Abstract
A brief personal perspective is provided for green fluorescent protein (GFP), covering the period 1994–2011. The topics discussed are primarily those in which my research group has made a contribution and include structure and function of the GFP polypeptide, the mechanism of fluorescence emission, excited state protein transfer, the design of ratiometric fluorescent protein biosensors and an overview of the fluorescent proteins derived from coral reef animals. Structure-function relationships in photoswitchable fluorescent proteins and nonfluorescent chromoproteins are also briefly covered.
Keywords: protein structure, crystallography, fluorescence spectroscopy, excited state proton transfer, chromophore, photochemistry
Introduction
Although the green fluorescent protein (GFP) has a scientific history predating 1960, I like to credit the February 11, 1994 issue of the journal Science (Fig. 1) as the beginning of the GFP revolution. The cover image,1 showing green-glowing sensory neurons in C. elegans, was instantly recognized as representing a major breakthrough for cell biology. That the image seems ho-hum today reflects on how dramatically GFP has changed our expectations of biological imaging.
Figure 1.

Cover of the February 11, 1994 issue of the journal Science (from Chalfie M, et al., Green fluorescent protein as a marker for gene expression, Science, 1994. 263. p. 802, reproduced with permission from AAAS.
Shortly thereafter, Milestone Zero for the revolution in GFP engineering was the more-or-less simultaneous publication of the crystal structure of wild type GFP from George Phillips' group2 and the popular mutant GFP/S65T†,‡ from mine.3 As a rather trivial example of engineering, the crystal structure of GFP/S65T permitted us to immediately create the popular yellow fluorescent protein YFP by making a single amino-acid substitution to wild-type GFP, T203Y.3 Given the unique structure and function of GFP, it is fitting that the icon of the GFP revolution, a ribbon diagram representing the “β-can” fold (Fig. 2) has also become the most easily recognized protein structure known.
Figure 2.

Schematic diagram showing the backbone fold of GFP, in this case roGFP2, an engineered version sensitive to the ambient thiol/disulfide equilibrium. The backbone is colored blue beginning at the N-terminal to red at the C-terminal of the chain. The chromophore and the introduced cysteines, which form a disulfide bond in the oxidized state (right hand side), are shown in ball and stick representation.
Fluorescent protein research has been the most rewarding and at times, the most exciting, of any topic in my entire research career. In this perspective, I provide a personal overview of the field. I regret that space limitations permit me to highlight only a few favorite topics, primarily those in which my research group has provided a contribution. Additional details on any topic mentioned can be found in the many cited reviews of GFP and its applications, beginning with Roger Tsien's comprehensive and still useful contribution.4 Several new reviews appeared following the 2008 award of the Nobel Prize in Chemistry to Martin Chalfie, Osamu Shimomura, and Roger Tsien for their pioneering work with GFP, and will be cited as appropriate.
Overall structure of GFP
Our crystals of GFP/S65T presented a number of technical problems, and the structure was slow to be resolved, but the effort was very rewarding. This is due in part to the unique molecular structure (Fig. 2), which seems remarkably well suited to its multiple tasks. The polypeptide backbone folds into a previously unobserved motif: a β-barrel of eleven strands, surrounding a central helix. Short distorted helical segments cap the barrel ends and help isolate the internal chromophore from solvent. An all-atom representation appears as a nearly perfect cylinder about 25 Å in diameter and 40 Å tall.
Despite its monolithic appearance, GFP can be expressed as two separate segments that nevertheless combine within cells to yield a functional fluorescent protein.5,6 This discovery has led to useful applications, for example in the two-hybrid scheme, the individual GFP segments are fused to two other proteins and the resulting genes expressed in parallel. Appearance of visible fluorescence suggests that the two fusion partners tightly interact in vivo. The amino acid sequence of GFP can also be circularly permuted, resulting in new N- and C-termini, without loss of function.7,8 Inspection of the fold suggests that it can be divided into sequential segments, for example, an N-terminal portion (roughly 1–80) consisting of three beta strands plus the central helix, and a larger C-terminal portion (roughly 81–238) that consists of eight beta strands arranged in a typical “Greek key” motif. However, to my knowledge, no one seems to have investigated whether isolated GFP segments have well-defined structures in solution.
Protein folding is essential for both chromophore formation and for fluorescence; see reviews.4,9,10 As is now well known, three amino acids within the central helix (serine 65, tyrosine 66, and glycine 67) rearrange covalently during the folding reaction, and in the presence of molecular oxygen become oxidized to form the chromophore. Denatured GFP is nonfluorescent, but fluorescence is regained upon renaturation.11 The isolated chromophore is also not fluorescent, either as the naked molecule12 or within the isolated hexapeptide.13 From these observations, it is clear that in addition to promoting the chemistry of fluorophore formation, a primary function of the polypeptide fold is to tightly restrain the chromophore. In this way, excitation energy is dissipated as light, rather than through nonradiative processes. Nature can have it both ways, though and in a later section, I'll discuss what makes “chromoproteins” nonfluorescent (although their purpose in Nature remains a mystery).
Chromophore structure and environment
All naturally occurring fluorescent protein chromophores are based on two rings linked by a double bond and in all known examples, are derived from the dipeptide tyrosine-glycine. The minimum energy configuration has the two rings coplanar, but both the cis (energetically favored in solution) and trans isomers of the central double bond are fluorescent. Within GFP, the chromophore is cis and approximately planar as expected [Fig. 3(a)],2,3 but in some other fluorescent proteins, the trans isomer predominates, e.g., in the red emitting eqFP611.14
Figure 3.
