Abstract
Sphingolipids are membrane constituents as well as signaling molecules involved in many essential cellular processes. Serine palmitoyltransferase (SPT) and sphingosine-1-phosphate lyase (SPL), both PLP (pyridoxal 5′-phosphate)-dependent enzymes, function as entry and exit gates of the sphingolipid metabolism. SPT catalyzes the condensation of serine and a fatty acid into 3-keto-dihydrosphingosine, whereas SPL degrades sphingosine-1-phosphate (S1P) into phosphoethanolamine and a long-chain aldehyde. The recently solved X-ray structures of prokaryotic homologs of SPT and SPL combined with functional studies provide insight into the structure–function relationship of the two enzymes. Despite carrying out different reactions, the two enzymes reveal striking similarities in the overall fold, topology, and residues crucial for activity. Unlike their eukaryotic counterparts, bacterial SPT and SPL lack a transmembrane helix, making them targets of choice for biochemical characterization because the use of detergents can be avoided. Both human enzymes are linked to severe diseases or disorders and might therefore serve as targets for the development of therapeutics aiming at the modulation of their activity. This review gives an overview of the sphingolipid metabolism and of the available biochemical studies of prokaryotic SPT and SPL, and discusses the major similarities and differences to the corresponding eukaryotic enzymes.
Keywords: sphingolipids, SPL, SPT, PLP, structure, function
Introduction
Sphingolipids are ubiquitous constituents of cell membranes, and their metabolites play the role of signaling molecules in eukaryotic cells.1–4 They are strictly found in eukaryotes, with the exception of some bacterial genera and viruses.5 Sphingolipids are composed of three modules: a sphingosine backbone called long-chain base (LCB), a polar head group, and an amine-linked fatty acid chain (Fig. 1). The common structure of the sphingosine backbone, 2-amino-1,3-diol-alkane, is unique in nature and possesses two chiral centers at carbon atoms 2 and 3. The naturally occurring diastereoisomers of sphingolipids have the (2d,3d) [or d(+)-erythro-] configuration.6 The sphingosine backbone and the fatty acid chain are variable in length, degree of unsaturation, and substitution. Sphingolipids can either be produced via the degradation of existing sphingolipids or de novo in a synthesis pathway initiated by serine palmitoyltransferase (SPT), an enzyme located in the endoplasmic reticulum (ER)2 (Figs. 2 and 3). SPT catalyzes the condensation of serine and palmitoyl-CoA to the first sphingolipid of the synthesis pathway, 3-keto-dihydrosphingosine (3-keto-dihydro-SPH)9 (Fig. 2). This reaction is the entry point of the sphingolipid metabolism and its rate-limiting step. The enzyme 3-keto-dihydro-SPH reductase (KDS) produces dihydrosphingosine (dihydro-SPH). N-Acylation of dihydro-SPH generates dihydroceramide (dihydro-CER),10 which is subsequently desaturated to form ceramide (CER),11 a sphingolipid metabolic hub. A second route for the generation of CER is provided by the breakdown of complex sphingolipids by specific hydrolases12 and of sphingomyelin (SM) by specific sphingomyelinases (SM'ases).13 CER, bound to CER transfer protein (CERT) or integrated to vesicles, is transported to the Golgi and serves as building block for glycosphingolipids (Glyco-SPHL),14 ceramide-1-phosphate (C1P),15–18 and SM19,20 (Figs. 2 and 3). CERT consists of ER- and Golgi membrane-interacting domains and a START (standing for steroidogenic acute regulatory protein-related lipid transfer) domain able to bind CER.21,22 SM and Glyco-SPHL are relocated to the plasma membrane (PM) by vesicular transport, whereas various Glyco-CER are specifically transported from the Golgi to the inner leaflet of the PM bound to glycolipid transfer protein (GLTP).23–25 CER deacylation by various ceramidases (CER'ase) produces the single-chain sphingolipid sphingosine (SPH).26 SPH is the substrate of sphingosine kinase (SK),27 which produces sphingosine-1-phosphate (S1P). S1P can be reversibly dephosphorylated by S1P phosphatase (S1PP)28 or irreversibly degraded by S1P lyase (SPL) in one long-chain aldehyde, hexadecenal (Hex), and one polar compound, phosphoethanolamine (PE)29 (Fig. 2). The cleavage of S1P catalyzed by SPL is the exit point of the sphingolipid metabolism. PE can serve as building block for the synthesis of certain phospholipids, for example, phosphatidylethanolamine (PtE),30 which might in turn be involved in further signaling events.31
Figure 1.
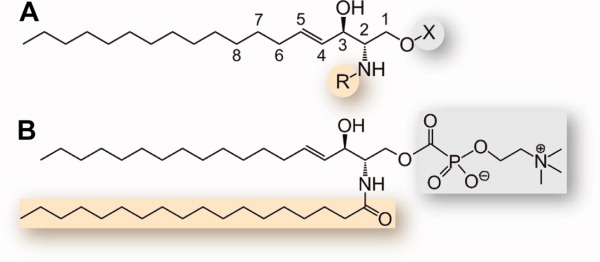
Chemical structure and diversity of sphingolipids. A: Sphingolipids d18:1Δ4trans. The module R is an amine-linked fatty acid in sphingomyelin (SM) and ceramide (CER) and an H in sphingosine (SPH) and sphingosine-1-phosphate (S1P), whereas X is either a phosphocholine in SM (see panel B), an H in CER and SPH, or a phosphate group in S1P. Phytosphingolipids carry a hydroxyl group at position 4, whereas 1-deoxy-sphingoid bases (DSBs) lack the hydroxyl at position 1. B: Sphingomyelin d18:1Δ4trans/18:0. For details about nomenclature, see Pata et al.4 Although the singular is used in the text, each class of sphingolipid is composed of a variety of molecules differing in length, degree of unsaturation, and substitution of the fatty acid and sphingolipid chains.
Figure 2.
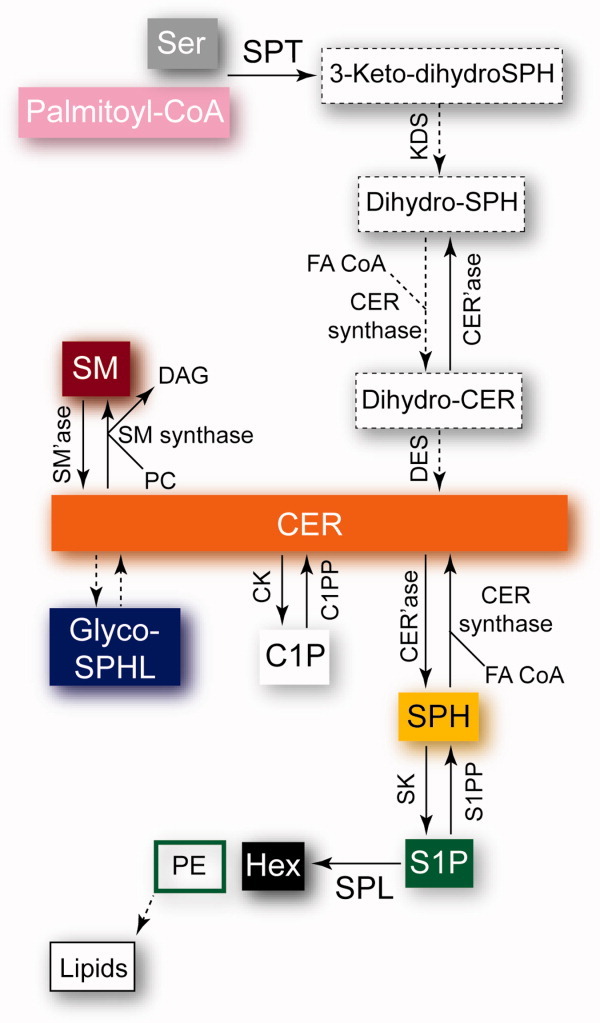
Overview of the sphingolipid metabolism. The direct pathway from entry to exit, that is, from SPT to SPL, goes from the gray and purple boxes until the black and green boxed rectangles. See text for abbreviations. FA CoA, fatty acyl-Coenzyme A; DES, dihydro-SPH desaturase; PC, phosphatidylcholine; DAG, diacylglycerol; CK, CER kinase; C1PP, C1P phosphatase. See Figure 3 for the cellular location of the reactions. Dashed boxes and arrows correspond to the respective dashed arrows in Figure 3.
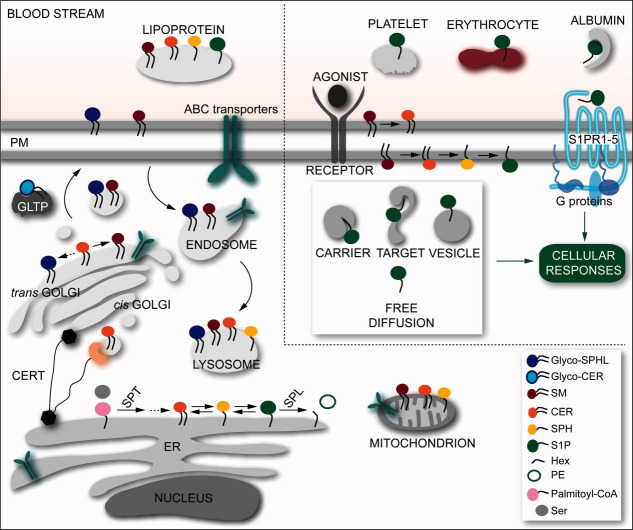
Figure 3.
Cellular locations of sphingolipids and S1P signaling modes. The color code for the various sphingolipids is the same as in Figure 2. The dashed arrows encompass reactions that are not displayed for clarity (see Fig. 2). The left part of the scheme illustrates the formation and degradation of sphingolipids in the ER, the subsequent transport of CER to the Golgi via CERT, the formation of Glyco-SPHL and SM therein, their transport to the PM via GLTP and vesicles, and their transport into the blood and their turnover via endosome. The ER- and Golgi membrane-interacting domains of CERT are represented as black hexagons, the CER-binding START is depicted in light orange, whereas the linkers between each domains are represented as black lines. In the cell, some sphingolipids also reside in the mitochondrion membrane. ABC transporters responsible for the export, import, or flipping of various sphingolipids were identified in different cellular compartments.7,8 The upper right part of the scheme depicts the extracellular transport of S1P in the blood, the stimulus-driven production of S1P (for example via binding of an antagonist to its cognate receptor), the putative intracellular transport modes of S1P, and the S1P extracellular signaling mode via the specific GPCRs S1PR1–5.
