR1. However, little is known about the catalytic-independent mechanisms of ADAR1 in mammalian cells. Likewise, the precise mechanism as to how ADAR1 impairs PKR and IRF3 activation is not resolved. Among the possibilities are the destabilization of dsRNA structures; dsRNA competition and sequestration; and altered protein-protein interactions. Nor is the mechanism resolved as to how PKR affects signaling to amplify IFNβ production, perhaps either by translational inhibition of suppressor proteins or by fulfilling an adaptor function during signaling. These remain important questions.
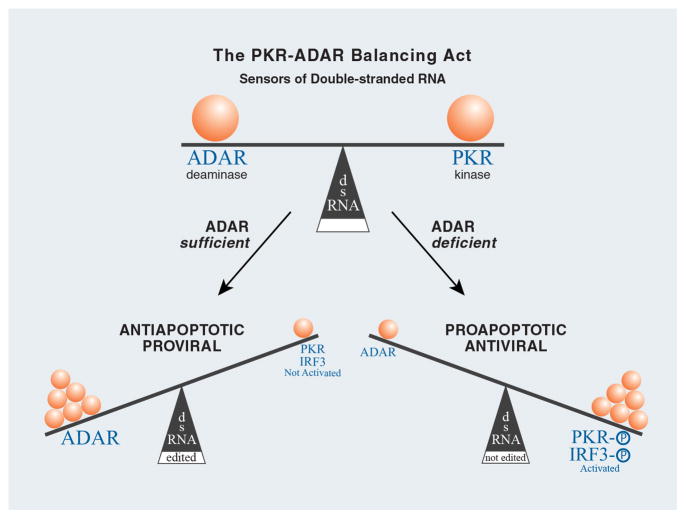
There are an increasing number of cellular sensors of foreign nucleic acids that have been identified which trigger the innate antiviral response, including RLRs and TLR3 for dsRNA. PKR and ADAR1 also are sensors of dsRNA (Fig. 3). Although both are IFN inducible proteins, constitutive levels of PKR and ADAR1 are typically present in most mammalian cells. A considerable body of evidence characterizes PKR function as antiviral and proapoptotic, whereas a growing body of evidence reveals ADAR1 function typically as proviral and antiapoptotic. A model that emerges is characterized by the actions of ADAR serving to balance those of PKR and other innate immune system responders (Fig. 3). Under spatiotemporal cellular conditions of ADAR sufficiency and PKR deficiency, PKR and IRF3 activities, for example, are minimized through functional inactivation of dsRNA by editing or sequestration. By contrast, under conditions of ADAR deficiency and PKR sufficiency, the activities of PKR and IRF3 are maximized. Future studies of the ADAR and PKR will provide opportunities to further test these notions, and no doubt will continue to provide us with surprises and new insights into biological functions of these IFN-inducible, dsRNA-binding enzymes.
Figure 3. Modulation of innate immune responses mediated by the relative balance between ADAR and PKR acting as RNA sensors.
Deamination of adenosine (A) to produce inosine (I) in duplex RNA structures catalyzed by ADAR leads to the nucleotide substitution of an I for an A in RNA. Because I base pairs with C instead of U, dsRNA with I:U mismatch base-pairs are less stable than A:U pairs. ADAR and PKR may also compete for dsRNA. ADAR, and A-to-I editing, may affect gene expression and function in virus-infected cells by a number of mechanisms including suppression of PKR and IRF3 activation.
Highlights.
PKR and ADAR1 as dsRNA sensors.
ADAR1 and PKR as opposing modulators of the interferon response.
ADAR1 suppresses PKR and IFNβ induction.
Proviral and cell protective functions of ADAR1.
Antiviral and Proapoptotic functions of PKR.
Acknowledgments
This work was supported in part by research grants AI-12520 and AI-20611 to CES from the National Institute of Allergy and Infectious Diseases, NIH, U.S. Public Health Service, and by a postdoctoral fellowship award to ZL from the Santa Barbara Foundation.