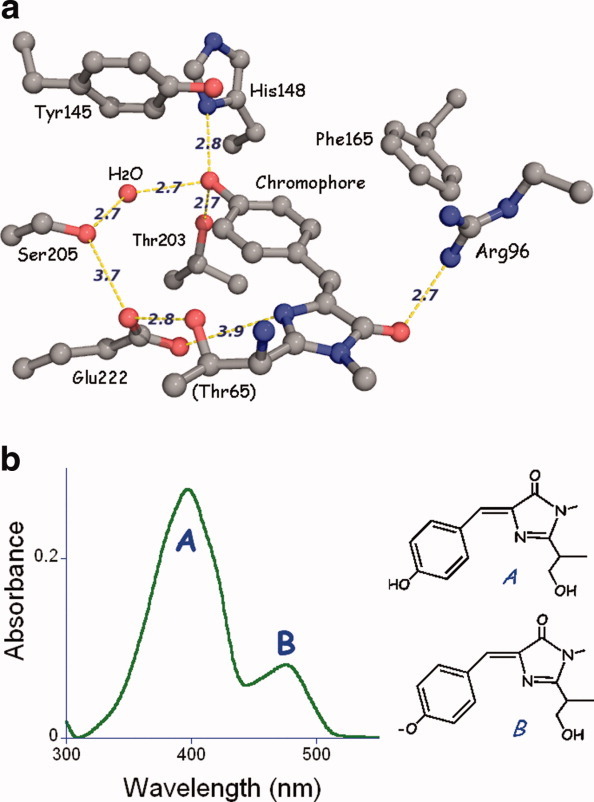
(a) Ball and stick representation of the GFP/S65T chromophore and its immediate environment. Nitrogen atoms are blue and oxygen is red. Hydrogen bonds and other potentially important interactions are shown as dashed lines and have the indicated lengths in Å. (b) Absorbance and excitation spectrum of wild-type GFP, together with a representation of the chromophore forms giving rise to the two peaks.
Light excitation promotes electrons to nonbonding orbitals, so in the minimum energy configuration of the first excited state of the chromophore, the two rings are approximately perpendicular.15 If such rearrangements are permitted by the environment, energy is usually dissipated by thermal rather than radiative processes. Consequently, the chromophore configuration and overall planarity are rigidly enforced by the surrounding protein matrix using a combination of hydrogen bonds and hydrophobic side chains, which in turn determine the shape of the enclosing cavity. Experimental evidence for the importance of a close cavity fit was provided quite early on by mutants at the Tyr 66 position (e.g., Phe, His, and Trp), all of which have substantially reduced brightness.16 However, brightness in these mutants can be improved by further mutagenesis of protein side chains lining the chromophore cavity. This result is consistent with the theoretical predictions that a loose cavity permits energy dissipation via internal conversion.17,18
In addition to the groups that determine cavity shape and polarity, two potentially charged side chains are absolutely conserved in all known fluorescent proteins. In GFP, these are Glu 222 and Arg 96 [Fig. 3(a)]. They are located on opposite edges of the chromophore, such that the polar side chains are close to the site where the chemistry takes place. Both groups are catalytic, that is, they have been determined to be very important but not absolutely essential for chromophore formation. In Figure 3a, some of the important interactions within the active site are shown, with key hydrogen bonds drawn as dashed lines. The chemistry of chromophore formation is now partially understood and quite complex. Rather than attempt to summarize what is known, I refer interested readers to two recent and comprehensive reviews19,20).
The GFP chromophore can exist as anionic or neutral, which explains the spectroscopic observation of two coincident absorbance and excitation bands [Fig. 3(b)]. The neutral protonated or A form absorbs in the ultraviolet (UV) at about 395 nm, whereas the anionic or B form absorbs at about 475 nm. Curiously, in wild-type GFP both bands are present at equilibrium in an A:B intensity ratio of about 6:1. This ratio is surprisingly insensitive to the environmental pH and salt concentration21 implying that in wild type, some as yet poorly understood mechanism protects the chromophore from changes in the solvent environment.
Point mutations within the chromophore cavity (and some without) readily affect the charge ratio, with two important examples being GFP/S65T16 (later with improved brightness as EGFP), showing only B band excitation, and the photoswitchable protein PA-GFP (GFP/T203H22,23), showing only A band excitation. A clear explanation for this change in behavior is provided by the crystal structures. The S65T mutation rearranges the hydrogen bond pattern of Glu222, suppressing its negative charge, in turn allowing the chromophore to become ionized.3 In PA-GFP, the exchange of His for Thr203 reduces hydrogen bonding opportunities to the chromophore hydroxyl group so that it preferentially remains neutral, with Glu222 carrying a negative charge. In PA-GFP, photoswitching is accomplished by brief exposure to UV light, during which Glu222 decarboxylates to become a neutral species, thus allowing the chromophore to become ionized.23
Mechanism of fluorescence emission and excited state proton transfer
Emission from the anionic chromophore is rather simply described as a transition from the first sin-glet excited state to ground (that is, a transition between the lowest unoccupied and highest occupied molecular orbital, see for example a recent study24). In most fluorescent proteins, protonation of the chromophore quenches fluorescence. The reason for this behavior is not completely understood, however in wild-type GFP and a few of its relatives, the chromophore is fluorescent in either protonation state (that is, upon either A or B band excitation). Comparison of the GFP chromophore with model compounds suggests that the excited state of the protonated chromophore (A*) should emit in the blue but instead, green emission is observed.21 The Boxer group25 used ultrafast fluorescence upconversion spectroscopy to show that upon excitation by 397-nm light, blue emission at 460 nm is observed as expected, but the blue emission decays with biphasic time constants of about 3 and 12 ps. The blue decay is matched by the rise in 510-nm green emission. Both rates are slowed by a factor of 5-6 upon substitution of exchangeable hydrogens with deuterium, suggesting that the A species is converted into an excited anionic intermediate I* by proton transfer. The I* state subsequently emits a green photon.
The proposed mechanism for A band emission, although incomplete, is generally accepted and is regarded as the first biologically relevant example of excited state proton transfer (ESPT) to be discovered. In solution, the chromophore pKa is about 8 but as found with tyrosine, the pKa drops to less than 1.0 upon excitation.26 The GFP chromophore is thus a strong photoacid.27 Detailed steps in the proton transfer process have been elucidated by several groups, demonstrating the existence of a photocycle with at least two excited state intermediates and multiple ground states (see for example28–32).