The specific inhibition or activation of the enzymes of the sphingolipid metabolism is crucial to control the concentration and the site of formation or degradation of sphingolipids, which play essential roles in living cells.32 Together with cholesterol and SM Glyco-SPHL form detergent-resistant membrane microdomains called “lipid rafts,” which are involved in the clustering of membrane proteins that are essential during signal transduction, in vesicular budding, and in the entry of pathogens.33 In addition, sphingolipids can directly interact with domains of membrane proteins, thereby modifying their activity.34 Moreover, some sphingolipids are “bioactive,” that is, a change in their concentration brings about functional consequences.1,3 The enzymes of the sphingolipid metabolism are biochemically interconnected, meaning that an increase in synthesis of one compound is related to a decrease in the other. Such interconnectivity increases the complexity of the signaling pathway and allows the cell to modulate the levels of a signaling molecule and its antagonist (Fig. 2). In addition, the location and transport of metabolites and enzymes, allowing pathways to operate in parallel, add another level of complexity (Fig. 3). Relative amounts of sphingolipids and imbalances in their metabolism have been univocally linked to hepatitis C,35 atopic dermatitis,36 rheumatoid arthritis,37 allergic response, anaphylaxis and asthma,38 multiple sclerosis, diabetes mellitus type I, psoriasis, Alzheimer's disease and atherosclerosis,39 cancer,3,36,40–42 and hereditary sensory and autonomic neuropathy type 1 (HSAN I).3 More specifically, Glyco-CER is involved in drug resistance.43 SPH modulates the activity of various kinases involved in the regulation of the cytoskeleton, endocytosis, cell cycle, and apoptosis,44 CER regulates stress-induced senescence45 and apoptosis,46 whereas C1P is involved in the inhibition of apoptosis,3 inflammation, vesicular trafficking, and phagocytosis.16,47 S1P is involved in the regulation of proliferation, cell growth, cell survival, cell migration, inflammation, angiogenesis, and resistance to apoptosis.40,48,49 It is a tumor-promoting agent antagonist to the cellular response primed by CER and SPH. Elevated amounts of S1P are found in cancer cells.50 Because of its physicochemical properties, micromolar concentrations of S1P can be found in the cytosol (Fig. 3), unlike the fully insoluble CER, SPH, and C1P.51 A recently identified intracellular target of nuclear S1P is histone deacetylases, suggesting that S1P is able to regulate gene transcription.52 In addition to its intracellular signaling mode, S1P can act extracellularly in an autocrine or a paracrine way by binding to five cognate receptors49,53,54 (Fig. 3). Consistently with its function in signaling, the concentration of S1P is tightly regulated by SK, S1PP, and SPL (Figs. 2 and 3).
SPL and SPT belong to the superfamily of pyridoxal 5′-phosphate (PLP)-dependent enzymes. PLP is covalently bound to the enzyme via a Schiff base between the aldehyde group of the PLP and the side-chain amine of a nearby lysine. Although they carry out two different reactions, SPL and SPT share a common fold, and their enzymatic mechanisms are in many aspects comparable. This article will review the recent advances in the structural and functional analysis of prokaryotic homologs of SPT and SPL, compare the two enzymes and relate them to their mammalian counterparts.
Structure and Function of SPL
The SPL gene has been first identified in budding yeast (Saccharomyces cerevisiæ),55 and the human gene was cloned in 2000.36,56 Yeast and mammalian SPLs are predicted single-pass ER membrane proteins with the transmembrane helix residing within the first fifth of the protein,57 but may localize to other organelles.36 Their N-terminus is situated in the ER lumen, where it undergoes glycosylation in yeast57 and their active site is exposed to the cytosol.58 Yeast SPL, called Dpl1p (dihydrosphingosine-1-phosphate lyase 1), is involved in resistance to heat stress59,60 and nutrient deprivation,61 in calcium homeostasis,62,63 as well as in endocytosis.64 In mammals, SPL plays a crucial role in the migration of T-cells from lymphoid tissues by maintaining the S1P gradient necessary for T-cells to egress from the lymph nodes into the blood stream.36,65 Similarly, SPL plays a central role in neutrophil egress from blood to tissues.66 Further, SPL has recently been confirmed as a central and crucial regulator of lipid homeostasis in mice liver.67 Disruption of SPL is linked to resistance to the chemotherapeutic agents etoposide and doxorubicin.68 SPL activity is also linked to an increase in CER levels in response to stress factors and is involved in apoptosis.69 The SPL gene is a transcriptional target of platelet-derived growth factor (PDGF),70 which regulates critical cellular events like cell growth, differentiation, and migration and is involved in cancer, inflammation, wound healing, and production of the extracellular matrix.71 The human SGPL1 gene is ubiquitously expressed with the exception of erythrocytes72 and platelets,73 and its expression is linked to tissues undergoing rapid cell turnover, like the intestine, colon, thymus, and liver.36,74 SGPL1 gene expression is downregulated in colon cancer,68,75 whereas it is high in enterocytes under normal conditions, where the protein is involved in metabolizing dietary sphingolipids.36,76 Notably, SPL is upregulated in certain malignant tissues, for example, in ovarian cancers77 as well as in the skin of patients suffering from atopic dermatitis,36,78 complicating the interpretation of its precise role in diseases. SPL has been recently directly linked to acute lung injury and might therefore be a target for the development of therapeutics.79 Additionally, SPL is involved in developmental events, especially in vascular maturation.36,74 Recently, the role of SPL in DNA repair and radioprotection was demonstrated.80
SPL is a member of the carbon–carbon lyase subclass of aldehyde-lyases (SPL, EC 4.1.2.27).36 SPL cleaves the amphiphilic substrate S1P between carbon atoms 2 and 3, yielding one hydrophobic and one hydrophilic product. Interestingly, Saba and coworkers have recently shown that the hydrophobic product of the cleavage of S1P, trans-2-hexadecenal, has a signaling function in mammalian cells.81 This indicates that the biological function of SPL is not restricted to the degradation of S1P but extends to the direct production of another bioactive lipid. This very important finding highlights the importance and the complexity of SPL in cell signaling events.
The full mechanism of accommodation and cleavage of the substrate and of release of the hydrophilic and hydrophobic products is not yet elucidated. The protein may contain a hydrophobic recognition site that interacts with the hydrophobic moiety of the substrate, whereas the hydrophilic head might be accommodated within the active site and can then be cleaved.6 Recent advances through biochemical and structural studies, however, have improved our understanding of the function of SPL,82 although biochemical studies are hindered by the poor solubility of phosphorylated sphingolipids, albeit zwitterionic in nature.6 Electrostatic and hydrophobic intermolecular interactions are presumably responsible for this property.83 Triton X-100 is so far considered the best detergent and is widely used to solubilize phosphorylated sphingolipids, because a number of other neutral and zwitterionic detergents have been reported to inhibit the enzyme,6,84 even though this needs to be confirmed in vitro. The pH optimum of SPL is in the range of 7.2–7.4. SPL shows a high level of specificity toward the stereochemistry of the substrate and cleaves exclusively the naturally occurring d-erythro (2d,3d configuration) sphingolipids.36 On the other hand, the enzyme exhibits very little specificity with respect to the chain length, degree of unsaturation, and substitution, as it cleaves S1P, dihydro-S1P, phyto-S1P (PS1P) [Fig. 1(A)], as well as methyl- or dimethyl-S1P.74,84,85 When the amino group of the substrate is modified, for example, by acetylation (N-acetyldihydro-S1P) or by methylation (N,N-dimethyl-S1P), the substrate is not cleaved.84 The classical PLP-dependent enzyme inhibitors semicarbazide, iodoacetamide, and N-methylmaleimide inhibit SPL to various extent, presumably by binding to either the cofactor, as shown for semicarbazide,82 or to a sulfhydryl group from a cysteine.74 Deoxypyridoxine also inhibits SPL, possibly by competing with PLP.6,36 There are known substrate analogs that inhibit SPL, for example, 1-deoxydihydrosphingosine-1-phosphonate,36,86 the threo isomer of dihydro-S1P,87 and 2-vinyldihydro-S1P.36,88 FTY720 (also called fingolimod) is an immunomodulatory drug phosphorylated by sphingosine kinase 289 that targets the S1P receptor 1.90,91 FTY720 inhibits SPL in vivo, when orally administered to mice.36 THI (2-acetyl-4-tetrahydroxybutyl-imidazole), found in the food colorant caramel colorant III, also inhibits SPL in vivo,36,65 although the mechanism of inhibition is not understood,74 as that of FTY720. A novel specific inhibitor of SPL, LX2931, is undergoing clinical trials.37,92 The divalent cations Ca2+ and Zn2+ have also been reported to inhibit the activity of SPL.93 It should be noted that biochemical results in the literature have been so far nearly invariably obtained using crude extracts or cell fractionation samples. Given the tight interconnectivity of S1P-related enzymes, this approach might introduce non-negligible bias.
Some bacteria, for example, Myxococcus xanthus and Symbiobacterium thermophilum, also carry a gene coding for a SPL. The structures of SPL from the thermophile Symbiobacterium thermophilum (StSPL) [Fig. 4(A)] and from an N-terminal truncation of Saccharomyces cerevisiae (Dpl1p) lacking the transmembrane helix reveal that the protein is a homodimer or a multiple thereof.82 Interestingly, the naturally occurring form of StSPL lacks a transmembrane domain and the recombinant protein is in vitro active, whereas the truncated Dpl1p fails to cleave S1P, dihydro-S1P, and PS1P in vitro.82 The cofactor occupancy is variable in several crystal forms of both SPLs. This indicates that the cofactor is labile in at least one subunit and that the two active sites might therefore carry out different functions. The very N-terminal parts of both proteins are flexible, suggesting that they might play a role in the activity or in the binding of other proteins, as is often the case for flexible regions.
Figure 4.