Abbreviations
- ADAR
adenosine deaminase acting on RNA
- dsRNA
double-stranded RNA
- IFN
interferon
- PKR
protein kinase regulated by RNA
- ssRNA
single-stranded RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the past two years, have been highlighted as:
*of special interest;
**of outstanding interest.
- 1.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 2.Tytell AA, Lampson GP, Field AK, Hilleman MR. Inducers of interferon and host resistance. 3. Double-stranded RNA from reovirus type 3 virions (reo 3-RNA) Proc Natl Acad Sci U S A. 1967;58:1719–1722. doi: 10.1073/pnas.58.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Field AK, Tytell AA, Lampson GP, Hilleman MR. Inducers of interferon and host resistance. II. Multistranded synthetic polynucleotide complexes. Proc Natl Acad Sci U S A. 1967;58:1004–1010. doi: 10.1073/pnas.58.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colby C, Morgan MJ. Interferon induction and action. Annu Rev Microbiol. 1971;25:333–360. doi: 10.1146/annurev.mi.25.100171.002001. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- 7.Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 8.O’Neill LA. DNA makes RNA makes innate immunity. Cell. 2009;138:428–430. doi: 10.1016/j.cell.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 9*.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. See annotation to ref. [10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. This paper like [9] shows that RNA pol III transcripts from AT-rich cytoplasmic DNA function as ligands for RIG-I. The authors provide evidence of a crosstalk between DNA and RNA sensing in antiviral innate immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samuel CE. Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc Natl Acad Sci U S A. 1979;76:600–604. doi: 10.1073/pnas.76.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 15.Sadler AJ, Williams BR. Structure and function of the protein kinase R. Curr Top Microbiol Immunol. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 16.Pindel A, Sadler A. The role of protein kinase R in the interferon response. J Interferon Cytokine Res. 2011;31:59–70. doi: 10.1089/jir.2010.0099. [DOI] [PubMed] [Google Scholar]
- 17.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George CX, Gan Z, Liu Y, Samuel CE. Adenosine deaminases acting on RNA, RNA editing, and interferon action. J Interferon Cytokine Res. 2011;31:99–117. doi: 10.1089/jir.2010.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuel CE. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411:180–193. doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wulff BE, Sakurai M, Nishikura K. Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat Rev Genet. 2011;12:81–85. doi: 10.1038/nrg2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogg M, Paro S, Keegan LP, O’Connell MA. RNA editing by mammalian ADARs. Adv Genet. 2011;73:87–120. doi: 10.1016/B978-0-12-380860-8.00003-3. [DOI] [PubMed] [Google Scholar]
- 22.Strobel SA, Cech TR, Usman N, Beigelman L. The 2,6-diaminopurine riboside.5-methylisocytidine wobble base pair: an isoenergetic substitution for the study of G. U pairs in RNA. Biochemistry. 1994;33:13824–13835. doi: 10.1021/bi00250a037. [DOI] [PubMed] [Google Scholar]
- 23.Serra MJ, Smolter PE, Westhof E. Pronounced instability of tandem IU base pairs in RNA. Nucleic Acids Res. 2004;32:1824–1828. doi: 10.1093/nar/gkh501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuel CE. ADARs: Viruses and Innate Immunity. Curr Top Microbiol Immunol. 2011 doi: 10.1007/82_2011_148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Wolff KC, Samuel CE. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology. 2010;396:316–322. doi: 10.1016/j.virol.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.George CX, Wagner MV, Samuel CE. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J Biol Chem. 2005;280:15020–15028. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- 30.George CX, Das S, Samuel CE. Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5′-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology. 2008;380:338–343. doi: 10.1016/j.virol.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fierro-Monti I, Mathews MB. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem Sci. 2000;25:241–246. doi: 10.1016/s0968-0004(00)01580-2. [DOI] [PubMed] [Google Scholar]
- 32.Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc Natl Acad Sci U S A. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol Cell Biol. 2001;21:7862–7871. doi: 10.1128/MCB.21.22.7862-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Samuel CE. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J Virol. 1996;70:1961–1968. doi: 10.1128/jvi.70.3.1961-1968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desterro JM, Keegan LP, Jaffray E, Hay RT, O’Connell MA, Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YG, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci U S A. 2003;100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 38**.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. This paper through inducible knockout of Adar1 shows that ADAR1 is an important innate immunity modulator in the mouse model, acting to suppress IFN signaling. The studies furthermore show that ADAR1 is important for the in vivo maintenance of hematopoietic stem cells and for successful embryogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 40**.XuFeng R, Boyer MJ, Shen H, Li Y, Yu H, Gao Y, Yang Q, Wang Q, Cheng T. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci U S A. 2009;106:17763–17768. doi: 10.1073/pnas.0903324106. This paper like [38] establishes an inducible mouse knockout of Adar1 and concludes that ADAR1 is important for the differentiation of hematopoietic progenitor cells in a manner that requires ADAR1 editing activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Ward SV, George CX, Welch MJ, Liou LY, Hahm B, Lewicki H, de la Torre JC, Samuel CE, Oldstone MB. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc Natl Acad Sci U S A. 2011;108:331–336. doi: 10.1073/pnas.1017241108. The authors describe a selective ADAR1-p150 knock-out mouse model and show that the interferon-inducible isoform of ADAR1 is essential for embryogenesis and cell protection against virus-induced cytotoxicity, whereas the constitutively expressed p110 is not sufficient. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuel CE. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 43.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 44.Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abraham N, Stojdl DF, Duncan PI, Methot N, Ishii T, Dube M, Vanderhyden BC, Atkins HL, Gray DA, McBurney MW, et al. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem. 1999;274:5953–5962. doi: 10.1074/jbc.274.9.5953. [DOI] [PubMed] [Google Scholar]
- 46.McCormack SJ, Thomis DC, Samuel CE. Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2 alpha protein kinase. Virology. 1992;188:47–56. doi: 10.1016/0042-6822(92)90733-6. [DOI] [PubMed] [Google Scholar]
- 47.Nanduri S, Rahman F, Williams BR, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su Q, Wang S, Baltzis D, Qu LK, Raven JF, Li S, Wong AH, Koromilas AE. Interferons induce tyrosine phosphorylation of the eIF2alpha kinase PKR through activation of Jak1 and Tyk2. EMBO Rep. 2007;8:265–270. doi: 10.1038/sj.embor.7400891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathews MB, Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nallagatla SR, Toroney R, Bevilacqua PC. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51**.McAllister CS, Toth AM, Zhang P, Devaux P, Cattaneo R, Samuel CE. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J Virol. 2010;84:380–386. doi: 10.1128/JVI.02630-08. Evidence is provided in this paper that PKR is required for maximized activation of IRF3 and IFNβ transcription via RIG-I signaling in response to viral infection. See also [55] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Schulz O, Pichlmair A, Rehwinkel J, Rogers NC, Scheuner D, Kato H, Takeuchi O, Akira S, Kaufman RJ, Reis e Sousa C. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. This paper describes a novel function of PKR in innate immune signaling, the regulation of IFNβ mRNA stability, besides translational control of IFNβ production through eIF2α phosphorylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilfoy FD, Mason PW. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:11148–11158. doi: 10.1128/JVI.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McAllister CS, Samuel CE. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J Biol Chem. 2009;284:1644–1651. doi: 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, et al. Hepatitis C virus controls interferon production through PKR activation. PLoS One. 2010;5:e10575. doi: 10.1371/journal.pone.0010575. See annotation to ref. [57] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. This publication together with [56] shows that the innate immune sensor PKR is subverted by HCV to impair development of the antiviral innate immune response at the translational level through activation of PKR and eIF2α phosphorylation, which is possible since HCV translation occurs independent of eIF2α and is hence not inhibited by PKR activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Sen A, Pruijssers AJ, Dermody TS, Garcia-Sastre A, Greenberg HB. The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF3. J Virol. 2011;85:3717–3732. doi: 10.1128/JVI.02634-10. This paper shows that PKR plays a crucial role in the RIG-I and MDA5-dependent IFNβ production during rotavirus infection as a positive regulator at a posttranscriptional stage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bischoff JR, Samuel CE. Mechanism of interferon action. Activation of the human P1/eIF-2 alpha protein kinase by individual reovirus s-class mRNAs: s1 mRNA is a potent activator relative to s4 mRNA. Virology. 1989;172:106–115. doi: 10.1016/0042-6822(89)90112-8. [DOI] [PubMed] [Google Scholar]