Structural basis for ESPT
The chief advantage of GFP/S65T is that it is excited primarily by blue light, making it more useful as a label.4 However at low pH, fluorescence is quenched and ESPT is not observed. Shortly after determining the structure of GFP/S65T, we initiated a collaboration with Titia Sixma's group to compare results with their structure of wild-type GFP, and proposed an atomic mechanism for the process. In this proposal, wild-type GFP has a built-in proton wire from the chromophore OH, through a water molecule and the hydroxyl of serine 205 to the acceptor Glu 222 (Fig. 4).33 Stoner-Ma et al.30 subsequently used ultrafast vibrational spectroscopy to confirm that Glu222 is the proton acceptor. Remarkably, the observed rate of protonation at Glu222 (t ∼ 15 ps) approximately matches the decay of blue fluorescence, suggesting that the proton transfer process is concerted (i.e., all three protons in Fig. 4 move simultaneously). Alternatively, proton transfer may be stepwise but the intermediate steps may take place too quickly for these techniques to resolve. Further studies of the proton transfer pathways in GFP are currently being conducted by us34 and by others,35 using a combination of mutagenesis, high resolution crystallography, and ultrafast spectroscopic techniques. GFP has turned out to be an outstanding system for study of the geometric factors controlling the rate of proton transfer, one of the most elementary reactions in biological chemistry.
Figure 4.

Proposed reaction scheme for ESPT in wild-type GFP. An ∼ 390 nm wavelength photon excites the system (upper right) and causes a series of proton transfer events to form the excited state anion (I*), left center, which subsequently emits a green photon (∼ 510 nm). A low frequency event is transformation of the ground state I to a metastable ground state anion, B (lower right).
Practical applications of ESPT: Genetically encoded biosensors
Cell biologists tend to think of fluorescent proteins as visible tags and overlook their potentially far more powerful applications as biosensors. With appropriate modifications, fluorescent proteins can be labels as well as report on conditions in the local cellular environment. Palmer et al.36 recently reviewed the many different types of fluorescent protein biosensors that have been realized and used.
A particularly attractive feature of GFP and some other fluorescent proteins is that one can take advantage of the charge state equilibrium of the chromophore to create ratiometric biosensors. Although in any indicator, the intensity of fluorescence emission obviously depends on the concentration of the indicator species, intensity of illumination, etc., for a two-state process with opposing changes in different parts of the spectrum, a simple ratio of two measurements removes or strongly reduces these effects. The theory and application of ratiometry has been discussed extensively for a class of fluorescent calcium indicators by Grynkiewicz et al.37
An obvious application would be a pH indicator, expressed within subcellular compartments. Miesenbock et al.38 first described pHfluorins, whose application utilizes a dual excitation scheme. For example, 510-nm emission is monitored as a function of excitation at 390 nm [I510390] and 475 nm [I510475]). The ratio I510390/I510475 will depend primarily on the local pH and be largely independent of pHfluorin concentration, path length and changes in illumination intensity.
In our lab, Hanson et al.39 later described dual emission pH sensors called deGFPs, which switch from blue emission (∼460 nm) at low pH to green (∼510 nm) at high pH. They are thus ratiometric by emission. Crystal structures obtained at different pH values revealed large structural rearrangements involving ESPT pathways, which in turn permitted a rational explanation for the phenomenon.39–41 Fluorescent protein pH indicators have so far seen few practical applications, possibly because most researchers are not fully aware of the possibilities they offer.
Two other important classes of genetically encoded biosensors include calcium indicators (for a recent review see Ref.42) and redox-sensitive GFPs, which report the local thiol-disulfide equilibrium.43,44 The most useful of these are ratiometric, that is, they respond to changes in the environment by opposing changes in spectroscopic signals.
Ratiometric redox-sensitive GFP indicators were first constructed by Hanson et al.44 In these indicators, surface-exposed cysteines were substituted on adjacent strands of the β-barrel, close to the chromophore, where disulfides could form (S147C/Q204C, see Fig. 2) or alternatively (N149C/S202C). Disulfide formation further constrains the β-barrel and reduces solvent accessibility of the chromophore, which results in readily detectible changes in the excitation spectrum. Reduction-oxidation sensitive GFPs (roGFPs) rapidly equilibrate with the local thiol-disulfide equilibrium and indicate the ambient potential by changes in the excitation spectrum as shown in Figure 5. Titrations against a redox buffer demonstrate that the transition is accurately modeled as two-state. Depending upon other modifications,45 redox indicators have been produced with midpoint potentials ranging from −220 to −290 mV at pH 7.0. Structural studies of roGFPs44.45 demonstrated the reversible formation of the disulfide and also revealed changes in the chromophore environment that permitted us to explain the observed changes in the protonation state of the chromophore. The more reducing indicators, such as roGFP1 and roGFP2, are well matched to the extremely reducing environments of the mitochondria and cytoplasm of mammalian cells (midpoint potentials at pH 7 in the range −270 to −290 mV), while the family roGFP1-iX is more suitable for applications in the endoplasmic reticulum (midpoint potentials ∼ −220 to −240 mV).
Figure 5.

(a) Detail of disulfide linkage and portion of the chromophore in the oxidized form of the redox biosensor roGFP2, showing a fragment of the final 2Fo-Fc electron density map contoured at 1 σ. (b) Graphical representation showing the effect of redox titration on the excitation spectrum of roGFP1.
These indicators have become quite popular and to date we have distributed vectors to over 350 laboratories. For recent reviews of genetically encoded redox-sensitive indicators, potential applications and some of the discoveries that have been made using them, see Refs. 46 and 47.