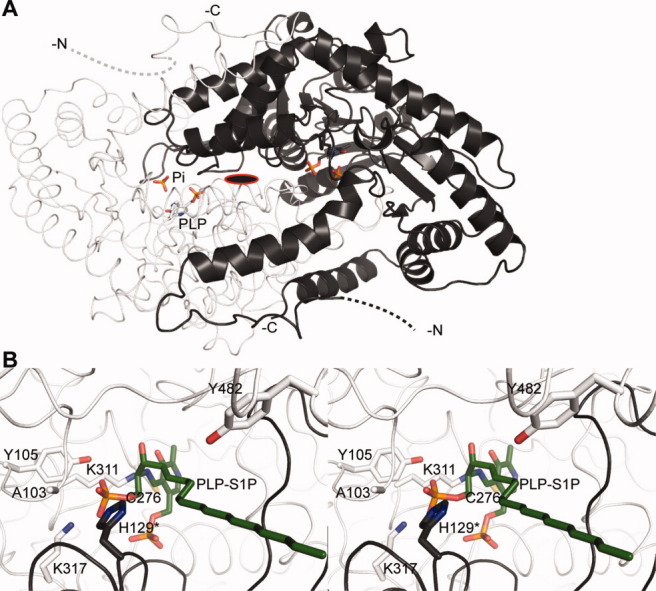
X-ray structure of StSPL. A: Dimeric structure of WT StSPL (PDB code 3MAD82). Subunit A is colored light gray, the subunit B dark gray, and represented in cartoon and ribbon, respectively. PLP and the covalently bound K311 as well as a phosphate ion appear in ball-and-stick in the color of the corresponding subunit and atom colors and are labeled in subunit A. Residues 1–57 are not visible and are represented by dashed lines in the color of the corresponding subunit. The N- and C-termini are indicated. The subunits are related by a twofold axis perpendicular to the plane of the sheet, indicated as a red-framed black ellipse. B: Stereo representation of the active site of subunit A showing the modeled external aldimine PLP-S1P in green (corresponding to step III in Figure 5). Residues important for activity appear in ball-and-stick in the color of the subunit. The asterisk denotes a residue belonging to the subunit B. Details about the modeling are given in Ref.82. This and the following structural figures were prepared with PyMOL.94
The presence of PLP in the active site allows for straightforward spectrophotometric characterization of wild type (WT) and mutants. Electron transfers as well as the microenvironment of PLP are reflected by enzyme-specific changes in the visible spectrum.95–101 WT StSPL features a broad peak at 420–460 nm and a shoulder at 360 nm.82 Upon addition of S1P d18:1Δ4trans, two transient new peaks at 403 and 420 nm appear. These peaks correlate with the enzymatic activity and were used to perform an analysis of StSPL mutants.82
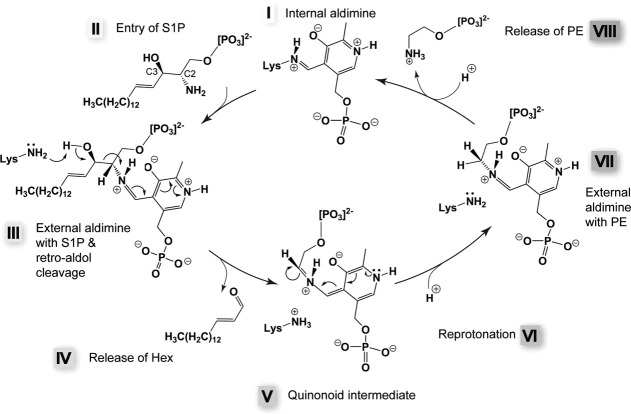
Already in 197488 and 1993,6 a reaction mechanism was proposed based on general knowledge of the biochemistry of PLP-dependent enzymes. Our recently published structural and functional investigations of purified SPLs82 shed a new light into the mechanism (Fig. 5). In the resting state of the enzyme, the PLP is covalently linked to the Lys residue 311, a complex called internal aldimine (step I). The amino group of the substrate replaces the lysine as PLP-binding partner (step III), and the covalent PLP-S1P adduct, named external aldimine [Fig. 4(B)], undergoes a retro-aldol cleavage by a yet unidentified base. The base was proposed to be C276 (in StSPL, corresponding to C317 in human SPL56), but our mutagenesis analysis did not confirm this finding.82 The formed adduct decomposes into a long-chain aldehyde that is released from the active site (step IV). A quinonoid intermediate (step V) is formed and a proton is stereospecifically incorporated (step VI). PE is then released (step VII) from the second external aldimine (step VI). Our mutagenesis analysis revealed that the highly conserved internal aldimine K311 as well as the neighboring K317 are strictly required for activity. Interestingly, Y482 located at the very C-terminus of the protein and covering the active site entrance is involved in enzymatic activity, suggesting an important role for the C-terminal part of SPL.82 Likewise, H129, belonging to the neighboring subunit, seems to be important for activity, because mutating it into Ala drastically impairs activity both in StSPL and Dpl1p.82 Mutating A103 into proline suppresses activity.82 Surprisingly, this residue is not conserved in human, because it is replaced by T148, suggesting that human SPL might feature a slightly different reaction mechanism or kinetic properties than StSPL and Dpl1p, at least concerning the role of this active-site loop. The same conclusion might be drawn for the inactive Y105F StSPL mutant compared to the corresponding Y174F Dpl1p mutant, which is partly active.82 Altogether, the recently published results highlight the dual role of lysines in catalysis and in the stabilization of reaction intermediates,82 as recently demonstrated for K88 of the PLP-dependent enzyme cystathionine β-synthase from Drosophila melanogaster.96
Figure 5.
Proposed SPL reaction mechanism.82 In the resting state, a lysine (311 in StSPL) forms a Schiff base with the aldehyde group of PLP (step I). The incoming substrate (S1P in the scheme, step II) triggers transaldimination and replaces the lysine as Schiff base partner of PLP. The resulting S1P-PLP complex is called external aldimine [step III, model presented in Figure 4(B), colored green]. After retro-aldol cleavage (step III), the long-chain aldehyde product (Hex in the scheme) leaves the active site (step IV) and a quinonoid intermediate is formed (step V). A stereospecific reprotonation (step VI) of this intermediate yields the second external aldimine (step VII). We propose that one of the two conserved lysines strictly required for activity carries out the retro-aldol cleavage in step III as well as the reprotonation (step VI). PE is then released (step VIII) and the internal aldimine is re-formed (step I).
Structure and Function of SPT
As for SPL, the effect of deleting the SPT gene was first studied in yeast.102 It clearly turned out that SPT is strictly required to produce sphingolipids and that it is crucial for sphingolipid homeostasis.103 Recently, yeast and human SPT has been shown to be negatively regulated by the yeast integral ER membrane proteins ORM1/2—ORMDL1/2/3 in human—by forming a complex with SPT and with the phosphoinositide phosphatase Sac1, termed SPOTS, and its formation is initiated by a yet unidentified sphingolipid intermediate. This provides a hint on the interconnectivity of the sphingolipid and the phosphoinositide metabolisms104,105 as well as on the regulation of the rate-limiting step of the sphingolipid metabolism.
In yeast and mammals, SPT is an ER membrane-associated complex.106 Human SPT is a heteromer composed of the SPTLC1 subunit and either SPTLC2 or 3.107 SPTLC1 and SPTLC2 share about 21% sequence identity and 45% similarity, whereas SPTLC2 and 3 share 68% identity and 84% similarity.107 SPTLC2 and 3 contain the PLP-binding residues and are therefore considered as the catalytic subunits.107,108 They have different acyl-CoA recognition affinities, increasing the variety of the sphingolipids produced. Importantly, despite its lack of cofactor, SPTLC1 is strictly required for activity.109 A so-called small subunit of the human SPT, named ssSPT, is responsible for maximal activity of the protein.110 The known isoforms, ssSPTa and ssSPTb, both activate a SPTLC dimer (either 1-2 or 1-3), with different preferences of acyl-CoA chain lengths.110
SPT (EC 2.3.1.50) is a member of the α-oxoamine synthase (AOS) subfamily.102,111,112 The enzyme catalyzes the formation of 3-keto-dihydro-SPH from serine and palmitoyl-CoA (Fig. 2), which is the rate-limiting step in sphingolipid synthesis. The bacteria Sphingomonas paucimobilis EY2395T, Sphingomonas wittichii RW1, Sphingobacterium multivorum, and the bacteria-predating bacterium Bdellovibrio stolpii carry SPT homologs that have recently been identified and characterized.113–117 Unlike Sphingomonas paucimobilis and Sphingomonas wittichii SPTs, which are cytosolic enzymes, the latter two SPTs are peripheral inner membrane proteins. Notably, the physiological function of the sphingolipids in bacteria is unknown.
The structure of Sphingomonas paucimobilis EY2395T SPT (SpSPT) was solved in 2007 at 1.3 Å resolution117 [Fig. 6(A)]. The first 20 and the last nine residues of the SpSPT sequence are not visible in the electron density map. Unlike eukaryotic SPTs, the enzyme is a homodimer with one active site per monomer. Each active site is composed of residues from both subunits. The role of the presumed Mg2+ ions sitting near the PLP [brown spheres in Figure 6(A)] remains to be clarified. Like StSPL, SpSPT produces an absorption spectrum characteristic of PLP-dependent enzymes, featuring a 416- to 426-nm and a 332- to 338-nm peak. The spectrum allows one to follow the equilibrium between the ketoenamine and enolimine tautomers of the Schiff base formed by PLP and K265 [Fig. 6(B)]. The ratio of those tautomers is modified upon addition of serine. A transient 505- to 515-nm peak, characteristic for a quinonoid intermediate, appears upon addition of the fatty acyl-CoA substrate.116,118,119 In 2009, a structural and functional study of Sphingobacterium multivorum SPT (SmSPT) was published (PDB code 3A2B).113 The structure, solved at 2.3 Å resolution, is similar to that of SpSPT, with minor differences in the N-terminal region.117 The structure of a third SPT from Sphingomonas wittichii RW1 (SwSPT) was solved in 2010 at 2.1 Å resolution by Campopiano and coworkers.114 SwSPT shares a similar fold with SpSPT and SmSPT, and its active site is nearly identical to that of SpSPT. Interestingly, the expression of the SwSPT gene seems to be coupled with a specific acyl carrier protein.114 This finding might provide new insight into the substrate accommodation into the active site of SPT.
Figure 6.
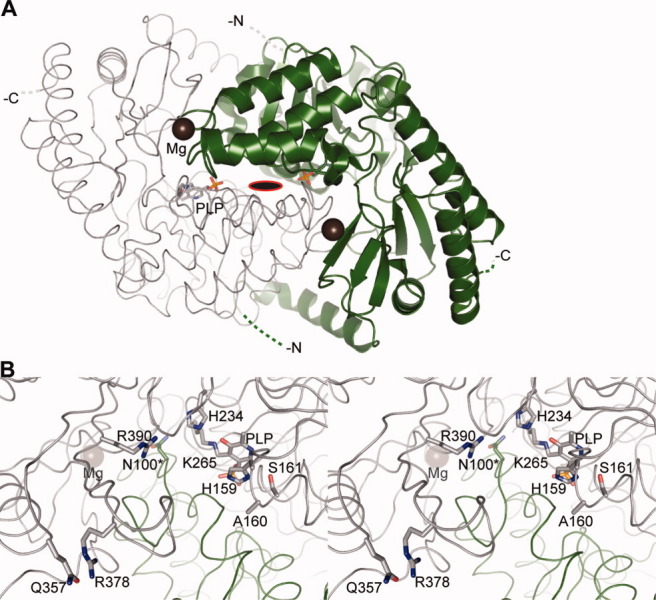
X-ray structure of SpSPT (PDB code 2JG2)117. A: Same as Figure 4(A) for holo-SpSPT. Subunit A is colored pale gray, whereas subunit B is dark green. The first 20 and the last nine residues of each subunit are represented by dashed lines. The bound Mg ions appear as brown spheres. B: Stereoview of the active site. The internal aldimine of subunit A as well as residues important for activity are shown as in Figure 4(B).