- 61.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 62.Gil J, Garcia MA, Gomez-Puertas P, Guerra S, Rullas J, Nakano H, Alcami J, Esteban M. TRAF family proteins link PKR with NF-kappa B activation. Mol Cell Biol. 2004;24:4502–4512. doi: 10.1128/MCB.24.10.4502-4512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies CC, Mak TW, Young LS, Eliopoulos AG. TRAF6 is required for TRAF2-dependent CD40 signal transduction in nonhemopoietic cells. Mol Cell Biol. 2005;25:9806–9819. doi: 10.1128/MCB.25.22.9806-9819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonnet MC, Daurat C, Ottone C, Meurs EF. The N-terminus of PKR is responsible for the activation of the NF-kappaB signaling pathway by interacting with the IKK complex. Cell Signal. 2006;18:1865–1875. doi: 10.1016/j.cellsig.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 65.Zhang P, Langland JO, Jacobs BL, Samuel CE. Protein kinase PKR-dependent activation of mitogen-activated protein kinases occurs through mitochondrial adapter IPS-1 and is antagonized by vaccinia virus E3L. J Virol. 2009;83:5718–5725. doi: 10.1128/JVI.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gelinas JF, Clerzius G, Shaw E, Gatignol A. ADAR1 enhancement of replication of RNA viruses via RNA editing and inhibition of PKR. J Virol. 2011 doi: 10.1128/JVI.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casey JL. Control of ADAR1 Editing of Hepatitis Delta Virus RNAs. Curr Top Microbiol Immunol. 2011 doi: 10.1007/82_2011_146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Toth AM, Li Z, Cattaneo R, Samuel CE. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem. 2009;284:29350–29356. doi: 10.1074/jbc.M109.045146. This study establishes that ADAR1 acts to suppress activation of proapoptotic and dsRNA-dependent activities associated with innate immunity including PKR and IRF3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nie Y, Hammond GL, Yang JH. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J Virol. 2007;81:917–923. doi: 10.1128/JVI.01527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Phuphuakrat A, Kraiwong R, Boonarkart C, Lauhakirti D, Lee TH, Auewarakul P. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J Virol. 2008;82:10864–10872. doi: 10.1128/JVI.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71*.Doria M, Neri F, Gallo A, Farace MG, Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 2009;37:5848–5858. doi: 10.1093/nar/gkp604. The authors describe that ADAR1 is beneficial for HIV-1 replication, by both editing-dependent and -independent mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doria M, Tomaselli S, Neri F, Ciafre SA, Farace MG, Michienzi A, Gallo A. ADAR2 editing enzyme is a novel human immunodeficiency virus-1 proviral factor. J Gen Virol. 2011;92:1228–1232. doi: 10.1099/vir.0.028043-0. [DOI] [PubMed] [Google Scholar]
- 73*.Clerzius G, Gelinas JF, Daher A, Bonnet M, Meurs EF, Gatignol A. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol. 2009;83:10119–10128. doi: 10.1128/JVI.02457-08. In this paper ADAR1 is shown to physically interact with PKR during HIV-1 infection and thereby inhibit PKR activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clerzius G, Gelinas JF, Gatignol A. Multiple levels of PKR inhibition during HIV-1 replication. Rev Med Virol. 2011;21:42–53. doi: 10.1002/rmv.674. [DOI] [PubMed] [Google Scholar]
- 75.Wang Y, Samuel CE. Adenosine deaminase ADAR1 increases gene expression at the translational level by decreasing protein kinase PKR-dependent eIF-2alpha phosphorylation. J Mol Biol. 2009;393:777–787. doi: 10.1016/j.jmb.2009.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.George CX, Samuel CE. Host Response to Polyomavirus Infection Is Modulated by RNA Adenosine Deaminase ADAR1 but Not by ADAR2. J Virol. 2011;85:8338–8347. doi: 10.1128/JVI.02666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77**.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. This study describes a screen of about 380 ISGs and shows that ADAR1 among the tested ISGs is a potent enhancer of viral replication, rather than inhibitor, for a broad variety of medically important RNA viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78**.Vitali P, Scadden AD. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat Struct Mol Biol. 2010;17:1043–1050. doi: 10.1038/nsmb.1864. The authors show that IU-mismatch pairs in dsRNA leads to suppression of IFN induction IRF3 activation and apoptosis, in agreement with the report [68] that ADAR1 suppresses PKR and IRF3 action. These studies underline the proviral, immune inhibitory and antiapoptotic activities of ADAR1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 81.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 84*.Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol. 2011;85:1581–1593. doi: 10.1128/JVI.02247-10. The paper shows that antiviral granules are formed during orthopoxvirus infection in a PKR-dependent manner and that these granules contribute to the cellular antiviral response by limiting viral replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang P, Jacobs BL, Samuel CE. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J Virol. 2008;82:840–848. doi: 10.1128/JVI.01891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86*.Qin Q, Carroll K, Hastings C, Miller CL. Mammalian Orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2{alpha} phosphorylation and PKR. J Virol. 2011 doi: 10.1128/JVI.01831-10. Evidence is provided that mammalian orthoreovirus disrupts stress granules at later stages of infection, which have been formed during earlier stages of infection. Furthermore, stress granules are required for a potent shutdown of translation despite the presence of phosphorylated eIF2α. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lindquist ME, Mainou BA, Dermody TS, Crowe JE., Jr Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology. 2011;413:103–110. doi: 10.1016/j.virol.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scadden AD. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol Cell. 2007;28:491–500. doi: 10.1016/j.molcel.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]