The fluorescent protein rainbow
Shortly after the gene for GFP was cloned, blue,16,48 cyan,16 and yellow3 variants were readily created by mutagenesis, but useful red mutants of GFP have not been discovered. The eventual discovery of an entire rainbow of fluorescent proteins in coral reef animals49 did not originate (as many people suppose) near a warm, sunny beach, but rather at a party in a Moscow apartment featuring a salt water aquarium. It seems that alcohol consumption encouraged speculation regarding the nature of the fluorescent pigmentation displayed by some sea anemones obtained at a nearby pet shop [M. V. Matz, personal communication] and the rest is history.
Members of this large protein family have easily recognizable sequence similarity and at last count, seven evolutionary classes are required to account for the diversity (see reviews20,50). These include five color classes (cyan, green, yellow, and two red classes), a class of colorless proteins found in the extracellular protein matrix of animals, including humans, that lack a chromophore entirely51 and a class of nonfluorescent chromoproteins52 that provide deep purple or blue coloration. More classes probably remain to be discovered, but so far, the evidence suggests that the GFP protein fold arose only once during the course of evolution.
I examined the molecular structure of the extracellular matrix protein nidogen,51 which is colorless and found in high concentrations just beneath our skin, and suspect that color could be restored by just 3-4 mutations. It is amusing to imagine that our evolutionary ancestors were once fluorescent! On the other hand in reef animals, as many as four different fluorescent proteins have been isolated from a single specimen,50 so it seems safe to conclude that fluorescent proteins play an important biological role. Some evidence suggests that this function is photoprotective53 and/or may be important for modulating light levels for photosynthetic symbionts.
Unfortunately, most of the reef-derived fluorescent proteins suffer from significant disadvantages with respect to applications. Most are obligate tetramers,54 even upon dilution to single molecules. This can be disastrous for labeling, because anything that is successfully labeled is often aggregated into higher order complexes. Furthermore, the red fluorescent proteins are slow to mature and frequently contain a large percentage of dead-end green fluorophores54 which complicates dual-label experiments with GFP. Finally, the red emitting proteins are often toxic to certain organisms, possibly because of reactive oxygen or free radical species that are produced during illumination55 (for an earlier review regarding applications in plants, see Ref.56). A dramatic but useful example of this side effect is provided by KillerRed, which can kill a host cell upon brief illumination.57
Fluorescent protein chromophores
Shortly after the discovery of DsRed, it was established58,59 that the red-shift in emission is due to oxidation of the protein backbone, which physically extends the chromophore conjugation by one double bond compared with GFP (Fig. 6). A hallmark of this modification is the appearance of multiple bands (usually three) on a denaturing SDS_PAGE gel, as heat and pH extremes readily lead to hydrolysis of the modified peptide bond adjacent to the chromophore.58 We described another interesting modification in yellow ZFP538 that added a third ring to the system.60 This was the first demonstration that an additional cyclization reaction could take place, but was slow to appear in print. The first manuscript was firmly rejected by Nature Structural Biology as most of the seven reviewers could not believe the result (now confirmed at high resolution61). Incidentally, our first submission on the structure of GFP/S65T was rejected by the editor of Science as one of the referees complained that the manuscript did not explain why light emission was important for the jellyfish.
Figure 6.

Schematic diagram showing the currently known types of naturally occurring fluorescent protein chromophores, colored roughly in accord with fluorescence emission. Note the two central single bonds characteristic of the most recently discovered type of chromophore, found in blue emitting mTagBFP.
GFP is capable of amazing chemistry! Many different chromogenic reactions have been observed to take place within GFP, and recent structural studies have expanded the repertoire of naturally-occurring chromophores to seven (summarized in Fig. 6). It seems likely that other chromophore modifications remain to be discovered.
In most reef-derived fluorescent proteins, an additional positively charged group, either Arg or Lys, is found adjacent to the chromophore and is required for development of red fluorescence. Lys 70 occupies this position in DsRed and replacement with anything other than Arg results in either colorless protein or formation of a green chromophore.54 In early work, it was assumed that a GFP-like chromophore was an intermediate along the pathway to development of the red chromophore, but more recent studies have required us to reject this hypothesis.20,62,63 However, none of the recently proposed mechanisms explains the requirement for Lys/Arg 70 in DsRed, so more work will be required.
Engineered monomeric fluorescent proteins
In reef-derived fluorescent proteins, the formation of tight oligomers is somehow coupled to chromophore development. Campbell et al.64 were able to produce a monomeric red fluorescent protein, but the effort required was heroic. Several rounds of directed evolution were required to first create from DsRed a weakly fluorescent dimer, the brightness of which had to be improved by cycles of semirandom mutagenesis and colony selection. The dimer was then broken into monomers by mutation at the subunit interface and again, brightness had to be improved by mutational cycles. In the final result, 33 individual mutations were incorporated into the parent gene to make mRFP1. It is unfortunately rather dim, with a quantum yield of about 0.25 (about 1/3 as efficient as DsRed or GFP).
Shaner et al. extended this effort by innovative use of directed evolution, somatic hypermutation and flow cytometry to select cells carrying color and brightness variants.65,66 This work resulted in “mFruits,” an invaluable collection of bright, photostable monomeric proteins ranging from yellow to deep red in emission  . We determined atomic resolution structures for several members of the family,67 which allowed us to rationalize the observed differences in emission and excitation wavelengths. One particularly interesting result of that study is the popular and brightly fluorescent mOrange, which was discovered to have another type of tricyclic chromophore (Fig. 6). In mOrange, the threonine Oγ of the chromogenic tripeptide cyclizes with a presumed acylimine in the backbone to generate an additional 2-hydroxy dihydrooxazole ring. A related three-ring chromophore, which develops from a Cys65-Tyr66-Gly67 tripeptide to form an additional 2-hydroxy-3-thiazoline ring, was later discovered in a variant of a naturally occurring orange fluorescent protein (mKO68).