Several compounds are known as potent inhibitors of SPT, for instance, sphingofungin B, myriocin, lipoxamycin, and viridofungin A,102 and are thought to mimic a reaction intermediate. In addition, l-cycloserine has recently been found to inhibit SpSPT.111
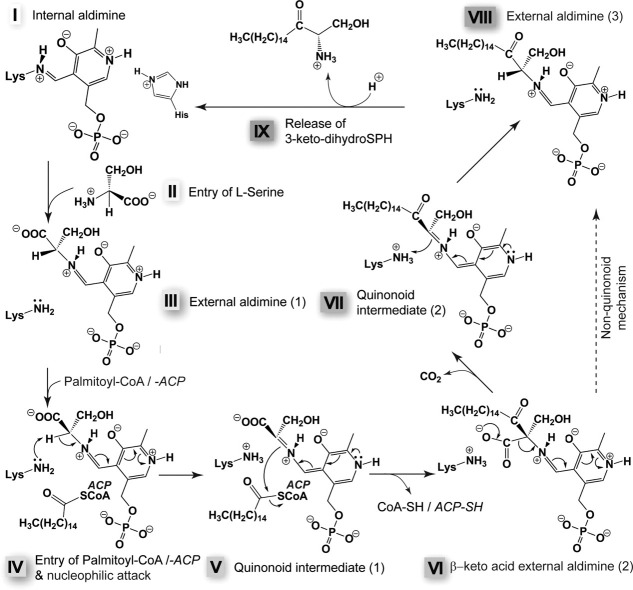
The reaction mechanism of SPT has been extensively studied, using, along with biochemical knowledge on the AOS family,112 the characteristic UV–visible spectra of the protein at various reaction stages of the WT and mutant forms.106,118–121 Briefly, the internal aldimine between PLP and K265 (in SpSPT) (Fig. 7, step I) undergoes transaldimination with l-serine to yield PLP-l-serine external aldimine (step III). Binding of the second substrate, a fatty acid conjugated to either coenzyme A (CoA) or to acyl-carrier protein (ACP), as recently suggested in Ref.114 (step IV), triggers a conformational change and ends up in α-deprotonation of serine. A first quinonoid intermediate is formed (step V). The resulting carbanion attacks the conjugated fatty acid to generate a condensation product (step VI), which is then decarboxylated to yield a second quinonoid intermediate (step VII). Protonation at Cα gives the external aldimine with the product, 3-keto-dihydro-SPH (step VIII), which is released from the active site (step IX)). The conserved residue H159, part of the HAS motif conserved throughout the AOS family,117 plays crucial roles in catalysis [Fig. 6(B)].118 It stacks against the pyridine ring of PLP, recognizes and anchors the carboxylate group of serine, regulates the conformation of the serine-PLP external aldimine to prevent unwanted reactions, and acts as acid catalyst in the condensation reaction.111 Another histidine, H234, plays a role in the reaction intermediate stabilization, as its position might suggest.111 In 2009, the structure of the external aldimine PLP-l-serine of SpSPT was reported,119 highlighting the crucial role of N100 from the neighboring subunit in binding the carboxylate of the external aldimine.119 Residue R378, the conformation of which is stabilized by a hydrogen bond with Q357,119 is involved in the stabilization of the quinonoid intermediate although it is not conserved in all SPTs.119 This residue, found in a mobile region of the protein, has two known conformations and is shown to play a crucial role in the binding of serine as well as of the inhibitor cycloserine.111 Interestingly, R390 of SpSPT located nearby is conserved in all SPTs and is strictly required for activity in Sphingomomas wittichii (R370).121 This recent finding shed a new light into the role of arginine in the reaction mechanism. As in Ref.96, the ɛ-amino group of K265 may play a critical role by abstracting the Cα proton in step IV and by stabilizing the carbanionic intermediate (step VI).
Figure 7.
Proposed SPT reaction mechanism.119,121 In the resting state, the PLP is linked to the enzyme via K265 (in SpSPT). Please note that the imidazole ring of H159 pi-stacks the pyridine ring of the PLP [see Fig. 6(B)]. For clarity, this residue is not shown in the subsequent steps (see comment below). The incoming amino acid (in this case serine) enters the active site (step II) and forms an external aldimine with PLP (step III). The external aldimine undergoes a nucleophilic attack, most likely by the internal aldimine-forming K265, after the second substrate, the acyl-CoA or the acyl-ACP (in this case palmitoyl), has entered the active site (step IV). The active site is expected to undergo conformational changes upon binding of the conjugated acyl. After the nucleophilic attack, a first quinonoid intermediate is formed (step V), the acyl carrier (either reduced CoA or ACP) leaves the active site, and a second external aldimine (with a β-keto acid) is formed (step VI). Decarboxylation of this intermediate produces the second quinonoid intermediate (step VII), which gets reprotonated to form the third external aldimine of the reaction cycle, between PLP and the reaction product (step VIII). The product 3-keto-dihydroSPH is then released (step IX). The proposed mechanism is based on the work of the Campopiano group.111,119,121 Importantly, H159 stabilizes by H-bond either the carboxyl group of the PLP-bound Ser (step III) or the carbonyl of the conjugated acyl (steps IV and V) and of the intermediates depicted in steps VI to VIII. For further explanations on the mechanism and especially the role of H159, please refer to the review by Ikushiro and Hayashi.106
SPT can metabolize other amino acids than serine.122 In the case where alanine is used in the condensation reaction, 1-deoxy-sphingoid bases (DSBs) are synthesized. DSBs lack the hydroxyl group at position 1 [Fig. 1(A)] and cannot be degraded by SPL. Accumulation of DSBs is toxic and was observed in cells expressing mutants of SPTLC1 and 2 linked to hereditary sensory and autonomic neuropathy type 1 (HSAN I) (Table I).108,123–125 These mutations most likely affect the residues involved in interaction with partner proteins (as hypothesized in Ref.108) or with the ssSPT subunit [Fig. 8(A)] or residues involved in PLP binding and substrate accommodation into the active site [Fig. 8(B), residues N100 and G268]. They result in a complete to partial loss of enzymatic activity. It turned out that some of these mutations perturbed the active site to facilitate the formation of the external aldimine with Ala.123,125,126 Interestingly, the accumulation of DSBs, rather than the reduced catalytic activity of SPT, seems to be the major factor responsible for HSAN I.122,126 In addition, elevated levels of DSBs were recently observed in patients suffering from diabetes mellitus.122 As alanine and serine derive from the glycolytic chain, this may suggest a link between the two pathologies. The corresponding HSAN I-causing mutations on the yeast SPT also result in reduced activity. However, the mutated subunit (2 or 3) is still able to form a heterodimer with subunit 1, indicating that the reaction mechanism is conserved from yeast to human.109 Moreover, SPTLC1 was shown to physically and functionally interact with ABCA1 and to inhibit the transport of cholesterol,127 providing evidence for coupling of the cholesterol and the sphingolipid metabolisms and a hint on a possible onset of atherosclerosis.
Table I.
Mutations of SPT Causing Hereditary Sensory and Autonomic Neuropathy Type 1 (HSAN I)
| Subunit | Human SPT | Yeast SPT | SpSPT corresponding residuea | Location in SpSPTa | References |
|---|---|---|---|---|---|
| SPTLC1 | C133W/Y | C180W | N100 | Active site | 110,111 |
| SPTLC1 | V144D | V191D | D121 | Surface exposed | 106,107,111 |
| SPTLC2 | V359M | N.R.b | V246 | Surface exposed | 112 |
| SPTLC2 | G382V | N.R.b | G268 | Active site | 112 |
| SPTLC2 | I504F | N.R.b | G385 | Surface exposed | 112 |
See Figure 8.
Not reported.
Figure 8.
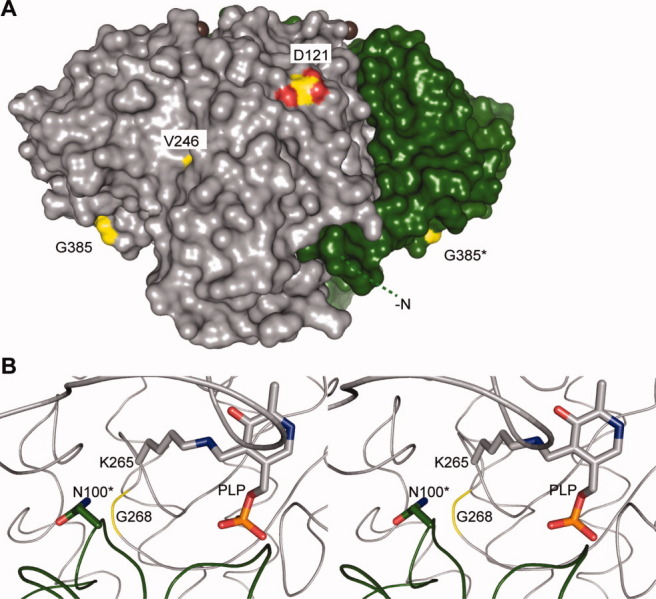
Hereditary sensory and autonomic neuropathy type 1 (HSAN I)–linked mutations on SpSPT. A: Surface representation of holo-SpSPT (PDB code 2JG2117). Subunit A is colored pale gray, whereas subunit B is dark green. The first 20 residues of subunit B are represented by a green dashed line. The residues of SpSPT corresponding to mutations linked to HSAN I are colored yellow and atom color. The bound Mg ions appear as brown spheres. Compared to Figure 6(A), the dimer was rotated 90° anticlockwise along an axis parallel to the plane of the sheet. This figure is linked to Table I. B: Stereoview of the active site. The residue G268 is colored yellow.
StSPL and SpSPT: Comparison
In addition to covalently linking PLP to the enzyme, K311 in StSPL and K265 in SpSPT most likely also play a catalytic role,82,106 mainly in the nucleophilic attack and protonation events. Likewise, H129 in StSPL and N100 in SpSPT, both located on an active site loop belonging to the neighboring subunit, are also key players in activity, demonstrating that the active form of both enzymes is a dimer or a multiple thereof.
The biological function of the encoding sphingolipid-related enzymes in bacteria and the origin of the corresponding genes are intriguing. The thermophile Symbiobacterium thermophilum has a significantly high number of genes acquired by horizontal transfer from different organisms.128 Notably, horizontal transfer of genes predicted to be involved in the biosynthesis of sphingolipids, including a putative SPT, has been reported between an algae and its DNA virus.129 The precise function of the proteins in the virus is unknown although they are thought to activate host cell death, thereby promoting the virus dissemination, to temporarily inhibit cell death to extend the length of infection, and to be involved in the virus recognition mechanism by the host cell by modulating the lipid profiles of the host/virus membranes.129 Indeed, in pathogenic bacteria and viruses, sphingolipid-related enzymes can mimic antigens during infection and escape from the immune system,5 modify the properties of the membrane to allow the entry of the pathogen,130 act as a virulence factor to promote infection, and interfere with the sphingolipid metabolism to promote infection,130 as found for Listeria ivanovii SM'ase131 and Pseudomonas aeroginosa CER'ase.132 In nonpathogenic bacteria such as Symbiobacterium thermophilum, it seems more likely that the main biological roles of StSPL are to degrade phospho-SPHL found in the environment and to contribute to the symbiotic relationship, similar to that between the symbiotic Bacteroides genus and a mammalian host in the intestine.5
The precise location of S1P and its mode of accommodation into the active site of SPL are still unknown, as are the exit modes of the hydrophilic product PE and of the hydrophobic product Hex. Likewise, the accommodation mode of the fatty acid into SPT and the exit path of the produced sphingolipid are still unknown. A conformational change is known to occur upon binding of the conjugated fatty acid in active site of SPT. In general, large conformational changes in SPL and SPT are expected to take place to accommodate and release molecules with different physicochemical properties. The enzymes lacking a transmembrane domain may interact with the membrane and accommodate their substrates or release their product via a transiently exposed hydrophobic stretch. Such a mechanism is known for Bacillus cereus and Listeria ivanovii SM'ases, two prokaryotic soluble homologs of membrane-bound eukaryotic SM'ases. There, a β-hairpin and a loop penetrate the lipid bilayer to accommodate the substrate in the active site.131,133 This phenomenon is not restricted to sphingolipid-related proteins. Other examples of peripheral membrane proteins include the SPL structural homolog E. coli GadB134,135 bound to the membrane at low pH, the hepatitis C virus protein N3S-4A,136 carnitine palmitoyltransferase 2,137 the peripheral membrane protein E. coli pyruvate oxidase,138 and A. aeolicus sulfide:quinone oxidoreductase.139 The N-terminal part of SPL and SPT varies across species, especially between prokaryotic and eukaryotic proteins,106,115 and may carry out species-specific functions. In addition, it would not be surprising if the flexible N-termini were found to initiate conformational changes before substrate accommodation and product release.
On the other hand, it should not be excluded that SPL and SPT may interact with a specific hydrophobic molecule carrier protein. Indeed, it was proposed that the expression of SPT is coupled with an acyl carrier protein in Sphingomonas witchii RW1.114
Mammalian Counterparts: Similarities and Differences
Although the respective active sites of SPL and SPT are most likely conserved from bacteria to human, a distinctive feature of eukaryotic SPLs and SPTs is the presence of transmembrane domains. In the absence of biophysical or structural data, the topology, oligomeric state, and relative orientation of the active site to the lipid bilayer remain elusive. Mammalian SPL is thought to be a homooligomer (Fig. 9), whereas SPT may be a heterooligomer. Hornemann et al. proposed that SPT forms a high-molecular-weight complex, most likely an octamer.140 Following this view, a possible octameric architecture is proposed in Figure 10. Given the well-described quaternary structure of PLP-dependent enzymes and the participation of both subunits to one active site, the oligomers are most likely formed by association of dimers. The third subunit of eukaryotic SPT (Fig. 10(B), red) adds another level of complexity to the challenge of defining the topology of the protein.
Figure 9.
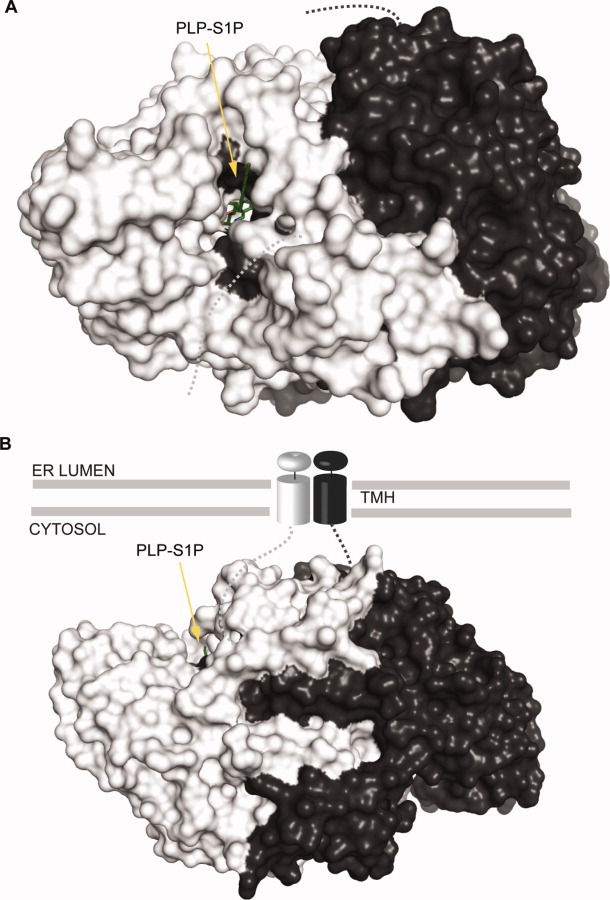
Putative membrane-bound SPL. Proposed topology of the membrane-bound mammalian and yeast SPL homodimer.82 StSPL containing a modeled PLP-S1P external aldimine is colored as in Figure 4 and displayed as surface. A: Compared to Figure 4(A), the protein was rotated 90° anticlockwise along an axis parallel to the plane of the sheet such that the putative active site entrance faces the reader. B: The protein is oriented as in Figure 4(A). TMH stands for transmembrane helix.
Figure 10.
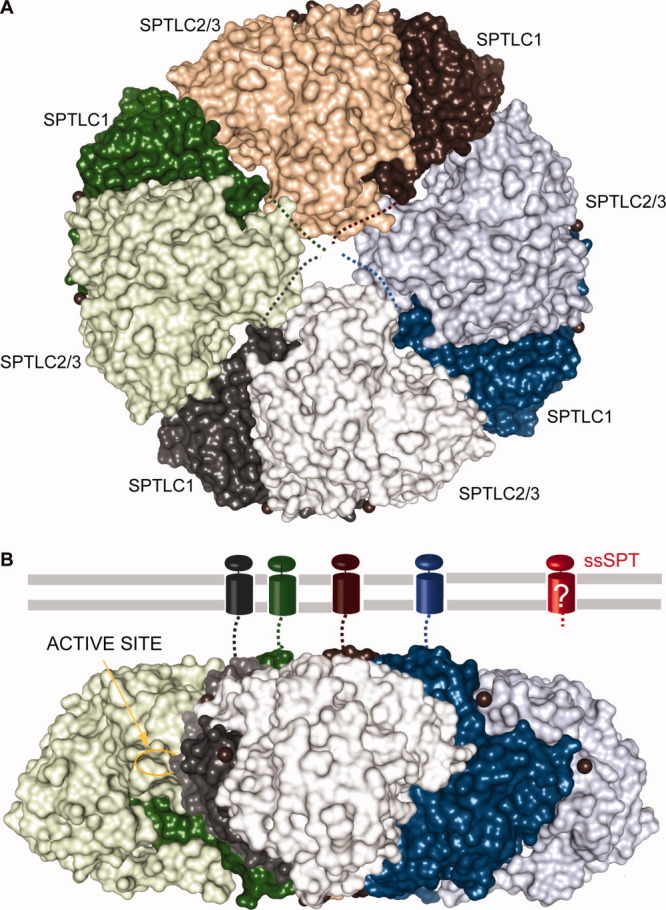
Putative membrane-bound SPT. Proposed topology of the membrane-bound mammalian and yeast SPT heteromer based on the assumption of Hornemann et al.140 SpSPT dimers in surface representation are colored green, brown, blue, and gray where SPTLC1 is colored with the darker shades and SPTLC2/3 with the darker shades. A: The orientation of the brown dimer corresponds to that of Figure 8(A), whereas the blue, gray, and green dimers are rotated by, respectively, 90, 180, and 270° in the plane of the sheet and assembled to form a disk. B: Lateral view of the octamer. Compared to the orientation in (A), the disk was rotated 90° anticlockwise along an axis parallel to the plane of the sheet. The four transmembrane helices of the SPTLC1 subunit are represented as in Figure 9. The active site of the green subunit SPTLC2/3 is indicated. Note that the topology of ssSPT, colored red, is unknown.
Another characteristic of eukaryotic SPT is the presence of PLP in only one subunit. This suggests that there is a single active site per dimer. Interestingly, it has been reported that the cofactor is labile in one subunit of StSPL and of a truncated form of Dpl1p.82 This may reflect regulation mechanisms mostly specific to eukaryotic enzymes. Further experimental data are needed to understand the differences in the two active sites and its consequences for the function of the enzymes.
Conclusion
The two PLP-dependent enzymes SPT and SPL play crucial roles in sphingolipid metabolism. Recent structural and functional analyses of prokaryotic SPL and SPT allowed describing the basic structure–function relationship of these enzymes. Nevertheless, important questions remain unanswered, such as the precise topology, the oligomeric state, the regulation mechanism of both enzymes, as well as the mode of substrate accommodation and product release in the prokaryotic as well as in the eukaryotic representatives. Finding answers to these questions is important for basic and applied research because entry and exit of the sphingolipid metabolism are targets of choice for drug design aimed at regulating the effects of given sphingolipid metabolites.39
Acknowledgments
The authors are grateful to the reviewer for the very valuable comments.
References
- 1.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 2.Bartke N, Hannun YA. Bioactive sphingolipids: metabolism and function. J Lipid Res. 2009;50:91–96. doi: 10.1194/jlr.R800080-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fyrst H, Saba JD. An update on sphingosine-1-phosphate and other sphingolipid mediators. Nat Chem Biol. 2010;6:489–497. doi: 10.1038/nchembio.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pata MO, Hannun YA, Ng CK. Plant sphingolipids: decoding the enigma of the Sphinx. New Phytol. 2009;185:611–630. doi: 10.1111/j.1469-8137.2009.03123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An D, Na C, Bielawski J, Hannun YA, Kasper DL. Membrane sphingolipids as essential molecular signals for Bacteroides survival in the intestine. Proc Natl Acad Sci USA. 2010;108:4666–4671. doi: 10.1073/pnas.1001501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Veldhoven PP, Mannaerts GP. Sphingosine-phosphate lyase. Adv Lipid Res. 1993;26:69–98. [PubMed] [Google Scholar]
- 7.Aye IL, Singh AT, Keelan JA. Transport of lipids by ABC proteins: interactions and implications for cellular toxicity, viability and function. Chem Biol Interact. 2009;180:327–339. doi: 10.1016/j.cbi.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher JI, Haber M, Henderson MJ, Norris MD. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer. 2010;10:523. doi: 10.1038/nrc2789. [DOI] [PubMed] [Google Scholar]
- 9.Linn SC, Kim HS, Keane EM, Andras LM, Wang E, Merrill AHJ. Regulation of de novo sphingolipid biosynthesis and the toxic consequences of its disruption. Biochem Soc Trans. 2001;29:831–835. doi: 10.1042/0300-5127:0290831. [DOI] [PubMed] [Google Scholar]
- 10.Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–25005. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- 11.Causeret C, Geeraert L, Van der Hoeven G, Mannaerts GP, Van Veldhoven PP. Further characterization of rat dihydroceramide desaturase: tissue distribution, subcellular localization, and substrate specificity. Lipids. 2000;35:1117–1125. doi: 10.1007/s11745-000-0627-6. [DOI] [PubMed] [Google Scholar]
- 12.Hakomori S. Traveling for the glycosphingolipid path. Glycoconj J. 2000;17:627–647. doi: 10.1023/a:1011086929064. [DOI] [PubMed] [Google Scholar]
- 13.Marchesini N, Hannun YA. Acid and neutral sphingomyelinases: roles and mechanisms of regulation. Biochem Cell Biol. 2004;82:27–44. doi: 10.1139/o03-091. [DOI] [PubMed] [Google Scholar]
- 14.Raas-Rothschild A, Pankova-Kholmyansky I, Kacher Y, Futerman AH. Glycosphingolipidoses: beyond the enzymatic defect. Glycoconj J. 2004;21:295–304. doi: 10.1023/B:GLYC.0000046272.38480.ef. [DOI] [PubMed] [Google Scholar]
- 15.Arana L, Gangoiti P, Ouro A, Trueba M, Gómez-Muñoz A. Ceramide and ceramide 1-phosphate in health and disease. Lipids Health Dis. 2010;9:1–12. doi: 10.1186/1476-511X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wijesinghe DS, Massiello A, Subramanian P, Szulc Z, Bielawska A, Chalfant CE. Substrate specificity of human ceramide kinase. J Lipid Res. 2005;46:2706–2716. doi: 10.1194/jlr.M500313-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Bornancin F. Ceramide kinase: the first decade. Cell Signal. 2010;23:999–1008. doi: 10.1016/j.cellsig.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Chalfant CE, Spiegel S. Sphingosine 1-phosphate and ceramide 1-phosphate: expanding roles in cell signaling. J Cell Sci. 2005;118:4605–4612. doi: 10.1242/jcs.02637. [DOI] [PubMed] [Google Scholar]
- 19.Milhas D, Clarke CJ, Hannun YA. Sphingomyelin metabolism at the plasma membrane: implications for bioactive sphingolipids. FEBS Lett. 2009;584:1887–1894. doi: 10.1016/j.febslet.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tafesse FG, Ternes P, Holthuis JC. The multigenic sphingomyelin synthase family. J Biol Chem. 2006;281:29421–29425. doi: 10.1074/jbc.R600021200. [DOI] [PubMed] [Google Scholar]
- 21.Hanada K, Kumagai K, Tomishige N, Kawano M. CERT and intracellular trafficking of ceramide. Biochim Biophys Acta. 2007;1771:644–653. doi: 10.1016/j.bbalip.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Kudo N, Kumagai K, Tomishige N, Yamaji T, Wakatsuki S, Nishijima M, Hanada K, Kato R. Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc Natl Acad Sci USA. 2008;105:488–493. doi: 10.1073/pnas.0709191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Structural basis for glycosphingolipid transfer specificity. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malinina L, Malakhova ML, Kanack AT, Lu M, Abagyan R, Brown RE, Patel DJ. The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure. PLoS Biol. 2006;4:e362. doi: 10.1371/journal.pbio.0040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Airenne TT, Kidron H, Nymalm Y, Nylund M, West G, Mattjus P, Salminen TA. Structural evidence for adaptive ligand binding of glycolipid transfer protein. J Mol Biol. 2006;355:224–236. doi: 10.1016/j.jmb.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 26.Xu R, Jin J, Hu W, Sun W, Bielawski J, Szulc Z, Taha T, Obeid LM, Mao C. Golgi alkaline ceramidase regulates cell proliferation and survival by controlling levels of sphingosine and S1P. FASEB J. 2006;20:1813–1825. doi: 10.1096/fj.05-5689com. [DOI] [PubMed] [Google Scholar]