. We determined atomic resolution structures for several members of the family,67 which allowed us to rationalize the observed differences in emission and excitation wavelengths. One particularly interesting result of that study is the popular and brightly fluorescent mOrange, which was discovered to have another type of tricyclic chromophore (Fig. 6). In mOrange, the threonine Oγ of the chromogenic tripeptide cyclizes with a presumed acylimine in the backbone to generate an additional 2-hydroxy dihydrooxazole ring. A related three-ring chromophore, which develops from a Cys65-Tyr66-Gly67 tripeptide to form an additional 2-hydroxy-3-thiazoline ring, was later discovered in a variant of a naturally occurring orange fluorescent protein (mKO68).
The research groups led by Campbell, Wiedenmann, and Miyawaki have utilized directed evolution approaches to produce bright, monomeric FPs with other valuable properties. For example, mTFP1 (teal,  )69 is optimized for use in FRET systems with a yellow or orange FP, while tandem dimers of EosFP offer high brightness and the capability of green to red photoconversion,70 albeit with larger molecular mass. Finally, mKeima71 offers an extremely large Stokes shift for dual color labeling, that is, it is excited in the blue at about 440 nm, but is red fluorescent (620 nm). We recently demonstrated that mKeima utilizes ESPT to achieve this large Stokes shift, but the proton transfer pathway is completely different from that found in GFP.72
)69 is optimized for use in FRET systems with a yellow or orange FP, while tandem dimers of EosFP offer high brightness and the capability of green to red photoconversion,70 albeit with larger molecular mass. Finally, mKeima71 offers an extremely large Stokes shift for dual color labeling, that is, it is excited in the blue at about 440 nm, but is red fluorescent (620 nm). We recently demonstrated that mKeima utilizes ESPT to achieve this large Stokes shift, but the proton transfer pathway is completely different from that found in GFP.72
How is one to choose from all of these different fluorescent proteins? Fortunately, a convenient and a thoughtful guide has been provided by Shaner et al.73 to assist in the selection of the appropriate fluorescent protein for a given application.
Nonfluorescent and reversibly photoswitchable proteins
Pocilloporin was the first example of a nonfluorescent GFP homolog (chromoprotein) to be described,74 although at the time of publication the authors had not yet recognized the relationship of pocilloporin to GFP. These proteins strongly absorb green or yellow light and thus appear purple or blue. They are potent pigments and often appear as decorations on coral reef denizens, such as on the tentacles of Anemonia sulcata. The latter observation led to the isolation of several other members of the group, for example asFP595.52
The crystal structures of pocilloporin, asFP595 and several other nonfluorescent chromoproteins have been determined.75–77 From these studies, it is clear that the lack of fluorescence is due to a highly twisted and bent chromophore, which in turn is enforced by the protein cavity. Such deformations are well known to reduce fluorescence quantum yield (QY, see e.g., Ref.78) but the effectiveness of this strategy is remarkable. An analog of the asFP595 chromophore has been synthesized79 and for the naked chromophore in solution, the QY is 10 times higher than the QY of the naturally occurring protein. This observation, combined with the abundance and diversity of nonfluorescent pigments argues for a strong evolutionary pressure in favor of efficient coloration, but against visible fluorescence in that context. I wonder why this should be so.
Mutants of asFP595 (also called the “kindling fluorescent protein” or KFP) are reversibly photoswitchable, in that exposure to intense green light can temporarily induce a weak orange fluorescent state that decays with time constants of seconds to minutes depending on the mutation.80,81 The process is reversible, as it is possible to switch the dark state back to the light state, usually by illumination with blue or ultraviolet light. The cycle may be repeated hundreds of times, suggesting that photoswitchable fluorescent proteins can be used for information storage at the single molecule level.
Studies of reversibly photoswitchable GFPs such as Dronpa and mTFP0.7 have revealed the basic mechanism behind photoswitching. In these proteins the “resting” state is brightly fluorescent, but under illumination they rapidly photobleach (become nonfluorescent). Upon exposure to blue or ultraviolet light, fluorescence is instantly restored. The structures of photoswitchable green fluorescent Dronpa82–84 and cyan fluorescent mTFP0.785 have been determined for both the light and dark states with strikingly similar results. In the fluorescent state, the chromophore is in both cases cis and planar, while in the dark state the chromophore is trans and highly twisted. Furthermore, in the dark state, the tyrosine moiety of the chromophore relocates to a rather hydrophobic environment and becomes protonated. In the protonated state, absorption of a ∼380 nm photon is expected to ionize the tyrosyl hydroxyl group and cause it to be expelled from the hydrophobic cavity, as is experimentally observed. The metastability of the photobleached state is explained by large structural changes within the protein cavity caused by the light-induced isomerization, which are in turn reversed by slow, thermally activated processes with energy barriers as large as 75 kJ/mole.85
Erasable labels have been proposed as tools to study motion of molecules within cells,70,86–88 while photoswitchable fluorescent proteins have been shown to be useful for superresolution light microscopy.88,89 These new techniques are extremely promising, but it is fair to say that no major discoveries in cell biology have yet been made using photoswitchable proteins. Perhaps it is too early to judge. On the other hand, one notes that GFP and its variants were discovered to blink and photoswitch as early as 1997.90 At that time, the effect was considered to be problematic, rather than potentially useful, so we are very definitely making progress.
Footnotes
The notation S65T indicates that serine 65 in wild-type was replaced by threonine in the mutant.
We chose to work on GFP/S65T for two reasons. First, GFP/S65T is primarily excited in the blue region of the visible spectrum, rather than in the UV as with wild-type and is therefore more useful for imaging applications. Second, several groups already had crystals of wild-type GFP and we felt that a comparison of wild-type versus mutant structures would immediately be useful.