- 27.Hait NC, Oskeritzian CA, Paugh SW, Milstien S, Spiegel S. Sphingosine kinases, sphingosine 1-phosphate, apoptosis and diseases. Biochim Biophys Acta. 2006;1758:2016–2026. doi: 10.1016/j.bbamem.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Johnson KR, Johnson KY, Becker KP, Bielawski J, Mao C, Obeid LM. Role of human sphingosine-1-phosphate phosphatase 1 in the regulation of intra- and extracellular sphingosine-1-phosphate levels and cell viability. J Biol Chem. 2003;278:34541–34547. doi: 10.1074/jbc.M301741200. [DOI] [PubMed] [Google Scholar]
- 29.Bandhuvula P, Saba JD. Sphingosine-1-phosphate lyase in immunity and cancer: silencing the siren. Trends Mol Med. 2007;13:210–217. doi: 10.1016/j.molmed.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Horibata Y, Hirabayashi Y. Identification and characterization of human ethanolaminephosphotransferase1. J Lipid Res. 2007;48:503–508. doi: 10.1194/jlr.C600019-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Kariya Y, Kihara A, Ikeda M, Kikuchi F, Nakamura S, Hashimoto S, Choi CH, Lee YM, Igarashi Y. Products by the sphingosine kinase/sphingosine 1-phosphate (S1P) lyase pathway but not S1P stimulate mitogenesis. Genes Cells. 2005;10:605–615. doi: 10.1111/j.1365-2443.2005.00862.x. [DOI] [PubMed] [Google Scholar]
- 32.Gangoiti P, Camacho L, Arana L, Ouro A, Granado MH, Brizuela L, Casas J, Fabriás G, Abad JL, Delgado A, Gómez-Muñoz A. Control of metabolism and signaling of simple bioactive sphingolipids: implications in disease. Prog Lipid Res. 2010;49:316–334. doi: 10.1016/j.plipres.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Ray PE. Shiga-like toxins and HIV-1 ‘go through’ glycosphingolipids and lipid rafts in renal cells. Kidney Int. 2009;75:1135–1137. doi: 10.1038/ki.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toledo MS, Suzuki E, Handa K, Hakomori S. Effect of ganglioside and tetraspanins in microdomains on interaction of integrins with fibroblast growth factor receptor. J Biol Chem. 2005;280:16227–16234. doi: 10.1074/jbc.M413713200. [DOI] [PubMed] [Google Scholar]
- 35.Ikeda H, Ohkawa R, Watanabe N, Nakamura K, Kume Y, Nakagawa H, Yoshida H, Okubo S, Yokota H, Tomiya T, Inoue Y, Nishikawa T, Ohtomo N, Tanoue Y, Koike K, Yatomi Y. Plasma concentration of bioactive lipid mediator sphingosine 1-phosphate is reduced in patients with chronic hepatitis C. Clin Chim Acta. 2010;411:765–770. doi: 10.1016/j.cca.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 36.Serra M, Saba JD. Sphingosine 1-phosphate lyase, a key regulator of sphingosine 1-phosphate signaling and function. Adv Enzyme Regul. 2009;50:349–362. doi: 10.1016/j.advenzreg.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagdanoff JT, Donoviel MS, Nouraldeen A, Carlsen M, Jessop TC, Tarver J, Aleem S, Dong L, Zhang H, Boteju L, Hazelwood J, Yan J, Bednarz M, Layek S, Owusu IB, Gopinathan S, Moran L, Lai Z, Kramer J, Kimball SD, Yalamanchili P, Heydorn WE, Frazier KS, Brooks B, Brown P, Wilson A, Sonnenburg WK, Main A, Carson KG, Oravecz T, Augeri DJ. Inhibition of sphingosine 1-phosphate lyase for the treatment of rheumatoid arthritis: discovery of (E)-1-(4-((1R,2S,3R)-1,2,3,4-tetrahydroxybutyl)-1H-imidazol-2-yl)ethanone oxime (LX2931) and (1R,2S,3R)-1-(2-(isoxazol-3-yl)-1H-imidazol-4-yl)butane-1,2,3,4-tetraol (LX2932) J Med Chem. 2010;53:8650–8662. doi: 10.1021/jm101183p. [DOI] [PubMed] [Google Scholar]
- 38.Price MM, Oskeritzian CA, Milstien S, Spiegel S. Sphingosine-1-phosphate synthesis and functions in mast cells. Future Lipidol. 2008;3:665–674. doi: 10.2217/17460875.3.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar A, Saba JD. Lyase to live by: sphingosine phosphate lyase as a therapeutic target. Expert Opin Ther Targets. 2009;13:1013–1025. doi: 10.1517/14728220903039722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- 41.Leong WI, Saba JD. S1P metabolism in cancer and other pathological conditions. Biochimie. 2010;92:716–723. doi: 10.1016/j.biochi.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oskouian B, Saba JD. Cancer treatment strategies targeting sphingolipid metabolism. Adv Exp Med Biol. 2010;688:185–205. doi: 10.1007/978-1-4419-6741-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gouaze-Andersson V, Cabot MC. Glycosphingolipids and drug resistance. Biochim Biophys Acta. 2006;1758:2096–2103. doi: 10.1016/j.bbamem.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Smith ER, Merrill AHJ, Obeid LM, Hannun YA. Effects of sphingosine and other sphingolipids on protein kinase C. Methods Enzymol. 2000;312:361–373. doi: 10.1016/s0076-6879(00)12921-0. [DOI] [PubMed] [Google Scholar]
- 45.Venable ME, Lee JY, Smyth MJ, Bielawska A, Obeid LM. Role of ceramide in cellular senescence. J Biol Chem. 1995;270:30701–30708. doi: 10.1074/jbc.270.51.30701. [DOI] [PubMed] [Google Scholar]
- 46.Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed cell death induced by ceramide. Science. 1993;259:1769–1771. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 47.Hinkovska-Galcheva V, Boxer LA, Kindzelskii A, Hiraoka M, Abe A, Goparju S, Spiegel S, Petty HR, Shayman JA. Ceramide 1-phosphate, a mediator of phagocytosis. J Biol Chem. 2005;280:26612–26621. doi: 10.1074/jbc.M501359200. [DOI] [PubMed] [Google Scholar]
- 48.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 49.Strub GM, Maceyka M, Hait NC, Milstien S, Spiegel S. Extracellular and intracellular actions of sphingosine-1-phosphate. Adv Exp Med Biol. 2010;688:141–155. doi: 10.1007/978-1-4419-6741-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson KR, Johnson KY, Crellin HG, Ogretmen B, Boylan AM, Harley RA, Obeid LM. Immunohistochemical distribution of sphingosine kinase 1 in normal and tumor lung tissue. J Histochem Cytochem. 2005;53:1159–1166. doi: 10.1369/jhc.4A6606.2005. [DOI] [PubMed] [Google Scholar]
- 51.García-Pacios M, Collado MI, Busto JV, Sot J, Alonso A, Arrondo JL, Goñi FM. Sphingosine-1-phosphate as an amphipathic metabolite: its properties in aqueous and membrane environments. Biophys J. 2009;97:1398–1407. doi: 10.1016/j.bpj.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivera J, Proia RL, Olivera A. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat Rev Immunol. 2008;8:753–763. doi: 10.1038/nri2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hla T, Brinkmann V. Sphingosine 1-phosphate (S1P): physiology and the effects of S1P receptor modulation. Neurology. 2011;76:S3–S8. doi: 10.1212/WNL.0b013e31820d5ec1. [DOI] [PubMed] [Google Scholar]
- 55.Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- 56.Van Veldhoven PP, Gijsbers S, Mannaerts GP, Vermeesch JR, Brys V. Human sphingosine-1-phosphate lyase: cDNA cloning, functional expression studies and mapping to chromosome 10q22(1) Biochim Biophys Acta. 2000;1487:128–134. doi: 10.1016/s1388-1981(00)00079-2. [DOI] [PubMed] [Google Scholar]
- 57.Mukhopadhyay D, Howell KS, Riezman H, Capitani G. Identifying key residues of sphinganine-1-phosphate lyase for function in vivo and in vitro. J Biol Chem. 2008;283:20159–20169. doi: 10.1074/jbc.M709753200. [DOI] [PubMed] [Google Scholar]
- 58.Ikeda M, Kihara A, Igarashi Y. Sphingosine-1-phosphate lyase SPL is an endoplasmic reticulum-resident, integral membrane protein with the pyridoxal 5′-phosphate binding domain exposed to the cytosol. Biochem Biophys Res Commun. 2004;325:338–343. doi: 10.1016/j.bbrc.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 59.Mao C, Saba JD, Obeid LM. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem J. 1999;342(Part 3):667–675. [PMC free article] [PubMed] [Google Scholar]
- 60.Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol. 1999;181:1134–1140. doi: 10.1128/jb.181.4.1134-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gottlieb D, Heideman W, Saba JD. The DPL1 gene is involved in mediating the response to nutrient deprivation in Saccharomyces cerevisiae. Mol Cell Biol Res Commun. 1999;1:66–71. doi: 10.1006/mcbr.1999.0109. [DOI] [PubMed] [Google Scholar]
- 62.Birchwood CJ, Saba JD, Dickson RC, Cunningham KW. Calcium influx and signaling in yeast stimulated by intracellular sphingosine 1-phosphate accumulation. J Biol Chem. 2001;276:11712–11718. doi: 10.1074/jbc.M010221200. [DOI] [PubMed] [Google Scholar]
- 63.Claas RF, ter Braak M, Hegen B, Hardel V, Angioni C, Schmidt H, Jakobs KH, Van Veldhoven PP, zu Heringdorf DM. Enhanced Ca2+ storage in sphingosine-1-phosphate lyase-deficient fibroblasts. Cell Signal. 2010;22:476–483. doi: 10.1016/j.cellsig.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 64.Grote E, Vlacich G, Pypaert M, Novick PJ. A snc1 endocytosis mutant: phenotypic analysis and suppression by overproduction of dihydrosphingosine phosphate lyase. Mol Biol Cell. 2000;11:4051–4065. doi: 10.1091/mbc.11.12.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwab SR, Pereira JP, Matloubian M, Xu Y, Huang Y, Cyster JG. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science. 2005;309:1735–1739. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 66.Allende ML, Bektas M, Lee BG, Bonifacino E, Kang J, Tuymetova G, Chen W, Saba JD, Proia RL. Sphingosine-1-phosphate lyase deficiency produces a pro-inflammatory response while impairing neutrophil trafficking. J Biol Chem. 2011;286:7348–7358. doi: 10.1074/jbc.M110.171819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bektas M, Allende ML, Lee BG, Chen W, Amar MJ, Remaley AT, Saba JD, Proia RL. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J Biol Chem. 2010;285:10880–10889. doi: 10.1074/jbc.M109.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Colié S, Van Veldhoven PP, Kedjouar B, Bedia C, Albinet V, Sorli SC, Garcia V, Djavaheri-Mergny M, Bauvy C, Codogno P, Levade T, Andrieu-Abadie N. Disruption of sphingosine 1-phosphate lyase confers resistance to chemotherapy and promotes oncogenesis through Bcl-2/Bcl-xL upregulation. Cancer Res. 2009;69:9346–9353. doi: 10.1158/0008-5472.CAN-09-2198. [DOI] [PubMed] [Google Scholar]
- 69.Reiss U, Oskouian B, Zhou J, Gupta V, Sooriyakumaran P, Kelly S, Wang E, Merrill AHJ, Saba JD. Sphingosine-phosphate lyase enhances stress-induced ceramide generation and apoptosis. J Biol Chem. 2004;279:1281–1290. doi: 10.1074/jbc.M309646200. [DOI] [PubMed] [Google Scholar]
- 70.Chen WV, Delrow J, Corrin PD, Frazier JP, Soriano P. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat Genet. 2004;36:304–312. doi: 10.1038/ng1306. [DOI] [PubMed] [Google Scholar]
- 71.Alvarez RH, Kantarjian HM, Cortes JE. Biology of platelet-derived growth factor and its involvement in disease. Mayo Clin Proc. 2006;81:1241–1257. doi: 10.4065/81.9.1241. [DOI] [PubMed] [Google Scholar]
- 72.Ito K, Anada Y, Tani M, Ikeda M, Sano T, Kihara A, Igarashi Y. Lack of sphingosine 1-phosphate-degrading enzymes in erythrocytes. Biochem Biophys Res Commun. 2007;357:212–217. doi: 10.1016/j.bbrc.2007.03.123. [DOI] [PubMed] [Google Scholar]
- 73.Yatomi Y, Igarashi Y, Yang L, Hisano N, Qi R, Asazuma N, Satoh K, Ozaki Y, Kume S. Sphingosine 1-phosphate, a bioactive sphingolipid abundantly stored in platelets, is a normal constituent of human plasma and serum. J Biochem. 1997;121:969–973. doi: 10.1093/oxfordjournals.jbchem.a021681. [DOI] [PubMed] [Google Scholar]
- 74.Fyrst H, Saba JD. Sphingosine-1-phosphate lyase in development and disease: sphingolipid metabolism takes flight. Biochim Biophys Acta. 2008;1781:448–458. doi: 10.1016/j.bbalip.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oskouian B, Sooriyakumaran P, Borowsky AD, Crans A, Dillard-Telm L, Tam YY, Bandhuvula P, Saba JD. Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is down-regulated in colon cancer. Proc Natl Acad Sci USA. 2006;103:17384–17389. doi: 10.1073/pnas.0600050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oskouian B, Saba J. Sphingosine-1-phosphate metabolism and intestinal tumorigenesis: lipid signaling strikes again. Cell Cycle. 2007;6:522–527. doi: 10.4161/cc.6.5.3903. [DOI] [PubMed] [Google Scholar]
- 77.Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema TRJ, Thiele JJ, Grindle SM, Bliss RL, Skubitz AP. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397–414. doi: 10.1016/S0002-9440(10)63306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seo EY, Park GT, Lee KM, Kim JA, Lee JH, Yang JM. Identification of the target genes of atopic dermatitis by real-time PCR. J Invest Dermatol. 2006;126:1187–1189. doi: 10.1038/sj.jid.5700234. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Y, Gorshkova IA, Berdyshev E, He D, Fu P, Ma W, Su Y, Usatyuk PV, Pendyala S, Oskouian B, Saba JD, Garcia JG, Natarajan V. Protection of LPS-induced murine acute lung injury by sphingosine-1-phosphate lyase suppression. Am J Respir Cell Mol Biol. 2011 doi: 10.1165/rcmb.2010-0422OC. Jun 9. [Epub ahead of print] doi: 10.1165/rcmb.2010-0422OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar A, Oskouian B, Fyrst H, Zhang M, Paris F, Saba JD. S1P lyase regulates DNA damage responses through a novel sphingolipid feedback mechanism. Cell Death Dis. 2011;2:e119. doi: 10.1038/cddis.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar A, Byun HS, Bittman R, Saba JD. The sphingolipid degradation product trans-2-hexadecenal induces cytoskeletal reorganization and apoptosis in a JNK-dependent manner. Cell Signal. 2011;23:1144–1152. doi: 10.1016/j.cellsig.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bourquin F, Riezman H, Capitani G, Grütter MG. Structural and functional studies of sphingosine-1-phosphate lyase, a key enzyme of sphigolipid metabolism. Structure. 2010;18:1054–1065. doi: 10.1016/j.str.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 83.Sasaki H, Arai H, Cocco MJ, White SH. pH dependence of sphingosine aggregation. Biophys J. 2009;96:72727–72733. doi: 10.1016/j.bpj.2008.12.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Veldhoven PP. Sphingosine-1-phosphate lyase. Methods Enzymol. 2000;311:244–254. doi: 10.1016/s0076-6879(00)11087-0. [DOI] [PubMed] [Google Scholar]
- 85.Stoffel W, LeKim D, Sticht G. Distribution and properties of dihydrosphingosine-1-phosphate aldolase (sphinganine-1-phosphate alkanal-lyase) Hoppe Seylers Z Physiol Chem. 1969;350:1233–1241. doi: 10.1515/bchm2.1969.350.2.1233. [DOI] [PubMed] [Google Scholar]
- 86.Stoffel W, Grol M. Chemistry and biochemistry of 1-desoxysphinganine 1-phosphonate (dihydrosphingosine-1-phosphonate) Chem Phys Lipids. 1974;13:372–388. doi: 10.1016/0009-3084(74)90011-5. [DOI] [PubMed] [Google Scholar]
- 87.Boumendjel A, Miller SP. Synthesis of sphingosine-1-phosphate and dihydrosphingosine-1-phosphate. J Lipid Res. 1994;35:2305–2311. [PubMed] [Google Scholar]
- 88.Stoffel W, Bauer E, Stahl J. The metabolism of sphingosine bases in Tetrahymena pyriformis. Sphingosine kinase and sphingosine-1-phosphate lyase. Hoppe Seylers Z Physiol Chem. 1974;355:61–74. doi: 10.1515/bchm2.1974.355.1.61. [DOI] [PubMed] [Google Scholar]
- 89.Paugh SW, Payne SG, Barbour SE, Milstien S, Spiegel S. The immunosuppressant FTY720 is phosphorylated by sphingosine kinase type 2. FEBS Lett. 2003;554:189–193. doi: 10.1016/s0014-5793(03)01168-2. [DOI] [PubMed] [Google Scholar]
- 90.Brinkmann V, Billich A, Baumruker T, Heining P, Schmouder R, Francis G, Aradhye S, Burtin P. Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat Rev Drug Discov. 2010;9:883–897. doi: 10.1038/nrd3248. [DOI] [PubMed] [Google Scholar]
- 91.Mullershausen F, Zecri F, Cetin C, Billich A, Guerini D, Seuwen K. Persistent signaling induced by FTY720-phosphate is mediated by internalized S1P1 receptors. Nat Chem Biol. 2009;5:428–434. doi: 10.1038/nchembio.173. [DOI] [PubMed] [Google Scholar]
- 92.Bagdanoff JT, Donoviel MS, Nouraldeen A, Tarver J, Fu Q, Carlsen M, Jessop TC, Zhang H, Hazelwood J, Nguyen H, Baugh SD, Gardyan M, Terranova KM, Barbosa J, Yan J, Bednarz M, Layek S, Courtney LF, Taylor J, Digeorge-Foushee AM, Gopinathan S, Bruce D, Smith T, Moran L, O'Neill E, Kramer J, Lai Z, Kimball SD, Liu Q, Sun W, Yu S, Swaffield J, Wilson A, Main A, Carson KG, Oravecz T, Augeri DJ. Inhibition of sphingosine-1-phosphate lyase for the treatment of autoimmune disorders. J Med Chem. 2009;52:3941–3953. doi: 10.1021/jm900278w. [DOI] [PubMed] [Google Scholar]
- 93.Van Veldhoven PP, Mannaerts GP. Subcellular localization and membrane topology of sphingosine-1-phosphate lyase in rat liver. J Biol Chem. 1991;266:12502–12507. [PubMed] [Google Scholar]
- 94.DeLano WL. The PyMOL molecular graphics system. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 95.Gut H, Dominici P, Pilati S, Astegno A, Petoukhov MV, Svergun DI, Grütter MG, Capitani G. A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase. J Mol Biol. 2009;392:334–351. doi: 10.1016/j.jmb.2009.06.080. [DOI] [PubMed] [Google Scholar]
- 96.Koutmos M, Kabil O, Smith JL, Banerjee R. Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine {beta}-synthase. Proc Natl Acad Sci USA. 2010;107:20958–20963. doi: 10.1073/pnas.1011448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Toney MD. Reaction specificity in pyridoxal phosphate enzymes. Arch Biochem Biophys. 2005;433:279–287. doi: 10.1016/j.abb.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 98.Metzler CM, Viswanath R, Metzler DE. Equilibria and absorption spectra of tryptophanase. J Biol Chem. 1991;266:9374–9381. [PubMed] [Google Scholar]
- 99.Ikushiro H, Hayashi H, Kawata Y, Kagamiyama H. Analysis of the pH- and ligand-induced spectral transitions of tryptophanase: activation of the coenzyme at the early steps of the catalytic cycle. Biochemistry. 1998;37:3043–3052. doi: 10.1021/bi971995+. [DOI] [PubMed] [Google Scholar]
- 100.Webster SP, Alexeev D, Campopiano DJ, Watt RM, Alexeeva M, Sawyer L, Baxter RL. Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies. Biochemistry. 2000;39:516–528. doi: 10.1021/bi991620j. [DOI] [PubMed] [Google Scholar]
- 101.Zhang J, Ferreira GC. Transient state kinetic investigation of 5-aminolevulinate synthase reaction mechanism. J Biol Chem. 2006;277:44660–44669. doi: 10.1074/jbc.M203584200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta. 2003;1632:16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 103.Huwiler A, Kolter T, Pfeilschifter J, Sandhoff K. Physiology and pathophysiology of sphingolipid metabolism and signaling. Biochim Biophys Acta. 2000;1485:63–99. doi: 10.1016/s1388-1981(00)00042-1. [DOI] [PubMed] [Google Scholar]
- 104.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci USA. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ikushiro H, Hayashi H. Mechanistic enzymology of serine palmitoyltransferase. Biochim Biophys Acta. doi: 10.1016/j.bbapap.2011.02.005. (in press). [Still in press doi: 10.1016/j.bbapap.2011.02.005] [DOI] [PubMed] [Google Scholar]