References
- 1.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802, 805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Yang F, Moss LG, Phillips GNJ. The molecular structure of green fluorescent protein. Nature Biotech. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 3.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 4.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J Am Chem Soc. 2000;122:5658–5659. [Google Scholar]
- 6.Kent KP, Childs W, Boxer SG. Deconstructing green fluorescent protein. J Am Chem Soc. 2008;130:9664–9665. doi: 10.1021/ja803782x. [DOI] [PubMed] [Google Scholar]
- 7.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topell S, Hennecke J, Glockshuber R. Circularly permuted variants of the green fluorescent protein. FEBS Lett. 1999;457:283–289. doi: 10.1016/s0014-5793(99)01044-3. [DOI] [PubMed] [Google Scholar]
- 9.Zimmer M. Green fluorescent protein (GFP): Applications, structure and related photophysical behavior. Chem Rev. 2002;102:759–781. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 10.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Bokman SH, Ward WW. Renaturation of Aequorea green-fluorescent protein. Biochem Biophys Res Commun. 1981;101:1372–1380. doi: 10.1016/0006-291x(81)91599-0. [DOI] [PubMed] [Google Scholar]
- 12.McCapra F, Razavi Z, Neary AP. The fluorescence of the chromophore of the green fluorescent protein of Aequorea and Renilla. J Chem Soc Chem Commun. 1988:12:790–791. [Google Scholar]
- 13.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 14.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. The 2.0A crystal structure of eqFP611, a far-red fluorescent protein from the sea anemone Entacmaea quadricolor. J Biol Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 15.Weber W, Helms V, McCammon JA, Langhoff PW. Shedding light on the dark and weakly fluorescent states of green fluorescent proteins. Proc Natl Acad Sci U S A. 1999;96:6177–6182. doi: 10.1073/pnas.96.11.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heim R, Prasher DC, Tsien RY. Wavelength mutations and postranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kummer AD, Kompa C, Lossau H, Pollinger-Dammer F, Michel-Beyerle ME, Silva CM, Bylina EJ, Coleman WJ, Yang MM, Youvan DC. Dramatic reduction in fluorescence quantum yield in mutants of green fluorescent protein due to fast internal conversion. Chem Phys. 1998;237:183–193. [Google Scholar]
- 18.Megley CM, Dickson LA, Maddalo SL, Chandler GJ, Zimmer M. Photophysics and dihedral freedom of the chromophore in yellow, blue, and green fluorescent protein. J Phys Chem B. 2009;113:302–308. doi: 10.1021/jp806285s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wachter RM. Chromogenic cross-link formation in green fluorescent protein. Acc Chem Res. 2007;40:120–127. doi: 10.1021/ar040086r. [DOI] [PubMed] [Google Scholar]
- 20.Wachter RM, Watkins JL, Kim H. Mechanistic diversity of red fluorescence acquisition by GFP-like proteins. Biochemistry. 2010;49:7417–7427. doi: 10.1021/bi100901h. [DOI] [PubMed] [Google Scholar]
- 21.Ward WW, Prentice HJ, Roth AF, Cody CW, Reeves SC. Spectral perturbations of the Aequorea green-fluorescent protein. Photochem Photobiol. 1982;35:803–808. [Google Scholar]
- 22.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 23.Henderson JN, Gepshtein R, Heenan JR, Kallio K, Huppert D, Remington SJ. Structure and mechanism of the photoactivatable green fluorescent protein. J Am Chem Soc. 2009;131:4176–4177. doi: 10.1021/ja808851n. [DOI] [PubMed] [Google Scholar]
- 24.Hasegawa J, Ise T, Fujimoto KJ, Kikuchi A, Fukumura E, Miyawaki A, Shiro Y. Excited states of fluorescent proteins, mKO and DsRed: Chromophore-protein electrostatic interaction behind the color variations. J Phys Chem B. 2010;114:2971–2979. doi: 10.1021/jp9099573. [DOI] [PubMed] [Google Scholar]
- 25.Chattoraj M, King BA, Bublitz GU, Boxer SG. Ultra-fast excited state dynamics in green fluorescent proteMultiple states and proton transfer. Proc Natl Acad Sci USA. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bent DV, Hayon E. Excited state chemistry of aromatic amino acids and related peptides. I. Tyrosine. J Am Chem Soc. 1975;97:2599–2606. doi: 10.1021/ja00843a002. [DOI] [PubMed] [Google Scholar]
- 27.Tolbert LM, Solntsev KM. Excited-state proton transfer: from constrained systems to “super” photoacids to superfast proton transfer. Acc Chem Res. 2002;35:19–27. doi: 10.1021/ar990109f. [DOI] [PubMed] [Google Scholar]
- 28.Kennis JT, Larsen DS, van Stokkum IH, Vengris M, van Thor JJ, van Grondelle R. Uncovering the hidden ground state of green fluorescent protein. Proc Natl Acad Sci U S A. 2004;101:17988–17993. doi: 10.1073/pnas.0404262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Thor JJ, Ronayne KL, Towrie M, Sage JT. Balance between ultrafast parallel reactions in the green fluorescent protein has a structural origin. Biophys J. 2008;95:1902–1912. doi: 10.1529/biophysj.108.129957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoner-Ma D, Jaye AA, Matousek P, Towrie M, Meech SR, Tonge PJ. Observation of excited-state proton transfer in green fluorescent protein using ultrafast vibrational spectroscopy. J Am Chem Soc. 2005;127:2864–2865. doi: 10.1021/ja042466d. [DOI] [PubMed] [Google Scholar]
- 31.Lossau H, Kummer A, Heinecke R, Poellinger-Dammer F, Kompa C, Bieser G, Jonsson T, Silva CM, Yang MM, Youvan DC, Michel-Beyerle ME. Time-resolved spectroscopy of wild-type and mutant Green Fluorescent Proteins reveals excited state deprotonation consistent with fluorophore-protein interactions. Chem Phys. 1996;213:1–16. [Google Scholar]