- 107.Hornemann T, Richard S, Rütti MF, Wei Y, von Eckardstein A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J Biol Chem. 2006;281:37275–37281. doi: 10.1074/jbc.M608066200. [DOI] [PubMed] [Google Scholar]
- 108.Hornemann T, Penno A, Richard S, Nicholson G, van Dijk FS, Rotthier A, Timmerman V, von Eckardstein A. A systematic comparison of all mutations in hereditary sensory neuropathy type I (HSAN I) reveals that the G387A mutation is not disease associated. Neurogenetics. 2009;10:135–143. doi: 10.1007/s10048-008-0168-7. [DOI] [PubMed] [Google Scholar]
- 109.Gable K, Han G, Monaghan E, Bacikova D, Natarajan M, Williams R, Dunn TM. Mutations in the yeast LCB1 and LCB2 genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J Biol Chem. 2002;277:10194–10200. doi: 10.1074/jbc.M107873200. [DOI] [PubMed] [Google Scholar]
- 110.Han G, Gupta SD, Gable K, Niranjanakumari S, Moitra P, Eichler F, Brown RHJ, Harmon JM, Dunn TM. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc Natl Acad Sci USA. 2009;106:8186–8191. doi: 10.1073/pnas.0811269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lowther J, Yard BA, Johnson KA, Carter LG, Bhat VT, Raman MC, Clarke DJ, Ramakers B, McMahon SA, Naismith JH, Campopiano DJ. Inhibition of the PLP-dependent enzyme serine palmitoyltransferase by cycloserine: evidence for a novel decarboxylative mechanism of inactivation. Mol Biosyst. 2010;6:1682–1693. doi: 10.1039/c003743e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mann S, Ploux O. Pyridoxal-5′-phosphate-dependent enzymes involved in biotin biosynthesis: structure, reaction mechanism and inhibition. Biochim Biophys Acta. doi: 10.1016/j.bbapap.2010.12.004. (in press). Still in press. doi: 10.1016/j.bbapap.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 113.Ikushiro H, Islam MM, Okamoto A, Hoseki J, Murakawa T, Fujii S, Miyahara I, Hayashi H. Structural insights into the enzymatic mechanism of serine palmitoyltransferase from Sphingobacterium multivorum. J Biochem. 2009;146:549–562. doi: 10.1093/jb/mvp100. [DOI] [PubMed] [Google Scholar]
- 114.Raman MC, Johnson KA, Clarke DJ, Naismith JH, Campopiano DJ. The serine palmitoyltransferase from Sphingomonas wittichii RW1: an interesting link to an unusual acyl carrier protein. Biopolymers. 2010;93:811–822. doi: 10.1002/bip.21482. [DOI] [PubMed] [Google Scholar]
- 115.Ikushiro H, Hayashi H, Kagamiyama H. Bacterial serine palmitoyltransferase: a water-soluble homodimeric prototype of the eukaryotic enzyme. Biochim Biophys Acta. 2003;1647:116–120. doi: 10.1016/s1570-9639(03)00074-8. [DOI] [PubMed] [Google Scholar]
- 116.Ikushiro H, Islam MM, Tojo H, Hayashi H. Molecular characterization of membrane-associated soluble serine palmitoyltransferases from Sphingobacterium multivorum and Bdellovibrio stolpii. J Bacteriol. 2007;189:5749–5761. doi: 10.1128/JB.00194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yard BA, Carter LG, Johnson KA, Overton IM, Dorward M, Liu H, McMahon SA, Oke M, Puech D, Barton GJ, Naismith JH, Campopiano DJ. The structure of serine palmitoyltransferase; gateway to sphingolipid biosynthesis. J Mol Biol. 2007;370:870–886. doi: 10.1016/j.jmb.2007.04.086. [DOI] [PubMed] [Google Scholar]
- 118.Shiraiwa Y, Ikushiro H, Hayashi H. Multifunctional role of his159in the catalytic reaction of serine palmitoyltransferase. J Biol Chem. 2009;284:15487–15495. doi: 10.1074/jbc.M808916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Raman MC, Johnson KA, Yard BA, Lowther J, Carter LG, Naismith JH, Campopiano DJ. The external aldimine form of serine palmitoyltransferase: structural, kinetic, and spectroscopic analysis of the wild-type enzyme and HSAN1 mutant mimics. J Biol Chem. 2009;284:17328–17339. doi: 10.1074/jbc.M109.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ikushiro H, Fujii S, Shiraiwa Y, Hayashi H. Acceleration of the substrate Calpha deprotonation by an analogue of the second substrate palmitoyl-CoA in serine palmitoyltransferase. J Biol Chem. 2008;283:7542–7553. doi: 10.1074/jbc.M706874200. [DOI] [PubMed] [Google Scholar]
- 121.Lowther J, Charmier G, Raman MC, Ikushiro H, Hayashi H, Campopiano DJ. Role of a conserved arginine residue during catalysis in serine palmitoyltransferase. FEBS Lett. 2011;585:1729–34. doi: 10.1016/j.febslet.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 122.Bertea M, Rütti MF, Othman A, Marti-Jaun J, Hersberger M, von Eckardstein A, Hornemann T. Deoxysphingoid bases as plasma markers in diabetes mellitus. Lipids Health Dis. 2010;9:1–7. doi: 10.1186/1476-511X-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gable K, Gupta SD, Han G, Niranjanakumari S, Harmon JM, Dunn TM. A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J Biol Chem. 2010;285:22846–22852. doi: 10.1074/jbc.M110.122259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eichler FS, Hornemann T, McCampbell A, Kuljis D, Penno A, Vardeh D, Tamrazian E, Garofalo K, Lee HJ, Kini L, Selig M, Frosch M, Gable K, von Eckardstein A, Woolf CJ, Guan G, Harmon JM, Dunn TM, Brown RHJ. Overexpression of the wild-type SPT1 subunit lowers desoxysphingolipid levels and rescues the phenotype of HSAN1. J Neurosci. 2009;29:14646–14651. doi: 10.1523/JNEUROSCI.2536-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rotthier A, Auer-Grumbach M, Janssens K, Baets J, Penno A, Almeida-Souza L, Van Hoof K, Jacobs A, De Vriendt E, Schlotter-Weigel B, Löscher W, Vondráček P, Seeman P, De Jonghe P, Van Dijck P, Jordanova A, Hornemann T, Timmerman V. Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. Am J Hum Genet. 2010;87:513–522. doi: 10.1016/j.ajhg.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Penno A, Reilly MM, Houlden H, Laurá M, Rentsch K, Niederkofler V, Stoeckli ET, Nicholson G, Eichler F, Brown RHJ, von Eckardstein A, Hornemann T. Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J Biol Chem. 2010;285:11178–11187. doi: 10.1074/jbc.M109.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tamehiro N, Zhou S, Okuhira K, Benita Y, Brown CE, Zhuang DZ, Latz E, Hornemann T, von Eckardstein A, Xavier RJ, Freeman MW, Fitzgerald ML. SPTLC1 binds ABCA1 to negatively regulate trafficking and cholesterol efflux activity of the transporter. Biochemistry. 2008;47:6138–6147. doi: 10.1021/bi800182t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nishida H, Yun CS. Phylogenetic and guanine-cytosine content analysis of Symbiobacterium thermophilum genes. Int J Evol Biol. 2010;2011:1–5. doi: 10.4061/2011/634505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Monier A, Pagarete A, de Vargas C, Allen MJ, Read B, Claverie JM, Ogata H. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009;19:1441–1449. doi: 10.1101/gr.091686.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heung LJ, Luberto C, Del Poeta M. Role of sphingolipids in microbial pathogenesis. Infect Immun. 2006;74:28–39. doi: 10.1128/IAI.74.1.28-39.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Openshaw AE, Race PR, Monzó HJ, Vázquez-Boland JA, Banfield MJ. Crystal structure of SmcL, a bacterial neutral sphingomyelinase C from Listeria. J Biol Chem. 2005;280:35011–35017. doi: 10.1074/jbc.M506800200. [DOI] [PubMed] [Google Scholar]
- 132.Inoue T, Okino N, Kakuta Y, Hijikata A, Okano H, Goda HM, Tani M, Sueyoshi N, Kambayashi K, Matsumura H, Kai Y, Ito M. Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase. J Biol Chem. 2009;284:9566–9577. doi: 10.1074/jbc.M808232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ago H, Oda M, Takahashi M, Tsuge H, Ochi S, Katunuma N, Miyano M, Sakurai J. Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus. J Biol Chem. 2006;281:16157–16167. doi: 10.1074/jbc.M601089200. [DOI] [PubMed] [Google Scholar]
- 134.Gut H, Pennacchietti E, John RA, Bossa F, Capitani G, De Biase D, Grutter MG. Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. EMBO J. 2006;25:2643–2651. doi: 10.1038/sj.emboj.7601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Capitani G, De Biase D, Aurizi C, Gut H, Bossa F, Grutter MG. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 2003;22:4027–4037. doi: 10.1093/emboj/cdg403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Brass V, Berke JM, Montserret R, Blum HE, Penin F, Moradpour D. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc Natl Acad Sci USA. 2008;105:14545–14550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rufer AC, Lomize AL, Benz J, Chomienne O, Thoma R, Hennig M. Carnitine palmitoyltransferase 2: analysis of membrane association and complex structure with a substrate analog. FEBS Lett. 2007;581:3247–3252. doi: 10.1016/j.febslet.2007.05.080. [DOI] [PubMed] [Google Scholar]
- 138.Neumann P, Weidner A, Pech A, Stubbs MT, Tittmann K. Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli. Proc Natl Acad Sci USA. 2009;105:17390–17395. doi: 10.1073/pnas.0805027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marcia M, Ermler U, Peng G, Michel H. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci USA. 2009;106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hornemann T, Wei Y, von Eckardstein A. Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochem J. 2007;405:157–164. doi: 10.1042/BJ20070025. [DOI] [PMC free article] [PubMed] [Google Scholar]