- 32.Agmon N. Proton pathways in green fluorescence protein. Biophys J. 2005;88:2452–2461. doi: 10.1529/biophysj.104.055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormo M, Remington SJ. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc Natl Acad Sci USA. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shu X, Leiderman P, Gepshtein R, Smith NR, Kallio K, Huppert D, Remington SJ. An alternative excited-state proton transfer pathway in green fluorescent protein variant S205V. Protein Sci. 2007;16:2703–2710. doi: 10.1110/ps.073112007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoner-Ma D, Jaye AA, Ronayne KL, Nappa J, Meech SR, Tonge PJ. An alternate proton acceptor for excited-state proton transfer in green fluorescent proterewiring GFP. J Am Chem Soc. 2008;130:1227–1235. doi: 10.1021/ja0754507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer AE, Qin Y, Park JG, McCombs JE. Design and application of genetically encoded biosensors. Trends Biotechnol. 2011;29:144–152. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grynkiewicz G, Martin P, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 38.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 39.Hanson GT, McAnaney TB, Park ES, Rendell M, Yarbrough DK, Chu S, Xi L, Boxer SG, Montrose MH, Remington SJ. Green fluorescent protein variants as ratiometric dual emission pH sensors. 1. Structural characterization and preliminary application. Biochemistry. 2002;41:15477–15488. doi: 10.1021/bi026609p. [DOI] [PubMed] [Google Scholar]
- 40.McAnaney TB, Park ES, Hanson GT, Remington SJ, Boxer SG. Green fluorescent protein variants as ratiometric dual emission pH sensors. 2. Excited-state dynamics. Biochemistry. 2002;41:15489–15495. doi: 10.1021/bi026610o. [DOI] [PubMed] [Google Scholar]
- 41.McAnaney TB, Shi X, Abbyad P, Jung H, Remington SJ, Boxer SG. Green fluorescent protein variants as ratiometric dual emission pH sensors: 3. Temperature dependence of proton transfer. Biochemistry. 2005;44:8701–8711. doi: 10.1021/bi050132a. [DOI] [PubMed] [Google Scholar]
- 42.Mank M, Griesbeck O. Genetically encoded calcium indicators. Chem Rev. 2008;108:1550–1564. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
- 43.Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson GT, Aggeler R, Oglesbee D, Canon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 45.Lohman JR, Remington SJ. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry. 2008;47:8678–8688. doi: 10.1021/bi800498g. [DOI] [PubMed] [Google Scholar]
- 46.Cannon MB, Remington SJ. Redox-sensitive green fluorescent proteprobes for intracellular redox responses. A review. Methods Mol Biol. 2008;476:51–65. doi: 10.1007/978-1-59745-129-1_4. [DOI] [PubMed] [Google Scholar]
- 47.Meyers A, Dick TP. Genetically encoded redox probes. Antioxidants Redox Signal. 2010;13:1–30. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 48.Wachter R, King BA, Heim R, Kallio K, Tsien RY, Boxer SG, Remington SJ. Crystal structure and photodynamic behavior of the blue emission variant Y66H/Y145F of green fluorescent protein. Biochemistry. 1997;36:9759–9765. doi: 10.1021/bi970563w. [DOI] [PubMed] [Google Scholar]
- 49.Matz MV, Arkady FF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nature Biotech. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 50.Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Hunt ME, Beltran-Ramirez V, Miller DJ, Wiedenmann J, Salih A, Matz MV. Diversity and evolution of coral fluorescent proteins. PLoS One. 2008;3:e2680. doi: 10.1371/journal.pone.0002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hopf M, Gohring W, Ries A, Timpl R, Hohenester E. Crystal structure and mutational analysis of a perlecan-binding fragment of nidogen-1. Nat Struct Biol. 2001;8:573–574. doi: 10.1038/89683. [DOI] [PubMed] [Google Scholar]
- 52.Lukyanov KA, Fradkov AF, Gurskaya NG, Matz MV, Labas YA, Savitsky AP, Markelov ML, Zaraisky AG, Zhao X, Fang Y, Tan W, Lukyanov SA. Natural animal coloration can be determined by a nonfluorescent green fluorescent protein homolog. J Biol Chem. 2000;275:25879–25882. doi: 10.1074/jbc.C000338200. [DOI] [PubMed] [Google Scholar]
- 53.Salih A, Larkum A, Cox G, Kuehl M, Hoegh-Guldberg O. Fluorescent pigments in corals are photoprotective. Nature. 2000;408:850–853. doi: 10.1038/35048564. [DOI] [PubMed] [Google Scholar]
- 54.Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J, Lin J, Zhou C, Deng X, Xia B. Cytotoxicity of red fluorescent protein DsRed is associated with the suppression of Bcl-xL translation. FEBS Lett. 2011;585:821–827. doi: 10.1016/j.febslet.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 56.Dixit R, Cyr R. Cell damage and reactive oxygen species production induced by fluorescence microscopy: effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J. 2003;36:280–290. doi: 10.1046/j.1365-313x.2003.01868.x. [DOI] [PubMed] [Google Scholar]
- 57.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 58.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;87:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He X, Bell AF, Tonge PJ. Synthesis and spectroscopic studies of model red fluorescent protein chromophores. Org Lett. 2002;9:1523–1526. doi: 10.1021/ol0200403. [DOI] [PubMed] [Google Scholar]
- 60.Remington SJ, Wachter RM, Yarbrough DK, Branchaud B, Anderson DC, Kallio K, Lukyanov KA. zFP538, a yellow fluorescent protein from Zoanthus, contains a novel three-ring chromophore. Biochemistry. 2005;44:202–212. doi: 10.1021/bi048383r. [DOI] [PubMed] [Google Scholar]
- 61.Pletneva NV, Pletnev SV, CD M, TT V, Popov VO, Martynov VI, Wlodawer A, Dauter Z, Pletnev VZ. Three-dimensional structure of yellow fluorescent protein zYFP538 from Zoanthus sp. at the resolution 1.8 angstrom. Bioorg Khim. 2007;33:421–430. doi: 10.1134/s1068162007040048. [DOI] [PubMed] [Google Scholar]
- 62.Subach OM, Malashkevich VN, Zencheck WD, MK S, Piatkevich KD, AS C, Verkhusha VV. Structural characterization of acylimine-containing blue and red chromophores in mTagBFP and TagRFP fluorescent proteins. Chem Biol. 2010;17:333–341. doi: 10.1016/j.chembiol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strack RL, Strongin DE, Mets L, Glick BS, Keenan RJ. Chromophore formation in DsRed occurs by a branched pathway. J Am Chem Soc. 2010;132:8496–8505. doi: 10.1021/ja1030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Structural basis for spectral variations in mFruits: Monomeric orange and red fluorescent proteins. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 68.Kikuchi A, Fukumura E, Karasawa S, Mizuno H, Miyawaki A, Shiro Y. Structural characterization of a thiazoline-containing chromophore in an orange fluorescent protein, monomeric Kusabira Orange. Biochemistry. 2008;47:11573–11580. doi: 10.1021/bi800727v. [DOI] [PubMed] [Google Scholar]
- 69.Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright, and photostable version of Clavularia cyan fluorescent protestructural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nienhaus GU, Nienhaus K, Holzle A, Ivanchenko S, Renzi F, Oswald F, Wolff M, Schmitt F, Rocker C, Vallone B, Weidemann W, Heilker R, Nar H, Wiedenmann J. Photoconvertible fluorescent protein EosFP: biophysical properties and cell biology applications. Photochem Photobiol. 2006;82:351–358. doi: 10.1562/2005-05-19-RA-533. [DOI] [PubMed] [Google Scholar]
- 71.Kogure T, Karasawa S, Arkai T, Saito K, Kinjo M, Miyawaki A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nature Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 72.Henderson JN, Osborne MF, Koon N, Gepshtein R, Huppert D, Remington SJ. Excited State Proton Transfer in the Red Fluorescent Protein mKeima. J Am Chem Soc. 2009;131:13212–13213. doi: 10.1021/ja904665x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 74.Dove S, Takabayashi M, Hoegh-Guldberg O. Isolation and characterization of the pink and blue pigments of Pocilloporid and Acroporid corals. Biol Bull. 1995;189:288–297. doi: 10.2307/1542146. [DOI] [PubMed] [Google Scholar]
- 75.Prescott M, Ling M, Beddoe T, Oakley AJ, Hoegh-Guldberg O, Devenish RJ, Rossjohn J. The 2.2 A crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation. Structure. 2003;11:275–284. doi: 10.1016/s0969-2126(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 76.Wilmann PG, Petersen J, Dvenish RJ, Prescott M, Rossjohn J. A polypeptide fragmentation within the chromophore revealed in the 2.1 A crystal structure of a nonfluorescent chromoprotein from anemonia sulcata. J Biol Chem. 2005;280:2401–2404. doi: 10.1074/jbc.C400484200. [DOI] [PubMed] [Google Scholar]
- 77.Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Lukyanov KA, Remington SJ. The kindling fluorescent protein from Anemonia sulcata: dark state structure at 1.38 Angstroms resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 78.Gepshtein R, Huppert D, Agmon N. Deactivation mechanism of the green fluorescent chromophore. J Phys Chem B. 2006;110:4434–4442. doi: 10.1021/jp0540095. [DOI] [PubMed] [Google Scholar]
- 79.Yampolsky IV, Remington SJ, Martynov VI, Potapov VK, Lukyanov S, Lukyanov KA. Synthesis and properties of the chromophore of asFP595 chromoprotein from Anemonia sulcata. Biochemistry. 2005;44:5788–5793. doi: 10.1021/bi0476432. [DOI] [PubMed] [Google Scholar]
- 80.Chudakov DM, Belousov VV, Zaraisky AG, Novoselov VV, Staroverov DB, Zorov DB, Lukyanov S, Lukyanov KA. Kindling fluorescent proteins for precise in vivo photolabeling. Nature Biotech. 2003;21:191–194. doi: 10.1038/nbt778. [DOI] [PubMed] [Google Scholar]
- 81.Chudakov DM, Foefanov AV, Mudrik NN, Lukyanov S, Lukyanov KA. Chromophore environment provides clue to “kindling fluorescent protein” riddle. J Biol Chem. 2003;278:7215–7219. doi: 10.1074/jbc.M211988200. [DOI] [PubMed] [Google Scholar]
- 82.Stiel AC, Trowitzsch S, Weber G, Andresen M, Eggeling C, Hell SW, Jakobs S, Wahl MC. 1.8 A bright-state structure of a reversibly photoswitchable fluorescent protein Dronpa guides the generation of fast switching mutants. Biochem J. 2007;402:35–42. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilmann PG, Turcic K, Battad JM, Wilce MC, Devenish RJ, Prescott M, Rossjohn J. The 1.7 A crystal structure of Dronpa: a photoswitchable green fluorescent protein. J Mol Biol. 2006;364:213–224. doi: 10.1016/j.jmb.2006.08.089. [DOI] [PubMed] [Google Scholar]
- 84.Andresen M, Stiel AC, Trowitzsch S, Weber G, Eggeling C, Wahl MC, Hell SW, Jakobs S. Structural basis for reversible photoswitching in Dronpa. Proc Natl Acad Sci U S A. 2007;104:13005–13009. doi: 10.1073/pnas.0700629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henderson JN, Ai HW, Campbell RE, Remington SJ. Structural basis for reversible photobleaching in a green fluorescent protein homologue. Proc Natl Acad Sci U S A. 2007;104:6672–6677. doi: 10.1073/pnas.0700059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 87.Chudakov DM, Verkhusha VV, Staroverov DB, Souslova EA, Lukyanov S, Lukyanov KA. Photoswitchable cyan fluorescent protein for protein tracking. Nature Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 88.Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci U S A. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 90.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:356–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]


